Abstract
Homogenous preparations of XenB of Pseudomonas putida, pentaerythritol tetranitrate reductase of Enterobacter cloacae, and N-ethylmaleimide reductase of Escherichia coli, all type II hydride transferases of the Old Yellow Enzyme family of flavoproteins, are shown to reduce the polynitroaromatic compound 2,4,6-trinitrotoluene (TNT). The reduction of this compound yields hydroxylaminodinitrotoluenes and Meisenheimer dihydride complexes, which, upon condensation, yield stoichiometric amounts of nitrite and diarylamines, implying that type II hydride transferases are responsible for TNT denitration, a process with important environmental implications for TNT remediation.
Although the polynitroaromatic 2,4,6-trinitrotoluene (TNT) is rather recalcitrant, a number of microorganisms have been described to be able to use this compound as a nitrogen source (reviewed in reference 4). The presence of the three electrophilic nitro substituents on the aromatic ring tends to inactivate its π system, thereby preventing oxidative attack by oxygenases that are frequently used by living organisms to catabolize aromatic compounds. Nevertheless, the nitro groups are easily reduced to hydroxylamino groups by nitroreductases (2, 8, 10, 15). Ammonium has been proposed to be released from these derivatives upon a Bamberger-type rearrangement (3, 6, 8). On the other hand, the aromatic ring of the polynitroaromatic compound is susceptible to nucleophilic attack by hydride ions to form Meisenheimer complex intermediates (13, 14). This type of ring reduction is catalyzed by reductases, which are referred to here as type II hydride transferases.
Our recent results show that the P. putida XenB protein, a type II hydride transferase of the Old Yellow Enzyme (OYE) family, reduces the nitro groups to produce hydroxylamines and also the aromatic ring to yield the transient production of the Meisenheimer monohydride complex (H−-TNT), which is further reduced to various isoforms of the Meisenheimer dihydride complex (2H−-TNT) (19). It is worth noting that nitrite is released in this reaction via the abiotic condensation of enzymatically produced hydroxylaminodinitrotoluenes and Meisenheimer dihydride complexes that yield diarylamines and stoichiometric amounts of nitrite, thereby making a mass balance possible for the first time (19).
Several enzymes of the OYE family of flavoproteins (17) with type II hydride transferase activity have been described, i.e., the pentaerythritol tetranitrate (PETN) reductase of Enterobacter cloacae PB2 (5), the N-ethylmaleimide (NEM) reductase of several Escherichia coli strains (6, 18), and xenobiotic reductase B (XenB) of Pseudomonas fluorescens I-C (9). However, the end products formed from TNT by these type II hydride transferases remained unclear, although in the case of the XenB enzyme of P. fluorescens, Pak et al. (9) previously proposed a putative biphenyl structure as an end product based on the molecular mass. A product with a molecular mass similar to that found by Pak et al. (9) was found in cell extracts of E. coli upon the transformation of TNT (11), but unequivocal identification was missing. In this study, we sought to identify and quantify the products for the PETN reductase of E. cloacae and the NEM reductase of E. coli and to discuss the implications of this group of enzymes for TNT bioremediation in general.
To purify the PETN reductase, the onr gene encoding the PETN reductase of E. cloacae was amplified by PCR using pONR1 (5) as a template and 5′-TTAGGATCCAAGCAAAAATGTCTCGCA-3′ and 5′-tttCTCGAGCAGTGAAGGGTAGTCGG-3′ (underlined characters in the tail of the former and latter primers represent sequences recognized by BamHI and XhoI, respectively). After digestion of the amplification product with BamHI and XhoI, the PCR product was ligated into the pET28b(+) vector (Novagen) that had been previously digested with the same restriction enzymes. For protein-His6 purification, the resulting plasmid was transformed into E. coli BL21(pLysS) cells (Novagen). Cultures were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and, after harvesting, disrupted by use of a French press (1,000 lb/in2) in a solution containing 25 mM sodium phosphate buffer (pH 7.0), 0.5 M NaCl, 5% (vol/vol) glycerol, and protease inhibitor cocktail (Complete; Roche). The His6-tagged protein was purified by nickel affinity chromatography and eluted with a continuous imidazole gradient, as described in the companion paper, in which the XenB protein of P. putida was purified to almost homogeneity (13). The NEM reductase of E. coli was also purified as a His6-tagged protein upon overexpression (6). XenB of P. putida, NEM reductase of E. coli, and PETN reductase of E. cloacae were all dialyzed against the same buffer, consisting of 25 mM sodium phosphate buffer (pH 7.5), 0.1 mM EDTA, 500 mM NaCl, 10% (vol/vol) glycerol, and 1 μM flavin mononucleotide.
