Abstract
Methanogens play a critical role in the decomposition of organics under anaerobic conditions. The methanogenic consortia in saturated wetland soils are often subjected to large temperature fluctuations and acidic conditions, imposing a selective pressure for psychro- and acidotolerant community members; however, methanogenic communities in engineered digesters are frequently maintained within a narrow range of mesophilic and circumneutral conditions to retain system stability. To investigate the hypothesis that these two disparate environments have distinct methanogenic communities, the methanogens in an oligotrophic acidic fen and a mesophilic anaerobic digester treating municipal wastewater sludge were characterized by creating clone libraries for the 16S rRNA and methyl coenzyme M reductase alpha subunit (mcrA) genes. A quantitative framework was developed to assess the differences between these two communities by calculating the average sequence similarity for 16S rRNA genes and mcrA within a genus and family using sequences of isolated and characterized methanogens within the approved methanogen taxonomy. The average sequence similarities for 16S rRNA genes within a genus and family were 96.0 and 93.5%, respectively, and the average sequence similarities for mcrA within a genus and family were 88.9 and 79%, respectively. The clone libraries of the bog and digester environments showed no overlap at the species level and almost no overlap at the family level. Both libraries were dominated by clones related to uncultured methanogen groups within the Methanomicrobiales, although members of the Methanosarcinales and Methanobacteriales were also found in both libraries. Diversity indices for the 16S rRNA gene library of the bog and both mcrA libraries were similar, but these indices indicated much lower diversity in the 16S digester library than in the other three libraries.
Methanogens are an integral part of carbon cycling on the planet, anaerobically catalyzing the production of methane primarily through carbon dioxide reduction with hydrogen or the conversion of methylated compounds such as acetate (64). These Archaea are essential community members in anaerobic digesters, which may be used to treat a variety of domestic, agricultural, and industrial wastes with concomitant energy production from the combustion of the methane-containing biogas. Anaerobic digestion is typically operated at circumneutral pH and mesophilic (30°C to 37°C) or thermophilic (55°C to 65°C) temperatures, with process stability decreasing dramatically outside of this range of conditions (5, 34, 38, 39). In these systems, methanogens are considered to be the most sensitive members of the methanogenic consortium to nonoptimal conditions (1, 21, 29), yet methanogenesis is known to proceed in both cold and acidic natural environments such as tundra (28, 63), Sphagnum-dominated peatlands (35, 49), and lake sediments (48, 55). Presumably, this difference in psychro- and acidotolerance can be attributed, in part, to differences in the methanogen communities, and a knowledge of these differences might lend insights into community-based strategies to increase digester stability with reduced chemical and energy inputs necessary to maintain narrow operating conditions.
Methanogens are difficult to isolate or culture under laboratory conditions, so communities are often examined through culture-independent techniques such as the amplification and sequencing of target DNA from environmental samples. The most widely used target for molecular analysis is the 16S rRNA genes, and a number of primers and probes have been developed specifically for methanogens or groups of methanogens (10, 44, 51, 52, 53, 57, 59, 65). To eliminate potential problems with nonspecific amplification, some researchers have developed primers for the gene sequence of the α-subunit of the methyl coenzyme M reductase (mcrA) (26, 43, 60). Mcr catalyzes the last step of methanogenesis and is conserved among all methanogens. Phylogenetic inference with mcrA sequences is similar to that obtained with 16S rRNA gene sequences, suggesting no lateral transfer (3, 43, 60). Moreover, Mcr is absent in all nonmethanogens, with the exception of the anaerobic methane-oxidizing Archaea, which are closely related to the methanogens (27). Due to the fact that methanogens may be examined exclusively from other bacteria present in an environment, mcrA has been increasingly used for phylogenetic analysis coupled with, or independent of, 16S rRNA genes.
