Abstract
Salmonella enterica serovar Heidelberg frequently causes food-borne illness in humans. There are few data on the prevalence, antimicrobial susceptibility, and genetic diversity of Salmonella serovar Heidelberg isolates in retail meats. We compared the prevalences of Salmonella serovar Heidelberg in a sampling of 20,295 meats, including chicken breast (n = 5,075), ground turkey (n = 5,044), ground beef (n = 5,100), and pork chops (n = 5,076), collected during 2002 to 2006. Isolates were analyzed for antimicrobial susceptibility and compared genetically using pulsed-field gel electrophoresis (PFGE) and PCR for the blaCMY gene. A total of 298 Salmonella serovar Heidelberg isolates were recovered, representing 21.6% of all Salmonella serovars from retail meats. One hundred seventy-eight (59.7%) were from ground turkey, 110 (36.9%) were from chicken breast, and 10 (3.4%) were from pork chops; none was found in ground beef. One hundred ninety-eight isolates (66.4%) were resistant to at least one compound, and 49 (16.4%) were resistant to at least five compounds. Six isolates (2.0%), all from ground turkey, were resistant to at least nine antimicrobials. The highest resistance in poultry isolates was to tetracycline (39.9%), followed by streptomycin (37.8%), sulfamethoxazole (27.7%), gentamicin (25.7%), kanamycin (21.5%), ampicillin (19.8%), amoxicillin-clavulanic acid (10.4%), and ceftiofur (9.0%). All isolates were susceptible to ceftriaxone and ciprofloxacin. All ceftiofur-resistant strains carried blaCMY. PFGE using XbaI and BlnI showed that certain clones were widely dispersed in different types of meats and meat brands from different store chains in all five sampling years. These data indicate that Salmonella serovar Heidelberg is a common serovar in retail poultry meats and includes widespread clones of multidrug-resistant strains.
Salmonella enterica is recognized as being one of the most common bacterial causes of food-borne diarrhea illness worldwide. It has been estimated that there are approximately 1.4 million cases of salmonellosis, resulting in 16,000 hospitalizations and 600 deaths each year in the United States (21). The majority of Salmonella infections are attributed to the ingestion of contaminated meat and eggs as well as fresh produce and seasonings (17). There has been increasing concern over the past 30 years regarding the worldwide emergence of multidrug-resistant phenotypes among Salmonella serovars, in particular Salmonella serovar Typhimurium and, more recently, Salmonella serovar Newport as well as several other serovars. The levels and extent of resistance vary in different regions and are influenced by antimicrobial use in humans and animals as well as geographical differences in the epidemiology of Salmonella.
In the United States and Canada, Salmonella serovar Heidelberg is among the most frequently isolated serovars both in clinical cases of salmonellosis and from retail meats and food animals. Salmonella serovar Heidelberg ranked first and fourth among serovars from food animals in 2002 and 2003, respectively (30), and ranked fifth and fourth among serovars from humans in 2003 and 2004, respectively (5, 6). Salmonella serovar Heidelberg was the most common serovar found in retail meats in both years (13, 34) and was found exclusively in poultry meats. The notion that poultry is the major reservoir of human infections in the United States and Canada is supported by case-control studies implicating table eggs (7, 11) and chicken meat (11) as being the main sources of Salmonella serovar Heidelberg infections.
Salmonella serovar Heidelberg is a major serovar only in the United States and Canada and is not among the top six serovars in other continents (25). Epidemic outbreaks of Salmonella serovar Heidelberg present a significant public health and economic burden in the region (1). Salmonella serovar Heidelberg has caused large outbreaks of food-borne illness in nursing homes, in hospitals, and within the community (4, 8, 19, 20). Antimicrobial drug resistance in this serovar is notable, where resistance to ceftiofur with decreased susceptibility to ceftriaxone has increased in recent years. These strains are also commonly resistant to streptomycin, tetracycline, sulfamethoxazole, chloramphenicol, and trimethoprim-sulfamethoxazole. Salmonella serovar Heidelberg is derived mainly from poultry products, where ceftiofur is used in day-old chicks to control early chick mortality due to Escherichia coli and Staphylococcus aureus. The occurrence of multiple drug resistance (MDR) in Salmonella serovar Heidelberg is particularly important because of the propensity of this serovar to produce severe extraintestinal infections (32) such as septicemia (12) and myocarditis (3). The objectives of this study were to determine the prevalences, antimicrobial susceptibility profiles, and genetic relatednesses of Salmonella serovar Heidelberg isolates from the National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) retail meat program in 2002 to 2006. Our overall goal was to gain a better understanding of the nature of MDR phenotypes disseminating in the U.S. retail meat supply.
MATERIALS AND METHODS
Retail meat sampling.
