Abstract
The Sinorhizobium meliloti ORFeome project cloned 6,314 open reading frames (ORFs) into a modified Gateway entry vector system from which the ORFs could be transferred to destination vectors in vivo via bacterial conjugation. In this work, a reporter gene destination vector, pMK2030, was constructed and used to generate ORF-specific transcriptional fusions to β-glucuronidase (gusA) and green fluorescent protein (gfp) reporter genes. A total of 6,290 ORFs were successfully transferred from the entry vector library into pMK2030. To demonstrate the utility of this system, reporter plasmids corresponding to 30 annotated sugar kinase genes were integrated into the S. meliloti SM1021 and/or SM8530 genome. Expression of these genes was measured using a high-throughput β-glucuronidase assay to track expression on nine different carbon sources. Six ORFs integrated into SM1021 and SM8530 had different basal levels of expression in the two strains. The annotated activities of three other sugar kinases were also confirmed.
Sinorhizobium meliloti forms a nitrogen-fixing symbiotic relationship with alfalfa by inducing root nodules on the host plant. Within the root nodule, the bacterium converts atmospheric dinitrogen into ammonia, a nitrogen compound alfalfa can assimilate, by using carbon-containing compounds provided by the plant to generate the energy and reductant needed for the nitrogenase reaction (19). Similar nitrogen-fixing symbiotic relationships with other legume hosts provide an important input of reduced nitrogen to natural and agricultural ecosystems.
The S. meliloti genome consists of a 3.7-Mb chromosome and two megaplasmids, pSymA (1.4 Mb) and pSymB (1.7 Mb), which collectively encode approximately 6,314 open reading frames (ORFs) (2, 8, 12, 27). Bioinformatic analysis has assigned many of these ORFs to functional categories based on similarities to genes for which there is more information, but there are limits to the precision of these assignments. Some of the S. meliloti ORFs have been characterized through genetic, biochemical, or physiological analysis, but a majority of the ORFs have not been examined directly.
Guided by the S. meliloti 1021 genome sequence (2, 8, 12), genome-scale tools to measure gene expression have been developed (3, 4, 10). Cowie et al. (9) constructed a reporter gene fusion library that contains 2,874 unique reporter fusions by insertion of random genomic DNA fragments into a vector. The resulting plasmids were then inserted into the genome of SM1021 via homologous recombination and resulted in transcriptional fusions or loss-of-function reporter insertions. There is limited utility in extending this approach, since fusions to unrepresented genes will be increasingly difficult to obtain using random DNA insertions. The S. meliloti ORFeome (17, 27) was designed to permit the rapid and cost-effective transfer of a comprehensive set of ORFs from a library of 6,314 entry clones into destination vectors. This ORFeome was constructed by bacteriophage lambda recombination, as implemented in the Invitrogen Gateway cloning system, using a modified entry vector that allowed in vivo transfer of ORFs to various destination vectors using bacterial mating (17, 27). This transfer of ORFs into destination vectors, and their subsequent integration into the SM1021 genome, was reported for a limited number of genes, but the application of this technique on a genome-level scale was not demonstrated (17).
In this work, we describe the construction of a reporter-type destination vector, pMK2030, which places the ORF upstream of the gfp gene, which codes for green fluorescent protein (GFP), and the gusA gene, which codes for β-glucuronidase (GUS). By using in vivo mating, 6,290 ORFs from the S. meliloti ORFeome library were transferred into pMK2030. The subsequent integration of these ORF-containing reporter plasmids into S. meliloti, by homologous recombination, places the reporter genes downstream of the native ORF, making their expression dependent on the natural promoter. Reporter gene activity was confirmed in S. meliloti, and a high-throughput liquid GUS assay was validated and allowed for large-scale screens of multiple ORFs. The expression levels of ORFs annotated as sugar kinases were analyzed by measuring the GUS activities of the resulting fusions under various growth conditions.
MATERIALS AND METHODS
Bacterial strains and media.
Strains used in this study (Table 1) were cultured at 33°C or 37°C (Escherichia coli) or 30°C (S. meliloti). The E. coli strains were cultured on Luria-Bertani (LB) broth or agar (26). E. coli containing the plasmid pRK2073 (Table 2) was cultured on M9 medium (26) supplemented with 1% vitamin-free Casamino Acids. For the E. coli strains, media were supplemented with antibiotics at the following final concentrations: tetracycline, 10 μg/ml (Tet10); rifampin, 100 μg/ml (Rif100); trimethoprim, 100 μg/ml (Tmp100); kanamycin, 40 μg/ml or 75 μg/ml (Kan40 or Kan75); chloramphenicol, 50 μg/ml (Cam50). Isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added to LB medium at a final concentration of 20 μg/ml (IPTG20) as indicated. S. meliloti strains were cultured on yeast mannitol broth (YMB) (28), Luria-Bertani magnesium calcium (LBMC) (23), or minimal ammonium (MNH4) medium (28) supplemented with 0.2% succinate, d-glucose, l-arabinose, d-raffinose, d-galactose, d-fructose, l-malate, d-mannitol, or glycerol. S. meliloti medium was supplemented with antibiotics at the following final concentrations: tetracycline, 1 μg/ml (Tet1); naladixic acid, 40 μg/ml (Nal40); cephalothin, 15 μg/ml (Ceph15).
TABLE 1.
E. coli and S. meliloti strains
| Strain | Genotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| DB3.1 | F−gyrA462 endA1 Δ(sr1-recA) mcrB mrr hsdS20(rB− mB−) supE44 ara-14 galK2 lacY1 proA2 rpsL20(Strr) xyl-5 Δleu mtl-1 | 5 |
| DB3.1 λpir | λpir lysogen of DB3.1 | 17 |
| DH5α | φ80lacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rk− mk+) supE44 relA1 deoR phoA Δ(lacZYA-argF)U169 | 15 |
| DH5α λpir | λpir lysogen of DH5α | 24 |
| HB101 | thi-1 hsdS20(rB− mB−) supE44 recA13 ara-14 leuB6 proA2 lacY1 galK2 rpsL20(Strr) xyl-5 mtl-1 | 6 |
| HB101R | Rifr derivative of HB101 | 17 |
| HB101R λpir | λpir lysogen of HB101R | This work |
| S. meliloti strains | ||
| SM1021 | SU47 str-21 | 21 |
| SM8530 | SM1021 expR+ | 14 |
| SM1021::pRG1SMxxxxx | SM1021 with pRG1 construct integrated into genome | This work |
| SM8530::pRG1SMxxxxx | SM8530 with pRG1 construct integrated into genome | This work |
TABLE 2.
Plasmids used and constructed in this work
| Plasmid | Description | Source or reference |
|---|---|---|
| pESMxxxxx | Kanr; entry vector with ORF inserted | 27 |
| pHRE1-Km | Donor of gfp and gusA genes | 25 |
| pKNOCK-Tc | Donor of RP4 oriT, R6K ori and tet | 1 |
| pMK2014 | Kanr; FRT-ccdB-Camr-FRT cassette | 17 |
| pMK2025 | pKNOCK-Tc with FRT cassette from pMK2014 | This work |
| pMK2030 | Tetr Camr; gfp and gusA reporter vector | This work |
| pMK2039 | pHRE1-Km missing SalI fragment | This work |
| pRG1SMxxxxx | Tetr; pMK2030 with ORF inserted | This work |
| pRK2013 | Kanr; helper plasmid for mobilization | 11 |
| pRK2073 | Tmpr; helper plasmid for mobilization | 20 |
| pXINT129 | Kanr; λ int and xis driven by Plac | 24 |
pMK2030 construction.