The increasing use of molecular-based community analysis has led to the identification of phylogenetic clusters of methanogens that are quite divergent in sequence from those of isolated, phenotypically described methanogens. Ecological indices such as the Shannon-Wiener or Simpson's indices and species accumulation curves, which are used to determine the extent of sampling of an environment, have been adapted for use in microbial gene surveys (30). The use of these ecological indices requires that genes or gene fragments be organized into operational taxonomic units (OTUs) representing a species or strain. Boone et al. (6) previously suggested a 16S rRNA gene sequence similarity of less than 98% as evidence for a separate methanogen species, which is slightly more restrictive than the minimum 97% sequence similarity suggested previously by Stackebrandt and Goebel (61) for sequences to be considered from the same species. Although these studies provide a basis for determining if gene sequences represent new species, little information is available about sequence similarities used to determine new taxonomic levels of methanogens above the species level. In addition, although many researchers use mcrA sequences alone or coupled with 16S rRNA gene sequences to examine methanogenic communities, little attention has been given to appropriate sequence similarity limits for mcrA in establishing a separate species or genus.
The hypothesis of this research is that the methanogen community of a mesophilic anaerobic digester is distinct from that of an acidic peat bog. The methanogen communities from these two environments were explored by creating 16S rRNA gene and mcrA clone libraries. To characterize the extent of community differences between these libraries, in silico analysis was performed on existing mcrA and 16S rRNA gene sequences for phenotypically characterized methanogens to describe the range of sequence similarity within currently accepted taxonomic levels.
MATERIALS AND METHODS
Site characteristics and sampling.
Bear Meadows Bog is an acidic, boreal peatland situated approximately 15 miles southeast of State College, PA. It is classified as a transitional bog, and vegetations consist predominantly of Sphagnum mosses, sedge grasses, and highbush blueberry, with peat approximately 1 m thick. The bog pH varies from 3.8 to 5.0, and bog waters contain little alkalinity, sulfate, or iron (J. F. Biddle, C. Turich, S. Brantley, and M. A. Bruns, presented at the American Geophysical Union, San Francisco, CA, 6 to 10 December 2002). Porewater methane reaches a maximum of 5,500 ppm at 30 cm below the water-sediment interface. Methane emitted from the bog has an isotopic signature of −60‰, suggesting a biogenic origin (Biddle et al., presented at the American Geophysical Union, San Francisco, CA, 6 to 10 December 2002). Temperatures in the region range from approximately −15°C to 35°C and precipitation are fairly evenly spaced throughout the year, with an average of 98 cm of rain and 115 cm of snow (www.nws.noaa.gov).
Cores were taken from two locations at Bear Meadows Bog covered with both Sphagnum mosses and sedges. Each core was collected in a series of three 6-in.-long Teflon sleeves (Ben Meadows Company, Janesville, WI) for a total of 1.5 feet extracted at each location. Immediately upon collection, the cores were placed into a sealed container with a GasPak (BBL Inc., Franklin Labs, NJ) to create an anaerobic environment (H2/CO2) for transport.
Anaerobic digester sludge was collected from the State College Wastewater Treatment Plant. The plant operates two fixed-cover primary digesters, which receive primary sludge and thickened secondary sludge and are constantly mixed and heated to 37°C. The pH is kept near neutral by adjusting the organic loading rate, and the solid retention time is 25 days. Collected sludge was transported anaerobically in a sealed container with a GasPak.
Construction of 16S rRNA gene and mcrA clone libraries.
Clone libraries were constructed for both methanogen-specific 16S rRNA genes and mcrA from the Bear Meadows Bog and anaerobic digester samples. DNA was extracted using a PowerSoil DNA extraction kit (MoBio Laboratories, Carlsbad, CA). For the bog peat cores, DNA extractions were performed on sediment taken from the central portions of the top and bottom of each of the three 6-in. core segments for a total of six DNA extractions from the bog peat. All of the DNA extractions from the bog peat were pooled before performing PCR for library construction.