NARMS retail meat monitoring is conducted in collaboration with FoodNet (http://www.cdc.gov/foodnet/). Each FoodNet site attempted to purchase 40 retail meats per month, comprising 10 samples each of chicken breast, ground turkey, ground beef, and pork chops from area grocery stores. Six FoodNet sites participated in 2002 (Connecticut, Georgia, Maryland, Minnesota, Oregon, and Tennessee), 8 sites participated in 2003 (with the addition of California and New York), and 10 sites participated in 2004 to 2006 (with the addition of Colorado and New Mexico). In 2002, Oregon collected retail meat samples from September to December only, and in 2003, Connecticut collected only 20 retail meats each month (5 of each meat type). Samples were kept cold during transport from the grocery store(s) to the laboratory.
Microbiological analysis.
In each laboratory, samples were refrigerated at 4°C and were processed no later than 96 h after purchase. Retail meat and poultry packages were kept intact until they were aseptically opened in the laboratory at the start of the examination. For chicken and pork samples, one cut of meat was examined. For ground product (beef and turkey), 25-g portions of product were used for culturing. Each sample was placed in separate sterile plastic bags with 250 ml of buffered peptone water, and the bags were vigorously shaken. Fifty milliliters of the rinse was mixed with 50 ml of double-strength lactose broth and incubated at 35°C for 24 h. From each culture grown overnight, 0.1 ml was transferred into 9.9 ml of Rappaport-Vassiliadis R10 broth. The tubes of Rappaport-Vassiliadis R10 medium were incubated in a water bath at 42°C for 16 to 20 h. One milliliter was transferred into 10 ml of prewarmed (35 to 37°C) M broth and incubated in a water bath at 35°C to 37°C for 6 to 8 h. From each M broth culture, 1 ml was heated at 100°C for 15 min, and the remaining portion was refrigerated. The heated portion from each culture was cooled to room temperature and tested using the Tecra Salmonella visual immunoassay kit (International BioProducts, Bothell, WA) or the Vidas Salmonella immunoassay kit (bioMérieux, Hazelwood, MO) according to the manufacturers' instructions. Positive Tecra or Vidas samples were streaked onto a xylose-lysine-deoxycholate agar plate for isolation and incubated at 35°C for 24 h. When Salmonella-like growth was observed, one well-isolated colony was streaked onto a trypticase soy agar plate supplemented with 5% defribrinated sheep blood (BBL, Becton, Dickinson, and Company, Sparks, MD) for isolation. Salmonella isolates were subsequently frozen at −60°C to −80°C in Brucella broth with 20% glycerol and shipped in cryovials on dry ice to the FDA Center for Veterinary Medicine. Upon arrival at the FDA Center for Veterinary Medicine, every isolate was streaked for purity onto a blood agar plate before being confirmed as Salmonella using the Vitek microbial identification system (bioMérieux, Hazelwood, MO). These isolates were further assayed for O and H antigens using either commercially available (Difco, Becton, Dickinson, and Company, Sparks, MD) or Centers for Disease Control and Prevention (CDC) antisera. Bacteria were stored in trypticase soy broth containing 15% glycerol at −80°C until use. All bacterial media were obtained from Difco, Becton, Dickinson, and Company, Sparks, MD, unless otherwise specified.
Antimicrobial susceptibility testing.
Salmonella isolates were assayed for susceptibility to a panel of 15 antimicrobials used by the NARMS program. Antimicrobial MICs of Salmonella isolates were determined via the Sensititre automated antimicrobial susceptibility system (Trek Diagnostic Systems, Westlake, OH) using cation-adjusted Mueller-Hinton broth, and results were read after 18 to 24 h of incubation at 36°C. MICs were interpreted according to CLSI standards where available (9, 10). The antimicrobial dilution ranges tested and the resistant breakpoints used were as follows: 0.5 to 64 and ≥64 μg/ml, respectively, for amikacin; 1 to 32 and ≥32 μg/ml, respectively, for amoxicillin-clavulanic acid; 1 to 32 and ≥32 μg/ml, respectively, for ampicillin; 0.5 to 32 and ≥32 μg/ml, respectively, for cefoxitin; 0.12 to 8 and ≥8 μg/ml, respectively, for ceftiofur; 0.5 to 64 and ≥64 μg/ml, respectively, for ceftriaxone; 2 to 32 and ≥32 μg/ml, respectively, for chloramphenicol; 0.0.15 to 4 and ≥4 μg/ml, respectively, for ciprofloxacin; 0.25 to 16 and ≥16 μg/ml, respectively, for gentamicin; 8 to 64 and ≥64 μg/ml, respectively, for kanamycin; 0.5 to 32 and ≥32 μg/ml, respectively, for nalidixic acid; 32 to 64 and ≥64 μg/ml, respectively, for streptomycin; 4 to 32 and ≥16 μg/ml, respectively, for tetracycline; 0.12 to 4 and ≥4 μg/ml, respectively, for trimethoprim-sulfamethoxazole; and 6 to 512 and ≥512 μg/ml, respectively, for sulfamethoxazole. Sulfamethoxazole was replaced with sulfisoxazole in 2004, and the same dilution ranges and breakpoints were used. For data reported here, these agents are collectively referred to as sulfonamides. Escherichia coli ATCC 25922, Escherichia coli ATCC 35218, Enterococcus faecalis ATCC 29212, Staphylococcus aureus ATCC 29213, and Pseudomonas aeruginosa ATCC 27853 were used as quality control organisms to ensure the validity of the susceptibility testing.