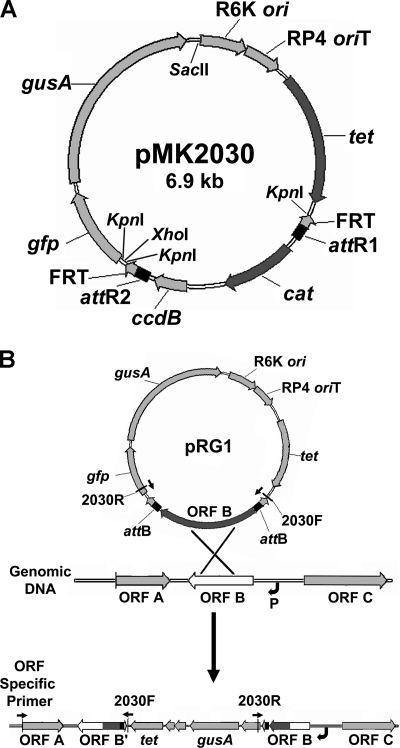
The reporter plasmid pMK2030 is composed of three DNA fragments originating from plasmids pMK2014, pKNOCK-Tc, and pHRE1-Km (Table 2; Fig. 1A). pMK2014 contributed a 1.8-kb FRT cassette, which contains the ccdB gene and a chloramphenicol acetyltransferase (cat) gene flanked by attR and FRT sequences. pMK2014 was digested with KpnI, and the FRT cassette was inserted into KpnI-digested pKNOCK-Tc to produce plasmid pMK2025. In addition to the FRT cassette, pMK2025 contains a tetracycline resistance gene (tet), RP4 oriT sequences for mobilization, and an oriV replication origin from plasmid R6K that is active only in strains containing the trans-acting R6K Pi replication protein. To obtain the reporter gene sequences, pHRE1-Km was digested with SalI and religated in order to remove a SalI fragment containing the kanamycin resistance gene, resulting in plasmid pMK2039. pMK2039 was then digested with both SacII and XhoI to release a SacII-XhoI gfp-gusA fragment, which was then ligated to pMK2025 DNA that had been cut with SacII and XhoI. The resulting plasmid was designated pMK2030 (Fig. 1A). pMK2030 is maintained in DB3.1 λpir (Table 1), which produces the R6K Pi protein.
FIG. 1.
(A) Map of reporter destination vector pMK2030. The restriction enzyme sites used in cloning the plasmid are marked on the interior of the diagram. The ccdB and cat genes between the attR1 and attR2 sites are replaced by ORF sequence from the entry vector during pentaparental mating. Because of the orientation of the ORF in the original construction, the gfp and gusA genes will always be downstream of the ORF after it is inserted by recombination. (B) Diagram of integration of pRG1 into S. meliloti genomic DNA. Single-crossover homologous recombination occurs between the ORF sequences in pRG1 and the genomic DNA, resulting in insertion of the pRG1 vector into the genomic DNA. Small black arrows indicate the direction of DNA amplification from primers 2030F, 2030R, and an ORF-specific primer. The curved black arrow indicates a DNA promoter region (P).
Generation of pRG1 constructs.
Pentaparental conjugation was used to transfer the cloned ORFs from the S. meliloti ORFeome (27) entry vectors (pEntry or pESMxxxxx) to the pMK2030 destination vector. SMxxxxx refers to the ORF ID number which originated from http://bioinfo.genopole-toulouse.prd.fr/annotation/iANT/bacteria/rhime/ and http://www.bioinformatics.wsu.edu/kahn/. The mating mix is composed of five E. coli strains (Tables 1 and 2): DH5α containing the entry vector (pESMxxxxx); DB3.1 (pRK2073) containing the mobilization helper plasmid; DH5α λpir containing pXINT129, from which the λ int and xis recombination factors are expressed; DB3.1 λpir (pMK2030); and HB101R λpir, the rifampin-resistant final host. To begin the conjugation, the pEntry isolates were transferred from an archival stock microplate into a 96-well deep well block (Qiagen, Valencia, CA) and filled with 1.3 ml LB-Kan75 by using a sterile 96-prong replicator tool (V & P Scientific Inc., San Diego, CA), and the 96 strains containing 96 different ORFs were grown overnight at 37°C with agitation. LB broth cultures (10 ml), with the appropriate antibiotic (Tables 1 and 2), of HB101R λpir and the pentaparental mating strains containing pMK2030, pXINT129, and pRK2073 were cultured separately overnight at 37°C with agitation. These strains were harvested by centrifugation at 4,000 × g for 5 min using a Sorvall RC 5B Plus centrifuge and SA-600 rotor and resuspended in 10 ml of sterile, chilled 0.85% NaCl in order to remove residual medium and antibiotics. This process was repeated twice. Five-ml aliquots of each washed culture were then combined in a 50-ml polypropylene tube and used as the four-parent mating mix. A 200-μl aliquot of each of the 96 pEntry clones was removed from the deep well block and placed in a 96-well microtiter plate. The cells were harvested by centrifugation at 885 × g at 4°C for 1 h by using a microtiter plate rotor (MTM 6.4) in a Jouan MR22i centrifuge. The supernatant was removed, and the cells were resuspended in 200 μl sterile, chilled 0.85% NaCl.
The four-parent mating mix was dispensed into a sterile, single-well, rectangular plate (OmniTray; Nalge Nunc, New York). A sterile 96-prong replicator was placed into the single-well plate containing the mating mix and then onto an OmniTray containing LB-IPTG20 agar. The replicator deposits approximately 10 μl of the mating mix onto the agar, and this was allowed to dry. The 96-prong replicator was flame sterilized, allowed to cool, and placed into the microtiter plate carrying the 96 pEntry vectors. The replicator was then placed onto the previously inoculated LB-IPTG20 agar, dispensing approximately 10 μl of each of the pEntry cultures on top of the spots containing the mating mix strains. The liquid was allowed to absorb into the agar and the plate was incubated for 8 h at 33°C. The resulting bacterial growth was removed with a sterile toothpick, streaked across the entire surface of an LB-Tet10Rif100 agar plate, and incubated overnight at 33°C in order to obtain individual colonies. The resulting transconjugants (about 200 to 400 colonies per plate) were screened by replica plating on LB-Tet10Rif100 and LB-Kan75 using a modified pin art toy (Toysmith, Auburn, WA). For each ORF, a single colony that grew on LB-Tet10Rif100, but not on LB-Kan75, was transferred again to LB-Tet10Rif100 and tested for kanamycin (LB-Kan75) and chloramphenicol (LB-Cam50) sensitivity, indicating that the pEntry plasmid was not present and that the Tetr pMK2030 derivative had lost the counterselection ccdB and Camr. If the first six colonies chosen for each ORF were not Kans Cams, 50 more colonies were screened before a remating was done. Once a colony was confirmed to have the correct antibiotic resistance properties, the size of the fragment inserted into pMK2030 was confirmed to be the same size as the ORF from the donor pEntry vector by PCR (see below). Plasmids containing inserts of the correct size were designated as pRG1 clones and given the construct name including the ORF ID number (pRG1SMxxxx). Freezer stocks with 20% dimethyl sulfoxide were made for all positive clones.
Conjugations that did not yield successful clones were remated using a modified version of the protocol. The parental strains were cultured in the same manner and media, except the pEntry strains were cultured in tubes of 3 ml LB-Kan75 instead of in a 96-well format. One-ml aliquots of all the strains were washed three times with 0.85% NaCl by using a microcentrifuge. Equal volumes of the five strains were combined, and 25 μl of the mating mix was placed on a sterile piece of nitrocellulose that was lying on LB-IPTG20 agar. The mating was incubated at 33°C for no more than 12 h, and then the nitrocellulose piece was transferred to a tube containing 1 ml of sterile water. The tube was vortexed to resuspend the cells and diluted 10-fold in sterile water. A 100-μl aliquot of the dilution was spread plated onto a LB-Tet10Rif100 plate and incubated at 33°C for 48 h. The resulting colonies were screened for kanamycin and chloramphenicol sensitivity as described above and confirmed by PCR (see below).
Integration of pRG1 clones into the S. meliloti genome.
