Abstract
In this study, variably absent or present (VAP) regions discovered through comparative genomics experiments were targeted for the development of a rapid, PCR-based method to subtype and fingerprint Escherichia coli O157:H7. Forty-four VAP loci were analyzed for discriminatory power among 79 E. coli O157:H7 strains of 13 phage types (PT). Twenty-three loci were found to maximize resolution among strains, generating 54 separate fingerprints, each of which contained strains of unique PT. Strains from the three previously identified major E. coli O157:H7 lineages, LSPA6-LI, LSPA6-LI/II, and LSPA6-LII, formed distinct branches on a dendrogram obtained by hierarchical clustering of comparative genomic fingerprinting (CGF) data. By contrast, pulsed-field gel electrophoresis (PFGE) typing generated 52 XbaI digestion profiles that were not unique to PT and did not cluster according to O157:H7 lineage. Our analysis identified a subpopulation comprised of 25 strains from a closed herd of cattle, all of which were of PT87 and formed a cluster distinct from all other E. coli O157:H7 strains examined. CGF found five related but unique fingerprints among the highly clonal herd strains, with two dominant subtypes characterized by a shift from the presence of locus fprn33 to its absence. CGF had equal resolution to PFGE typing but with greater specificity, generating fingerprints that were unique among phenotypically related E. coli O157:H7 lineages and PT. As a comparative genomics typing method that is amenable for use in high-throughput platforms, CGF may be a valuable tool in outbreak investigations and strain characterization.
Escherichia coli O157:H7 is one of the most significant human pathogens, responsible for outbreaks worldwide of food- and waterborne illnesses that range from diarrhea to the hemolytic uremic syndrome (15, 45, 46, 50). Two major lineages of this pathogen that differ in both genotype and phenotype have been identified using octamer-based genome scanning, with strains of lineage I more frequently linked to human illness (26). The lineage-specific polymorphism assay (LSPA6) is based on polymorphisms in six genetic loci (53) and was developed as a means to quickly identify strain lineage in a single PCR. As food production and distribution becomes increasingly centralized, outbreaks of infection associated with this pathogen from fresh produce and meats have an increased risk of being widely disseminated and infecting multiple populations from a single source, making the ability to quickly and effectively determine epidemiologically related strains of paramount importance to public health (10, 12).
Bacteriophage typing was among the first subtyping schemes developed to characterize E. coli O157:H7 isolates (1). While the method is still commonly used to screen E. coli O157:H7 isolates during outbreak investigations, the common occurrence of certain phage types (PTs), along with the low number of PTs overall, limits its resolution and frequently results in the classification of outbreak- and non-outbreak-related E. coli O157:H7 isolates within the same PT.
DNA-based subtyping methods, which rely on the unique genetic fingerprint of each bacterial strain, allow a higher level of discrimination among E. coli O157:H7 isolates than is possible with bacteriophage typing (51, 54). Among the DNA-based methods commonly used to genotype bacteria are multilocus sequence typing (MLST), amplified fragment length polymorphism (AFLP) analysis, ribotyping, PCR-restriction fragment length polymorphism (PCR-RFLP) analysis, multilocus variable-number tandem repeat analysis (MLVA), and the current standard, pulsed-field gel electrophoresis (PFGE).
MLST has been shown to be of inferior resolution to other genotyping methods such as PFGE because the small number of housekeeping genes analyzed by MLST do not contain sufficient variation to be useful as the sole epidemiological typing method (5, 33). Some groups have suggested AFLP as an alternative to PFGE (21, 56), and although AFLP could discriminate O157:H7 strains from non-O157:H7 strains, it was found to lack within-serogroup resolution compared to PFGE (18, 19). Likewise, ribotyping alone does not offer the discriminatory power needed to effectively fingerprint E. coli O157:H7 strains, although it has been successfully used in conjunction with PFGE to offer additional discrimination (3, 38). PCR-RFLP based on virulence regions (such as the regulatory region of Stx phage) has been used to unambiguously determine clonality among E. coli O157:H7 strains but lacks the resolution to differentiate among strains that are temporally or geographically distant (42, 43). The analysis of tandem duplication in the genome with MLVA has been useful in characterizing clonal organisms such as E. coli O157:H7 (20) with greater discriminatory power than PFGE (32, 36), although its usefulness in higher-level analyses, such as lineage typing, has not been determined. Unfortunately, MLVA requires the PCR products be sequenced or analyzed using capillary electrophoresis to accurately determine the number of tandem repeats (20), although estimation of size based on traditional agarose gels has also been used (24).
PFGE is currently considered the “gold standard” of DNA-based subtyping methods and has been shown to discriminate between O157:H7 and non-O157:H7 isolates, among isolates from different geographic regions, between outbreak- and non-outbreak-related isolates, and to identify outbreak sources (4, 6, 7). Although PFGE has been an extremely powerful tool in E. coli O157:H7 genotyping, it requires complex computer-dependent graphical analysis using dedicated software and does not always produce consistent results, particularly when the analyses are conducted in different laboratories (35). In response to these challenges, the U.S. Centers for Disease Control and Prevention has attempted to improve the reproducibility of PFGE by developing PulseNet as a means of interlaboratory standardization and sharing of PFGE data.
While all of these subtyping methods are useful, none produces a stand-alone method for subtyping E. coli O157:H7 isolates and many indicate seemingly contradictory levels of diversity. Genomic variation in E. coli has been found in large part to be due to genomic islands and phage-related areas that often contain virulence-associated genes, which arise through horizontal gene transfer (17). Examples include the phage-encoded Shiga toxins (31, 37), the locus of enterocyte effacement pathogenicity island (29), and other genomic islands containing virulence-related factors, such as fimbriae (41) and iron uptake systems (9, 34). Recently a “seropathotype” designation based on the association between the occurrence and severity of human disease and certain serogroups has been proposed. It was found that those strains more virulent to humans also contain specific genes within the virulence-associated genomic island OI#122 (23). Genotyping methods, such as octamer-based genome scanning, have also shown that certain lineages of E. coli O157:H7 appear to be more commonly associated with human disease than others, suggesting differences in phenotype and possibly virulence within this serotype (26, 57). As all molecular subtyping methods are based on heterogeneity among DNA sequences, the ideal scheme would be based on comparative genomic analysis of the entire genome sequences of all isolates. However, the practical considerations of whole-genome sequencing do not as of yet make the analysis of multiple E. coli O157:H7 isolates feasible for rapid typing of outbreak strains.
Our previous work with microarray-based comparative genomics and subtractive hybridization led to the identification of genetic regions that are variably absent or present (VAPs) among E. coli O157:H7 strains (48, 55). In this study, VAPs were used to create a PCR-based comparative genomic fingerprinting (CGF) method capable of identifying taxonomically unique subtypes and epidemiologically related strains.
MATERIALS AND METHODS
Selection of loci.
Thirty-four E. coli O157:H7 strains which had previously been characterized by microarray-based comparative genome hybridization (CGH) were analyzed for genes that were variably absent or present (55). The microarray data contained 1,751 VAP loci, and those with binary log ratio distributions were selected for further examination, as they were expected to increase the probability that a locus was present or absent rather than heterogeneous in sequence. From the set of loci consisting of binary log ratio distributions, those that gave the greatest strain discrimination were selected preferentially over those that had very little discriminatory power. Thus, if the presence and absence of two loci were identical among all strains, only one was kept. No knowledge of lineage or other phylogenetic relationship was used in the selection of loci. A set of 34 loci were chosen that were distributed across the entire E. coli genome and offered the greatest combined resolution among the strains, without reference to any other typing method.