Detection of blaCMY.
The presence of blaCMY was determined by PCR using previously published methods (35). DNA template was prepared using MoBio Ultraclean DNA isolation kits (MoBio Laboratories, Inc., Carlsbad, CA). Amplifications were carried out using 200 ng of template, 250 μM each deoxynucleoside triphosphate, 1.5 mM MgCl2, 50 pmol of primers, and 1 U of Amplitaq Gold Taq polymerase (Applied Biosystems, Foster City, CA). The amplified products were separated by gel electrophoresis on 1.0% agarose gels stained with ethidium bromide. The gels were visualized under UV light. The DNA sequence was determined for a subset of amplicons using the ABI 3730 automated sequencer and analyzed using the National Center for Biotechnology Information's BLAST network service.
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed to determine genomic DNA fingerprinting profiles of Salmonella serovar Heidelberg according to the protocol developed by the CDC (23) using Salmonella serovar Braenderup H9812 as the control strain. Agarose-embedded DNA was digested with 50 U of XbaI or BlnI (Boehringer Mannheim, Indianapolis, IN) for at least 4 h in a water bath at 37°C. The restriction fragments were separated by electrophoresis in 0.5× TBE buffer (Invitrogen, Carlsbad, CA) at 14°C for 18 h using a Chef Mapper electrophoresis system (Bio-Rad, Hercules, CA) with pulse times of 2.16 to 63.8 s. Isolates presenting DNA smears were retested using plugs digested with XbaI or BlnI and electrophoresis buffer containing 50 μM thiourea in 0.5× TBE buffer. The gels were stained with ethidium bromide, and DNA bands were visualized with UV transillumination (Bio-Rad). PFGE results were analyzed using BioNumerics software (Applied Maths, Kortrijk, Belgium), and banding pattern similarities were compared using two-enzyme analysis with a 1.5% band position tolerance (27). All PFGE profiles generated from this study were submitted to the PulseNet national database located at the CDC for comparison with isolates from clinical salmonellosis cases.
Statistical analysis.
Trends were analyzed using the Cochran-Armitage test for trend, and z statistics were calculated (18). Tests that are statistically significant at the level of an α value of 0.05 were considered to indicate an increasing or decreasing trend in prevalence over the 5 years of sampling.
RESULTS
Prevalence of Salmonella serovar Heidelberg in retail meats.
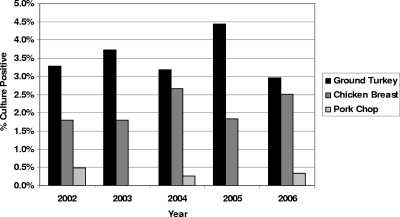
A total of 20,295 retail meat samples, comprised of chicken breasts (n = 5,075), ground turkey (n = 5,044), ground beef (n = 5,100), and pork chops (n = 5,076), were collected during 2002 to 2006 for the NARMS retail meat surveillance program (Table 1). Salmonella serovar Heidelberg was the most frequently isolated serovar in all 5 years. A total of 298 isolates were recovered, representing 21.6% (298/1,380) of all Salmonella serovars. Among the 298 isolates, 35 were isolated in 2002, 48 were isolated in 2003, 71 were isolated in 2004, 75 were isolated in 2005, and 69 were isolated in 2006. The annual prevalence of Salmonella serovar Heidelberg from 2002 to 2006 was not significantly different within any given meat type (P = 0.44 for all meats combined). Figure 1 shows the annual prevalence of Salmonella serovar Heidelberg from raw meats. Poultry meat was the dominant source of Salmonella serovar Heidelberg, accounting for 96.6% of all isolates, with 59.7% (n = 178) from ground turkey and 36.9% (n = 110) from chicken breast (Fig. 1). Only 3.4% (n = 10) of all isolates were from pork chops, and none was found in ground beef, where Salmonella spp. are rarely isolated.
TABLE 1.
Number of meat samples analyzed by yeara
| Yr | No. of meat samples analyzed
|
||||
|---|---|---|---|---|---|
| Ground turkey | Chicken breast | Pork chop | Ground beef | Total | |
| 2002 | 642 | 616 | 613 | 642 | 2,513 |
| 2003 | 857 | 897 | 899 | 880 | 3,533 |
| 2004 | 1,165 | 1,172 | 1,176 | 1,186 | 4,699 |
| 2005 | 1,195 | 1,194 | 1,196 | 1,196 | 4,781 |
| 2006 | 1,185 | 1,196 | 1,192 | 1,196 | 4,769 |
| Total | 5,044 | 5,075 | 5,076 | 5,100 | 20,295 |
The total number of meat specimens analyzed increased as the number of participating test sites expanded from 5 in 2002 to 10 in 2006 (14).