A fragment of the 16S rRNA genes was amplified with A21f, an Archaea-specific forward primer (16), and Eury498 (8), a reverse primer developed from a Euryarchaeota-specific probe. PCR conditions were based on those reported previously by Girguis et al. (23). The mcrA sequences were amplified with three different forward primers, ME1 (26), ML-f (43), and mlas (5′-GGTGGTGTMGGDTTCACMCARTA-3′), which is a truncated version of forward primer ML-f, designed for a better melting temperature match with the reverse primer, a lower 3′ stability to improve specificity, and two additional degeneracies to improve coverage (31). The ML primer set is reported to have wide coverage of methanogen groups, detecting members of the Methanosarcinaceae, Methanosaetaceae, Methanobacteriales (both mcrA and mrtA), Methanococcales, and the uncultured groups fen cluster, rice cluster I, and MCR-2 (17, 18, 32, 43, 58). The ME primer set also captures a wide range of methanogens (2, 19, 20) and has been reported to capture methanogen groups related to the fen cluster not detected by the ML primer set, although the ME primers are reported to have difficulty amplifying members of the Methanosarcinaceae (33). We thought that by using both forward primers ML and ME, we would capture a greater representation of the methanogen diversities at these sites. All three forward primers were paired with the same reverse primer, mcrA-rev (5′-CGTTCATBGCGTAGTTVGGRTAGT-3′), which is a consensus sequence of three previously reported reverse primers for mcrA (26, 43, 60). Reaction conditions were the same for all three mcrA-targeted primer sets and included an initial denaturation step at 95°C for 3 min, followed by five cycles of denaturation at 95°C for 30 s, annealing at 48°C for 45 s, and extension at 72°C for 30 s, with a ramp rate of 0.1°C/s from the annealing to the extension temperature. These initial five cycles were followed with 30 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 45 s, and extension at 72°C for 30 s, followed by a final extension step at 72°C for 10 min.
PCR products were ligated into vector pCR 2.1 using a TA cloning kit, and the ligation products were used to transform Escherichia coli Top10 cells according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Ampicillin- and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-amended LB agar was used for blue-white screening of transformants, which were subject to whole-cell PCR directly to amplify the plasmid insert. PCR products were sent to the Nucleic Acid Facility at Penn State University for sequencing, and sequencing electropherograms were examined for accuracy using Sequence Scanner v1.0 (Applied Biosystems, Inc., Foster City, CA).
Determination of sequence similarity cutoffs for methanogen taxonomic levels.
16S rRNA and mcrA gene sequences from isolated methanogens were analyzed to determine how to group clone sequences from the libraries into appropriate OTUs. A current list of methanogens with recognized taxonomic standing is maintained by the Subcommittee on Taxonomy of Methanogens of the International Committee on Systematics of Prokaryotes (http://www.the-icsp.org/taxa/methanogenslist.htm). Using this accepted taxonomic structure, corresponding 16S rRNA gene and mcrA sequences for these methanogens were obtained from GenBank, when available. Sequences were eliminated from the analysis if they were too short or contained several undetermined bases (>2% of the sequence). Sequences belonging to Methanocalculus sp. were eliminated from the analysis, as the placement of this group is still under study by the International Committee on Systematics of Prokaryotes. Sequences for Methanospirillum hungatei and Methanopyrus kandleri were also eliminated from the analysis, as they each represent the only sequence for their genus and family. Sequences were truncated from bp 114 to 1435 (E. coli numbering) for 16S rRNA genes and bp 1014 to 1423 (Methanobacterium thermautotrophicum numbering) for mcrA. These truncations were necessary to ensure that all sequences were of the same length prior to the analysis. The sequences were aligned with ClustalW (http://www.ebi.ac.uk/clustalw/), and Mega3 (36) was used to create distance matrices based on the Jukes-Cantor algorithm. Sequences were grouped together by taxonomic level, and Mega3 was used to calculate the mean sequence similarities within each taxonomic unit and between taxonomic units. For instance, within the genus Methanobacterium, the average sequence similarity was calculated based on comparisons of each species to every other species in that genus. To calculate the percent similarity within the family Methanobacteriaceae, the mean similarity was calculated based on the similarities between each species in this family and every other species in a separate genus of this family.
