Abstract
In Hypocrea jecorina, Xyr1 (xylanase regulator 1) is the main transcription activator of hydrolase-encoding genes, such as xyn1, xyn2, bxl1, cbh1, cbh2, egl1, and bgl1. Even though Xyr1 mediates the induction signal for all these genes derived from various inducing carbon sources and compounds, xyr1 transcription itself is not inducible by any of these substances. However, cultivation on glucose as the carbon source provokes carbon catabolite repression of xyr1 transcription mediated by Cre1. In addition, xyr1 transcription is repressed by the specific transcription factor Ace1. Moreover, Xyr1 is permanently available in the cell, and no de novo synthesis of this factor is needed for a first induction of xyn1 transcription. The constitutive expression of xyr1 leads to a significant elevation/deregulation of the xyn1, xyn2, and bxl1 transcription compared to what is seen for the parental strain. Overall, the corresponding xylanolytic enzyme activities are clearly elevated in a constitutively xyr1-expressing strain, emphasizing this factor as an auspicious target for genetically engineered strain improvement.
Hypocrea jecorina (anamorph Trichoderma reesei [22]) is a filamentous ascomycete which abundantly occurs in nature wherever biomass is obtainable. Hydrolases secreted by this fungus are used in a broad range of industrial applications covering, e.g., pulp and paper (4, 32, 53), food and feed (10, 24, 52), and textile (20, 23, 33) industries as well as biofuels and bioenergy (14, 17, 36). The set of cellulolytic and xylanolytic enzymes produced by H. jecorina includes cellobiohydrolases (EC 3.2.1.91; reference 48, e.g.); endo-β-1,4-glucanases (EC 3.2.1.4; reference 34, e.g.); 1,4-β-glucosidases (EC 3.2.1.21; references 9 and 39, e.g.); two major endo-β-1,4-xylanases, XYNI and XYNII (EC 3.2.1.8) (49); and one β-xylosidase, BXLI (EC 3.2.1.37) (16).
In Aspergillus, the xylanolytic and cellulolytic systems are strictly coregulated via the inducer d-xylose (references 11 and 15, e.g.), but the corresponding enzymes in H. jecorina hydrolytic systems are differentially regulated. In particular, genes encoding xylanolytic enzymes are induced by their respective degradation and/or transglycosylation products of xylan, e.g., xyn1 (xylanase I-encoding) by d-xylose (26, 56), xyn2 (xylanase II-encoding) by xylobiose and sophorose (18, 29), and bxl1 (β-xylosidase 1-encoding) by xylobiose (29). In addition, lactose has been identified as a potent inducing molecule (29, 45). Transcriptional activation by Xyr1 (xylanase regulator 1) is common to all these genes (43, 45).
xyr1 encodes a zinc binuclear cluster protein binding to a GGCTAA-like motif arranged as an inverted repeat in the xyn1 and xyn2 promoters (37, 43). The motif closely resembles the consensus sequence for the binding of the Aspergillus niger XlnR transactivator (51). XlnR not only is a central regulator protein responsible for the activation of more than 10 genes involved in the degradation of xylan and cellulose but also contributes to the regulation of d-xylose metabolism (11, 15, 50). The functional domains of XlnR include a N-terminal zinc binuclear cluster domain, a central coiled-coil domain probably involved in nuclear localization, and a C-terminal activation domain (reviewed in reference 44).
Beyond Xyr1/XlnR-mediated induction, the carbon catabolite repressor Cre1/CreA has been described for both organisms as a wide domain repressor of particular hydrolase-encoding genes (references 7, 8, 19, and 26, e.g.). However, in H. jecorina, only one endoxylanase-encoding gene, xyn1, is under direct Cre1 control (19, 26). The other, xyn2, is not directly regulated by Cre1 (57). In addition, the isolation of the two transcription factors Ace1 (activator of cellulases 1) and Ace2 (activator of cellulases 2), potentially involved in the regulation of hydrolase formation in H. jecorina, have been reported (2, 38). Ace1 (1) antagonizes Xyr1 function by competing for one of its binding sites in the xyn1 promoter (37). Ace2 was shown to contact the Xyr1-binding sites in the xyn2 promoter and is needed for basal transcription (54) as well as for the continuous extension of xyn2 expression (46). Ace2 is not involved in the xyn1 transcription (2, 37). It was proposed that these transcription factors are narrow domain regulators, which modulate the general activator Xyr1 (43, 46). Up to now, no mechanisms involving respective orthologous regulators have been identified in the expression of Aspergillus hydrolases.
Little is known about the transcriptional regulation of the xyr1 and xlnR genes themselves. The absence of the H. jecorina Ace2 leads to higher xyr1 transcript levels under inducing conditions (46). Further, Cre1-mediated derepression is involved in the transcriptional regulation of xyr1 on lactose in H. jecorina (45). In Aspergillus nidulans, the monitoring of an xlnR::goxC reporter system indicated that the xlnR promoter is repressed by glucose via CreA (47). Investigations of a Cre1/CreA-mediated carbon catabolite repression (CCR) of xyr1 and xlnR genes at the mRNA level are lacking in industrially important strains of H. jecorina and A. niger.
Herein, we examine aspects of the transcriptional regulation of the H. jecorina xyr1 gene.
MATERIALS AND METHODS
Strains and growth conditions.
H. jecorina (T. reesei) QM9414 (ATCC 26921, a cellulase hyperproducing mutant derived from wild-type strain QM6a [28], termed herein the “parental strain”), Rut-C30 (a Cre1-negative strain; ATCC 56765) (31), and CK11 (a cre1 retransformation strain of Rut-C30) (6) were used throughout this study. An H. jecorina xyr1 deletion strain (Δxyr1 strain) (43) was transformed with the xyr1 structural gene fused to the nag1 promoter of Hypocrea atroviridis (Trichoderma atroviride) (leading to a constitutive expression in the new transformant strain [A. R. Mach-Aigner and R. L. Mach, unpublished data]). All strains were maintained on malt agar.
For replacement experiments, mycelia were precultured in 1-liter Erlenmeyer flasks on a rotary shaker (250 rpm) at 30°C for 18 h in 250 ml of Mandels-Andreotti (MA) medium (27) containing 1% (wt/vol) glycerol as the sole carbon source. Conidia (final concentration, 108 per liter) were used as the inoculum. Pregrown mycelia were washed and thereafter equal amounts were resuspended in MA medium containing 1% (wt/vol) glucose or d-xylose as the sole carbon source. Mycelia were also replaced on MA medium without a carbon source (control) or on medium without a carbon source but supplemented with 1.5 mM xylobiose as an inducer molecule. Incubation was continued for 8 h, and 15-ml samples were taken after 3, 5, and 8 h. In certain replacement experiments on d-xylose, the cultures were supplemented with 5 mM cycloheximide (Sigma, Steinheim, Germany) to inhibit translation (5, 41).
Direct cultivations were performed in 1-liter Erlenmeyer flasks with 200 ml MA medium containing 1% (wt/vol) oat spelt xylan (Sigma) and inoculated with 108 conidia per liter (final concentration). Incubation was performed for 72 h at 30°C and 250 rpm, and 15-ml samples were taken after 0, 12, 24, 36, 48, 60, and 72 h.
Vector construction and fungal transformation.