The VAP loci identified by our CGH work consisted of only O157:H7 lineage I open reading frames (55). In order to remove potential bias in subsequent analyses, 10 lineage II-specific VAP loci identified by subtractive hybridization experiments (48) were added to the 34 identified through microarray-based CGH, and this final set of 44 were used in testing. Although 10 loci were lineage II specific, no effort was made to preferentially keep them; all loci were treated equally, and those that offered the greatest discrimination were kept.
Isolation of DNA.
The E. coli strains used in this study were obtained from a variety of human, bovine, and environmental sources (Table 1). Growth of the bacteria was carried out in 10 ml of brain heart infusion broth, at 37°C for ∼16 h in a shaking incubator (200 rpm). The cultures were centrifuged at 8,000 rpm for 10 min, and the bacterial pellet was dissolved in 15 ml of 10 mM NaCl, 20 mM Tris-HCl (pH 8.0), 1 mM EDTA, 100 μg/ml proteinase K, and 0.5% sodium dodecyl sulfate. This suspension was incubated at 50°C for 2 h and extracted with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1). Following centrifugation for 10 min at 8,000 rpm, the upper phase was removed and precipitated by adding 0.1 volume of 3 M NaO-acetate (pH 5.2) and 2 volumes of 99% ethanol. The DNA precipitate was then spooled out of solution using a sterile glass rod, washed with 70% ethanol, and dissolved in 3 ml of Tris-EDTA (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) buffer.
TABLE 1.
The E. coli O157:H7 strains studied and their source, date of isolation, phage type, LSPA6 lineage, XbaI PFGE digestion pattern, and CGF profile
| Name | Source | Date isolated | Phage type | LSPA6 lineage | PFGE XbaI | CGF profile |
|---|---|---|---|---|---|---|
| R1388 | Human | 1996 | 1 | I/II | X01.0091 | CGF55 |
| 59243 | Human | 10-Jun-95 | 2 | I/II | X01.0102 | CGF53 |
| 71074 | Human | 19-Jul-95 | 2 | I/II | X01.0110 | CGF52 |
| 09601Fe046.1 | Bovine | NAa | 2 | I/II | X01.0274 | CGF52 |
| Zap0046 | Bovine | NA | 2 | I/II | X01.0209 | CGF54 |
| S2302-1 | Human | 1996 | 14 | I | X01.0002 | CGF18 |
| S3722 | Human | 1997 | 14 | I | X01.0156 | CGF40 |
| S2628 | Human | 1998 | 14 | I | X01.0002 | CGF26 |
| 97701 | Human | 1995 | 14 | I | X01.0143 | CGF41 |
| F5 | Bovine | 8-Aug-95 | 14 | I | X01.0109 | CGF42 |
| F1299 | Bovine | 26-Aug-96 | 14 | I | X01.0528 | CGF34 |
| TS-97 | Bovine | 11-Oct-01 | 14 | I | X01.0109 | CGF19 |
| LRH73 | Human | 7-Dec-99 | 14 | I | X01.0119 | CGF31 |
| 23339 | Human | 1996 | 14 | I | X01.0003 | CGF38 |
| F30 | Bovine | 8-Aug-95 | 14 | I | X01.0056 | CGF23 |
| LN637-4 | Environmental | 2001 | 14 | I | X01.0074 | CGF24 |
| LS110 | Bovine | 22-Aug-01 | 14 | I | X01.0132 | CGF39 |
| AA1002-1 | Environmental | 10-Jun-02 | 14 | I | X01.0176 | CGF23 |
| 93111 | Human | 1993 | 14 | I | X01.0001 | CGF50 |
| EDL933 | Human | 1982 | 21 | I | X01.0138 | CGF21 |
| LRH13 | Human | 1-Nov-96 | 23 | II | X01.0095 | CGF10 |
| F12 | Bovine | 8-Aug-95 | 23 | II | X01.0095 | CGF8 |
| F1081 | Bovine | 25-Jun-96 | 23 | II | X01.0097 | CGF5 |
| F1305 | Bovine | 26-Aug-96 | 23 | II | X01.0093 | CGF11 |
| R1797 | Human | 1996 | 23 | II | X01.0527 | CGF12 |
| LS68 | Bovine | 2001 | 23 | II | X01.0049 | CGF9 |
| FRIK920 | Bovine | NA | 23 | II | X01.0160 | CGF2 |
| FRIK1999 | Bovine | NA | 23 | II | X01.0162 | CGF1 |
| EC19920026 | Bovine | NA | 23 | II | X01.0046 | CGF7 |
| 63154 | Human | 25-Jun-95 | 31 | I | X01.0002 | CGF20 |
| 8-1360-1 | Human | NA | 31 | I | X01.0191 | CGF47 |
| 58212 | Human | 1996 | 31 | I | X01.0144 | CGF20 |
| F2 | Bovine | 8-Aug-95 | 31 | I | X01.0192 | CGF49 |
| F744 | Bovine | 18-Mar-96 | 31 | I | X01.0047 | CGF48 |
| F1095 | Bovine | 25-Jun-96 | 31 | I | X01.0002 | CGF22 |
| R1195 | Human | 1-Jul-95 | 31 | I | X01.0002 | CGF47 |
| F1103 | Bovine | 25-Jun-96 | 32 | I | X01.0150 | CGF44 |
| Sakai | Human | ATCC | 32 | I | X01.0139 | CGF27 |
| AA1000-2 | Environmental | 10-Jun-02 | 32 | I | X01.0174 | CGF43 |
| M01MD3265 | NA1 | NA | 32 | I | X01.0154 | CGF46 |
| APF59-3 | Environmental | 3-Jun-04 | 32 | I | X01.0247 | CGF25 |
| 70490 | Human | 9-Jul-95 | 34 | I | X01.0002 | CGF36 |
| F732 | Bovine | 18-Mar-96 | 34 | I | X01.0088 | CGF45 |
| AA619-2 | Environmental | 4-Jul-01 | 34 | II | X01.0092 | CGF13 |
| LS236 | Bovine | 10-Oct-01 | 34 | I | X01.0008 | CGF37 |
| AA995-2 | Environmental | 10-Jun-02 | 34 | II | X01.0173 | CGF15 |
| FRIK1985 | Bovine | NA | 45 | II | X01.0161 | CGF4 |
| FRIK1990 | Bovine | NA | 54 | II | X01.0111 | CGF16 |
| FRIK2001 | Bovine | NA | 54 | II | X01.0163 | CGF6 |
| EC970520 | NA1 | 12-Dec-02 | 67 | II | X01.0525 | CGF3 |
| 12491 | Human | 03-Feb-95 | 74 | II | X01.0526 | CGF14 |
| E2328 | Human | 12-Jan-95 | 87 | I | X01.0062 | CGF29 |
| F1082 | Bovine | 25-Jun-96 | 87 | I | X01.0062 | CGF32 |
| H432 | Bovine | 21-May-96 | 87 | I | X01.0062 | CGF29 |
| H435 | Bovine | 21-May-96 | 87 | I | X01.0062 | CGF29 |
| H451 | Bovine | 21-May-96 | 87 | I | X01.0080 | CGF33 |
| H453 | Bovine | 21-May-96 | 87 | I | X01.0062 | CGF29 |
| H454 | Bovine | 21-May-96 | 87 | I | X01.0062 | CGF29 |
| H568 | Bovine | 17-Jun-96 | 87 | I | X01.0062 | CGF29 |
| H571 | Bovine | 17-Jun-96 | 87 | I | X01.0062 | CGF29 |
| H572 | Bovine | 17-Jun-96 | 87 | I | X01.0062 | CGF29 |
| H573 | Bovine | 17-Jun-96 | 87 | I | X01.0062 | CGF29 |
| H574 | Bovine | 17-Jun-96 | 87 | I | X01.0063 | CGF35 |
| H2160 | Bovine | 24-Mar-97 | 87 | I | X01.0065 | CGF28 |
| H2161 | Bovine | 24-Mar-97 | 87 | I | X01.0065 | CGF28 |
| H2163 | Bovine | 24-Mar-97 | 87 | I | X01.0065 | CGF29 |
| H2164 | Bovine | 24-Mar-97 | 87 | I | X01.0343 | CGF29 |
| H2176 | Bovine | 24-Mar-97 | 87 | I | X01.0086 | CGF29 |
| H2704 | Bovine | 9-May-97 | 87 | I | X01.0522 | CGF30 |
| H2718 | Bovine | 9-May-97 | 87 | I | X01.0086 | CGF29 |
| H2723 | Bovine | 9-May-97 | 87 | I | X01.0065 | CGF29 |
| H2727 | Bovine | 9-May-97 | 87 | I | X01.0065 | CGF29 |
| H2731 | Bovine | 9-May-97 | 87 | I | X01.0065 | CGF29 |
| H4420 | Bovine | 23-Dec-97 | 87 | I | X01.0086 | CGF30 |
| EC980120 | Bovine | 23-Dec-97 | 87 | I | X01.0086 | CGF30 |
| EC980121 | Bovine | 23-Dec-97 | 87 | I | X01.0086 | CGF30 |
| EC980122 | Bovine | 23-Dec-97 | 87 | I | X01.0086 | CGF30 |
| EC980125 | Bovine | 23-Dec-97 | 87 | I | X01.0086 | CGF30 |
| OK1 | Human | 1996 | Atypical | I | X01.0529 | CGF51 |
| K12 MG1655 | ATCC | NA | NA | NA | NA | CGF17 |
NA, not available.