Phylogenetic and diversity analyses of bog and digester communities.
The program DOTUR (54) was used to group clone sequences into OTUs based upon decreasing levels of similarity using the furthest-neighbor algorithm, which states that any two sequences in a group defined by a percent similarity are at least that similar to one another. The input files for DOTUR were distance matrices generated by Mega3 using a Jukes-Cantor algorithm. EstimateS (version 7.5; R. K. Colwell [http://purl.oclc.org/estimates]) was used to calculate Coleman rarefaction curves, Shannon-Wiener and Simpson indices, and Chao1 and abundance-based coverage estimator (ACE) values. For the 16S rRNA gene libraries, statistics were conducted for OTUs of 98 and 96% similarity, respectively, representing similarity within a methanogen species suggested previously by Boone et al. (6) and similarity within a methanogen genus as determined by this study. For the mcrA libraries, statistics were conducted for OTUs of 89% similarity, representing the average mcrA sequence similarity of methanogens within a genus, as there is no standard mcrA divergence for determining a new methanogen species. For each run, 100 randomizations were made with sample replacement. To construct phylogenetic trees, clones were grouped with DOTUR by family-level associations as determined in this study. One representative sequence was chosen from each clone group, and these clone sequences were aligned with ClustalW, and phylogenetic trees were created in Mega3 using the maximum parsimony method with clone-neighbor interchange search level 1 and bootstrapped with 100 trials. Trees for both the 16S rRNA genes and mcrA were rooted with Methanopyrus kandleri as the outgroup.
Nucleotide sequence accession numbers.
Sequences derived from the analysis were deposited in GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html) under accession numbers DQ680352 to DQ680670 and DQ781023 to DQ781063.
RESULTS
Sequence similarities for methanogen taxonomic levels.
The mean percent similarities of methanogen 16S rRNA gene sequences were calculated for 15 genera containing at least two sequences and eight families representing four orders of methanogens (Table 1). Results obtained using a Kimura two-parameter algorithm (data not shown) were nearly identical to those obtained with the Jukes-Cantor algorithm. The average sequence similarity of species within a genus was 96.0% with a range of 88.2 to 100%. The average sequence similarity within a family was 94.3% when considering all methanogen families and was 93.5% for families containing at least two genera. The mean percent similarities of mcrA sequences were calculated for 15 genera and eight families of methanogens (Table 2). The average percent similarity of mcrA sequences within a genus was 88.9%, which ranged from 69.3 to 100%. The average similarity overall within a family was 83.5% and was 79.0% for families containing at least two genera.
TABLE 1.
Sequence percent similarity values within the methanogens for 16S rRNA genesa
| Family | Mean % similarity | Genus | No. of isolates in analysis | Range of % similarity (mean) | GenBank accession no. |
|---|---|---|---|---|---|
| Methanobacteriaceae | 92.8 | Methanobacterium | 11 | 92.1-99.8 (95.4) | M36508, AY386124, DQ649335, AY350742, M59124, AF233586, AF095261, AF028690, AF093061, DQ649330, AF095264 |
| Methanobrevibacter | 9 | 88.2-97.1 (94.0) | AY196666, AF242652, AB065294, U62533, U41095, U82322, U55239, U55237, U55240 | ||
| Methanosphaera | 1 | NA | M59139 | ||
| Methanothermobacter | 5 | 98.6-99.9 (99.3) | X68720, X99046, X99047, X99048, AB104858, | ||