FIG. 1.
Proportion of retail meats that were culture positive for Salmonella serovar Heidelberg in 2002 to 2006. The total numbers of meat samples tested each year from 2002 to 2006 are shown in Table 1. No Salmonella serovar Heidelberg isolates were recovered from ground beef during the sampling interval.
Antimicrobial susceptibility profiles.
One hundred ninety-nine of 298 isolates (67%) were resistant to at least one antimicrobial agent, with 16.4% (n = 49) of the isolates being resistant to at least five antimicrobials. Six isolates (3.0%), all recovered from ground turkey, were resistant to at least nine antimicrobials, including beta-lactams, aminoglycosides, chloramphenicol, sulfamethoxazole, and tetracycline. For chicken and turkey isolates combined, resistance to tetracycline was most common (39.9%), followed by streptomycin (37.8%), sulfamethoxazole (27.7%), gentamicin (25.7%), kanamycin (21.5%), ampicillin (19.8%), amoxicillin-clavulanic acid (10.4%), cefoxitin (9.0%), and ceftiofur (9.0%). Rare isolates were resistant to chloramphenicol (1%), nalidixic acid (1.0%), and trimethoprim-sulfamethoxazole (0.7%). All isolates were susceptible to amikacin, ceftriaxone, and ciprofloxacin.
In general, isolates from ground turkey were more frequently resistant than were those from chicken (Fig. 2). Analysis for trends in susceptibility from 2002 to 2006 showed that for all isolates combined, streptomycin resistance decreased from 65.7% to 33.3% (P = 0.05). In Salmonella serovar Heidelberg isolates from chicken meat, kanamycin resistance declined from 36.4% to 0% (P = 0.0002) (Fig. 2). Other aminoglycosides decreased, but the trends were not statistically significant. These changes in aminoglycoside resistance were due largely to the high resistance levels observed in the inaugural year of testing (Fig. 2). In addition to this class of compounds, tetracycline resistance in chicken isolates decreased significantly from 45.5% in 2002 to 3.3% in 2006 (P = 0.004).
FIG. 2.
Rates of antimicrobial resistance of Salmonella serovar Heidelberg isolates from ground turkey (A) and chicken breast (B) in 2002 to 2006. Antimicrobial resistance rates are presented for tetracycline (Tet), sulfonamides (Sul), streptomycin (Str), kanamycin (Kan), gentamicin (Gen), ampicillin (Amp), amoxicillin-clavulanate (Amc), ceftiofur (Tio), cefoxitin (Fox), chloramphenicol (Chl), and nalidixic acid (Nal). Trimethoprim-sulfamethoxazole resistance (not shown) was detected in only two chicken breast isolates in 2006. All isolates were susceptible to amikacin, ceftriaxone, and ciprofloxacin.
Compared with 2002 levels, chicken isolates showed increasing resistance to ceftiofur, cefoxitin, and amoxicillin-clavulanate, rising from 0% to 10% (P = 0.36). Ceftiofur resistance was not detected in ground turkey isolates in 2003, compared with 17.1% in 2006 (P = 0.009). Among the 56 ampicillin-resistant isolates from all sources, 26 showed resistance to ceftiofur, 6 of which displayed the MDR-AmpC phenotype (33). Twenty isolates displayed the amoxicillin, cefoxitin, ceftiofur, and ampicillin (A2C-AMP) resistance pattern. As expected, the 26 isolates that showed resistance to ceftiofur also showed decreased susceptibility to ceftriaxone (8 to 32 μg/ml), with 17 originating from ground turkey and 9 originating from chicken breast. All 26 isolates were positive by PCR for the blaCMY cephamycinase gene. There was no resistance to beta-lactams among the 10 pork isolates, where 5 were tetracycline resistant and 7 were resistant to sulfonamides (data not shown).
PFGE profiles.