PCR.
PCRs were carried out in a reaction volume of 50 μl containing 1× PCR buffer (Qiagen), 1 mM each deoxynucleoside triphosphate (Invitrogen), 0.2 μM each primer (Alpha DNA), 1 U Taq DNA polymerase (Qiagen), and distilled H2O to fill the remaining volume. Amplification was performed using either a GeneAmp PCR System 9700 (Applied Biosystems) or a Mastercycler epGradient (Eppendorf), with an initial denaturing step of 95°C for 5 min followed by 30 cycles of 95°C for 30 s, an annealing step of 20 s at the appropriate temperature and an extension step of 1 min/kb expected product size at 72°C, and completed with a final extension of 72°C for 5 min. Visualization of PCRs was carried out following agarose gel electrophoresis. Briefly, 8 μl of PCR mixture combined with 2 μl loading dye (0.25% [wt/vol] bromophenol blue, 40% [wt/vol] sucrose) were run on a 1% (wt/vol) agarose gel stained with ethidium bromide for 40 min at 110 V and visualized under UV light.
Phage, pulsed-field gel electrophoresis, and lineage typing.
Bacteriophage typing was carried out as described by Ahmed and colleagues and extended by Khakhria and colleagues (1, 25). PFGE was carried out at the Laboratory for Foodborne Zoonoses in Guelph, Ontario, according to the Centers for Disease Control and Prevention manual standard 1-day protocol, as previously described (40). Lineage typing was carried out using the E. coli O157:H7 lineage-specific polymorphism assay (53).
Construction of dendrograms.
Results of the PCR amplifications were converted to binary values (0 for absence, 1 for presence) and clustered using Bionumerics version 5.1 (Applied Maths) with the simple matching distance metric and the average linkage method of clustering. The dendrogram was rooted with K-12 strain MG1655.
PFGE banding patterns were analyzed with Bionumerics version 5.1 (Applied Maths) and clustered by UPGMA. The dendrogram was created using the Dice similarity coefficient with an optimization of 1.5% and a tolerance of 1.5%.
RESULTS
Forty-four loci that were previously found to be variously absent or present in genomic islands of E. coli O157:H7 and which were distributed throughout the entire genome (Tables 2 and 3) were selected for initial testing and assessed by PCR assay across 79 E. coli O157:H7 isolates and the laboratory strain K-12 MG1655 (Table 1). Of the initial 44 loci targeted by PCR, 13 generated nonbinary data (e.g., multiple bands or bands of various sizes). The remaining 31 loci produced binary results and generated 54 unique fingerprints. Each of the 31 loci was assessed for its ability to discriminate among strains. It was found that 23 loci differentiated strains that only differed at one, two, or three loci. The remaining eight loci only offered additional discrimination between strains where four or more loci already differed and did not contribute additional fingerprints (data not shown); exclusion of these loci from the final set maintained the resolution of the 54 unique fingerprints (Table 4).
TABLE 2.
The E. coli O157:H7 lineage II-specific VAP loci initially testeda
| Region | Function |
|---|---|
| A01 | EC93 contact-dependent inhibition (cdiB) gene |
| A05 | Bacteriophage P2 genes J and I, encoding baseplate assembly proteins |
| A06b | E. coli Sakai intergenic region between ECs1159 and ECs1160 |
| A07 | E. coli Sakai ORF ECs1928, encoding a hypothetical protein, adjacent to ECs1997, encoding a putative filament protein |
| A12 | DNA sequence without significant homology to annotated DNA sequences |
| B01 | Shigella flexneri bacteriophage V ORFs orf39, orf40, and dam, encoding a replication protein, a hypothetical protein, and a DNA adenine methylase |
| B02 | Bacteriophage P2 genes J and I, encoding baseplate assembly proteins |
| B05 | ColE6-CT14 colicin plasmid, colicin E6 gene |
| B13 | DNA sequence without significant homology to annotated DNA sequences |
| B18 | pColD-157 plasmid, colicin D-157 activity protein |
Identified by Steele et al. (48).
TABLE 3.