To insert the nag1::xyr1 gene fusion (see above) into the genome of the H. jecorina xyr1 deletion strain (the Δxyr1 strain), plasmid pARS3 was constructed as follows. The pZEGA3 plasmid (55), obtained from our department stock, was digested by XbaI/NsiI, yielding a 4.5-kb linear fragment containing the H. atroviridis nag1 promoter and the H. jecorina cbh2 terminator. Religation of this fragment using a stuffer (gained from annealing the oligonucleotides XbaIAcc65I and Acc65INotI) (Table 1) led to the vector pARS4 bearing an additional Acc65I and a NotI restriction site. An iCycler (Bio-Rad, Hercules, CA) was used to run 30 cycles of 15 s at 95°C, 15 s at 59°C, and 3 min at 72°C applying a high-fidelity polymerase (Fermentas, St. Leon-Rot, Germany) to introduce an Acc65I and a NotI restriction site flanking the xyr1 structural gene. All primers used are listed in Table 1. The 2.9-kb PCR product was cloned into the pARS4 to create pARS3 bearing the pnag1::xyr1::tcbh2 expression cassette. Retransformation of the Δxyr1 strain was performed according to an optimized protocol described in reference 13 and applying cotransformation of pARS3 and pAN7 (35), bearing the hph gene (encoding hygromycin B phosphotransferase) of Escherichia coli. All vector constructs were verified via DNA sequencing.
TABLE 1.
Primers and probes used throughout the study
| Name | Sequence (5′-3′) | Employmenta |
|---|---|---|
| KpnIxyr1f | ATATAGGTACCATGTTGTCCAATCCTCTCCGTC | Construction of pARS3 |
| NotIxyr1r | TATATGCGGCCGCTTAGAGGGCCAGACCGGTTCC | Construction of pARS3 |
| Acc65INotI | GCGGCCGCGGTACC | Construction of pARS4 |
| XbaIAcc65I | CTAGGGTACCGCGGCCGCTGCA | Construction of pARS4 |
| Taqman xyn1 FAM | CGTCCAACCAACGCCCACAACAA | xyn1 real-time PCR |
| Taqxyn1f | CAGCTATTCGCCTTCCAACAC | xyn1 real-time PCR |
| Taqxyn1Dr | GAGGAGTCCTCCTACGCAGAA | xyn1 real-time PCR |
| Taqxyn1r | CCAAAGTTGATGGGAGCAGAA | xyn1 real-time PCR |
| Taqman xyn2 FAM | CTGCCATCCCTTGCCGCC | xyn2 real-time PCR |
| Taqxyn2Dr | GGTAAGGGTAGGTAGTCTTACTTGTTC | xyn2 real-time PCR |
| Taqxyn2r | CCGAGAAGTTGATGACCTTGTTC | xyn2 real-time PCR |
| Taqxyn2f | GGTCCAACTCGGGCAACTTT | xyn2 real-time PCR |
| Taqman bxl1 FAM | CGACCTCGTTCCCCATGCCCATCCTCACTACG | bxl1 real-time PCR |
| bxl1f | GCCAACTTCGCCACCAAGG | bxl1 real-time PCR |
| bxl1r | CGGCAATCTGGTGGATCAATGTG | bxl1 real-time PCR |
Employment of the oligonucleotides during the study is given.
Southern blot analysis.
Fungal genomic DNA was isolated as described previously (12). Southern hybridization was carried out as described by Sambrook and coworkers (40). Chromosomal DNA of the nag1::xyr1 transformant strains (nx strains) was digested with EcoRV. Hybridization was performed with a 1.5-kb [α-32P]dCTP-labeled SacII fragment of pARS3.
Parallel DNA and RNA extraction, reverse transcription, and real-time PCR analysis.
Harvested mycelia were homogenized in 1 ml peqGOLD TriFast DNA/RNA/protein purification system (PEQLAB Biotechnologie, Erlangen, Germany) using a FastPrep FP120 BIO101 ThermoSavant cell disrupter (Qbiogene, Carlsbad, CA). DNA and RNA were simultaneously isolated in a two-step process according to the manufacturer's instructions.
Synthesis of cDNA from mRNA was carried out applying the RevertAid H minus first-strand cDNA synthesis kit (Fermentas) according to the manufacturer's instructions.
All PCRs were performed in an iCycler iQ real-time detection system (Bio-Rad). The software of the iCycler (iCycler iQ, Optical System Software, version 3.0a; Bio-Rad) was used to compile PCR protocols and define plate setups. All PCR reactions were accomplished in triplicate in 25-μl reaction mixtures including 1× iQ Supermix (Bio-Rad), 0.1 μM corresponding dual-labeled probe (MWG Biotech, Ebersberg, Germany), 0.1 μM forward primer, 0.1 μM reverse primer, and as a template DNA or cDNA (100-fold diluted). Primers and probes are given in Table 1. Each run included a blank (sterile bidistilled water instead of sample) and a no-amplification control (sodium dodecyl sulfate [final concentration, 0.01%] added to the reaction mixture). The respective PCR protocols were followed: 3 min of initial denaturation at 95°C, followed by 45 cycles of 15 s at 95°C, 15 s at 60°C (for real-time PCR of xyr1) or 59°C (for real-time PCR of xyn1, xyn2, and bxl1), and 15 s at 72°C. The threshold level was set automatically to noise-to-signal ratio conditions by the Optical System Software. Calculation of amounts of mRNA (cDNA) per gene dose was performed as previously described (43). Results of transcription analysis are given in relative amounts of mRNA (cDNA) per gene dose. These amounts are always related to one reference sample within a certain experiment; consequently, the comparison of ratios is valid only within one experimental setup.
Enzyme assays.
Endo-β-1,4-xylanase activity was measured applying Xylazyme AX tablets (Megazyme, Wicklow, Ireland) according to the manufacturer's instructions. One unit of activity is defined as the amount of enzyme required to release 1 micromole of reducing xylose sugar equivalents per minute under the defined assay conditions (40°C, 10 min).
β-Xylosidase activity was measured by using 4-nitrophenyl-β-d-xylopyranoside as a substrate dissolved in a 50 mM sodium citrate buffer, pH 5.0. The enzyme assay was performed at 50°C as previously described (21).
Determination of fungal growth on xylan.
Harvested mycelia were suspended in 1 ml of 0.1 N NaOH in a reaction tube with screw cap (2 ml; Brand GmbH+Co KG, Wertheim, Germany). Glass beads (a mixture of 0.37-g beads [diameter, 0.1 to 0.01 mm], 0.25-g beads [diameter, 1 mm], and one glass bead [diameter, 3 mm]) (Braun Biotech International GmbH, Melsungen, Germany) were added. This suspension was solubilized in a Teflon homogenizer (FastPrep 120; BIO 101 Savant Instruments, Hobrook, NY). After incubation at room temperature for 3 hours, the samples were centrifuged at 14,000 rpm and 4°C for 10 min. Next, protein concentration was determined via Quick Start Bradford protein assay according to the manufacturer's guidelines (Bio-Rad).
Statistical analyses of real-time PCR data.
Data were analyzed by applying the Friedman test (nonparametric analyses of the variance of related samples); two consecutive tests were performed either including or excluding the most differing parameter group (e.g., replacement to glucose-containing medium). Because of the consecutive dual test approach, the level of significance was corrected according to the Bonferroni method.