| Methanococcaceae | 93.4 | Methanococcus | 4 | 91.4-95.9 (93.5) | M36507, U39016, U38486, U38461, |
| Methanothermococcus | 2 | (95.0) | M59128, AB057722, | ||
| Methanocaldococcaceae | 95.6 | Methanocaldococcus | 5 | 95.7-98.8 (96.8) | M59126, AF056938, AF547621, AF025822, AF051404 |
| Methanotorris | 1 | NA | AB100884 | ||
| Methanomicrobiaceae | 92.7 | Methanomicrobium | 1 | NA | M59142 |
| Methanoculleus | 6 | 95.7-99.9 (97.3) | AB065298, AB038795, M59134, Y16382, AB065297, AF531178 | ||
| Methanofollis | 3 | 95.7-99.7 (97.3) | AF095272, AY186542, Y16428, | ||
| Methanogenium | 3 | 90.0-98.4 (93.2) | M59130, AJ862839, DQ177345 | ||
| Methanolacinia | 1 | NA | AY196678 | ||
| Methanoplanaceae | 94.4 | Methanoplanus | 2 | (94.4) | M59143, U76631, |
| Methanocorpusculaceae | 99.9 | Methanocorpusculum | |||
| Methanosarcinaceae | 92.9 | Methanosarcina | 4 | 99.7-100.0 (99.9) | AY260435, AF095266, AY260436, AF095268 |
| Methanococcoides | 8 | 93.7-99.1 (96.6) | AY196682, AE010299, AY663809, DQ058823, AB065296, AJ012742, U89773, M59140 | ||
| Methanohalobium | 2 | (97.6) | M59127, X65537 | ||
| Methanohalophilus | 1 | NA | U20149 | ||
| Methanolobus | 1 | NA | M59133 | ||
| Methanomethylovorans | 4 | 96.6-98.2 (97.4) | U20148, U20152, U20154, U20155, | ||
| Methanimicrococcus | 1 | NA | AF120163 | ||
| Methanosaetaceae | 92.5 | Methanosaeta | 2 | 92.5 | M59146, AB071701 |
| Mean % similarity | 94.3 | 96.0 | |||
| With multiple genera | 93.5 |
Ranges and means were calculated with distance matrices created with the Jukes-Cantor algorithm. NA, not applicable.
TABLE 2.
Sequence percent similarity values within the methanogens for mcrAa
| Family | Mean % similarity | Genus | No. of isolates in analysis | Range of % similarity (mean) | GenBank accession no. |
|---|---|---|---|---|---|
| Methanobacteriaceae | 71.7 | Methanobacterium | 4 | 69.3-75.8 (73.1) | AF414050, AY386125, AF313806, AY289750 |
| Methanobrevibacter | 4 | 77.2-90.0 (82.6) | AF414046, AF414035, DQ251045, DQ251046 | ||
| Methanosphaera | 1 | NA | AF414047 | ||
| Methanothermobacter | 4 | 97.2-100.0 (98.6) | U10036, AY303950, AY289752, AY289748 | ||
| Methanothermaceae | 98.0 | Methanothermus | 2 | (98.0) | J03375, AY289747 |
| Methanococcaceae | 83.2 | Methanococcus | 4 | 76.0-92.7 (83.3) | M16893, AY354034, BX957223, X07793 |
| Methanothermococcus | 2 | 85.9 | AF414048, AY354033 | ||
| Methanocaldococcaceae | 81.4 | Methanocaldococcus | 2 | 82.5 | AF414040, AY354035 |
| Methanotorris | 1 | NA | AF414039 | ||
| Methanomicrobiaceae | 79.5 | Methanomicrobium | 1 | NA | AF414044 |
| Methanoculleus | 5 | 89.3-100.0 (92.1) | AF414036, AB288270, NZAASI01000002, AF313804, DQ229156 | ||
| Methanofollis | 1 | NA | AF414041 | ||
| Methanogenium | 4 | 82.8-93.8 (86.9) | DQ229157, DQ229158, DQ229159, DQ229160 | ||
| Methanocorpusculaceae | 94.9 | Methanocorpusculum | 3 | 93.8-97.1 (94.9) | AY260445, AF414049, AY260441 |
| Methanosarcinaceae | 79.4 | Methanosarcina | 7 | 88.3-99.3 (91.8) | Y10058, AE010299, AY260443, AF414043, U22248, U22250, U22251 |
| Methanococcoides | 2 | 96.0 | U22235, U22234 | ||
| Methanohalobium | 1 | NA | U22236 | ||
| Methanohalophilus | 3 | 95.0-99.0 (96.7) | U22237, U22259, U22239 | ||
| Methanolobus | 5 | 87.4-96.3 (91.6) | U22244, U22257, U22242, U22243, U22245 | ||
| Methanomethylovorans | 1 | NA | AY260442 | ||
| Methanosalsum | 1 | NA | U22252 | ||
| Methanosaetaceae | 79.6 | Methanosaeta | 2 | 79.6 | AF313802, CP000477 |
| Mean % similarity | 83.5 | 88.9 | |||
| With multiple genera | 79.0 |
Ranges and means were calculated using distance matrices created with the Jukes-Cantor algorithm. NA, not applicable.