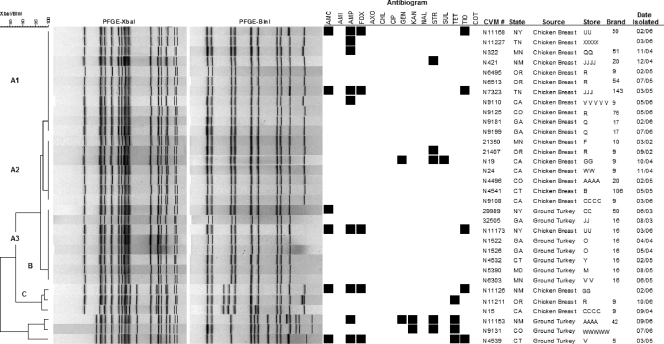
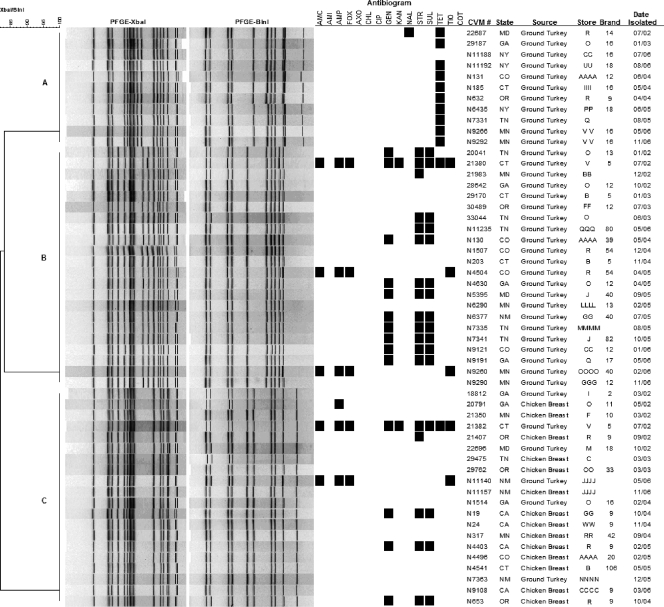
To assess genetic relatedness, PFGE was used to analyze all 298 Salmonella serovar Heidelberg isolates. A total of 61 PFGE patterns were generated with XbaI, and 107 patterns were generated with XbaI plus BlnI using the combined banding patterns. Figures 3 and 4 show PFGE data for a subset of poultry isolates and dendrograms generated from the combination of banding patterns obtained using both enzymes. These isolates are shown to demonstrate the added discriminatory power of two-enzyme analysis (Fig. 3) and the temporal and geographical dispersal of common clones in poultry products (Fig. 4).
FIG. 3.
PFGE patterns and antimicrobial resistance profiles of selected clones of Salmonella serovar Heidelberg. For the susceptibility results, a black box indicates resistance to a particular antimicrobial. AMC, amoxicillin-clavulanic acid; AMI, amikacin; AMP, ampicillin; FOX, cefoxitin; AXO, ceftriaxone; CHL, chloramphenicol; CIP, ciprofloxacin; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; STR, streptomycin; SUL, sulfonamides; TET, tetracycline; TIO, ceftiofur; COT, trimethoprim-sulfamethoxazole; CVM, FDA Center for Veterinary Medicine.
FIG. 4.
PFGE and antimicrobial resistance profiles of select Salmonella serovar Heidelberg clones. For the susceptibility results, a black box indicates resistance to a particular antimicrobial. AMC, amoxicillin-clavulanic acid; AMI, amikacin; AMP, ampicillin; FOX, cefoxitin; AXO, ceftriaxone; CHL, chloramphenicol; CIP, ciprofloxacin; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; STR, streptomycin; SUL, sulfonamides; TET, tetracycline; TIO, ceftiofur; COT, trimethoprim-sulfamethoxazole; CVM, FDA Center for Veterinary Medicine.
In Fig. 3, the combined-pattern analysis reveals the value of a second enzyme to increase discriminatory power. Using XbaI and BlnI combination analysis, cluster A was subdivided into clusters A1, A2, and A3. Clusters A1 and A2 were composed of 18 chicken breast isolates recovered from different states over a period of 5 years, which had the same BlnI pattern but two different XbaI patterns. Conversely, the eight isolates in cluster A3 had indistinguishable XbaI patterns but different BlnI patterns than did cluster A1. Seven of the isolates in cluster A3 were recovered from ground turkey, and one was recovered from chicken breast. Cluster B contained three chicken isolates, which displayed indistinguishable XbaI patterns but different BlnI patterns showing one-band differences from each other. Similar observations were seen in cluster C (Fig. 3).
Figure 4 shows three Salmonella serovar Heidelberg clusters (clusters A, B, and C) based on PFGE patterns with both enzymes. These data show that some clones can be widely spread among different meat products sold in different retail outlets under different brands over time. There were some general correlations between PFGE groupings and antimicrobial resistance phenotypes. For example, 10/11 isolates in cluster A displayed resistance to tetracycline, while the majority of isolates in cluster B demonstrated resistance to gentamicin, streptomycin, and trimethoprim-sulfamethoxazole. The 20 isolates in cluster C showed mixed resistant profiles; however, no resistant profiles were shared between isolates recovered from ground turkey and chicken breast (Fig. 4).
PFGE subtyping also showed that some clones were restricted to specific meat types, while others were widespread in both chicken and turkey meats. For example, cluster C in Fig. 3 and clusters A and B in Fig. 4 were composed solely of isolates recovered from ground turkey from different stores, meat brands, and states over the 5-year testing period, whereas clusters A1, A2, and B in Fig. 3 included only chicken isolates. In contrast, cluster A3 in Fig. 3 and cluster C in Fig. 4 contained isolates originating from both ground turkey and chicken breast.