RESULTS
Transcription of xyr1 is not activated via a specific inducer molecule.
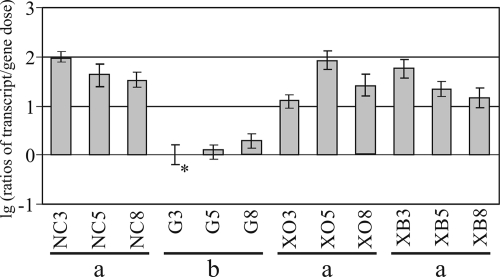
In H. jecorina, the expression hydrolase-encoding genes such as xyn1, xyn2, bxl1, cbh1 (cellobiohydrolase 1), cbh2 (cellobiohydrolase 2), egl1 (endoglucanase 1), and bgl1 (β-glucosidase 1) is regulated by the general activator Xyr1, independent of the carbon source or inducer (43, 45). Nevertheless, different expression/induction patterns for these genes have been observed in earlier studies (29, 56). Consequently, we examined if the transcription of this regulatory protein itself responds to certain induction signals or differs on various carbon sources. After precultivation, the H. jecorina parental strain was transferred to medium without a carbon source or to medium containing 1% (wt/vol) glucose or d-xylose or 1.5 mM xylobiose and incubated for 3, 5, or 8 h. After parallel DNA and RNA extraction followed by cDNA synthesis, the transcript levels were analyzed via real-time PCR. Statistical analyses revealed that similar levels of xyr1 transcript formation were observed for the xyn1-inducing carbon source d-xylose and for the xyn2-inducing substance xylobiose. These transcript levels never exceeded those observed on medium without a carbon source. Significantly less transcript accumulated on medium containing glucose (Fig. 1). Changes of xyr1 transcript levels after transfer to d-xylose or xylobiose are probably due to differences in the experimental setup, as d-xylose is applied as the carbon source, whereas xylobiose is applied as an inducer molecule. These data strongly indicate the influence of a glucose repression but not of an induction mechanism on xyr1 transcription. Consequently, the regulation of xyr1 transcription in a carbon catabolite-dependent manner was examined next.
FIG. 1.
Transcription analysis of carbon source-dependent regulation of xyr1 transcription. Strain QM9414 was precultured on glycerol and thereafter transferred to MA medium containing 1% (wt/vol) glucose (G) or xylose (XO) as the sole carbon source, that without a carbon source (NC), or that supplemented with 1.5 mM xylobiose (XB) and incubated for 3, 5, or 8 h as indicated. Transcriptional analysis of xyr1 was performed via real-time PCR applying a dually labeled probe after parallel extraction of RNA and DNA followed by cDNA synthesis. Ratios of amounts of mRNA (cDNA) per gene dose are given in a decade logarithmic scale (lg). The values are means of results from five independent biological replicates. Error bars indicate standard deviations. The asterisk indicates the reference sample. Groups a and b represent samples exhibiting differences with (a to b) or without (a to a) statistical significance.
Transcription of xyr1 is regulated via Cre1-mediated CCR.
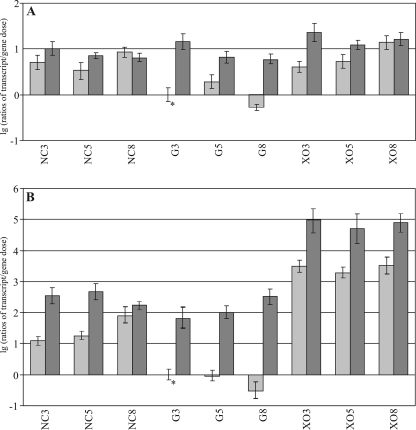
To investigate whether the reduced transcript level of xyr1 on glucose is due to Cre1-mediated CCR, we cultured a Cre1-negative strain (Rut-C30). To prove that the observed effect is due to a lack of Cre1 formation, a corresponding cre1 retransformation strain of Rut-C30 (CK11) was also cultivated. Equal amounts of mycelia of both strains pregrown on glycerol were transferred into media supplemented with 1% (wt/vol) glucose or d-xylose or without any carbon source. A transcript analysis of xyr1 in Rut-C30 revealed similar levels for all conditions tested (Fig. 2A), indicating a general release from Cre1-dependent CCR (i.e., derepression). Because xyn1 is known as a gene being subject to CCR by Cre1 (26), we examined xyn1 transcription in parallel as a reference. In strict accordance with the observed xyr1 transcript levels, xyn1 transcription on glucose is as high as on medium without a carbon source, if cultivation is in a Cre1-negative background (Fig. 2B). But in contrast to xyr1, xyn1 transcription is inducible by d-xylose, which does coincide with former observations (26, 37, 43) (Fig. 2A and B). An analogous analysis of the xyr1 and xyn1 transcript formation in the cre1 retransformation strain CK11 showed a restoration of a glucose-dependent repression (Fig. 2A and B). This was also observed for strain QM9414, thus confirming that the reduced transcript levels on glucose can be ascribed to a Cre1-mediated CCR. Furthermore, we observed a significantly reduced derepression (transferred to the absence of a carbon source) and induction (transferred to xylose as the carbon source) of xyn1 transcription in CK11 compared to what was seen for Rut-C30 (Fig. 2B). A possible explanation for these findings is that the restoration of cre1 expression in CK11 probably acts as a negative regulator even under derepressing and inducing conditions. As a summary, we conclude that the transcriptional regulation of xyr1 is subject to a Cre1-dependent CCR/derepression but not to an induction mechanism.
FIG. 2.
Transcription analysis of carbon source-dependent regulation of xyr1 transcription. Strain Rut-C30 (dark gray) and strain CK11 (light gray) were precultured on glycerol and thereafter transferred to MA medium containing 1% (wt/vol) glucose (G) or xylose (XO) as the sole carbon source or that without a carbon source (NC) and incubated for 3, 5, or 8 h as indicated. Transcriptional analysis of xyr1 (A) and xyn1 (B) was performed via real-time PCR applying a dual-labeled probe after parallel extraction of RNA and DNA, followed by cDNA synthesis. Ratios of amounts of mRNA (cDNA) per gene dose are given in a decade logarithmic scale (lg). The values are means of results from seven independent biological replicates. Error bars indicate standard deviations. The asterisk indicates the reference sample.
Ace1 antagonizes xyr1 transcription.
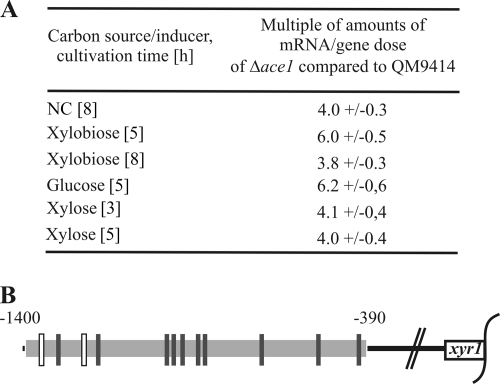
Since the influence of Cre1 could be demonstrated in this study and an elevated level of xyr1 transcript in an ace2 deletion strain was reported previously we consequently investigated if the repressing transcription factor Ace1 also negatively affects transcription of xyr1. Therefore, the QM9414 parental strain and an ace1 deletion strain (the Δace1 strain) were pregrown and thereafter replaced to MA medium containing glucose or d-xylose as a carbon source or xylobiose as an inducing substance or else with no carbon source as a control. Interestingly, xyr1 transcription levels were 3.8 to 6.4 times elevated in the Δace1 strain (Fig. 3A). Accordingly, we performed an in silico analysis of the xyr1 promoter region (−1 to −1400 bp) for potential regulatory binding motives. We identified 2 Ace1-binding sites (AGGCA [38]) and 10 Cre1-binding sites (SYGGRG [42]) (4 of them arranged as a repeat) in a region 1,400 bp upstream of the xyr1 structural gene (Fig. 3B). Although there is no biochemical evidence confirming the binding of any of these sites, we speculate that Ace1 acts, next to Cre1, as a repressor of xyr1 transcription itself.