Triparental matings were used to transfer the pRG1 constructs into S. meliloti strains SM1021 and SM8530 (Table 1). The strains used in the triparental matings were E. coli HB101 λpir containing the pRG1 clones, E. coli DB3.1 containing the helper plasmid pRK2013, and SM8530 or SM1021 as recipients (Tables 1 and 2). S. meliloti strains were cultured in YMB for 48 h at 30°C, and the E. coli strains were cultured at 37°C overnight in LB-Kan40 for pRK2013 and LB-Tet10 for pRG1 clones. All cultures were washed and resuspended with chilled, sterile 0.85% NaCl equal to the original volume of cells washed. Cells were mixed in a 4:1:1 ratio of SM1021 or SM8530, pRK2013, and pRG1SMxxxxx, respectively, in a total volume of 100 μl. For high-throughput matings, the cells were combined in wells of a 96-well plate and then stamped onto LBMC agar using a 96-prong replicator tool. The mating mixtures were incubated at 30°C for 48 h and then transferred with the replicator tool to LBMC-Tet1Nal40Ceph15 in order to kill the E. coli cells. The colonies that grew after incubation for 48 h at 30°C were then streaked for single colonies on individual LBMC-Tet1Nal40Ceph15 plates, and these plates were incubated for 96 h at 30°C. Isolated colonies were further purified by patching (50 isolates/plate) onto MNH4-Succ-Tet1 and incubated for 72 h at 30°C. Of the bacteria that grew, two isolates for each ORF were three-phase streaked onto MNH4-Succ-Tet1, allowed to grow at 30°C for 72 h, and then screened by PCR for recombination of the pRG1 plasmid into the chromosome (see below). Small-scale triparental matings, and remates of previously failed pRG1 integrations, were essentially done as the high-throughput matings with a few changes. A 25-μl aliquot of the mating mixture was placed on a sterile nitrocellulose piece lying on LBMC agar. After 48 h, the nitrocellulose piece was placed in a tube containing 1 ml of sterile water and the cells were resuspended by vortexing. The cell suspension was diluted 1:100 and 1:50 in sterile water, and 100 μl of each dilution was spread plated on LBMC-Tet1Nal40Ceph15 agar and incubated at 30°C for 72 h. The resulting colonies were then streaked on MNH4-Succ-Tet1 and screened by PCR.
Colony screening by PCR.
Cell lysis PCR was performed to confirm that the ORF inserted into the pRG1 constructs during the pentaparental mating was the same size as the ORF carried in the respective pEntry construct. The primers attBUF (5′-AGGCTTAGGAGGCTCTTCAATG-3′) and attBUR (5′-ACTTTGTACAAGAAAGCTGGGTTC-3′) flank the attB sites in pRG1 (Fig. 1) and amplify the region containing the ORF inserts. The PCR conditions were as previously described (27). Colonies resulting from the triparental matings that grew on MNH4-Succ-Tet1 agar were screened by PCR to confirm integration of the pRG1 construct into the SM1021 or SM8530 genome. This was accomplished by demonstrating the linkage of an adjacent gene to the pMK2030 sequence. Figure 1B shows the location of the primers used in this screen. The direction of the ORF of interest (ORF B′ in Fig. 1B) with respect to the upstream ORF (ORF A in Fig. 1B) determined which primer set was used. One primer was specific for the ORF upstream of the target and was previously used to amplify the ORF from the genome (27). The sequences for the gene-specific primers can be found in the S. meliloti ORFeome database at http://www.bioinformatics.wsu.edu/kahn. If the ORF was transcribed from the top (5′) strand, 2030R (5′-GCATCACCTTCACCCTCTCC-3′) was used as the second primer; if the ORF was transcribed from the bottom (3′) strand, 2030F (5′-AAGCTGACGCGTCCTCGGTA-3′) was used (Fig. 1B). The PCR was performed on colonies using a cell lysis protocol as described by Schroeder et al. (27).
β-Glucuronidase assays to quantify gene expression.
The standard GUS assay (22) measuring p-nitrophenyl-β-d-glucoronide (PNPG) hydrolysis was adapted for use in a 96-well microtiter plate format. Cultures were started from 96-well freezer stock plates by stamping with the 96-prong replicator tool onto YMB (for SM1021 strains) or LBMC (for SM8530 strains). The plates were incubated at 30°C for 48 h, and then a sterile replicator tool was used to inoculate 96-well, deep well blocks that contained 1.3 ml of MNH4 medium supplemented with one of nine carbon sources (see “Bacterial strains and media” above). The blocks were then covered with AirPore tape sheets (Qiagen, Valencia, CA) and placed on a platform shaker for 48 h at room temperature (22°C). Fifty-microliter aliquots of the cultures were then transferred to new deep well blocks containing 1.3 ml of the corresponding medium, covered with AirPore strips, and incubated for 48 h at room temperature on a platform shaker. Aliquots of 200 μl of cells were transferred into a microtiter plate from each block, and the cell density (the optical density at 600 nm [OD600]) was determined. Absorbance was measured using a Molecular Devices SpectraMax 250 plate reader and SoftMax Pro version 1.2.0 software. Aliquots of 100 μl of these cells were then transferred to a new microtiter plate in which the assay was carried out. Eight μl of the assay buffer (125 mM sodium phosphate dibasic, pH 7.0, 25 mM Na2H2EDTA, 0.25% Sarkosyl, 0.25% Triton X-100, and 25 mM 2-mercaptoethanol) was added to the cells, followed by 20 μl of 10 mM PNPG. Samples were incubated at room temperature for 70 min, and 100 μl of 0.5 M sodium carbonate was added to stop the reactions. Readings of the OD415 and OD600 were recorded. Two replications, of three samples for each strain, were performed. The activity of the GUS enzyme was calculated using the equation from Miller (22), the average blank value (uninoculated media) was subtracted, and the average of the six values for each strain was calculated. The Miller calculation coefficient that corrects for cell debris within the OD415 reading of the assay was used to normalize the data between medium types. The data were normalized by using coefficient values that gave Miller unit values of 19 to 21 for the wild-type SM1021 and SM8530 strains; the resulting values are listed for each medium type (Table 3). Biochemical pathway information was researched using the MetaCyc website (http://www.metacyc.org).
TABLE 3.
Correction factors for minimal ammonia media used in the GUS assay
| C source | Correction for Miller calculationa
|
|
|---|---|---|
| SM1021 | SM8530 | |
| Arabinose | 1.50 | 1.15 |
| Fructose | 1.50 | 0.97 |
| Galactose | 1.51 | 1.20 |
| Glucose | 1.60 | 1.17 |
| Glycerol | 1.25 | 1.20 |
| Malate | 1.45 | 1.13 |
| Mannitol | 1.44 | 1.05 |
| Raffinose | 1.56 | 1.25 |
| Succinate | 1.55 | 1.30 |
In the calculation method used by Miller, there is a term used to correct for the contribution to absorbance at 420 nm for the culture density, measured at 600 nm. This correction factor differed for the various media, and use of a single correction term was not adequate.
Plant inoculation and staining of nodules for GUS expression.
Alfalfa (Medicago sativa cv. Champ) seed was sterilized, planted, and inoculated as previously described (29). The nodules were harvested after 4 weeks and cut in half lengthwise by using a scalpel. The cut nodules were stained for GUS activity as described by Jefferson et al. (18). Nodules were photographed using an Olympus BH-2 light microscope and a Nikon Coolpix 990 digital camera after 3 and 20 h of staining. Nodules infected with wild-type SM1021 or SM8530 never showed evidence of GUS activity after 20 h.
Nucleotide sequence accession number.
The sequence of pMK2030 has been deposited in GenBank under accession no. EU825945.
RESULTS AND DISCUSSION
pMK2030 reporter construct.