The E. coli O157:H7 lineage I VAP loci initially tested, the primers targeting them, and their locations in the sequenced genomes of K-12 MG1655 and E. coli O157:H7 strains EDL933 and Sakai
| Primera | K-12 ORF | EDL933 ORF | Sakai ORF | Genomic island | Function |
|---|---|---|---|---|---|
| fprn01 | B0546 | NAb | NA | KI#37 | Putative ARAC-type regulatory protein; ybcM [B] |
| fprn02 | B1201 | NA | NA | KI#74 | Putative sensor-type regulator |
| fprn03 | B2082-B2083 | NA | NA | KI#103 | Hypothetical protein |
| fprn04 | B2083 | NA | NA | KI#103 | Prophage P2 ogr protein and hypothetical protein |
| fprn05 | B2361 | NA | NA | KI#121 | Hypothetical protein |
| fprn06 | NA | NA | ECs4943-ECs4944 | S-loop#274 | Putative regulatory protein; putative DNA-binding protein |
| fprn07 | NA | NA | ECs4955-ECs4956 | S-loop#274 | Hypothetical proteins |
| fprn08 | NA | Z0020 | ECs0020 | OI#1/S-loop#1 | Hypothetical protein |
| fprn09 | NA | Z0309 | ECs0275 | OI#8/S-loop#16 | Similar to regulatory protein Cro (phage lambda) |
| fprn10 | NA | NA | ECs1184 | S-loop#69 | Hypothetical protein |
| fprn11 | NA | Z1203 | ECs1386 | OI#43/S-loop#72 | Hypothetical protein |
| fprn12 | NA | Z1770 | ECs1506 | OI#50/S-loop#77 | Hypothetical protein |
| fprn13 | NA | Z1963 | ECs1695 | OI#55/S-loop#83 | Hypothetical protein |
| fprn14 | NA | Z2400 | ECs1941 | S-loop#91 | Putative transcriptional regulator |
| fprn15 | NA | Z2386-Z2387 | ECs1954 | S-loop#93 | Hypothetical protein |
| fprn16 | NA | Z3358 | ECs2990 | OI#93/S-loop#153 | Putative prophage repressor CI |
| fprn17 | NA | Z3939 | ECs3507 | OI#108/S-loop#186 | Hypothetical protein |
| fprn18 | NA | Z4188 | ECs3724 | OI#115/S-loop#197 | Type III secretion protein EpaQ |
| fprn20 | NA | NA | ECs4954 | S-loop#274 | Hypothetical protein |
| fprn21 | NA | Z5690 | ECs5072 | OI#167/S-loop#276 | Putative carbohydrate ABC transport system permease |
| fprn22 | B1289 | NA | NA | KI#87 | Hypothetical protein |
| fprn23 | NA | Z1204 | ECs1387 | OI#43/S-loop#72 | Hypothetical protein |
| fprn24 | NA | Z1447 | NA | OI#45 | Putative repressor protein CI of bacteriophage BP-933W |
| fprn25 | NA | Z1481-Z1483 | ECs1225-ECs1228 | OI#45/S-loop#69 | Hypothetical protein and putative tail fiber protein |
| fprn26 | NA | Z1835 | ECs1574 | OI#51/S-loop#78 | Putative integrase |
| fprn27 | NA | Z1841 | ECs1579-ECs1580 | OI#51/S-loop#78 | Hypothetical protein |
| fprn28 | NA | Z1963-Z1965 | ECs1695-ECs1698 | OI#55/S-loop#83 | Hypothetical protein; putative ferric enterobactin transport ATP-binding protein; putative ABC transporter permease protein; putative ATP-binding protein of ABC transporter |
| fprn29 | NA | Z2090 | ECs2209 | OI#57/S-loop#108 | Putative repressor protein encoded within prophage CP-933O |
| fprn30 | B1519 | Z2186 | ECs2126 | NA | Putative enzyme |
| fprn31 | NA | Z2311-Z2312 | ECs2016-ECs2017 | OI#66/S-loop#98 | Hypothetical protein |
| fprn32 | NA | Z2322 | ECs2007 | OI#67/S-loop#95 | Putative outer membrane protein |
| fprn33 | NA | Z2969 | ECs2620 | OI#76/S-loop#119 | Putative transcriptional regulator |
| fprn34 | NA | Z4092 | ECs3637 | OI#112/S-loop#194 | Hypothetical membrane protein |
| fprn35 | NA | Z5885 | ECs5249 | OI#172/S-loop#286 | Putative resolvase |
Forward and reverse primer sequences are given in Table 4.
NA, not applicable.
TABLE 4.
The 23 CGF primers and the conditions for their amplification
| Primer | Expected size (bp) | Annealing temp (°C) | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|---|---|
| fprn02 | 413 | 55.0 | ACGGGCAATCACTGAACTC | TGACGGCGATATAAGGACC |
| fprn30 | 614 | 55.0 | TCGCCATTACCAACTGAC | GATAACGGGCATCGACTC |
| fprn03 | 347 | 57.0 | CAATGTATCGCTGTACCGACTC | GCAGGACTGGACAATCACTAAG |
| fprn05 | 422 | 55.0 | GCCGGTCATCTTTCTCAC | GCGCATTTGCGTAACTCC |
| fprn10 | 408 | 55.0 | TTGAGGCCGAACGTAAAC | CGCCTCTTAACGGTAGTC |
| fprn11 | 442 | 55.0 | GCGGATGAATCCCTCAATCG | CCAGCAGGAACATCGCATAG |
| fprn13 | 208 | 55.0 | ACCGCCCTGGATATCTTC | ACGTCAGCTGGAGTCTTG |
| fprn28 | 954 | 57.0 | CCGCAGCCCATATCGAATAAGG | TCATGCGTCTCTGGGTTACTCC |
| fprn14 | 150 | 55.0 | CTCGTCCAGAAGGTCAATGATG | CCTCGCCGAGAGAAATTAAG |
| fprn15 | 648 | 55.0 | GGTCGCATGCCTTAGATAACTC | TTGGTGGTCCGATGCTTGAAG |
| fprn32 | 2210 | 55.0 | CGTTGCGGTTTGTGTATC | CTGCGGTAGTGAATAACG |
| fprn29 | 1002 | 55.0 | CCATCGTCAACCTCATC | GCTCCGTAAGTCAATGG |
| fprn33 | 359 | 55.0 | TGTCACCTCCCTACTATCAC | TGCCATGAGCGACATTG |
| fprn16 | 512 | 55.0 | GAAAGGTTAGCGCGGATG | TCAGCCAAACGTCTCTTC |
| fprn34 | 1007 | 55.0 | AACCGGGACATTAACGAC | ACGAAGAGCGAGAATGAG |
| fprn18 | 101 | 55.0 | TTCACGCCAAATGGCAAC | TCTGCAGGACCTATTGCAGTAG |
| fprn06 | 807 | 57.0 | ACCCGCTTTAGATTCTACCG | AAAGAGCGCAGTGATAAGCC |
| fprn07 | 926 | 57.0 | GCAGGACTGCCAGAAAGTTAAG | CGCCGATTTCCCTAAAGGTAAG |
| fprn35 | 489 | 55.0 | ACCAGCGCATGGAACTTG | GCGTCGCTCTTGAAACAC |
| fprn24 | 691 | 55.0 | CACTGGTTACCAGCTATG | CGCAAGCCTGTAAAGAAG |
| A01a | 432 | 60.8 | ACCAAGGCATCCCCCGTGTGAA | ATAATCCGCTGGGGCTGGCTGAC |
| B01a | 316 | 60.7 | AAGCCCCCACGTAATTCCCTGACA | CATTTCCCGCGCCTGACTGAGA |
| A12a | 446 | 55.9 | GGGCGGACTTTGTTTGGTTGAA | GCCTGGCGGAAATGGACTGTAT |
Identified by Steele et al. (48).
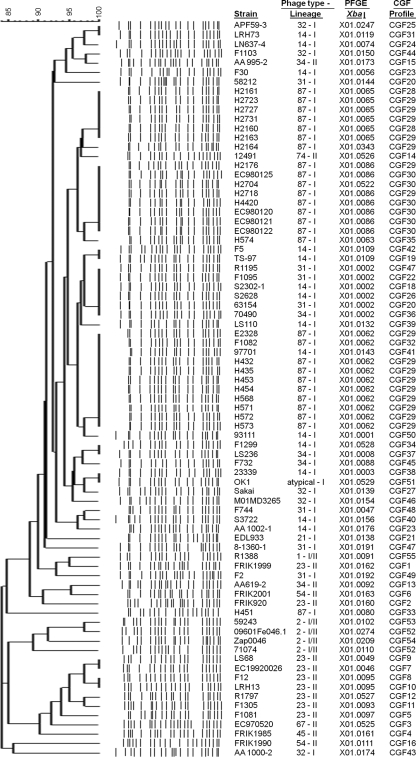
Phage typing, PFGE analysis, and lineage typing of the 79 E. coli O157:H7 strains resolved 13 separate PTs, 52 unique PFGE profiles, and 58 LSPA6-LI, 5 LSPA6-LI/II, and 16 LSPA6-LII strains. The 23-locus binary fingerprint of every strain was hierarchically clustered using Bionumerics version 5.1 (Fig. 1). As can be seen, at a 60% similarity cutoff, four unique clusters of strains were observed. One cluster contained the non-O157:H7 strain K-12 MG1655, which was used as an outgroup, while the three remaining clusters corresponded to the three major O157:H7 lineages, LSPA6-LI, LSPA6-LI/II, and LSPA6-LII. The first cluster contained the 16 LSPA6-LII strains of PT23, PT34, PT45, PT54, PT67, and PT74; the second contained the 58 LSPA6-LI strains of PT14, PT21, PT31, PT32, PT34, and PT87; the third contained the 5 LSPA6-LI/II strains of PT1 and PT2. Every PT was exclusive to a cluster, except for PT34, which was observed in both LSPA6-LI and LSPA6-LII clusters. When strains were grouped according to PFGE banding pattern (Fig. 2), the LSPA6 lineage groupings were no longer absolute. LSPA6-LI strain AA1000-2 grouped with LSPA6-LII strains, LSPA6-LI/II strain R1388 grouped apart from the other four LSPA6-LI/II strains, and LSPA6-LII strains AA995-2 and 12491 grouped among the LSPA6-LI strains.