FIG. 3.
(A) Transcript analysis of the xyr1 gene in the QM9414 parental strain and the ace1 deletion strain (Δace1 strain). Both strains were precultured in MA medium containing glycerol and thereafter transferred to MA medium containing 1% (wt/vol) of a carbon source or 1.5 mM xylobiose as the inducer and incubated for 3, 5, or 8 h as indicated. Time points chosen for this experiment correspond to the strongest regulatory effects observed for strain QM9414. Results are given in multiple amounts of mRNA per gene dose of Δace1 strain compared to QM9414. The values are means of results from three independent biological replicates with standard deviations indicated. Note that values gained from different inducers/carbon sources are not comparable. (B) In silico analysis of the xyr1 promoter (−1 to −1400 bp upstream the structural gene) for identification of binding sites of the transcription regulators Cre1 (SYGGRG [black vertical bars]) and Ace1 (AGGCA [white vertical bars]).
De novo synthesis of Xyr1 is not essential for an initial induction of xyn1.
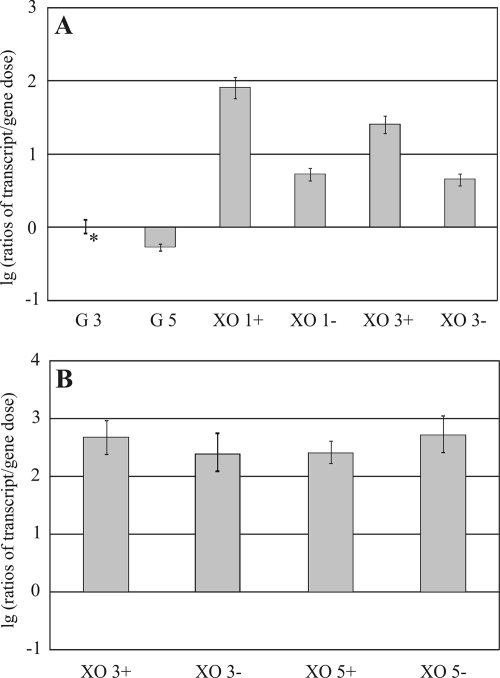
On one hand, xyr1 transcription is subject to CCR mediated via Cre1; on the other hand, it was recently demonstrated that Xyr1 even under repressing conditions (glucose-grown cultures) binds at least to the upstream part of its corresponding regulatory element (GGCTAA motif) in the xyn1 promoter (37). Together, these observations appear contradictory and underscore the question of the role of Xyr1 under repressing conditions. Furthermore, we questioned whether the de novo synthesis of Xyr1 is necessary for an initial induction of xyn1 transcription. To answer this, the parental strain was replaced either to d-xylose either with or without a translation inhibitor (5 mM cycloheximide, previously proven as an effective translation inhibitor in filamentous fungi [references 5 and 41, e.g.]) or else to glucose. First, we found again that the transcription of the xyr1 gene is strongly reduced on glucose compared with what was seen for d-xylose. In all cases, transcription affected the translation inhibitor. However, mRNA (cDNA) isolated from d-xylose-grown cultures with translation inhibitor never dropped below the levels observed for glucose without inhibitor (Fig. 4A). One interpretation of these observations is the release from Cre1-mediated CCR on d-xylose on the one hand; on the other hand, the absence of de novo-synthesized Cre1 may cause elevated transcript levels of xyr1 when the translation inhibitor is added. These findings suggest that protein synthesis is not needed for a low constitutive xyr1 transcript level. It is firmly established that the induction of xyn1 expression is strictly dependent on Xyr1 (43). Therefore, xyn1 expression is an ideal model to investigate whether de novo synthesis of Xyr1 is essential for an initial induction of xyn1 transcription. As can be inferred from Fig. 4B, the xyn1 transcript levels are not significantly altered, independent of cycloheximide supplementation. Thus, even though the addition of a translation inhibitor prevents de novo synthesis of Xyr1, xyn1 transcription, which is strictly dependent on Xyr1, is induced. We assume that a low constitutive level of Xyr1 is present under all growth conditions.
FIG. 4.
Influence of a translation inhibitor on transcription of xyr1 and xyn1 in H. jecorina. Strain QM9414 was precultured on glycerol and thereafter transferred to MA medium containing 1% (wt/vol) glucose (G) or xylose (XO) as the sole carbon source supplemented with (+) or without (−) 5 mM cycloheximide. Sampling and transcript analysis were performed after 1 and 3 h for xyr1 and after 3 and 5 h for xyn1. This time-staggered sampling should allow analysis of the influence of de novo-synthesized Xyr1 on xyn1 transcription. Transcriptional analysis of xyr1 (A) and xyn1 (B) was performed via real-time PCR applying a dually labeled probe after parallel extraction of RNA and DNA, followed by cDNA synthesis. Ratios of amounts of mRNA (cDNA) per gene dose are given in a decade logarithmic scale (lg). The results are means of results from three independent biological replicates. Error bars indicate standard deviations. The asterisk indicates the reference sample.
Insertion of a constitutively expressed copy of xyr1 into H. jecorina.
To create a strain with a constitutively expressed xyr1 (GenBank accession no. AF479644), the respective structural gene was put under the control of the nag1 promoter of H. atroviridis. Transformation of this nag1::xyr1 fusion into the genome of an H. jecorina Δxyr1 strain (43) was carried out via cotransformation using the plasmid pARS3, bearing the fusion and the vector pAN7 (35) and conferring hygromycin B resistance. This yielded approximately 15 mitotically stable transformants, of which 10 were further characterized (nx strains). Southern blot analysis in all cases revealed the insertion of the nag1::xyr1 construct as homologous single-copy integration at the xyr1 locus, replacing the former amdS-based deletion cassette (data not shown). Prescreening of the transformants in liquid culture with xylan as the sole carbon source showed similar growth rates and xylanase activities for all of them (data not shown). Transcript analysis of three randomly chosen strains (nx 7, nx 10, and nx 11) via real-time PCR indicated the restoration of xyr1 transcript formation for all of them (see Fig. S1 in the supplemental material). The nx transformant strains showed growth rates on malt extract medium that were similar to those of the QM9414 parental strain or the Δxyr1 strain (see Fig. S1 in the supplemental material). However, the conidiospores are white to brownish instead of dark green and conidiation is slightly delayed in the nx strains compared with the parental strain (see Fig. S1 in the supplemental material). One of the transformant strains (nx7 strain) was randomly chosen to perform further analyses.