A new destination vector, pMK2030, was designed to use the S. meliloti ORFeome library (27) for constructing reporter fusions. The S. meliloti ORFeome contains PCR-amplified ORFs inserted into pMK2010, a modified entry plasmid vector (17) that can use bacteriophage λ integrase and excisionase-mediated recombination to transfer cloned DNA between this vector and vectors with compatible att sites. pMK2030 (Fig. 1A) was constructed with attR sites to allow the transfer of ORFs from the attL-containing pEntry constructs using a pentaparental mating system. In pMK2030, the ccdB and cat genes are located between the attR sites. CcdB, which inhibits DNA gyrase and is toxic to most E. coli strains, allows for counterselection against pMK2030 vectors that have not recombined. E. coli DB3.1 (Table 1) is resistant to CcdB and was used to maintain pMK2030. The cat gene will also be removed in the attL-attR recombination, providing a chloramphenicol-sensitive phenotype for a recombined plasmid. Promoterless reporter genes gusA and gfp are located downstream of the attR sites in pMK2030. When the insert containing pMK2030 plasmid is recombined into the S. meliloti genome, by selecting for tetracycline resistance carried on pMK2030, the placement of the reporter genes connects their expression to the promoter controlling the native ORF. pMK2030 contains an R6K origin of replication (ori). This allows for replication in E. coli strains containing the R6K pir gene, but not in most other bacteria, including S. meliloti. pMK2030 must be maintained in an E. coli strain that has a λ prophage integrated into the chromosome, in this case DB3.1 λpir (Table 1). An RP4 origin of transfer (oriT), which allows for the mobilization of the plasmid during conjugation, is also present on pMK2030 (Fig. 1A). Inverted FRT sites allow for pMK2030 to be used to generate ORF deletions in S. meliloti (17).
Recombination of ORFs into pMK2030.
The standard Invitrogen Gateway method for transferring an ORF from an entry vector into a destination vector involves an LR clonase reaction to catalyze recombination between attL and attR sites in vitro. This is time-consuming and expensive because of the need for DNA purification, the reaction itself, and the subsequent transformation. A pentaparental mating protocol was developed to allow the recombination between attL and attR sites to be carried out in vivo (17). Using pentaparental conjugation, the ORFs were transferred from pEntry clones into pMK2030. ORFs from 6,290 (99.6%) of the 6,314 pEntry clones were successfully transferred into the pMK2030 construct in E. coli HB101R λpir. The constructs are collectively referred to as pRG1 and pRG1SMxxxxx for individual clones. The 24 clones that did not successfully transfer to pRG1, most of which are annotated as hypothetical proteins, were remated multiple times and appear to be recalcitrant to transfer. A list of the 24 ORFs that are not in the pRG1 library can be found at http://www.bioinformatics.wsu.edu/kahn/under the “pRG1 Library Information” link. The reporter vector pMK2030 accepts the transfer of specific ORFs from the pEntry ORFeome library via pentaparental mating and allowed for the directed construction of fusions representing most of the genes in S. meliloti. This approach supplements the S. meliloti-compatible reporter vector described by Cowie et al. (9), which is based on the cloning of random DNA fragments and therefore is unable to capture all of the target genes without specific constructions. Twelve of the ORFs not represented in the pRG1 library are present in the library constructed by Cowie et al. (9).
Integration of pRG1 constructs into S. meliloti SM8530.
The pRG1 construct must be recombined into the S. meliloti genome before expression data can be measured, since the expression of the gfp and gusA reporter genes depends on transcription initiating upstream of the inserted ORF. To date, 1,071 of the pRG1 constructs have been integrated into the genome of SM8530 by using triparental matings. After the pRG1 constructs integrate by homologous recombination, two functional copies of the ORF are present, flanking the integrated plasmid (Fig. 1B). Integration of pRG1 results in the loss of native regulation of downstream genes in an operon, but low-level constitutive expression of the downstream genes does occur via readthrough from the tetracycline (tet) promoter in pRG1. Addition of the appropriate tet repressor to a pRG1 strain represses the tet promoter and leads to expression of downstream genes that can be induced by the addition of dihydrotetracycline (S. Clark and M. L. Kahn, unpublished data).
The original library was constructed with DNA sequences from SM1021, but the pRG1 clones were integrated into SM8530. SM8530 is a derivative of SM1021 that has a functional expR gene as the result of the excision of a natural insertion sequence. ExpR is a LuxR transcriptional regulator that mediates quorum sensing (23). ExpR regulates at least 200 genes, including many involved in exopolysaccharide production, motility, and chemotaxis (13, 16). Since ExpR regulates genes of many key processes, including some involved in symbiosis, we decided to construct the reporter library in the SM8530 background. The pRG1 constructs are maintained in an E. coli library, so the utility of the reporter constructs is that they can be used with any SM1021 derivative and in other strains with close DNA sequence identity.
The efficiency of the triparental mating used to integrate pRG1s into SM8530 is low. The reduced efficiency is caused by the low recombination frequency in S. meliloti. Of the 1,615 ORF integrations attempted in the triparental mating, only 66.3% of the ORFs were successfully integrated into the genome of SM8530. A list of pRG1 constructs successfully integrated into SM8530 can be found at http://www.bioinformatics.wsu.edu/kahn/ via the “SM8530 Integrant Library Information” link. However, not all successful pRG1 integrations were obtained on the first mating attempt. After one mating attempt, 935 ORFs (57.9%) were positive for pRG1 integration into SM8530. The unintegrated pRG1s were remated, and 136 (8.4%) showed a successful integration. If the pRG1 still did not integrate, a modified triparental mating method was attempted, yet 543 pRG1s (33.6%) failed to integrate into SM8530. At this time it is unclear why these pRG1 constructs do not yield S. meliloti integrants. Analysis of the 1,615 attempted pRG1 integrations determined that the average ORF length (1,030 bp) of the 57.9% that integrated the first time was significantly larger (Student's t test, P < 0.005) than the average length (750 bp) of the 33.6% that did not recombine.
The decreased efficiency of homologous recombination could also have been due to errors in the ORF sequence of the pRG1 clones. Although only a representative sample of the original pEntry clones of the ORFeome were sequenced (27), we are confident that there are very few mutations in the ORFeome. The sequences of the pSymA clones (pESMaxxxx) were verified using custom, high-density oligonucleotide arrays from NimbleGen Systems, Inc. Using the NimbleGen genome-wide mutation mapping technology, pooled DNA from 1,221 pEntry clones, containing pSymA sequences, was hybridized with a chip containing the sequence of pSymA tiled in 29 nucleotide oligomers with 7-base overlaps. Mismatches within the ORF sequences were detected by altered hybridization ratios of the sample DNA to the oligomers. Within the pEntry clones of the ORFeome, 96.6% of the pSymA ORFs were mutation free (data not shown). Therefore, DNA sequence mutations cannot explain the much higher frequency (33.6%) of failed triparental matings.
GUS activity in pRG1 integrants.
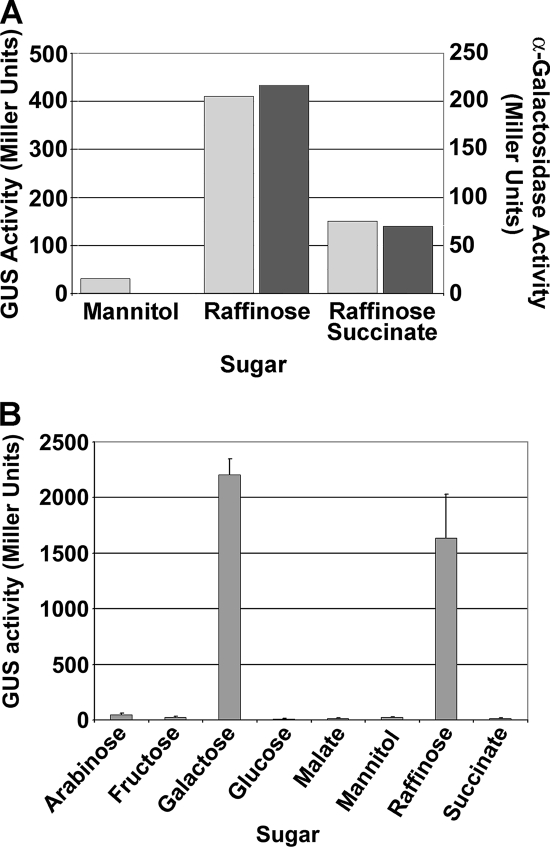
The ultimate goal with the S. meliloti ORFeome is to create a tool that allows for “off-the-shelf” genetics. Off-the-shelf genetics refers to a genome-scale set of individual ORFs in plasmids and strains that contain reporter genes or deletions that have been previously constructed and are ready for use at any time, which reduces the preparation time needed to do an experiment. The pEntry library of the ORFeome is not useful for analyzing genetics in S. meliloti, but once the pRG1 library is integrated into S. meliloti, off-the-shelf genetics using reporter genes is then possible. With this system, the expression level of specific genes can be measured using GusA or GFP reporters which are under the control of native promoters. In order to test whether the pRG1 constructs provided ORF-specific expression of the reporter genes, a reporter fusion was made to the α-galactosidase gene (agaL1; SMb21648) and integrated into the genome of SM1021. AgaL1 is not expressed on mannitol but is induced by raffinose, and this induction is subject to catabolite repression by succinate (7). Figure 2A shows that the pattern of GUS expression on these compounds correlates with the induction of α-galactosidase activity in SM1021. When the GUS assay was adapted for the 96-well plate format, the agaL1 GUS activity was highly induced by raffinose and also by galactose, but minimal activity was detected for the other six sugars (Fig. 2B). These data indicate that the gusA reporter gene is expressed in an ORF-specific manner and that the level of GUS expression provides a quantitative representation of the ORF expression level. It was confirmed that the gfp reporter gene was functional by visualization of a SM1021::pRG1SMa0815 (nifA) reporter strain-infected root nodule (E. Daghighi, S. N. Yurgel, and M. L. Kahn, unpublished data).