FIG. 1.
The dendrogram created when the 23-locus binary fingerprint of each of the 79 E. coli O157:H7 strains and non-O157:H7 strain K-12 MG1655 were clustered using Bionumerics version 5.1. A black square represents the presence of a locus and a white square represents the absence of a locus, as determined by PCR. Sequence data for EDL933 indicate that locus fprn16 of OI#93/S-loop#153 is homologous to that of Sakai; however, repeated efforts to amplify fprn16 in EDL933 were unsuccessful. Similarly, previous work with the Sakai strain used in this study has shown a deletion of fprn29 in OI#57/S-loop#108.
FIG. 2.
The dendrogram created when the XbaI PFGE banding pattern of each of the 79 E. coli strains studied were clustered by UPGMA using the Dice coefficient in Bionumerics version 5.1, with an optimization of 1.5% and a tolerance of 1.5%.
Strains with the same CGF profile were found to share the same PT, a trend that was observed for all seven CGF profiles represented by multiple strains in the data set (CGF20, CGF23, CGF28, CGF29, CGF30, CGF47, and CGF52). By contrast, PFGE profile X01.0002 contained strains of PT14, PT31, and PT34.
In order to determine whether CGF could distinguish epidemiologically relevant subpopulations, our data set included 25 PT87 isolates that had been obtained from a closed herd of cattle at the Animal Diseases Research Institute (ADRI) in Lethbridge, Alberta, Canada, over a period of 18 months. Five separate isolates were obtained from each of five sampling dates from 21 May 1996 through 23 Dec 1997. As Fig. 1 shows, a discrete branch in the dendrogram representing strains with fingerprints having greater than 91% identity (i.e., fewer than three differences in a CGF profile) contained all PT87 strains in the data set and included the 25 isolates from the ADRI cattle herd. The cattle strains could be further subtyped into two major groups based on a 100% similarity threshold, one of which (CGF29) consisted of strains positive for locus fprn33 isolated between 21 May 1996 and 9 May 1997. The other cluster (CGF30) contained strains found to be negative for locus fprn33, including all five isolates from 23 Dec 1997 and one from the prior sampling date of 9 May 1997. This suggests a shift in the clonal architecture of the O157:H7 herd strains over time. Microarray analysis of three PT87 strains showed that the phage-related OI#76/S-loop#119, in which fprn33 is located, was completely absent in fprn33-negative strain H4420 but fully intact in fprn33-positive strains E2328 and F1082 (55). PFGE typing was able to distinguish the isolates from 23 Dec 1997, one isolate from 9 May 1997, and one isolate from 24 March 1997 as a separate type, X01.0086, and delineated seven separate PFGE types among the herd strains, but as Fig. 2 shows the herd strains did not form a discrete cluster when grouped by PFGE banding pattern.
DISCUSSION
The rapid progress in bacterial genome sequencing has allowed the development of high-resolution subtyping methods that can provide inter- and intraspecies bacterial genome comparisons. This study capitalized on prior comparative genomics hybridization experiments that examined the genomes of 34 E. coli O157:H7 strains and regions of variability therein, which allowed the development of a PCR-based typing system able to group strains into phenotypically related lineage-specific types, to determine those that were epidemiologically linked, and to offer discrimination between related and nonrelated isolates (48, 55). A study by Wick et al. using a smaller set of strains found most of these same regions to be divergent and phage related (52). The CGF loci used in this study were selected to capture the diversity in the E. coli O157:H7 population, not to reproduce the dendrogram created using microarray data from the original strains used in the study by Zhang et al. (55). It is therefore interesting to see that the relationships observed among strains with the microarray data have been maintained in the CGF-based dendrogram. We therefore conclude that CGF data provide a meaningful snapshot of the genetic diversity of the E. coli O157:H7 population and the genotypes of specific strains. This CGF method relies on 23 loci that are either present or absent and thus in theory offers 223 (8,388,608) possible fingerprints.
On the basis of octamer-based genome scanning, E. coli O157:H7 strains have been split into two distinct clonal lineages, lineage I and lineage II, that differ in genotype and host ecology. Lineage II strains are less frequently associated with human disease due either to inefficient transmission from bovine sources or lack of virulence to humans (26). The work of Zhang and colleagues identified an intermediate lineage that shared characteristics with both octamer-based genome scanning O157:H7 lineages (55). Those intermediary strains were designated lineage I/II and corresponded to LSPA6 type 211111 (53). All strains in this study grouped according to LSPA6 lineage, and each lineage formed its own branch of the dendrogram at a similarity threshold of 60%. It is worth noting that CGF data could be used to cluster strains into meaningful groups concordant with broad subtypes, such as LSPA6 lineage, and narrow subtypes, such as PT, respectively, depending on the percent similarity threshold value that was used, while targeting none of the loci used in LSPA6 typing. We have recently published a study in which a strong relationship between PT and LSPA6 lineage genotype was shown to exist among E. coli O157:H7 strains from Canada (57). In that study we found that PT23, PT45, PT54, PT67, and PT74 were LSPA6-LII specific, that LSPA6-LII strains were significantly less likely to be isolated from humans than LSPA6-LI strains, and that these LSPA6-specific groups were phenotypically meaningful in traits such as toxin production and antimicrobial resistance. Thus, CGF appears to produce genotypically and phenotypically relevant subtyping data.
CGF was able not only to offer groupings of similar strains but also to provide discrimination within the groups, both of which are traits essential for a typing system (8). CGF analysis of our data set generated 49 distinct fingerprints among 54 unrelated strains and three highly related fingerprints among the 25 strains from a closed herd of cattle. Strains isolated from the same feedlot or farm have been shown to be highly clonal, with a few dominant subtypes frequently detected and other subtypes occasionally or sporadically detected (13, 16, 27). CGF analysis revealed two dominant subtypes (CGF29 and CGF30) and three sporadic subtypes (CGF28, CGF33, and CGF35) among the herd strains. The most common dominant subtype (CGF29) was exhibited by 15 of 20 herd strains isolated from 21 May 1996 through 9 May 1997, while five strains obtained during this sampling period each showed a one-locus difference with respect to CGF29. One of these variants, strain H2704 (isolated on 9 May 1997) and all five strains isolated in the subsequent sampling period (23 December 1997) shared the same fingerprint (CGF30) consisting of an absent locus fprn33 and comprised a second dominant subtype, suggesting a clonal shift in the dominant subtype of the herd strains. Locus fprn33 encodes a putative transcriptional regulator in the phage-related OI#76/S-loop#119, which also contains genes encoding hypothetical and tail fiber proteins. Microarray data showed that this entire genomic region was lost in a PT87 strain negative for fprn33 (55). It therefore seems that some loci are very stable over time and allow the differentiation of strains that diverged in the distant past, such as those specific to lineage, while others are highly unstable and allow the differentiation of closely related strains, such as those of the ADRI cattle herd. This is consistent with other findings that have found stability in genetic elements, such as the presence of stx2c being strongly associated with lineage II strains (56), and the fact that some phage-related genomic regions may prevent the incorporation of other genetic elements through changes in phage receptors or immunity to superinfection by other bacteriophages (14, 22). As many of the CGF loci are phage related, and some PTs have been shown to be lineage specific (57), such phage-related genetic elements may play a role in the stability of clusters identified by CGF. The PT87 herd isolates did not share a CGF fingerprint with other strains, and two dominant and three sporadic subtypes could be distinguished within them, suggesting that the method may be useful in separating and typing E. coli O157:H7 outbreak isolates.