Constitutive expression of xyr1 causes elevated xylanase formation.
To investigate effects of constitutively expressed, deregulated (CCR-independent) xyr1 expression, we precultivated the parental strain and the nx7 transformant strain on glycerol and thereafter transferred them into media containing glucose, d-xylose, xylobiose, or no carbon source. Real-time reverse transcription-PCR analysis of xyr1 revealed similar transcript levels under all conditions tested for the nx7 strain, contrasting with the glucose repression of this gene seen for the parental strain (Table 2). It can be speculated that release from CCR in the nx7 transformant strain is due to the fact that the nag1 promoter is not regulated by Cre1 (25). Whereas the xyr1 gene is constitutively expressed in the mutant strain, no transcript formation of the xyn1 gene was observed under glucose-repressing conditions for QM9414. These observations are consistent with the known regulatory mechanisms of the parental strain (26, 37, 43) (Table 2). However, xyn1 transcript accumulation under d-xylose inducing conditions dramatically increased in the nx7 transformant strain (Table 2). The basal level of xyn2 transcription, described previously for the parental strain (43, 54), could also be detected in the transformant strain (Table 2). In contrast to the transcription of xyn1, the induction of the xyn2 gene via xylobiose was weaker at early time points in the nx7 transformant strain but later exceeded the levels of the parental strain (Table 2). To complete the analysis of all genes coding for xylan backbone-degrading enzymes in H. jecorina, the bxl1 transcript formation was examined. Again, higher levels of transcript were observed for the transformant strain (Table 2).
TABLE 2.
Transcription levels of the xyr1, xyn1, xyn2, and bxl1 genes
| Gene | Carbon source/inducer (h of cultivation) | Amt of mRNA (cDNA) per gene dose fora:
|
|
|---|---|---|---|
| QM9414 | nx7 | ||
| xyr1 | NC (3) | 22.9 ± 2 | 18.1 ± 2 |
| Xylobiose (3) | 12.7 ± 1 | 18.3 ± 2 | |
| Glucose (3) | 1.0 ± 0 | 32.4 ± 3 | |
| Xylose (3) | 11.6 ± 1 | 27.4 ± 2 | |
| xyn1 | Glucose (3) | ND | ND |
| Glucose (5) | ND | ND | |
| Glucose (8) | ND | ND | |
| Xylose (3) | ND | 7,652 ± 399 | |
| Xylose (5) | 1.0 ± 0.0 | 388 ± 29 | |
| Xylose (8) | 12 ± 2 | 416 ± 21 | |
| xyn2 | NC (3) | 157 ± 10 | 81 ± 3 |
| NC (5) | 18 ± 1 | 9 ± 0 | |
| NC (8) | 9 ± 0 | 1 ± 0 | |
| Xylobiose (3) | 20,613 ± 978 | 5,934 ± 289 | |
| Xylobiose (5) | 54,104 ± 2,536 | 1,897 ± 91 | |
| Xylobiose (8) | 1,072 ± 59 | 3,906 ± 175 | |
| bxl1 | NC (3) | 1.0 ± 0.0 | 8.9 ± 0.5 |
| NC (5) | 0.3 ± 0.0 | 0.2 ± 0.0 | |
| NC (8) | 0.3 ± 0.0 | 1.2 ± 0.0 | |
| Xylobiose (3) | 7,179 ± 315 | 11,474 ± 491 | |
| Xylobiose (5) | 5,056 ± 242 | 5,331 ± 258 | |
| Xylobiose (8) | 460 ± 24 | 10,450 ± 507 | |
Values are means ± standard deviations from three independent experiments. ND, no detection.
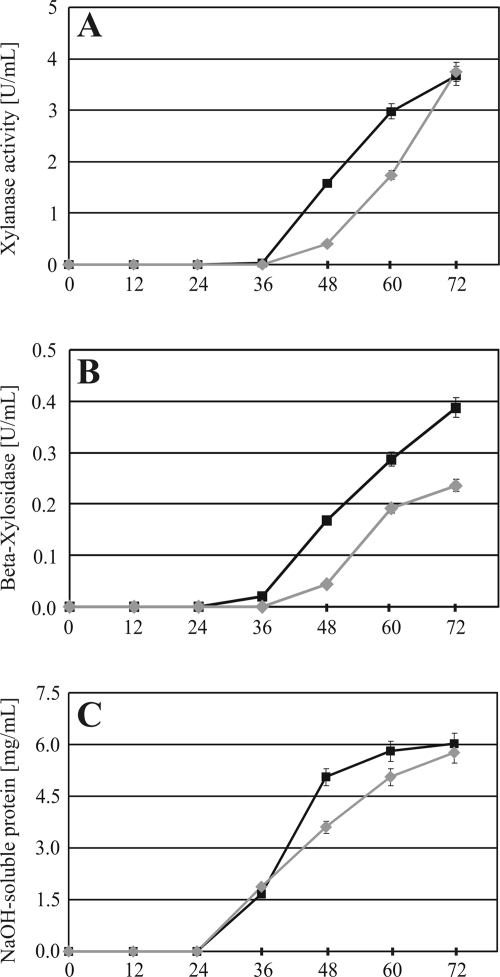
To evaluate the impact of the increased mRNA levels on xylanolytic enzyme activity, the parental strain and the nx7 transformant strain were cultivated on medium containing xylan. We observed endo-β-1,4-xylanase activity at an earlier culture stage (Fig. 5A) and β-xylosidase activity was clearly elevated (Fig. 5B) in the nx7 transformant strain, thus confirming the results of the transcript analysis. Soluble protein measurements showed that growth characteristics slightly differed in the strains tested (Fig. 5C), most likely due to the delayed xylanase production in the parental strain. Complete clearing (i.e., enzymatic degradation of the insoluble xylan compounds) of the xylan-containing medium by the nx7 transformant strain was visible after approximately 50 h of cultivation, whereas the parental strain could not clear the medium during the duration of the experiment. To summarize, these findings strongly support the above-suggested postulate that Xyr1 expression alone is not sufficient for xylanase induction and that posttranslational modifications are needed to convert this transcription factor into its active form. However, a deregulated expression of xyr1 (i.e., release of xyr1 from CCR) strongly elevates the production of the xylanolytic enzyme activity of H. jecorina.
FIG. 5.
Analysis of xylanase enzyme activity formation. Endo-β-1,4-xylanases (A) and β-xylosidase (B) activities in culture supernatants of the QM9414 parental strain (gray diamonds) and the nx7 mutant strain (black squares) (A to C). Detection of enzyme activity formation of both strains was accomplished after 0, 12, 24, 36, 48, 60, and 72 h of cultivation in shake flasks on 1% (wt/vol) xylan after inoculation with spores. One unit of activity is defined as the amount of enzyme required to release 1 micromole of reducing xylose sugar equivalents or 4-nitrophenyl residues per minute under the defined assay conditions. (C) Determination of biomass formation in medium containing insoluble xylan by measurement of NaOH-soluble protein. All data are means of results from three independent biological replicates. Error bars indicate standard deviations.
DISCUSSION
In recent years, the isolation of a main hydrolase regulatory protein (Xyr1/XlnR) has been reported for several fungal species, including those of Aspergillus (XlnR [51]), Hypocrea (Xyr1 [43]), and Fusarium (Xyr1 [3]). Although its central role in the transcriptional activation of cellulolytic as well as xylanolytic enzyme-encoding genes has been demonstrated, investigations on the regulation of xyr1 expression itself remained incomplete.