FIG. 2.
AgaL1 GUS activity. A. AgaL1 GUS activity compared to α-galactosidase activity. Gray bars, GUS activity from the reporter gene gusA; black bars, α-galactosidase activity from AgaL1. B. AgaL1 GUS activity in the high-throughput assay. AgaL1 is induced by raffinose and galactose. The data were reproducible in a 96-well format.
To test the usefulness of the integrated pRG1 reporter constructs, we chose to analyze putative sugar and sugar alcohol kinase genes (Table 4). Since S. meliloti appears to lack phosphotransferase transport systems, these kinases are key for bringing imported carbohydrates into metabolism. The S. meliloti genome contains 36 genes predicted to code for these kinases. However, these have not been studied extensively, and while bioinformatics analysis is thought to do a good job of assigning proteins to families based on sequence conservation related to enzyme mechanism, it is not as good at predicting the substrate specificity of these enzymes. Gene expression patterns, or the phenotypes of mutants, can help in identifying the correlation between sugars and enzymes. The pRG1 integrants for the sugar kinases were used in an attempt to identify substrates that induced expression of these ORFs. Using MNH4 media supplemented with nine different carbon sources, the expression levels of the kinase genes were measured in GUS assays. These assays were performed in a 96-well format to test the reproducibility of the data in a high-throughput manner. In the future, once all the pRG1 constructs are integrated into S. meliloti, the whole ORFeome could be screened in a similar fashion.
TABLE 4.
pRG1 constructs integrated into S. meliloti strains SM1021 and/or SM8530
| ORF ID | Gene | Present in strain
|
Annotation | |
|---|---|---|---|---|
| SM1021 | SM8530 | |||
| Control | ||||
| SMb21648 | agaL1 | X | α-Galactosidase (melibiase) | |
| Sugar kinases | ||||
| SMa0137 | X | Putative kinase/esterase | ||
| SMa0514 | idnK | X | Gluconate kinase | |
| SMa0705 | dgoK2 | X | X | 2-Dehydro-3-deoxygalactonokinase |
| SMb20307 | X | X | Putative dihydroxyacetone kinase | |
| SMb20313 | X | X | Dihydroxyacetone kinase | |
| SMb20489 | X | X | Carbohydrate kinase | |
| SMb20767 | dak | X | X | Putative dihydroxyacetone kinase |
| SMb20852 | X | X | Putative sugar kinase | |
| SMb21009 | glpK | X | Probable glycerol kinase | |
| SMb21022 | X | Probable sugar kinase | ||
| SMb21119 | gntK | X | X | Putative gluconokinase |
| SMb21184 | ackA2 | X | Putative acetate kinase (acetokinase) | |
| SMb21217 | X | X | Putative sugar kinase | |
| SMb21373 | X | X | Putative sugar kinase | |
| SMb21374 | X | Putative sugar kinase, PfkB family | ||
| SMc00473 | X | X | Carbohydrate kinase | |
| SMc00881 | dgoK1 | X | 2-Dehydro-3-deoxygalactonokinase | |
| SMc01103 | rbsK | X | Ribokinase | |
| SMc01165 | iolC | X | X | Sugar kinase |
| SMc01531 | kdgK | X | X | 2-Dehydro-3-deoxygluconokinase |
| SMc01618 | X | Carbohydrate kinase | ||
| SMc01623 | eriA | X | Erythritol kinase | |
| SMc02164 | frk | X | X | Fructokinase |
| SMc02335 | X | X | Pentose kinase | |
| SMc02341 | X | Sugar kinase | ||
| SMc02846 | X | X | Sugar kinase | |
| SMc03138 | X | Sugar kinase | ||
| SMc03164 | xylB | X | X | Xylulose kinase |
| SMc03817 | X | Sugar kinase | ||
| SMc04005 | pykA | X | X | Pyruvate kinase II |
The pRG1 plasmids corresponding to 30 predicted sugar and sugar alcohol kinases were integrated into the genomes of S. meliloti SM1021 and/or SM8530. Twenty-five of the pRG1 constructs successfully integrated into SM1021, 22 into SM8530, and 17 of the ORFs were in both SM1021 and SM8530 (Table 4). Integrants were not obtained in either S. meliloti strain for ORFs SMb20314, SMb20497, SMb21462, SMc01503, SMc02334, and SMc04213. GUS activity was measured for the sugar kinase reporter strains cultured in MNH4 media supplemented with 0.2% of arabinose, fructose, galactose, glucose, malate, mannitol, raffinose, succinate, or glycerol as the carbon source. The assays were carried out in a high-throughput manner using 96-well microtiter plates with two replications, each with three wells for each strain. The activities in hydrolyzing the GUS substrate PNP-gluc are reported as Miller units. Values obtained for the two replications on each of the carbon sources were plotted against each other, giving a regression with r2 values of 0.92 for SM1021 (n = 655) and 0.88 for SM8530 (n = 559).
GUS activity was measured, and the ratio of activity in the fusion strain to the background activity of the parental strain (SM1021 or SM8530) not containing a fusion plasmid was calculated. About one-third of the sugar kinase ORFs assayed in SM1021 were not expressed (0 to 2×) on any of the carbon sources, but only 16% of the sugar kinases were not expressed in SM8530 (Table 5). The general upregulation in SM8530 is consistent with previous findings that over 200 ORFs are upregulated in an ExpR+ background (13, 16). The percentages of low-level (2 to 4×) and high-level (>12×) sugar kinase expression in SM1021 and SM8530 were similar (Table 5), but in the moderate expression range (4 to 12×) 32% of the tested ORFs were expressed in SM8530, and 18% in SM1021. Interestingly enough, none of the sugar kinase ORFs from this study had been shown to be regulated by ExpR in previous microarray studies (13, 16). This may be due to the type of medium used to culture the cells for the microarray analysis. These data demonstrate that the high-throughput GUS assay can be used to monitor global gene expression of the pRG1 reporters in different parental backgrounds.
TABLE 5.
Patterns of expression and overexpression of sugar kinase fusions in SM1021 and SM8530 backgrounds
| C source | % of ORFs with indicated fold increase in expression over backgrounda
|
|||||||
|---|---|---|---|---|---|---|---|---|
| SM1021 (n = 25)
|
SM8530 (n = 22)
|
|||||||
| 0-2× | 2-4× | 4-12× | >12× | 0-2× | 2-4× | 4-12× | >12× | |
| Arabinose | 36 | 24 | 20 | 20 | 18 | 27 | 37 | 18 |
| Fructose | 28 | 32 | 16 | 24 | 13 | 32 | 32 | 23 |
| Galactose | 28 | 32 | 16 | 24 | 9 | 36 | 32 | 23 |
| Glucose | 32 | 32 | 24 | 12 | 14 | 32 | 36 | 18 |
| Glycerol | 52 | 12 | 20 | 16 | 14 | 32 | 36 | 18 |
| Malate | 44 | 20 | 20 | 16 | 14 | 36 | 32 | 18 |
| Mannitol | 24 | 40 | 16 | 20 | 18 | 23 | 32 | 27 |
| Raffinose | 36 | 28 | 20 | 16 | 18 | 37 | 27 | 18 |
| Succinate | 32 | 36 | 12 | 20 | 27 | 32 | 23 | 18 |
| Overallb | 35 | 28 | 18 | 19 | 16 | 32 | 32 | 20 |
The ratio of activity in the fusion strain to the “background” activity of the parental strain (SM1021 or SM8530) not containing a fusion plasmid was calculated, and the percentage of ORF fusions in each activity range was tabulated. Bold numbers indicate data of interest that are discussed further in the text.