While PFGE is currently the “gold standard” of typing and has been shown to differentiate between strains of the same PT, it requires highly trained staff and extensive standardization to generate accurate results that can be compared between laboratories (2, 30, 35, 39). Figure 2 demonstrates that the tolerance levels required to compare PFGE patterns across multiple gels create clusters of identical strains that may appear slightly different to the naked eye. Moreover, temporally and geographically unrelated strains are occasionally given the same typing designation by PFGE (36). This phenomenon was evident in our data where strains TS-97 and F5 were indistinguishable by PFGE despite being isolated over 6 years apart, whereas CGF typing identified five loci where these two strains differed. Conversely, three lineage II strains isolated from the province of Alberta over 4 years (AA619-2, 12491, and AA995-2) were shown to be genetically similar and clustered together with CGF, while PFGE typing did not group the strains and placed the lineage II strains AA995-2 and 12491 among those of lineage I. This suggests that while PFGE is more appropriate for identifying the source of an outbreak, phage typing appears to give more taxonomically relevant data. CGF combines the benefits of both, being able to group strains into taxonomically meaningful subtypes as well as discriminating between and within the groupings. As has been shown, PFGE type X01.0002 in this study was found to contain unrelated strains of various PTs, making it unsuitable for higher-level discrimination among E. coli O157:H7 strains. Additionally, when the strains from this study were clustered by PFGE pattern, although no PFGE type was observed in more than one lineage, the lineage designations were no longer specific to a grouping of strains. The low specificity of PFGE has led to the recommendation that PCR-based methods be used over PFGE in typing studies (8, 44).
As has been shown, every CGF profile was specific to a PT; excluding the PT87 herd strains there were four instances where CGF was incapable of discriminating between two strains. The question of whether seemingly unrelated strains, such as F30 and AA1002-1, both of CGF23 and isolated nearly 7 years apart, are actually similar or if CGF simply lacks the resolution to properly discern them was tested. Microarray data from the work of Zhang et al. (55) were examined where the strains 71074 and 09601Fe046.1, both of CGF52, had been previously analyzed. It was found that the genomic content of strains 71074 and 09601Fe046.1 differed from each other by 0.96% when clusters of two or more open reading frames were examined; the next most closely related strain, Zap0046, differed from 71074 and 09601Fe046.1 by 1.14% and 1.53%, respectively (data not shown). The CGF52 strains 71074 and 09601Fe046.1 were thus found to be extremely similar by microarray analysis and grouped together when the microarray data were hierarchically clustered among 31 E. coli O157:H7 strains of various PTs and lineage. Therefore, although the epidemiological data may not show an obvious linkage between strains of the same CGF type, the genomes of the strains examined showed that they were clearly related. The resolution of CGF is therefore such that confidence can be taken in the relatedness of strains grouped together, even in the absence of epidemiological data, a task for which PFGE proved unsuitable in the analysis of our strain collection.
CGF offers high reproducibility among replicates, as the analysis involves only the presence or absence of single bands, with no judgment of band size or pattern. Such ease of data acquisition allows immediate and accurate interlaboratory reproducibility and exchange. CGF does not require expensive sequencing equipment or analysis software, such as is required for MLST, MLVA, or PFGE, which lends itself to immediate implementation by any PCR-capable laboratory; the loci may also be easily transferable to higher-throughput platforms, such as single-tube suspension microarrays.
Further, the 23-locus fingerprint offers a glimpse of the genetic structure of the E. coli O157:H7 isolate being typed, not just anonymous bands generated from enzyme recognition sites, the sequences of a few loci, or the number of repeats of a small segment of DNA. Because the presence of specific genomic islands has been linked to increased virulence of strains of several bacterial species (17, 23), and as every locus in the fingerprint targets a region of a given genomic island, it is possible that a strain could be identified as “more” or “less” pathogenic to humans based on its CGF subtype.
Recently, Manning et al. (28) used single-nucleotide polymorphism (SNP) analysis to identify a unique group (clade 8) of hypervirulent E. coli O157:H7 strains in the United States. In our study CGF was shown to distinguish among E. coli O157:H7 strains with a high and low propensity for human infection, so it would be interesting to see if this hypervirulent clade of the organism could also be identified with this genotyping approach. Unfortunately, restrictions in the international transfer of pathogenic microorganisms have made it difficult to develop a common set of reference E. coli O157:H7 strains for use in genotyping studies. However, the availability of nucleotide sequence data for an increasing number of E. coli O157:H7 strains in GenBank may allow such a typing set to be available in silico.
This study presents the successful development of a high-resolution molecular typing method that exploits data obtained through comparative genomics-based population studies. A similar methodology has recently been described for typing Streptococcus pneumoniae (11), Campylobacter jejuni (49), and Escherichia coli strains (47), suggesting that this approach is likely to be useful in the development of next-generation genotyping methods. Subsequent iterations of VAP analysis targeting additional loci will be derived from new E. coli O157:H7 sequence data as they become available. There are 16 sequencing projects currently under way on this pathogen available at the NCBI “genomes in progress” web page. At the very least, insight into the presence or absence of the 23 loci is a starting point for further genetic analysis.
Conclusion.
CGF was shown to be superior to both phage typing and the current typing standard, PFGE, as it offers comparable resolution between typed strains and much greater specificity. It also benefits from being a simple and fast PCR-based assay generating binary results and one that requires minimal training for both the actual testing and the data analysis, simplifying interlaboratory comparisons. Since CGF is a molecular typing method developed from whole-genome comparisons and O157:H7 lineage-specific sequences, it targets regions found to be most variable among strains, and thus most amenable for use in typing. Furthermore, the subtype groupings given by CGF appear to be phenotypically meaningful, while those given by PFGE appear not to be. In the future, as new sequence data become available, additional variable loci should be evaluated for their use in high-resolution typing of this pathogen. CGF could be an alternative or adjunct to PFGE typing, and its performance should be evaluated against other emerging molecular typing methods, such as MLVA.
Acknowledgments
We thank the Canadian Food Inspection Agency for use of research facilities at the Animal Diseases Research Institute, Lethbridge, Alberta. Additional thanks are extended to Clayton Ross of Lethbridge College for experimental work and Irene Yong and Shelley Frost from the Laboratory for Foodborne Zoonoses in Guelph, Ontario, for typing results.
This research was funded by the Public Health Agency of Canada.
Footnotes
Published ahead of print on 12 September 2008.