We demonstrate here that in contrast to its regulation targets (e.g., xyn1, xyn2, bxl1, cbh1, cbh2, egl1, and bgl1), the regulation of xyr1 expression itself is subject only to a repression/derepression mechanism. These findings are in strict accordance with the in silico analysis of the region upstream of xyr1, where no Xyr1-binding sites (with the potential for autoregulation) could be identified. On the other hand, the presence of 10 Cre1 sites, 4 of them arranged as tandem repeats, gave a first indication for a Cre1-dependent regulation of the transcription of xyr1. In Hypocrea, Cre1 sites are functional in vivo only if they are arranged as either inverted (xyn1 [26]) or tandem (cbh1 [19]) repeats. Performing transcription studies of the xyr1 gene in a Cre1-negative background provided an in vivo proof of the CCR of xyr1 transcription, finally supported by the fact that a complementation of the Cre1-defective strain led to a restoration of the glucose repression of xyr1. Upon the transfer of these two strains to an xyn1-inducing carbon source (i.e., d-xylose), no increase of xyr1 transcription above the derepressed level was observed. These findings again support the model of a repression/derepression regulatory mechanism. It is noteworthy that a Cre1-dependent regulation of xyr1 transcription can now also explain the Cre1-related expression of hydrolytic enzyme-encoding genes in Hypocrea, which do not bear functional Cre1 binding sites in their own promoters (e.g., cbh2 [57] and xyn2 [56]). At first glance, these findings point to a simple model of Xyr1 de novo synthesis-mediated gene regulation. However, two previous findings contradict such a simple regulatory explanation: (i) in vivo footprinting analyses of the xyn1 and xyn2 promoters revealed binding of Xyr1 even under repressing conditions (37, 46); and (ii) the constitutive level of xyn2 transcription, also present under CCR conditions, is strictly dependent on Xyr1 (46). These results are a clear indication of the presence of Xyr1 also under repressing conditions. Xyr1 is present either at a concentration that is inappropriate to confer induction (still consistent with a de novo synthesis model) or in an inactive form, most probably lacking an activating posttranslational modification, or else in a combination of both. Consequently, following this working hypothesis, we investigated the necessity of a de novo synthesis of Xyr1 for an initial induction of xylanase formation. Using cycloheximide, a translation inhibitor (5, 41), we demonstrated that the induction of xyn1 transcription occurs independent of protein synthesis. This observation is in good agreement with the above-mentioned binding of Xyr1 to its respective elements even under repressing conditions. As even low levels of Xyr1 are sufficient to mediate induction, the hypothesis of a posttranslational modification has to be favored and will be a focus of future investigations.
Having these results in hand, we questioned if the amount of disposable Xyr1 influences the expression of the hydrolase-encoding genes in Hypocrea. To study this, the xyr1 structural gene was placed under the control of the H. atroviridis promoter of nag1, which is constitutively expressed in H. jecorina. The entire gene locus of xyr1 was replaced by a single copy of this construct. A comparison of the transcription of xyr1 in this strain (nx7) with the parental strain under derepressing as well as inducing conditions clearly demonstrated that levels of transcript formation are similar in both strains. Under CCR conditions, the xyr1 mRNA is detectable for the nx7 strain only at levels comparable to those seen for inducing conditions. However, the expression of xyr1 during CCR does not lead to any xylanase-encoding transcript formation on glucose, which is again perfectly consistent with the above-suggested posttranslational activation of Xyr1. Investigating the transcription patterns of the three major xylanase-encoding genes (xyn1, xyn2, and bxl1) revealed a strong influence of the deregulated/constitutive expression of xyr1 under inducing conditions. The transcription of xyn1 (up to several hundredfold) and to a lesser extent of bxl1 is highly increased during all time points tested. xyn2 mRNA formation in the nx7 strain lacks the characteristic induction peak following an addition of the inducer molecule observed for the parental strain. However, as for the other xylanase-encoding genes, no downregulation of the transcription of xyn2 can be seen for the nx7 strain, whereas it can be observed in the parental strain. A possible explanation for this different change in the transcription profile of the xyn2 gene may be the fact that, in addition to Xyr1, the transcription factor Ace2, which is not overexpressed in the nx7 strain, is required for a full induction (46). Accordingly, Ace2 is not involved in the transcriptional regulation of the other xylanolytic enzyme-encoding genes of H. jecorina (46). It should be noted that the measurements of corresponding enzyme activities are perfectly congruent with the transcription analyses. These findings are consistent with recently published data for an A. nidulans strain expressing xlnR under the A. nidulans gpdA promoter, which showed higher transcript levels of xlnA and xlnB and partly higher levels of xlnD in a Northern blot analysis. However, also in this case, all xylanase-encoding genes require d-xylose for the induction of their transcription (47).
Ace1 was previously described as a repressing transcription factor of hydrolases in Hypocrea (1) and, more recently, it has been demonstrated that Ace1 directly competes with Xyr1 for one of its binding elements in the xyn1 promoter (37). In this study, we provide evidence that Ace1 also negatively acts on the transcriptional regulation of xyr1, which is consistent with the presence of two Ace1-binding sites in the respective upstream regulatory sequence. Thus, in addition to the above-described Cre1-dependent CCR of xyr1 and xyn1 transcription, a second negative regulatory effect mediated by Ace1 acts on this regulation cascade. Such a regulatory mechanism can be understood as a “double-double” lock system (named accordingly to the double-lock regulation of the alcA gene in Aspergillus nidulans, described as a CreA-mediated double repression of alcR and alcA [30]) of the xyn1 promoter. Such a tight “shut-off-and-on” mechanism of this particular promoter is in agreement with transcription analyses allowing no detection of transcript formation (neither of the xyn1 gene nor of heterologously expressed genes [26]) when the fungus is grown on glucose as the sole carbon source. In addition, the xyn1 gene is highly inducible via the cheap carbon source d-xylose, even in the presence of glucose (26). This highlights the xyn1 promoter as a precisely regulatable expression system for Hypocrea and other fungal species (e.g., Fusarium species [K. Brunner, R. Mitterbauer, G. Adam, and R. L. Mach, unpublished data]).
To summarize, Xyr1 is the major regulator of cellulolytic and xylanolytic enzyme formation in H. jecorina, which is itself regulated on a transcriptional level by a repression/derepression mechanism. Since these studies are done with the cellulase-hyperproducing strains QM9414 and Rut-C30, the mechanisms/phenomena cannot be assumed to be operative in wild-type QM6a. Nevertheless, in QM9414 deregulating the repression of xyr1 expression already leads to a significant increase of hydrolase production, which is of course still dependent on the presence of the respective inducer molecule. The identification of a possible activating posttranslational modification of Xyr1 will probably have the potential to completely deregulate hydrolase production. Accordingly, this should work out independent of the presence of an inducer when xyr1 is expressed under a constitutive promoter. This working hypothesis demands further investigations as it is hopefully a major key for strain improvement via genetic engineering, not only for Hypocrea, but also for other hydrolase-producing fungi.