DISCUSSION
The retail meat component of the NARMS is designed to be a sentinel monitoring program to assess the status of antimicrobial resistance among select food-borne bacteria originating from select foods of animal origin. We report 5 years of PFGE typing and antimicrobial susceptibility data for the most common Salmonella serovar (22%), Salmonella serovar Heidelberg, from a sampling of 20,295 retail raw meats. NARMS surveillance for the sampling interval from 2002 to 2006 indicated that nearly all (96.6%) Salmonella serovar Heidelberg isolates were recovered from poultry meats: 59.7% from ground turkey (n = 178) and 36.9% from chicken breast (n = 110), with only 3.4% (n = 10) of isolates from pork chop and none from ground beef. This finding is consistent with those of the U.S. Food Safety Inspection Service, which reported that Salmonella serovar Heidelberg isolates were identified mainly in poultry, representing one of the five most prevalent serovars each year in both birds and meat products, and rarely recovered from other food animal sources (29). Thus, the preponderance of Salmonella serovar Heidelberg isolates in turkey meat from this study relative to those of other serovars most likely reflects its presence in live animals, where it is known to be acquired at various stages in the production cycle (22, 28).
One hundred ninety-nine of 298 isolates (67%) were resistant to at least one antimicrobial agent, with 16.4% (n = 49) being resistant to at least five antimicrobials. When data from 2002 and 2006 were compared, several changes in the ratio of resistant isolates were observed (Fig. 2), but only declining kanamycin and tetracycline resistance in chicken isolates exhibited statistical significance. The most extensive resistance patterns were among turkey isolates, with some showing resistance to nine or more antimicrobials. These findings are consistent with what is seen in Enterococcus and E. coli isolates tested in the NARMS program, where resistance levels and MDR phenotypes are most extensive among turkey meat isolates (14).
In salmonellae, resistance to quinolones and cephalosporins is of primary medical concern. Nalidixic acid resistance remained at low levels in Salmonella serovar Heidelberg, peaking in 2006 at 3.3% in chicken breast and in 2002 at 4.8% in ground turkey. No resistance to ciprofloxacin was detected in Salmonella serovar Heidelberg isolates or in any other salmonellae from NARMS retail meat to date (14). Resistance to trimethoprim-sulfamethoxazole and chloramphenicol remained low year to year in Salmonella serovar Heidelberg, as was noted previously for other serovars (14).
While we do not have access to data for antimicrobial use in food animal production, the increasing resistance to beta-lactam compounds indicates considerable reliance on these agents in poultry production during the sampling years. Among food isolates of Salmonella spp. in the United States, the acquisition of ceftiofur resistance is strongly associated with MDR and the presence of a plasmid-borne blaCMY-2 gene (31), which is widely dispersed geographically among various members of the Enterobacteriaceae. In addition to ceftiofur resistance, blaCMY-2 confers decreased susceptibility to ceftriaxone along with resistance to ampicillin, amoxicillin-clavulanate, cephalothin, and cefoxitin. It is commonly present on an IncA/C plasmid backbone with numerous other resistance genes (31) but may be carried on other plasmid backbones (16, 35) with various codeterminants. In our isolates of ceftiofur-resistant Salmonella serovar Heidelberg, the MDR-AmpC phenotype was present in 5/17 ground turkey isolates and 1/9 chicken breast isolates. Previous work showed that for all six of these isolates, the blaCMY gene was carried on an IncA/C backbone (31). The remaining ceftiofur-resistant isolates exhibited only the A2C-AMP patterns.
In Canada, the A2C-AMP patterns became predominant in serovar Heidelberg compared with other serovars, representing the most frequently found resistance pattern (24). Surveillance data revealed substantial increases in resistance to cephalosporins among Salmonella serovar Heidelberg isolates from humans and chicken in 2003 to 2004 in Quebec (26). In the latter half of 2003, approximately 30% of isolates from humans from Quebec were resistant to ceftiofur. The prevalence gradually increased over 2004 to approximately 48% in the second quarter of 2005. This increase was accompanied temporally by an increase in ceftiofur resistance in Salmonella serovar Heidelberg isolates from retail chicken from Quebec (25) and Ontario. An investigation into possible risk factors showed that ceftiofur was commonly used in chicken hatcheries to control E. coli infections. A voluntary suspension of ceftiofur use in February 2005 was followed temporally by a dramatic decline in the prevalence of ceftiofur resistance in Salmonella serovar Heidelberg isolates from humans, chicken at abattoir, and chicken at retail in Ontario (2). Concern about increasing cephalosporin resistance prompted the FDA to propose an extralabel prohibition for cephalosporin use in food animals in the United States (15).
PFGE using XbaI and BlnI digestion resulted in a total of 107 patterns for the 298 strains examined. Data suggest different patterns of dissemination in food production. Clones from ground turkey (clusters A and B) (Fig. 4) isolated from 2002 to 2006 from different stores in different states suggest contamination from a common source, perhaps at a grinding facility separate from the vendor location, which serves multiple commercial brands and retail outlets. Cluster A also showed a highly consistent antimicrobial susceptibility pattern, with 9/11 isolates being resistant to tetracycline only, adding strength to the notion of a common source. Susceptibility patterns also show evidence of subpopulations, presumably signifying the acquisition of different antimicrobial resistance plasmids. For example, nearly all the strains in cluster B displayed one of three resistance patterns, including the MDR-AmpC phenotype. Similarly, with chicken breast isolates (clusters A1 and A2) (Fig. 3), clonal strains present in the same commercial brand in multiple U.S. states at different times suggest contamination from a production facility associated with same brand. Finding the same PFGE clone over several years is evidence that some strains are better adapted to persist in an animal host environment or survive food processing treatments. Regardless, their presence in different meats from different locations over time implies that Salmonella contamination of food can occur at different stages of meat production and that some clones were likely disseminated from farms.