REFERENCES
- 1.Ahmed, R., C. Bopp, A. Borczyk, and S. Kasatiya. 1987. Phage-typing scheme for Escherichia coli O157:H7. J. Infect. Dis. 155:806-809. [DOI] [PubMed] [Google Scholar]
- 2.Aires-de-Sousa, M., K. Boye, H. de Lencastre, A. Deplano, M. C. Enright, J. Etienne, A. Friedrich, D. Harmsen, A. Holmes, X. W. Huijsdens, A. M. Kearns, A. Mellmann, H. Meugnier, J. K. Rasheed, E. Spalburg, B. Strommenger, M. J. Struelens, F. C. Tenover, J. Thomas, U. Vogel, H. Westh, J. Xu, and W. Witte. 2006. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44:619-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avery, S. M., E. Liebana, C. Reid, M. J. Woodward, and S. Buncic. 2002. Combined use of two genetic fingerprinting methods, pulsed-field gel electrophoresis and ribotyping, for characterization of Escherichia coli O157 isolates from food animals, retail meats, and cases of human disease. J. Clin. Microbiol. 40:2806-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, J. G. Wells, B. P. Bell, K. D. Greene, J. Lewis, and P. M. Griffin. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beutin, L., S. Kaulfuss, S. Herold, E. Oswald, and H. Schmidt. 2005. Genetic analysis of enteropathogenic and enterohemorrhagic Escherichia coli serogroup O103 strains by molecular typing of virulence and housekeeping genes and pulsed-field gel electrophoresis. J. Clin. Microbiol. 43:1552-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Böhm, H., and H. Karch. 1992. DNA fingerprinting of Escherichia coli O157:H7 strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 30:2169-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bopp, D. J., B. D. Sauders, A. L. Waring, J. Ackelsberg, N. Dumas, E. Braun-Howland, D. Dziewulski, B. J. Wallace, M. Kelly, T. Halse, K. A. Musser, P. F. Smith, D. L. Morse, and R. J. Limberger. 2003. Detection, isolation, and molecular subtyping of Escherichia coli O157:H7 and Campylobacter jejuni associated with a large waterborne outbreak. J. Clin. Microbiol. 41:174-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burucoa, C., V. Lhomme, and J. L. Fauchere. 1999. Performance criteria of DNA fingerprinting methods for typing of Helicobacter pylori isolates: experimental results and meta-analysis. J. Clin. Microbiol. 37:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calderwood, S. B., and J. J. Mekalanos. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 169:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooley, M., D. Carychao, L. Crawford-Miksza, M. T. Jay, C. Myers, C. Rose, C. Keys, J. Farrar, and R. E. Mandrell. 2007. Incidence and tracking of Escherichia coli O157:H7 in a major produce production region in California. PLoS ONE 2:e1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dagerhamn, J., C. Blomberg, S. Browall, K. Sjöström, E. Morfeldt, and B. Henriques-Normark. 2008. Determination of accessory gene patterns predicts the same relatedness among strains of Streptococcus pneumoniae as sequencing of housekeeping genes does and represents a novel approach in molecular epidemiology. J. Clin. Microbiol. 46:863-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erickson, M. C., and M. P. Doyle. 2007. Food as a vehicle for transmission of Shiga toxin-producing Escherichia coli. J. Food Prot. 70:2426-2449. [DOI] [PubMed] [Google Scholar]
- 13.Faith, N. G., J. A. Shere, R. Brosch, K. W. Arnold, S. E. Ansay, M. S. Lee, J. B. Luchansky, and C. W. Kaspar. 1996. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl. Environ. Microbiol. 62:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fogg, P. C. M., S. M. Gossage, D. L. Smith, J. R. Saunders, A. J. McCarthy, and H. E. Allison. 2007. Identification of multiple integration sites for Stx-phage φ24b in the Escherichia coli genome, description of a novel integrase and evidence for a functional anti-repressor. Microbiology 153:4098-4110. [DOI] [PubMed] [Google Scholar]
- 15.Friesema, I., B. Schimmer, O. Stenvers, A. Heuvelink, E. de Boer, K. van der Zwaluw, C. de Jager, D. Notermans, I. van Ouwerkerk, R. de Jonge, and W. van Pelt. 2007. STEC O157 outbreak in the Netherlands, September-October 2007. Euro. Surveill. 12:E071101.1. [DOI] [PubMed] [Google Scholar]
- 16.Gannon, V. P. J., T. A. Graham, R. King, P. Michel, S. Read, K. Ziebell, and R. P. Johnson. 2002. Escherichia coli O157:H7 infection in cows and calves in a beef cattle herd in Alberta, Canada. Epidemiol. Infect. 129:163-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 18.Hahm, B., Y. Maldonado, E. Schreiber, A. K. Bhunia, and C. H. Nakatsu. 2003. Subtyping of foodborne and environmental isolates of Escherichia coli by multiplex-PCR, REP-PCR, PFGE, ribotyping and AFLP. J. Microbiol. Methods 53:387-399. [DOI] [PubMed] [Google Scholar]
- 19.Heir, E., B. A. Lindstedt, T. Vardund, Y. Wasteson, and G. Kapperud. 2000. Genomic fingerprinting of Shiga toxin-producing Escherichia coli (STEC) strains: comparison of pulsed-field gel electrophoresis (PFGE) and fluorescent amplified-fragment-length polymorphism (FAFLP). Epidemiol. Infect. 125:537-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyytiä-Trees, E., S. C. Smole, P. A. Fields, B. Swaminathan, and E. M. Ribot. 2006. Second generation subtyping: a proposed Pulsenet protocol for multiple-locus variable-number tandem repeat analysis of Shiga toxin-producing Escherichia coli O157 (STEC O157). Foodborne Pathog. Dis. 3:118-131. [DOI] [PubMed] [Google Scholar]
- 21.Iyoda, S., A. Wada, J. Weller, S. J. Flood, E. Schreiber, B. Tucker, and H. Watanabe. 1999. Evaluation of AFLP, a high-resolution DNA fingerprinting method, as a tool for molecular subtyping of enterohemorrhagic Escherichia coli O157:H7 isolates. Microbiol. Immunol. 43:803-806. [DOI] [PubMed] [Google Scholar]
- 22.Kameyama, L., L. Fernández, J. Calderón, A. Ortiz-Rojas, and T. A. Patterson. 1999. Characterization of wild lambdoid bacteriophages: detection of a wide distribution of phage immunity groups and identification of a Nus-dependent, nonlambdoid phage group. Virology 263:100-111. [DOI] [PubMed] [Google Scholar]
- 23.Karmali, M. A., M. Mascarenhas, S. Shen, K. Ziebell, S. Johnson, R. Reid-Smith, J. Isaac-Renton, C. Clark, K. Rahn, and J. B. Kaper. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with Vero cytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41:4930-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawamori, F., M. Hiroi, T. Harada, K. Ohata, K. Sugiyama, T. Masuda, and N. Ohashi. 2008. Molecular typing of Japanese Escherichia coli O157:H7 isolates from clinical specimens by multilocus variable-number tandem repeat analysis and PFGE. J. Med. Microbiol. 57:58-63. [DOI] [PubMed] [Google Scholar]
- 25.Khakhria, R., D. Duck, and H. Lior. 1990. Extended phage-typing scheme for Escherichia coli O157:H7. Epidemiol. Infect. 105:511-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, J., J. Nietfeldt, and A. K. Benson. 1999. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc. Natl. Acad. Sci. USA 96:13288-13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeJeune, J. T., T. E. Besser, D. H. Rice, J. L. Berg, R. P. Stilborn, and D. D. Hancock. 2004. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover. Appl. Environ. Microbiol. 70:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manning, S. D., A. S. Motiwala, A. C. Springman, W. Qi, D. W. Lacher, L. M. Ouellette, J. M. Mladonicky, P. Somsel, J. T. Rudrik, S. E. Dietrich, W. Zhang, B. Swaminathan, D. Alland, and T. S. Whittam. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. USA 105:4868-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neely, M. N., and D. I. Friedman. 1998. Arrangement and functional identification of genes in the regulatory region of lambdoid phage H-19B, a carrier of a Shiga-like toxin. Gene 223:105-113. [DOI] [PubMed] [Google Scholar]