Supplementary Material
Acknowledgments
This study was supported by a grant from the Austrian Fonds zur Förderung Wissenschaftlicher Forschung (P 20192-B03) and by a grant from the Vienna University of Technology, DemoTech-Innovative Project, both given to R.L.M., which are gratefully acknowledged.
Footnotes
Published ahead of print on 12 September 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aro, N., M. Ilmén, A. Saloheimo, and M. Penttilä. 2003. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl. Environ. Microbiol. 69:56-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aro, N., A. Saloheimo, M. Ilmen, and M. Penttila. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276:24309-24314. [DOI] [PubMed] [Google Scholar]
- 3.Brunner, K., A. M. Lichtenauer, K. Kratochwill, M. Delic, and R. L. Mach. 2007. Xyr1 regulates xylanase but not cellulase formation in the head blight fungus Fusarium graminearum. Curr. Genet. 52:213-220. [DOI] [PubMed] [Google Scholar]
- 4.Buchert, J., T. Oksanen, J. Pere, M. Siika-aho, A. Suurnäkki, and L. Viikari. 1998. Applications of Trichoderma reesei enzymes in the pulp and paper industry, p. 343-357. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma and Gliocladium, vol. 2. Taylor & Francis, Ltd., London, United Kingdom. [Google Scholar]
- 5.Cybis, J., and P. Weglenski. 1972. Arginase induction in Aspergillus nidulans. The appearance and decay of the coding capacity of messenger. Eur. J. Biochem. 30:262-268. [DOI] [PubMed] [Google Scholar]
- 6.Cziferszky, A., R. L. Mach, and C. P. Kubicek. 2002. Phosphorylation positively regulates DNA binding of the carbon catabolite repressor Cre1 of Hypocrea jecorina (Trichoderma reesei). J. Biol. Chem. 277:14688-14694. [DOI] [PubMed] [Google Scholar]
- 7.de Graaff, L. H., H. C. van den Broeck, A. J. van Ooijen, and J. Visser. 1994. Regulation of the xylanase-encoding xlnA gene of Aspergillus tubigensis. Mol. Microbiol. 12:479-490. [DOI] [PubMed] [Google Scholar]
- 8.de Vries, R. P., J. Visser, and L. H. de Graaff. 1999. CreA modulates the XlnR-induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res. Microbiol. 150:281-285. [DOI] [PubMed] [Google Scholar]
- 9.Fowler, T., and R. D. Brown, Jr. 1992. The bgl1 gene encoding extracellular beta-glucosidase from Trichoderma reesei is required for rapid induction of the cellulase complex. Mol. Microbiol. 6:3225-3235. [DOI] [PubMed] [Google Scholar]
- 10.Galante, Y. M., R. Monteverdi, S. Inama, C. Caldini, A. De Conti, V. Lavelli, and F. Bonomi. 1993. New applications of enzymes in wine making and olive oil production. Ital. Biochem. Soc. Trans. 4:34. [Google Scholar]
- 11.Gielkens, M. M., E. Dekkers, J. Visser, and L. H. de Graaff. 1999. Two cellobiohydrolase-encoding genes from Aspergillus niger require d-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl. Environ. Microbiol. 65:4340-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruber, F., J. Visser, C. P. Kubicek, and L. H. De Graaff. 1990. Cloning of the Trichoderma reesei pyrG gene and its use as a homologous marker for a high-frequency transformation system. Curr. Genet. 18:447-451. [DOI] [PubMed] [Google Scholar]
- 13.Gruber, F., J. Visser, C. P. Kubicek, and L. H. de Graaff. 1990. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 18:71-76. [DOI] [PubMed] [Google Scholar]
- 14.Hahn-Hägerdal, B., M. Galbe, M. F. Gorwa-Grauslund, G. Liden, and G. Zacchi. 2006. Bio-ethanol—the fuel of tomorrow from the residues of today. Trends Biotechnol. 24:549-556. [DOI] [PubMed] [Google Scholar]
- 15.Hasper, A. A., J. Visser, and L. H. de Graaff. 2000. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates d-xylose reductase gene expression. Mol. Microbiol. 36:193-200. [DOI] [PubMed] [Google Scholar]
- 16.Herrmann, M. C., M. Vrsanska, M. Jurickova, J. Hirsch, P. Biely, and C. P. Kubicek. 1997. The beta-d-xylosidase of Trichoderma reesei is a multifunctional beta-d-xylan xylohydrolase. Biochem. J. 321:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Himmel, M. E., S. Y. Ding, D. K. Johnson, W. S. Adney, M. R. Nimlos, J. W. Brady, and T. D. Foust. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804-807. [DOI] [PubMed] [Google Scholar]
- 18.Hrmova, M., P. Biely, and M. Vrsanska. 1986. Specificity of cellulase and β-xylanase induction in Trichoderma reesei QM 9414. Arch. Microbiol. 144:307-311. [Google Scholar]
- 19.Ilmén, M., C. Thrane, and M. Penttilä. 1996. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol. Gen. Genet. 251:451-460. [DOI] [PubMed] [Google Scholar]
- 20.Koo, H., M. Ueda, T. Wakida, Y. Yoshimura, and T. Igarashi. 1994. Cellulase treatment of cotton fabrics. Text. Res. J. 64:70-74. [Google Scholar]
- 21.Kristufek, D., S. Zeilinger, and C. P. Kubicek. 1995. Regulation of β-xylosidase formation by xylose in Trichoderma reesei. Appl. Microbiol. Biotechnol. 42:713-717. [Google Scholar]
- 22.Kuhls, K., E. Lieckfeldt, G. J. Samuels, W. Kovacs, W. Meyer, O. Petrini, W. Gams, T. Borner, and C. P. Kubicek. 1996. Molecular evidence that the asexual industrial fungus Trichoderma reesei is a clonal derivative of the ascomycete Hypocrea jecorina. Proc. Natl. Acad. Sci. USA 93:7755-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, A., M. Lepola, and C. Purtell. 1994. Enzyme finishing of man-made cellulosic fabrics. Text. Chem. Color. 26:25-28. [Google Scholar]
- 24.Lanzarini, G., and P. G. Pifferi. 1989. Enzymes in the fruit juice industry, p. 189-222. In C. Cantarelli and G. Lanzarini (ed.), Biotechnology applications in beverage production. Elsevier Science, London, United Kingdom.