The average of each expression range for all carbon sources was calculated to compare the global change in gene expression between SM1021 and SM8530.
Specific gene expression differences between the SM1021 and SM8530 backgrounds can also be determined. There are three annotated dihydroxyacetone kinases: SMb20307, SMb20313, and SMb20767. Dihydroxyacetone kinases phosphorylate dihydroxyacetone during glycerol degradation. The resulting product, dihydroxyacetone phosphate, is an intermediate in glycolysis that can be converted to pyruvate, with the net generation of one ATP. SMb20313 showed low or no GUS expression on all of the compounds tested and similar expression in both SM1021 and SM8530 (Table 6). These data indicate that SMb20313 is not highly expressed by any of the carbon sources evaluated in either strain, suggesting that it is not the primary dihydroxyacetone kinase used under these growth conditions. However, there was a striking difference in GUS expression by SMb20307 and SMb20767 (dak) fusions between SM1021 and SM8530 when cultured with all carbon sources. SMb20307 was expressed on average 6.2-fold in SM1021, while in SM8530 this ORF was expressed 32.5-fold. For SMb20767, the expression was reversed with expression in SM1021 being 33.8-fold and in SM8530 5.7-fold. The high GUS expression levels, regardless of carbon source, of SMb20767 in SM1021 and SMb20307 in SM8530 appear to be strain dependent. This suggests that ExpR may have a role in regulating SMb20307 in SM8530. Four other ORFs exhibited different expression levels in SM8530 compared to SM1021 on all carbon sources. SMc03164 (xylB), which is involved in xylose degradation, is expressed on average 25.7 times more than background in SM8530 on all carbon sources, compared to 2.7× in SM1021 (Table 6). Xylose degradation results in d-xylulose-5-phosphate, which is a substrate in the nonoxidative branch of the pentose phosphate pathway, which produces glyceraldehyde-3-P and fructose-6-P. SMb20489, SMc01531, and SMc01165 have medium expression in SM8530 and low expression in SM1021. SMb20489 is annotated as a carbohydrate kinase and SMc01165 (iolC) as a sugar kinase. SMc01531 (kdgK) is annotated as a 2-dehydro-3-deoxygluconokinase, which is the final enzyme in the β-d-glucuronide and d-galacturonate degradation pathways. Again, the differential GUS expression of the above ORFs between SM1021 and SM8530 could be due to the functional ExpR gene in SM8530. SM8530 has increased exopolysaccharide production (9, 12), and this may be linked to the upregulation of sugar kinases.
TABLE 6.
GUS expression from ORF reporters in SM1021 and SM8530a
| Strain and ORF ID | Fold increase above background (normalized)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Arabinose | Fructose | Galactose | Glucose | Glycerol | Malate | Mannitol | Raffinose | Succinate | |
| SM1021 | |||||||||
| SMa0705 | 1.29 | 1.35 | 1.25 | 1.40 | 0.00 | 1.18 | 1.35 | 1.07 | 1.18 |
| SMb20307 | 5.80 | 6.76 | 6.73 | 6.33 | 4.72 | 5.30 | 6.93 | 7.21 | 5.79 |
| SMb20313 | 2.05 | 2.23 | 2.68 | 2.05 | 0.73 | 1.95 | 2.38 | 1.90 | 2.25 |
| SMb20489 | 1.72 | 2.64 | 2.00 | 2.18 | 0.62 | 1.86 | 2.24 | 2.43 | 1.79 |
| SMb20767 | 31.33 | 35.99 | 40.30 | 29.13 | 31.76 | 31.15 | 39.24 | 30.88 | 33.97 |
| SMb20852 | 1.96 | 2.25 | 2.43 | 1.76 | 0.55 | 1.74 | 2.17 | 1.76 | 1.89 |
| SMb21119 | 5.48 | 5.65 | 5.27 | 4.15 | 2.35 | 4.71 | 5.08 | 3.99 | 4.00 |
| SMb21217 | 1.41 | 1.37 | 1.41 | 1.37 | 1.29 | 1.30 | 1.62 | 1.37 | 1.55 |
| SMb21373 | 1.37 | 1.46 | 1.78 | 1.45 | 0.68 | 1.26 | 1.61 | 1.41 | 1.33 |
| SMc00473 | 4.09 | 4.59 | 4.44 | 3.56 | 3.09 | 3.36 | 3.84 | 4.09 | 3.77 |
| SMc01165 | 2.43 | 1.93 | 2.18 | 2.08 | 1.51 | 2.12 | 2.63 | 2.26 | 2.45 |
| SMc01531 | 1.51 | 1.65 | 1.77 | 1.53 | 2.96 | 1.57 | 1.70 | 1.45 | 1.72 |
| SMc02164 | 5.95 | 14.52 | 6.28 | 4.90 | 5.28 | 5.16 | 9.23 | 5.59 | 6.50 |
| SMc02335 | 2.18 | 2.32 | 2.37 | 2.14 | 0.62 | 1.98 | 2.50 | 2.62 | 2.02 |
| SMc02846 | 15.02 | 13.45 | 14.45 | 10.52 | 9.24 | 12.13 | 14.35 | 11.98 | 14.10 |
| SMc03164 | 2.86 | 3.06 | 3.15 | 2.42 | 1.26 | 2.60 | 3.38 | 2.92 | 2.73 |
| SMc04005 | 13.31 | 22.23 | 16.58 | 16.36 | 21.01 | 12.07 | 19.30 | 15.96 | 14.89 |
| SM8530 | |||||||||
| SMa0705 | 1.51 | 1.88 | 1.45 | 1.60 | 1.58 | 1.63 | 1.33 | 1.40 | 0.86 |
| SMb20307 | 37.23 | 34.71 | 37.35 | 27.32 | 41.98 | 29.14 | 34.09 | 26.08 | 24.53 |
| SMb20313 | 2.97 | 2.81 | 3.32 | 3.14 | 2.83 | 2.26 | 3.02 | 2.63 | 2.27 |
| SMb20489 | 5.01 | 5.62 | 5.72 | 4.29 | 5.59 | 3.42 | 5.94 | 4.19 | 3.89 |
| SMb20767 | 6.59 | 6.11 | 6.27 | 6.20 | 6.15 | 4.55 | 6.70 | 4.25 | 4.65 |
| SMb20852 | 3.02 | 3.17 | 2.85 | 3.13 | 2.88 | 2.55 | 3.00 | 2.43 | 2.15 |
| SMb21119 | 4.43 | 4.82 | 4.78 | 4.85 | 5.76 | 4.93 | 4.03 | 3.76 | 3.93 |
| SMb21217 | 1.92 | 2.07 | 2.26 | 2.10 | 2.44 | 2.21 | 1.89 | 1.91 | 1.25 |
| SMb21373 | 1.70 | 1.97 | 2.34 | 1.82 | 1.96 | 1.95 | 1.86 | 1.80 | 1.10 |
| SMc00473 | 6.32 | 5.85 | 5.77 | 5.34 | 7.00 | 4.32 | 5.30 | 4.45 | 4.48 |
| SMc01165 | 11.04 | 8.61 | 12.08 | 11.13 | 10.23 | 9.04 | 12.53 | 7.31 | 10.15 |
| SMc01531 | 5.47 | 4.68 | 5.38 | 5.96 | 6.27 | 4.19 | 7.03 | 4.60 | 4.88 |
| SMc02164 | 8.96 | 23.90 | 8.52 | 9.62 | 10.27 | 9.70 | 15.87 | 10.48 | 7.73 |
| SMc02335 | 2.58 | 2.45 | 2.75 | 2.69 | 2.76 | 2.48 | 2.31 | 2.31 | 1.73 |
| SMc02846 | 22.90 | 18.29 | 17.48 | 18.39 | 26.85 | 14.25 | 43.25 | 12.75 | 13.96 |
| SMc03164 | 25.02 | 23.76 | 30.19 | 30.11 | 26.13 | 20.54 | 26.66 | 27.75 | 20.72 |
| SMc04005 | 24.65 | 36.15 | 27.40 | 35.36 | 35.08 | 12.58 | 99.53 | 24.78 | 13.34 |
Bold numbers indicate data of interest that are discussed further in the text.