First of all, we determined the kinetic parameters of each enzyme in assays with appropriate amounts of NADPH and TNT with 0.25 μM enzyme (based on flavin concentrations) in 50 mM potassium phosphate buffer (pH 7.0) at 25°C. The kinetic parameters were determined by measuring the changes in TNT concentration over time after high-performance liquid chromatography (HPLC) separation of the products rather than using the more typical spectrophotometrical monitoring of NADPH oxidation. This was done in this way because the NEM reductase exhibits high oxidase activity in the absence of TNT (18). For HPLC, a Waters-Alliance chromatograph equipped with a photodiode array detector (model 2996) and a 5-μm C8 reversed-phase column (Novapak C8, 150 by 3.9 mm; Waters S.A., Barcelona, Spain) were used. Samples (25 μl) were run for 20 min in a 35% (vol/vol) methanol water solution at an 0.85-ml/min flow rate with the detector set at 230 nm. From Lineweaver-Burk plots, we determined Km and Vmax values. The Km values for TNT for the three enzymes ranged from 15 to 64 μM (Table 1), whereas the Km values for NADPH were in the range of between 13 and 117 μM (Table 1). Consistently, XenB showed the highest Vmax, followed by PETN reductase (Table 1). The different TNT reduction rates of each enzyme were reflected in the results from time course experiments (see Fig. S1 in the supplemental material).
TABLE 1.
Comparison of enzymatic activities of type II hydride transferasesa
| Enzyme | Avg Km (μM TNT) ± SE | Avg Km (μM NADPH) ± SE | Avg Vmax (μM/min·mg protein) ± SE |
|---|---|---|---|
| XenB | 48 ± 3 | 13 ± 5 | 105 ± 2 |
| PETN reductase | 64 ± 4 | 117 ± 32 | 37 ± 0.5 |
| NEM reductase | 15 ± 3 | 27 ± 3 | 8 ± 0.5 |
Enzymatic parameters were assayed by monitoring consumed TNT. Data represent the results of at least three different experiments. The error is given as the standard error at 95% confidence.
A second series of assays was directed to determine the nature of the products of the reaction mediated by each enzyme and their stoichiometry. To this end, we used TNT-saturated buffers and an NADPH regeneration system, as described previously (19). Briefly, 1 μM enzyme was added to 0.1 mM NADPH in 50 mM potassium phosphate buffer (pH 7) previously saturated with TNT. For NADPH recycling, 5 units of NADP+-dependent secondary alcohol dehydrogenase from Thermoanaerobium brockii (Sigma) and 2% (vol/vol) isopropanol (previously saturated with TNT) were added to the in vitro reaction mixture. The reaction took place at 25°C for 8 h, after which the entire reaction mixture was dissolved in an equal volume of acetonitrile to ensure the dissolution of any poorly soluble diarylamine that may be formed. Both TNT and the evolved products were monitored by HPLC; the above-described HPLC method was used to quantify TNT and its amino- and hydroxylamine derivatives, while a second method was used to separate nitrite, the isoforms of TNT hydride adducts, and the diaryl adducts from each other. In this method, the mobile phase was 5 mM tetrabutylammonium phosphate (Fluka) at pH 7.0 and acetonitrile-water. Samples (25 μl) were run at a flow rate of 0.85 ml/min, and the detector was set at 210, 230, and 450 nm. The method consisted of a linear gradient from 0% to 25% (vol/vol) acetonitrile during 20 min and from 25% to 62% acetonitrile for another 23 min. The column was then reequilibrated with 100% 5 mM tetrabutylammonium solution for another 9 min. For 2-hydroxylamino-4,6-dinitrotoluene, 4-hydroxylamino-2,6-dinitrotoluene, 2-amino-4,6-dinitrotoluene, and 4-amino-2,6-dinitrotoluene, reference standards were obtained from AccuStandard (New Haven, CT). For Meisenheimer dihydride complexes, diarylamines and diaryl hydroxylamines standards were prepared and verified as described previously (19).