- 25.Mach, R. L., C. K. Peterbauer, K. Payer, S. Jaksits, S. L. Woo, S. Zeilinger, C. M. Kullnig, M. Lorito, and C. P. Kubicek. 1999. Expression of two major chitinase genes of Trichoderma atroviride (T. harzianum P1) is triggered by different regulatory signals. Appl. Environ. Microbiol. 65:1858-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mach, R. L., J. Strauss, S. Zeilinger, M. Schindler, and C. P. Kubicek. 1996. Carbon catabolite repression of xylanase I (xyn1) gene expression in Trichoderma reesei. Mol. Microbiol. 21:1273-1281. [DOI] [PubMed] [Google Scholar]
- 27.Mandels, M. 1985. Applications of cellulases. Biochem. Soc. Trans. 13:414-416. [DOI] [PubMed] [Google Scholar]
- 28.Mäntylä, A. L., K. H. Rossi, S. A. Vanhanen, M. E. Penttilä, P. L. Suominen, and K. M. Nevalainen. 1992. Electrophoretic karyotyping of wild-type and mutant Trichoderma longibrachiatum (reesei) strains. Curr. Genet. 21:471-477. [DOI] [PubMed] [Google Scholar]
- 29.Margolles-Clark, E., M. Ilmén, and M. Penttilä. 1997. Expression patterns of ten hemicellulase genes of the filamentous fungus Trichoderma reesei on various carbon sources. J. Biotechnol. 57:167-179. [Google Scholar]
- 30.Mathieu, M., and B. Felenbok. 1994. The Aspergillus nidulans CREA protein mediates glucose repression of the ethanol regulon at various levels through competition with the ALCR-specific transactivator. EMBO J. 13:4022-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montenecourt, B. S., and D. E. Eveleigh. 1979. Production and characterization of high yielding cellulase mutants of Trichoderma reesei. TAPPI J. 28:101-108. [Google Scholar]
- 32.Noé, P., J. Chevalier, F. Mora, and J. Comtat. 1986. Action of xylanases on chemical pulp fibers. Part II. Enzymatic beating. J. Wood Chem. Technol. 6:167-184. [Google Scholar]
- 33.Pedersen, G. P., G. A. Screws, and D. A. Cereoni. 1992. Biopolishing of cellulosic fabrics. Can. Text. J. December:31-35. [Google Scholar]
- 34.Penttila, M., P. Lehtovaara, H. Nevalainen, R. Bhikhabhai, and J. Knowles. 1986. Homology between cellulase genes of Trichoderma reesei: complete nucleotide sequence of the endoglucanase I gene. Gene 45:253-263. [DOI] [PubMed] [Google Scholar]
- 35.Punt, P. J., R. P. Oliver, M. A. Dingemanse, P. H. Pouwels, and C. A. van den Hondel. 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56:117-124. [DOI] [PubMed] [Google Scholar]
- 36.Ragauskas, A. J., C. K. Williams, B. H. Davison, G. Britovsek, J. Cairney, C. A. Eckert, W. J. Frederick, Jr., J. P. Hallett, D. J. Leak, C. L. Liotta, J. R. Mielenz, R. Murphy, R. Templer, and T. Tschaplinski. 2006. The path forward for biofuels and biomaterials. Science 311:484-489. [DOI] [PubMed] [Google Scholar]
- 37.Rauscher, R., E. Würleitner, C. Wacenovsky, N. Aro, A. R. Stricker, S. Zeilinger, C. P. Kubicek, M. Penttilä, and R. L. Mach. 2006. Transcriptional regulation of xyn1, encoding xylanase I, in Hypocrea jecorina. Eukaryot. Cell 5:447-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saloheimo, A., N. Aro, M. Ilmén, and M. Penttilä. 2000. Isolation of the ace1 gene encoding a Cys(2)-His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J. Biol. Chem. 275:5817-5825. [DOI] [PubMed] [Google Scholar]
- 39.Saloheimo, M., J. Kuja-Panula, E. Ylösmäki, M. Ward, and M. Penttila. 2002. Enzymatic properties and intracellular localization of the novel Trichoderma reesei β-glucosidase BGLII (Cel1A). Appl. Environ. Microbiol. 68:4546-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 41.Strauss, J., H. K. Horvath, B. M. Abdallah, J. Kindermann, R. L. Mach, and C. P. Kubicek. 1999. The function of CreA, the carbon catabolite repressor of Aspergillus nidulans, is regulated at the transcriptional and post-transcriptional level. Mol. Microbiol. 32:169-178. [DOI] [PubMed] [Google Scholar]
- 42.Strauss, J., R. L. Mach, S. Zeilinger, G. Hartler, G. Stoffler, M. Wolschek, and C. P. Kubicek. 1995. Cre1, the carbon catabolite repressor protein from Trichoderma reesei. FEBS Lett. 376:103-107. [DOI] [PubMed] [Google Scholar]
- 43.Stricker, A. R., K. Grosstessner-Hain, E. Würleitner, and R. L. Mach. 2006. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and d-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 5:2128-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stricker, A. R., R. L. Mach, and L. H. de Graaff. 2008. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei). Appl. Microbiol. Biotechnol. 78:211-220. [DOI] [PubMed] [Google Scholar]
- 45.Stricker, A. R., M. G. Steiger, and R. L. Mach. 2007. Xyr1 receives the lactose induction signal and regulates lactose metabolism in Hypocrea jecorina. FEBS Lett. 581:3915-3920. [DOI] [PubMed] [Google Scholar]
- 46.Stricker, A. R., P. Trefflinger, N. Aro, M. Penttilä, and R. L. Mach. 2008. Role of Ace2 (activator of cellulases 2) within the xyn2 transcriptosome of Hypocrea jecorina. Fungal Genet. Biol. 45:436-445. [DOI] [PubMed] [Google Scholar]
- 47.Tamayo, E. N., A. Villanueva, A. A. Hasper, L. H. de Graaff, D. Ramon, and M. Orejas. 2008. CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet. Biol. 45:984-993. [DOI] [PubMed] [Google Scholar]
- 48.Teeri, T., I. Salovouri, and J. Knowles. 1983. The molecular cloning of the major cellulase gene from Trichoderma reesei. Biotechnology 1:696-699. [Google Scholar]
- 49.Törrönen, A., A. Harkki, and J. Rouvinen. 1994. Three-dimensional structure of endo-1,4-beta-xylanase II from Trichoderma reesei: two conformational states in the active site. EMBO J. 13:2493-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Peij, N. N., M. M. Gielkens, R. P. de Vries, J. Visser, and L. H. de Graaff. 1998. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl. Environ. Microbiol. 64:3615-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Peij, N. N., J. Visser, and L. H. de Graaff. 1998. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol. Microbiol. 27:131-142. [DOI] [PubMed] [Google Scholar]
- 52.Walsh, G. A., R. F. Power, and D. R. Headon. 1993. Enzymes in the animal-feed industry. Trends Biotechnol. 11:424-430. [DOI] [PubMed] [Google Scholar]
- 53.Welt, T., and R. J. Dinus. 1995. Enzymatic deinking—a review. Prog. Pap. Recycl. 4:36-47. [Google Scholar]
- 54.Würleitner, E., L. Pera, C. Wacenovsky, A. Cziferszky, S. Zeilinger, C. P. Kubicek, and R. L. Mach. 2003. Transcriptional regulation of xyn2 in Hypocrea jecorina. Eukaryot. Cell 2:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeilinger, S., C. Galhaup, K. Payer, S. L. Woo, R. L. Mach, C. Fekete, M. Lorito, and C. P. Kubicek. 1999. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genet. Biol. 26:131-140. [DOI] [PubMed] [Google Scholar]
- 56.Zeilinger, S., R. L. Mach, M. Schindler, P. Herzog, and C. P. Kubicek. 1996. Different inducibility of expression of the two xylanase genes xyn1 and xyn2 in Trichoderma reesei. J. Biol. Chem. 271:25624-25629. [DOI] [PubMed] [Google Scholar]
- 57.Zeilinger, S., M. Schmoll, M. Pail, R. L. Mach, and C. P. Kubicek. 2003. Nucleosome transactions on the Hypocrea jecorina (Trichoderma reesei) cellulase promoter cbh2 associated with cellulase induction. Mol. Genet. Genomics 270:46-55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.