- 32.Noller, A. C., M. C. McEllistrem, A. G. F. Pacheco, D. J. Boxrud, and L. H. Harrison. 2003. Multilocus variable-number tandem repeat analysis distinguishes outbreak and sporadic Escherichia coli O157:H7 isolates. J. Clin. Microbiol. 41:5389-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noller, A. C., M. C. McEllistrem, O. C. Stine, J. G. J. Morris, D. J. Boxrud, B. Dixon, and L. H. Harrison. 2003. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogura, Y., T. Ooka, Asadulghani, J. Terajima, J. Nougayrède, K. Kurokawa, K. Tashiro, T. Tobe, K. Nakayama, S. Kuhara, E. Oswald, H. Watanabe, and T. Hayashi. 2007. Extensive genomic diversity and selective conservation of virulence-determinants in enterohemorrhagic Escherichia coli strains of O157 and non-O157 serotypes. Genome Biol. 8:R138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pei, Y., J. Terajima, Y. Saito, R. Suzuki, N. Takai, H. Izumiya, T. Morita-Ishihara, M. Ohnishi, M. Miura, S. Iyoda, J. Mitobe, B. Wang, and H. Watanabe. 2008. Molecular characterization of enterohemorrhagic Escherichia coli O157:H7 isolates dispersed across Japan by pulsed-field gel electrophoresis and multiple-locus variable-number tandem repeat analysis. Jpn. J. Infect. Dis. 61:58-64. [PubMed] [Google Scholar]
- 37.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pradel, N., Y. Bertin, C. Martin, and V. Livrelli. 2008. Molecular analysis of Shiga toxin-producing Escherichia coli strains isolated from hemolytic uremic syndrome patients and dairy samples in France. Appl. Environ. Microbiol. 74:2118-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preston, M. A., W. Johnson, R. Khakhria, and A. Borczyk. 2000. Epidemiologic subtyping of Escherichia coli serogroup O157 strains isolated in Ontario by phage typing and pulsed-field gel electrophoresis. J. Clin. Microbiol. 38:2366-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.PulseNetUSA. 2004. One-day (24-28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7, non-typhoidal Salmonella serotypes, and Shigella sonnei by pulsed field gel electrophoresis (PFGE). PulseNet PFGE manual 5.1-5.3:1-13. [Google Scholar]
- 41.Shen, S., M. Mascarenhas, R. Morgan, K. Rahn, and M. A. Karmali. 2005. Identification of four fimbria-encoding genomic islands that are highly specific for verocytotoxin-producing Escherichia coli serotype O157 strains. J. Clin. Microbiol. 43:3840-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shima, K., J. Terajima, T. Sato, K. Nishimura, K. Tamura, H. Watanabe, Y. Takeda, and S. Yamasaki. 2004. Development of a PCR-restriction fragment length polymorphism assay for the epidemiological analysis of Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 42:5205-5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shima, K., N. Yoshii, M. Akiba, K. Nishimura, M. Nakazawa, and S. Yamasaki. 2006. Comparison of PCR-RFLP and PFGE for determining the clonality of enterohemorrhagic Escherichia coli strains. FEMS Microbiol. Lett. 257:124-131. [DOI] [PubMed] [Google Scholar]
- 44.Shima, K., Y. Wu, N. Sugimoto, M. Asakura, K. Nishimura, and S. Yamasaki. 2006. Comparison of a PCR-restriction fragment length polymorphism (PCR-RFLP) assay to pulsed-field gel electrophoresis to determine the effect of repeated subculture and prolonged storage on RFLP patterns of Shiga toxin-producing Escherichia coli O157:H7. J. Clin. Microbiol. 44:3963-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siegler, R. L. 1995. The hemolytic uremic syndrome. Pediatr. Clin. North Am. 42:1505-1529. [DOI] [PubMed] [Google Scholar]
- 46.Sigmundsdottir, G., A. Atladottir, H. Hardardottir, E. Gudmundsdottir, M. Geirsdottir, and H. Briem. 2007. STEC O157 outbreak in Iceland, September-October 2007. Euro. Surveill. 12:E071101.2. [DOI] [PubMed] [Google Scholar]
- 47.Srinivasan, U., L. Zhang, A. M. France, D. Ghosh, W. Shalaby, J. Xie, C. F. Marrs, and B. Foxman. 2007. Probe hybridization array typing: a binary typing method for Escherichia coli. J. Clin. Microbiol. 45:206-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steele, M., K. Ziebell, Y. Zhang, A. Benson, R. Johnson, E. Taboada, and V. Gannon. 2007. Distribution of OBGS lineage I and lineage II conserved regions among non-O157 E. coli serotypes, abstr. B-285/145. Abstr. 107th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 49.Taboada, E. N., J. M. Mackinnon, J. Johnson, M. J. Roberts, S. Ross, W. O. S. Mauro, A. Ratansi, J. Yan, J. A. Lorentz, J. Thomas, K. Rahn, and V. P. J. Gannon. 2007. The use of high-throughput comparative genomics-based molecular typing enhances cluster detection in epidemiological studies of Campylobacter jejejuni. Campylobacter Helicobacter-Related Organisms 2007 Meet., 2 to 5 September 2007, Rotterdam, The Netherlands. Zoonoses Public Health 54(Suppl. 1):20-0008. [Google Scholar]
- 50.Tsai, T., W. Luo, F. Wu, and T. Pan. 2005. Molecular subtyping for Escherichia coli O157:H7 isolated in Taiwan. Microbiol. Immunol. 49:579-588. [DOI] [PubMed] [Google Scholar]
- 51.van Belkum, A., P. T. Tassios, L. Dijkshoorn, S. Haeggman, B. Cookson, N. K. Fry, V. Fussing, J. Green, E. Feil, P. Gerner-Smidt, S. Brisse, and M. Struelens. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl. 3):1-46. [DOI] [PubMed] [Google Scholar]
- 52.Wick, L. M., W. Qi, D. W. Lacher, and T. S. Whittam. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 187:1783-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, Z., J. Kovar, J. Kim, J. Nietfeldt, D. R. Smith, R. A. Moxley, M. E. Olson, P. D. Fey, and A. K. Benson. 2004. Identification of common subpopulations of non-sorbitol-fermenting, β-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl. Environ. Microbiol. 70:6846-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaidi, N., K. Konstantinou, and M. Zervos. 2003. The role of molecular biology and nucleic acid technology in the study of human infection and epidemiology. Arch. Pathol. Lab. Med. 127:1098-1105. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, Y., C. Laing, M. Steele, K. Ziebell, R. Johnson, A. K. Benson, E. Taboada, and V. P. J. Gannon. 2007. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics 8:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao, S., S. E. Mitchell, J. Meng, S. Kresovich, M. P. Doyle, R. E. Dean, A. M. Casa, and J. W. Weller. 2000. Genomic typing of Escherichia coli O157:H7 by semi-automated fluorescent AFLP analysis. Microbes Infect. 2:107-113. [DOI] [PubMed] [Google Scholar]
- 57.Ziebell, K., M. Steele, Y. Zhang, A. Benson, E. Taboada, C. Laing, S. McEwan, B. Ciebin, R. Johnson, and V. Gannon. 2008. Genotypic characterization and prevalence of virulence factors among Canadian Escherichia coli O157:H7 strains. Appl. Environ. Microbiol. 74:4314-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]