SMc02164 (frk), a fructokinase, was highly expressed (>12×) by fructose in SM1021 and by both fructose and mannitol in SM8530 (Table 6). Mannitol is the reduced sugar alcohol corresponding to fructose; mannitol-1-phosphate is oxidized to fructose-6-phosphate by a dehydrogenase. The induction by fructose in both strains supports the annotation of SMc02164 as a functional fructokinase. Two ORFs were expressed at high levels in both SM1021 and SM8530. The first, SMc02846, which is annotated as a sugar kinase, is expressed on average 20.9-fold over background in SM8530 and 12.8-fold in SM1021 (Table 6). The second, SMc04005 (pykA), which encodes pyruvate kinase II, was highly expressed in both strains on all carbon sources (Table 6). It was not surprising to observe the high expression, since PykA converts pyruvate into phosphoenolpyruvate, which is needed for mixed acid fermentation, glycolysis, and anaerobic respiration. Also, many of the observed sugar kinases that were expressed are involved in pathways where pyruvate is an end product. The increased production of pyruvate would result in the high expression of PykA. Two annotated 2-dehydro-3-deoxygalactonokinases (SMa0705 and SMc00881) were also evaluated for GUS expression on the nine carbon sources. 2-Dehydro-3-deoxygalactonokinase phosphorylates 2-dehydro-3-deoxy-d-galactonate during d-galactonate degradation, and the final products, pyruvate and glyceraldehyde-3-P, are substrates in numerous biochemical reactions. SMa0705 (dgoK2) was not expressed in SM1021 or SM8530, indicating the gene may not be functional or is not expressed under the tested conditions (Table 6). Data for SMc00881 (dgoK1) were only obtained for SM1021 (Table 7) and indicated that dgoK1 is highly induced by raffinose (12.2×) and galactose (25.9×) and moderately induced by the other carbon sources. The induction by raffinose and galactose of dgoK1 is logical, since d-galactonate degradation occurs after galactose degradation and raffinose can be degraded into galactose.
TABLE 7.
GUS expression from ORF reporters in SM1021 or SM8530a
| Strain and ORF ID | Fold increase above background (normalized)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Arabinose | Fructose | Galactose | Glucose | Glycerol | Malate | Mannitol | Raffinose | Succinate | |
| SM1021 | |||||||||
| SMb21009 | 2.51 | 3.25 | 3.20 | 3.97 | 22.68 | 2.66 | 2.74 | 2.83 | 3.57 |
| SMb21022 | 1.56 | 1.55 | 1.55 | 1.44 | 0.43 | 1.24 | 1.38 | 1.26 | 1.45 |
| SMb21184 | 19.67 | 25.47 | 22.13 | 16.47 | 24.47 | 19.11 | 22.95 | 17.69 | 20.74 |
| SMb21374 | 1.44 | 1.45 | 1.65 | 1.52 | 0.29 | 1.45 | 1.63 | 1.70 | 1.39 |
| SMc00881 | 9.19 | 7.83 | 25.85 | 6.81 | 7.13 | 6.19 | 6.43 | 12.15 | 8.34 |
| SMc01103 | 14.57 | 13.07 | 13.10 | 9.11 | 8.30 | 11.87 | 14.05 | 9.09 | 12.25 |
| SMc03138 | 2.00 | 2.14 | 2.07 | 1.89 | 1.00 | 1.76 | 2.34 | 1.84 | 2.03 |
| SMc03817 | 2.82 | 2.84 | 2.65 | 2.63 | 1.52 | 2.48 | 2.27 | 2.16 | 2.71 |
| SM8530 | |||||||||
| SMa0137 | 4.74 | 5.10 | 4.95 | 5.23 | 5.17 | 4.62 | 5.51 | 3.44 | 3.38 |
| SMa0514 | 1.37 | 1.66 | 1.50 | 1.54 | 1.61 | 1.54 | 0.98 | 1.28 | 0.79 |
| SMc01618 | 2.82 | 2.24 | 2.87 | 2.82 | 3.12 | 2.47 | 3.75 | 2.74 | 2.03 |
| SMc01623 | 2.28 | 2.50 | 2.24 | 2.37 | 2.78 | 2.04 | 4.78 | 2.26 | 2.24 |
| SMc02341 | 2.87 | 2.99 | 2.94 | 2.80 | 3.14 | 2.75 | 3.30 | 2.30 | 1.98 |
Bold numbers indicate data of interest that are discussed further in the text.
Three other sugar kinase genes, represented only by fusions in SM1021, were found to be expressed. SMc01103 (rbsK), a ribokinase, was moderately to highly expressed on all of the carbon sources (Table 7). d-Ribose-5-phosphate, the product from a ribokinase reaction, is a substrate in numerous biochemical pathways, such as the pentose phosphate pathway. A putative acetokinase, SMb21184 (ackA2), was highly expressed on all the carbon sources (Table 7). This enzyme catalyzes reactions in threonine degradation and pyruvate fermentation. A gene involved in glycerol degradation was induced by glycerol in SM1021 (Table 7). SMb21009 (glpK), annotated as a probable glycerol kinase, is strongly induced by glycerol in SM1021.
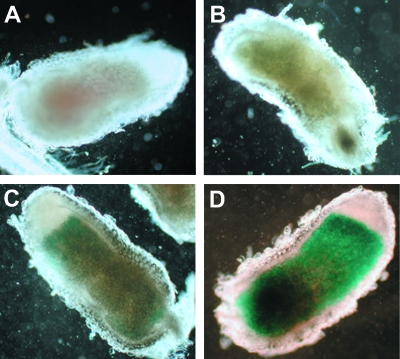
GUS activity can also be assessed in root nodules of plants infected with the reporter strains. Inoculating reporter strains onto alfalfa allows the detection of gene activity during symbiosis. Fourteen of the SM1021 sugar kinase reporter strains (Table 8) were inoculated onto alfalfa and induced nodules which were stained for GUS activity. Stained nodules were qualitatively analyzed after 3 and 20 h of staining (Table 8). For genes that are highly expressed, GUS staining was observed at the 3-h time point (Fig. 3D). If nodules remained colorless after 20 h, the ORF was considered to be not expressed (Fig. 3B). Usually, even if the gene was expressed at a low level, there was medium to dark staining after 20 h. Only six of the sugar kinase reporter strain-induced nodules showed any GUS staining after 3 h (Table 8). Seven more reporter strain nodules exhibited staining after 20 h (Table 8). Generally, the GUS activity in nodules correlated with the GUS activity seen in assays with cell cultures, as observed for SMb20307 (Fig. 3C; Table 6), which was consistently expressed at a low level in SM1021, and SMb20767, which was reliably expressed at a high level (Fig. 3D; Table 6). Unfortunately, the carbon sources in the nodule are not defined, and since a limited number of carbon sources were tested in the cell assays, any observed differences were most likely due to the presence of other carbon sources. One ORF (SMa0705) that did not have any activity in the liquid GUS assays was not expressed in nodules at either time point (Fig. 3B).
TABLE 8.
GUS activity in stained nodules from SM1021 reporter strain-infected alfalfa
| ORF ID | GUS staining in nodules ata:
|
|
|---|---|---|
| 3 h | 20 h | |
| SMa0705 | − | − |
| SMb20307 | + | ++++ |
| SMb20489 | − | ++ |
| SMb20767 | +++ | ++++ |
| SMb20852 | − | ++ |
| SMb21009 | − | ++++ |
| SMb21022 | − | ++++ |
| SMb21184 | ++ | ++++ |
| SMb21373 | − | ++ |
| SMc00881 | + | ++++ |
| SMc02335 | ++ | +++ |
| SMc02846 | ++ | ++++ |
| SMc03138 | − | +++ |
| SMc03164 | − | ++++ |
−, no staining; +, faint staining; ++, light staining; +++, medium staining; ++++, dark staining.