Figure 4 shows that certain clones were dispersed in both chicken and turkey and different meat brands from different store chains in all five sampling years. Some clonal clusters fitting this description (data not shown) included isolates from pork. A PFGE cluster comprised of Salmonella serovar Heidelberg isolates from chicken, turkey, and pork was sold by five vendors in three states from 2002 to 2006. As with other clusters, these isolates showed very similar antimicrobial resistance patterns. Identical strains that were isolated from ground turkey and pork chops were also from the same store during the same month, suggesting that Salmonella spreads by cross-contamination at the retail outlet.
Salmonella serovar Heidelberg is common only in the United States and Canada, where it has been associated with more invasive infections than those caused by other common serovars (12, 32). Continuous monitoring of its prevalence and resistance in the food supply, along with other Salmonella serotypes of public health importance, will significantly enhance the surveillance of Salmonella infections and future outbreak investigations.
Footnotes
Published ahead of print on 29 August 2008.
REFERENCES
- 1.Barnass, S., M. O’Mahony, P. N. Sockett, J. Garner, J. Franklin, and S. Tabaqchali. 1989. The tangible cost implications of a hospital outbreak of multiply-resistant Salmonella. Epidemiol. Infect. 103:227-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulianne, M. 2005. Results from a large poultry study in Québec examining antimicrobial use and antimicrobial resistance. In Agriculture's Role in Managing Antimicrobial Resistance Conference 2005, Toronto, Canada.
- 3.Burt, C. R., J. C. Proudfoot, M. Roberts, and R. H. Horowitz. 1990. Fatal myocarditis secondary to Salmonella septicemia in a young adult. J. Emerg. Med. 8:295-297. [DOI] [PubMed] [Google Scholar]
- 4.CDC. 1986. Salmonella heidelberg outbreak at a convention—New Mexico. MMWR Morb. Mortal. Wkly. Rep. 35:91. [PubMed] [Google Scholar]
- 5.CDC. 2007. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates final report, 2003. CDC, Atlanta, GA.
- 6.CDC. 2007. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates final report, 2004. CDC, Atlanta, GA.
- 7.Chittick, P., A. Sulka, R. V. Tauxe, and A. M. Fry. 2006. A summary of national reports of foodborne outbreaks of Salmonella Heidelberg infections in the United States: clues for disease prevention. J. Food Prot. 69:1150-1153. [DOI] [PubMed] [Google Scholar]
- 8.Choi, M., T. T. Yoshikawa, J. Bridge, A. Schlaifer, D. Osterweil, D. Reid, and D. C. Norman. 1990. Salmonella outbreak in a nursing home. J. Am. Geriatr. Soc. 38:531-534. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; fifteenth informational supplement. CLSI document M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Clinical and Laboratory Standards Institute. 2008. Method for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard, sixth edition. CLSI document M7-A6. Clinical and Laboratory Standards Institute, Wayne, PA.
- 11.Currie, A., L. MacDougall, J. Aramini, C. Gaulin, R. Ahmed, and S. Isaacs. 2005. Frozen chicken nuggets and strips and eggs are leading risk factors for Salmonella Heidelberg infections in Canada. Epidemiol. Infect. 133:809-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demczuk, W., R. Ahmed, D. Woodward, C. Clark, and F. Rodgers. 2000. Laboratory surveillance for enteric pathogens in Canada, 2000 annual summary. National Laboratory for Enteric Pathogens, National Microbiology Laboratory, Health Canada, Canadian Science Centre for Human and Animal Health, Winnipeg, Manitoba, Canada.
- 13.FDA. 2006. National Antimicrobial Resistance Monitoring System—Enteric Bacteria (NARMS): 2003 Executive Report. FDA, Bethesda, MD.
- 14.FDA. 2008. National Antimicrobial Resistance Monitoring System—Enteric Bacteria (NARMS): retail meat isolates final report, 2005. FDA, Bethesda, MD. http://www.fda.gov/cvm/2005NARMSAnnualRpt.htm.
- 15.Federal Register. 2008. Cephalosporin drugs. Extralabel animal drug use; order of prohibition. Fed. Regist. 73:38110-38113. [Google Scholar]
- 16.Giles, W. P., A. K. Benson, M. E. Olson, R. W. Hutkins, J. M. Whichard, P. L. Winokur, and P. D. Fey. 2004. DNA sequence analysis of regions surrounding blaCMY-2 from multiple Salmonella plasmid backbones. Antimicrob. Agents Chemother. 48:2845-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez, T. M., Y. Motarjemi, S. Miyagawa, F. K. Kaferstein, and K. Stohr. 1997. Foodborne salmonellosis. World Health Stat. Q. 50:81-89. [PubMed] [Google Scholar]
- 18.Kuzma, J. W., and S. E. Bohnenblust. 1998. Basic statistics for the health sciences. Mayfield Publishing Co., Mountain View, CA.