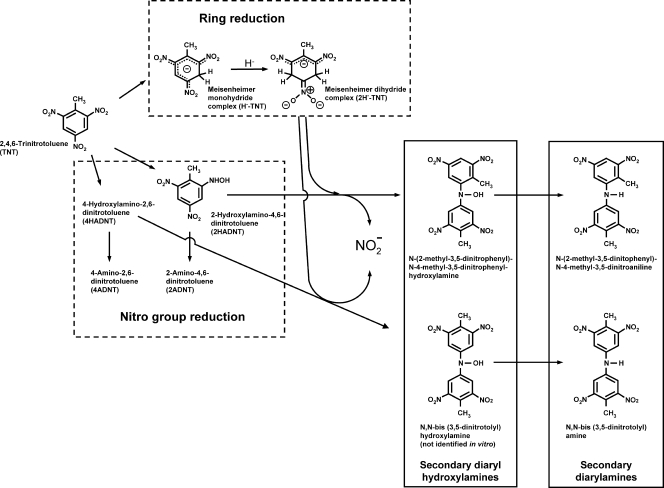
All three enzymes produced the same reactive intermediates as well as nitrite and the secondary diarylamines (see Fig. 2). Five Meisenheimer dihydride complex isoforms were found to be produced with TNT as a substrate with all three type II hydride transferases used in this study. In agreement with data described previously by Ziganshin et al. (20), we found that when each dihydride isoform was separated and isolated by ion pair HPLC, in time, they turned into the other isoforms in the eluent buffer without releasing nitrite. This points toward the existence of an equilibrium that is obviously pH dependent. When the NEM reductase was used in the assays, we observed a large amount of secondary diaryl hydroxylamine production as well (see Fig. S1 in the supplemental material). This intermediate was previously detected only in chemical condensation reactions (19), thereby confirming the reaction scheme proposed in Fig. 1. All three type II hydride transferases were also found to produce larger amounts of N-(2-methyl-3,5-dinitrophenyl)-4-methyl-3,5-dinitroaniline than the symmetric diarylamine, suggesting that, initially, more 2-hydroxylamino-4,6-dinitrotoluene is produced than 4-hydroxylamino-2,6-dinitrotoluene.
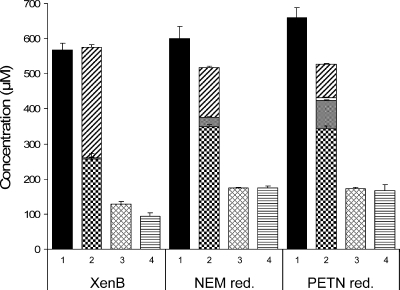
FIG. 2.
Molar relationship between the TNT consumed (solid bar) (1), the total TNT-derived products formed (Meisenheimer dihydride isoforms [hatched bars], amino dinitrotoluenes [open bars], hydroxylaminodinitrotoluenes [shaded bars], and stochiometric diaryl adduct production [checkered bars]) (2), diaryl adducts produced (cross-hatched bars) (3), and nitrite released (striped bars) (4) with XenB, NEM reductase (NEM red.), and PETN reductase (PETN red.) in vitro after 8 h. Error bars indicate standard errors at a 95% confidence interval of three separate experiments.
FIG. 1.
Pathways leading to the products detected in vitro in TNT biotransformations with the type II hydride transferases.
Previously, due to the lack of knowledge about the end products of TNT biotransformation, mass balance studies were largely impossible. However, with the resolution of the main denitration mechanism and the production of end products in sufficient amounts to be used as standards for quantification (19), mass balances can now be calculated. Therefore, we were able to obtain the mass balance of the TNT biotransformation catalyzed by each of the type II hydride transferases tested by relating the sum of the formed products to the amount of TNT consumed in the in vitro reactions.