FIG. 3.
GUS expression in nodules. A. SM1021-induced nodules showed no GUS staining after 20 h. B. Nodules resulting from SM1021::pRG1SMa0705 (dgoK2) also showed no activity after 20 h. C. SM1021::pRG1SMb20307-induced nodules had faint GUS staining after 3 h. D. Nodules from SM1021::pRG1SMb20767 (dak) had medium GUS staining after 3 h.
Conclusions.
The construction and integration of the pRG1 library allow for “off-the-shelf” genomics in S. meliloti. Currently, 1,071 pRG1 clones have been integrated into SM8530 and are ready to use in GUS and GFP assays. However, since the 6,290 pRG1 constructs are harbored in E. coli, they can be easily transferred and integrated into SM1021, SM8530, and possibly other closely related rhizobia strains. As a result, this system reduces the amount of time needed to create a reporter fusion from scratch and allows for the quick acquisition of gene expression data which are representative of the target gene expression. High-throughput GUS assays can be done with the integrated reporters, allowing for large-scale analyses of groups of genes under various growth conditions, as shown in Table 5. The data obtained from the GUS assays can be used like microarray data to measure global changes in expression between strains or growth conditions and may be more accurate, since actual gene expression is measured instead of RNA production. More sugar kinase genes were moderately expressed in SM8530 than in SM1021, a result that may be due to the expression of ExpR in SM8530 (13, 16). Six ORFs had noticeable differences in expression between SM1021 and SM8530, which again could be due to the functional ExpR. Changes in the expression of individual genes, with different carbon sources and strains, were also measured, and the GUS activity confirmed the annotated function and substrate specificity in some cases. SMc02164 (frk), SMb21009 (glpK), and SMc00881 (dgoK1) were found to be specifically induced by fructose, glycerol, and galactose, respectively. Even though a relatively small number of ORFs were evaluated (30 versus 6,290), with a limited set of carbon substrates, useful data were obtained and demonstrate the utility of having an “off-the-shelf” genomics tool.
Acknowledgments
We extend special thanks to Jenny Kleene, Casey Thomas, and Elizabeth Daghighi for their technical support. No robots were used in the development of the expression library of S. meliloti, but a group of Washington State University students were important at various stages of the work, including Nadine Santos, Jody Thomas, Rebecca Flannigan, Malia Haddock, Michelle Mace, Jennifer Topham, Kelsey Wertzler, Logan Smith, Sarah Chavez, Harvey Doty, Marcie Tonnemaker, Wendy Cheung, Melissa Nguyen, Helen Yi, Austin Harvie, Jason Neal-McKinney, Kristi Podelnyk, Paul Guyett, and Ryan Loosveldt, and were critical to the completion of this project.
This project was supported by a grant from the Microbial Genetics Program at the U.S. National Science Foundation (MCB-0133258).
Footnotes
Published ahead of print on 12 September 2008.
REFERENCES
- 1.Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26:824-828. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. W. Hyman, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, C. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K.-C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 98:9883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, M. J., C. J. Toman, R. F. Fisher, and S. R. Long. 2004. A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. USA 101:16636-16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, A., H. Bergès, E. Krol, C. Bruand, S. Rüberg, D. Capela, E. Lauber, E. Meilhoc, F. Ampe, F. J. de Bruijn, J. Fourment, A. Francez-Charlot, D. Kahn, H. Küster, C. Liebe, A. Pühler, S. Weidner, and J. Batut. 2004. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant-Microbe Interact. 17:292-303. [DOI] [PubMed] [Google Scholar]
- 5.Bernard, P., K. E. Kezdy, L. van Melderen, J. Steyaert, L. Wyns, M. L. Pato, P. N. Higgins, and M. Couturier. 1993. The F plasmid CcdB protein induces efficient ATP-dependent DNA cleavage by gyrase. J. Mol. Biol. 234:534-541. [DOI] [PubMed] [Google Scholar]
- 6.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 7.Bringhurst, R. M., and D. J. Gage. 2002. Control of inducer accumulation plays a key role in succinate-mediated catabolite repression in Sinorhizobium meliloti. J. Bacteriol. 184:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dréano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. M. Pohl, D. Portetelle, A. Pühler, B. Purnelle, U. Ramsperger, C. Renard, P. Thébault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowie, A., J. Cheng, C. D. Sibley, Y. Fong, R. Zaheer, C. L. Patten, R. M. Morton, G. B. Golding, and T. M. Finan. 2006. An integrated approach to functional genomics: construction of a novel reporter gene fusion library for Sinorhizobium meliloti. Appl. Environ. Microbiol. 72:7156-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djordjevic, M. A. 2004. Sinorhizobium meliloti metabolism in the root nodule: a proteomic perspective. Proteomics 4:1859-1872. [DOI] [PubMed] [Google Scholar]
- 11.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F.-J. Vorhölter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Pühler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, M., H. Chen, A. Eberhard, M. R. Gronquist, J. B. Robinson, B. G. Rolfe, and W. D. Bauer. 2005. sinI- and expR-dependent quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 187:7931-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glazebrook, J., and G. C. Walker. 1989. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661-672. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 16.Hoang, H. H., A. Becker, and J. E. González. 2004. A LuxR homolog ExpR, in combination with the Sin quorum sensing system, plays a central role in Sinorhizobium meliloti gene expression. J. Bacteriol. 186:5460-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.House, B. L., M. W. Mortimer, and M. L. Kahn. 2004. New recombination methods for Sinorhizobium meliloti genetics. Appl. Environ. Microbiol. 70:2806-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jefferson, R. A., T. A. Kavanagh, and M. W. Bevan. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in plants. EMBO J. 6:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, K. M., H. Kobayashi, B. W. Davies, M. E. Taga, and G. C. Walker. 2007. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5:619-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leong, S. A., G. S. Ditta, and D. R. Helinski. 1983. Heme biosynthesis in Rhizobium: identification of a cloned gene coding for δ-aminolevulinic acid synthetase from Rhizobium meliloti. J. Biol. Chem. 257:8724-8730. [PubMed] [Google Scholar]
- 21.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and M. Ausubel. 1982. Physical and genetic characteristics of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 23.Pellock, B. J., M. Teplitski, R. P. Boinay, W. D. Bauer, and G. C. Walker. 2002. A LuxR homolog controls production of symbiotically active extracellular polysaccharide II by Sinorhizobium meliloti. J. Bacteriol. 184:5067-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Platt, R., C. Drescher, S.-K. Park, and G. J. Phillips. 2000. Genetic system for reversible integration of DNA constructs and lacZ gene fusions into the Escherichia coli chromosome. Plasmid 43:12-23. [DOI] [PubMed] [Google Scholar]
- 25.Ramos, H. J. O., L. D. B. Roncato, L. D. B. Roncato-Maccari, E. M. Souza, J. R. L. Soares-Ramos, H. Hungria, and F. O. Pedrosa. 2002. Monitoring Azospirillum-wheat interactions using the gfp and gusA genes constitutively expressed from a new broad-host range vector. J. Biotechnol. 97:243-252. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Schroeder, B. K., B. L. House, M. W. Mortimer, S. N. Yurgel, S. C. Maloney, K. L. Ward, and M. L. Kahn. 2005. Development of a functional genomics platform for Sinorhizobium meliloti: construction of an ORFeome. Appl. Environ. Microbiol. 71:5858-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Somerville, J. E., and M. L. Kahn. 1983. Cloning of the glutamine synthetase I gene from Rhizobium meliloti. J. Bacteriol. 156:168-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yurgel, S. N., and M. L. Kahn. 2005. Sinorhizobium meliloti dctA mutants with partial ability to transport dicarboxylic acids. J. Bacteriol. 187:1161-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]