- 19.Layton, M. C., S. G. Calliste, T. M. Gomez, C. Patton, and S. Brooks. 1997. A mixed foodborne outbreak with Salmonella heidelberg and Campylobacter jejuni in a nursing home. Infect. Control Hosp. Epidemiol. 18:115-121. [DOI] [PubMed] [Google Scholar]
- 20.Lyons, R. W., C. L. Samples, H. N. DeSilva, K. A. Ross, E. M. Julian, and P. J. Checko. 1980. An epidemic of resistant Salmonella in a nursery. Animal-to-human spread. JAMA 243:546-547. [PubMed] [Google Scholar]
- 21.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nayak, R., and T. Stewart-King. 2008. Molecular epidemiological analysis and microbial source tracking of Salmonella enterica serovars in a preharvest turkey production environment. Foodborne Pathog. Dis. 5:115-126. [DOI] [PubMed] [Google Scholar]
- 23.Panhotra, B. R., A. K. Saxena, and A. M. Al-Arabi Al-Ghamdi. 2004. Emerging nalidixic acid and ciprofloxacin resistance in non-typhoidal Salmonella isolated from patients having acute diarrhoeal disease. Ann. Saudi Med. 24:270-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Public Health Agency of Canada. 2007. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2005. Public Health Agency of Canada, Guelph, Ontario, Canada. http://www.phac-aspc.gc.ca/cipars-picra/2005-eng.php.
- 25.Public Health Agency of Canada. 2007. Salmonella Heidelberg—ceftiofur-related resistance in human and retail chicken isolates. Public Health Agency of Canada, Guelph, Ontario, Canada. http://www.phac-aspc.gc.ca/cipars-picra/heidelberg/pdf/heidelberg_e.pdf.
- 26.Reference deleted.
- 27.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, S. B. Hunter, B. Swaminathan, and T. J. Barrett. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59-67. [DOI] [PubMed] [Google Scholar]
- 28.Santos, F. B., D. H. Dsouza, L. Jaykus, P. R. Ferket, and B. W. Sheldon. 2007. Genotypes, serotypes, and antibiotic resistance profiles of Salmonella isolated from commercial North Carolina turkey farms. J. Food Prot. 70:1328-1333. [DOI] [PubMed] [Google Scholar]
- 29.USDA. 2008. Serotypes profile of Salmonella isolates from meat and poultry products January 1998 through December 2007. Food Safety Inspection Service, USDA, Washington, DC. http://www.fsis.usda.gov/PDF/Serotypes_Profile_Salmonella_Tables_&_Figures.pdf.
- 30.USDA. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS). USDA, Washington, DC. http://www.ars.usda.gov/Main/docs.htm?docid=6750&page=4.
- 31.Welch, T. J., W. F. Fricke, P. F. McDermott, D. G. White, M. L. Rosso, D. A. Rasko, M. K. Mammel, M. Eppinger, M. J. Rosovitz, D. Wagner, L. Rahalison, J. E. Leclerc, J. M. Hinshaw, L. E. Lindler, T. A. Cebula, E. Carniel, and J. Ravel. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS ONE 2:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilmshurst, P., and H. Sutcliffe. 1995. Splenic abscess due to Salmonella Heidelberg. Clin. Infect. Dis. 21:1065. [DOI] [PubMed] [Google Scholar]
- 33.Zaidi, M. B., V. Leon, C. Canche, C. Perez, S. Zhao, S. K. Hubert, J. Abbott, K. Blickenstaff, and P. F. McDermott. 2007. Rapid and widespread dissemination of multidrug-resistant blaCMY-2 Salmonella Typhimurium in Mexico. J. Antimicrob. Chemother. 60:398-401. [DOI] [PubMed] [Google Scholar]
- 34.Zhao, S., P. F. McDermott, S. Friedman, J. Abbott, S. Ayers, A. Glenn, E. Hall-Robinson, S. K. Hubert, H. Harbottle, R. D. Walker, T. M. Chiller, and D. G. White. 2006. Antimicrobial resistance and genetic relatedness among Salmonella from retail foods of animal origin: NARMS retail meat surveillance. Foodborne Pathog. Dis. 3:106-117. [DOI] [PubMed] [Google Scholar]
- 35.Zhao, S., D. G. White, P. F. McDermott, S. Friedman, L. English, S. Ayers, J. Meng, J. J. Maurer, R. Holland, and R. D. Walker. 2001. Identification and expression of cephamycinase blaCMY genes in Escherichia coli and Salmonella isolates from food animals and ground meat. Antimicrob. Agents Chemother. 45:3647-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]