Figure 2 shows that almost all of the TNT consumed in the reaction can be accounted for in the formed products if the concentration of the adducts is multiplied by 2, taking into account that two TNT molecules are necessary to form one adduct molecule. The condensation of the reactive intermediates was relatively slow (19), which explains why in some reactions the hydroxylaminodinitrotoluene intermediates that were formed were not fully transformed after 8 h. According to the reaction scheme shown in Fig. 1, the amount of formed adducts should be equal to the amount of released nitrite. Figure 2 shows that, indeed, almost equimolar amounts of adducts and nitrite were formed by each of the three type II hydride transferases under study.
The fact that all three of the type II hydride transferases that we tested produced the same end products suggests that the condensation reaction that produces diaryl adducts is, indeed, initiated by the three type II hydride transferases of the OYE family studied here. This can also be the case for other type II hydride transferases since Pak et al. (9), with XenB of P. fluorescens, and Stenuit et al. (11), with cell extracts of E. coli, previously described compounds with the same molecular masses as those exhibited by the secondary diarylamines whose structures were unequivocally resolved by nuclear magnetic resonance and Fourier transform infrared spectroscopy in our previous study (19). Williams et al. (18) did not identify end products with the PETN reductase of E. cloacae or with the NEM reductase of E. coli, probably because they used three- to fourfold-lower TNT concentrations in their system, which made peak detections more difficult.
Homologs belonging to the OYE family of flavoproteins are found throughout prokaryotic species but also in plants, fungi, and nematodes (17). Similarity searches in databases using BLAST (1) with type II hydride transferase sequences (i.e., XenB, NEM reductase, and PETN reductase) revealed that many bacteria could harbor OYE homologs with type II hydride activity (data not shown). This could have implications for TNT remediation technologies, as it suggests that TNT denitration via diarylamine formation could be a relatively common process in nature. If so, the presence of this ability in indigenous microbial populations could account for the high capacity of TNT degradation in some soils and why field studies with TNT-degrading strain P. putida JLR11 showed little additional effects in the rhizoremediation of TNT-contaminated soils compared to uninoculated controls, in contrast with the results found under laboratory conditions (12). More importantly, if TNT denitration is indeed common in the environment, the recalcitrance and toxicity of diarylamines, as well as its reactivity with organic/inorganic material (7, 16), will need to be elucidated. We have found that poorly soluble diarylamines are stable in aqueous solution for months and that these were not further reduced by XenB in vitro. These observations and its nitroaromatic nature suggest that diarylamines probably represent the final recalcitrant end products hitherto not identified in environmental samples. Future studies regarding TNT biodegradation should therefore also focus on the degradation of diarylamines.
Supplementary Material
Acknowledgments
This study was supported by a grant from the European Commission (MADOX QLRT-2001-00345) and a grant from Proyecto de Excelencia CIV344 from the Junta de Andalucia (Spain).
We thank Neil Bruce for providing pONR1. We also thank José A. Paz and David Martín for technical assistance, Lourdes Sánchez from the Servicio de Instrumentación Científica of the Estación Experimetal del Zaidín for help with HPLC analysis, and M. Mar Fandila and Carmen Lorente for secretarial assistance.
Footnotes
Published ahead of print on 12 September 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caballero, A., J. J. Lázaro, J. L. Ramos, and A. Esteve-Nuñez. 2005. PnrA, a new nitroreductase-family enzyme in the TNT-degrading strain Pseudomonas putida JLR11. Environ. Microbiol. 7:1211-1219. [DOI] [PubMed] [Google Scholar]
- 3.Caballero, A., and J. L. Ramos. 2006. A double mutant of Pseudomonas putida JLR11 deficient in the synthesis of the nitroreductase PnrA and assimilatory nitrite reductase NasB is impaired for growth on 2,4,6-trinitrotoluene (TNT). Environ. Microbiol. 8:1306-1310. [DOI] [PubMed] [Google Scholar]
- 4.Esteve-Nuñez, A., A. Caballero, and J. L. Ramos. 2001. Biological degradation of 2,4,6-trinitrotoluene. Microbiol. Mol. Biol. Rev. 65:335-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.French, C. E., S. Nicklin, and N. C. Bruce. 1998. Aerobic degradation of 2,4,6-trinitrotoluene by Enterobacter cloacae PB2 and by pentaerythritol tetranitrate reductase. Appl. Envrion. Microbiol. 64:2864-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.González-Pérez, M. M., P. van Dillewijn, R.-M. Wittich, and J. L. Ramos. 2007. Escherichia coli has multiple enzymes that attack TNT and release nitrogen for growth. Environ. Microbiol. 9:1535-1540. [DOI] [PubMed] [Google Scholar]
- 7.Heiss, G., and H.-J. Knackmuss. 2002. Bioelimination of nitroaromatic compounds: immobilization versus mineralization. Curr. Opin. Microbiol. 5:282-287. [DOI] [PubMed] [Google Scholar]
- 8.Hughes, J. B., C. Wang, K. Yesland, A. Richardson, R. Bhadra, G. Bennet, and F. Rudolph. 1998. Bamberger rearrangement during TNT metabolism by Clostridium acetobutylicum. Environ. Sci. Technol. 32:494-500. [Google Scholar]
- 9.Pak, J. W., K. L. Knoke, D. R. Noguera, B. G. Fox, and G. H. Chambliss. 2000. Transformation of 2,4,6-trinitrotoluene by purified xenobiotic reductase B from Pseudomonas fluorescens I-C. Appl. Environ. Microbiol. 66:4742-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smets, B. F., H. Yin, and A. Esteve-Nuñez. 2007. TNT biotransformation: when chemistry confronts mineralization. Appl. Microbiol. Biotechnol. 76:267-277. [DOI] [PubMed] [Google Scholar]
- 11.Stenuit, B., L. Eyers, R. Rozenberg, J. L. Habib-Jiwan, and S. N. Agathos. 2006. Aerobic growth on 2,4,6-trinitrotoluene (TNT) as sole nitrogen source by Escherichia coli and evidence of TNT denitration with whole cells and cell-free extracts. Appl. Environ. Microbiol. 72:7945-7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Dillewijn, P. A. Caballero, J. A. Paz, M. M. González-Pérez, J. M. Oliva, and J. L. Ramos. 2007. Bioremediation of 2,4,6-trinitrotoluene under field conditions. Environ. Sci. Technol. 41:1378-1383. [DOI] [PubMed] [Google Scholar]
- 13.van Dillewijn, P., A. Caballero, R. Wittich, and J. L. Ramos. 2008. Subfunctionality of enzymatic activities in hydride transferases of the Old Yellow Enzyme family of flavoproteins of Pseudomonas putida. 74:6703-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vorbeck, C., H. Lenke, P. Fischer, J. C. Spain, and H.-J. Knackmuss. 1998. Initial reductive reactions in aerobic microbial metabolism of 2,4,6-trinitrotoluene. Appl. Environ. Microbiol. 64:246-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watrous, M. M., S. Clark, R. Kutty, S. Huang, F. B. Rudolph, J. B. Hughes, and G. N. Bennet. 2003. 2,4,6-Trinitrotoluene reduction by an Fe-only hydrogenase in Clostridium acetobutylicum. Appl. Environ. Microbiol. 69:1542-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss, M., R. Geyer, R. Russow, H. H. Richnow, and M. Kästner. 2004. Fate and metabolism of [15N]2,4,6-trinitrotoluene in soil. Environ. Toxicol. Chem. 23:1852-1860. [DOI] [PubMed] [Google Scholar]
- 17.Williams, R. E., and N. C. Bruce. 2002. ‘New uses for an Old Enzyme’—the Old Yellow Enzyme family of flavoenzymes. Microbiology 148:1607-1614. [DOI] [PubMed] [Google Scholar]
- 18.Williams, R. E., D. A. Rathbone, N. S. Scrutton, and N. C. Bruce. 2004. Biotransformation of explosives by the Old Yellow Enzyme family of flavoproteins. Appl. Environ. Microbiol. 7:3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wittich, R. M., A. Haidour, P. van Dillewijn, and J. L. Ramos. 2008. OYE flavoprotein reductases initiate the condensation of TNT-derived intermediates to secondary diarylamines and nitrite. Environ. Sci. Technol. 42:734-739. [DOI] [PubMed] [Google Scholar]
- 20.Ziganshin, A. M., R. Gerlach, T. Borch, A. V. Naumov, and R. P. Naumova. 2007. Production of eight different hydride complexes and nitrite release from 2,4,6-trinitrotoluene by Yarrowia lipolytica. Appl. Environ. Microbiol. 73:7898-7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.