Collector's curves and diversity indices of bog and digester communities.
A total of 97 methanogen 16S rRNA gene sequences were obtained, 47 from the bog sediment and 50 from the digester sludge. A total of 259 mcrA sequences were obtained, 105 from the bog sediment and 154 from the digester sludge. For the 105 mcrA sequences derived from the bog, 35, 38, and 32 sequences were obtained with ME, ML, and mlas as the forward primer, respectively. For the 154 mcrA sequences obtained from the digester sludge, 21, 72, and 61 sequences were obtained with ME, ML, and mlas as the forward primers, respectively.
The 16S rRNA genes and mcrA libraries were analyzed by constructing species accumulation curves and calculating diversity indices. Minimum sequence similarities of 98 and 96% were used to group 16S rRNA gene sequences into OTUs, as these values represented the suggested sequence similarity within a methanogen species (6) and the calculated average sequence similarity of species within a genus from this study (Tables 1 and 2). Sequences for mcrA generated from all three forward primers were analyzed together, but sequences identified as the isoenzyme mrtA were excluded from this analysis. Only members of the Methanobacteriales and the Methanococcales possess the Mrt operon (21), so these sequences were excluded to avoid a biased overrepresentation of species from these orders in the library. At a sequence similarity of 98%, the 16S rRNA gene libraries were grouped into 26 and 16 clades for the bog and digester, respectively. At a sequence similarity of 96%, the 16S rRNA gene libraries were grouped into 19 and 7 clades for the bog and digester, respectively. At a sequence similarity of 89%, mcrA sequences were grouped into 24 clades each for the bog and digester libraries.
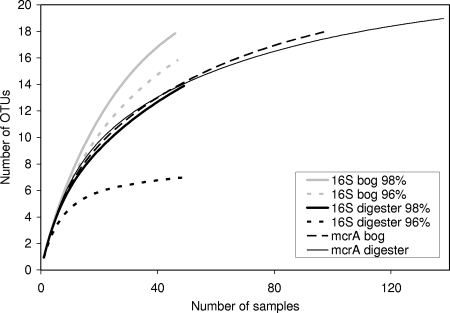
Based on these clone groupings, Coleman rarefaction curves, which represent how extensively the genetic diversity at a site has been sampled, were constructed for both environments and gene targets (Fig. 1). The diversity indices Chao1, ACE, Shannon-Wiener index (H′), and Simpson's index (D) were calculated using EstimateS (Table 3). The values for Chao1 and ACE represent the expected number of OTUs present in an environment if sampling were complete (30). The Shannon-Wiener index (H′) and Simpson's index are measures of species richness and the evenness of distribution of species within a community, and both increase with increasing genetic diversity at a site. Comparing the 16S rRNA gene libraries, diversity was lower and dominance was greater in the digester than in the bog library at both the 96 and 98% similarity cutoffs for OTU groupings (Table 3). As expected, for both 16S rRNA gene libraries, the rarefaction curves show a flattening with a cutoff of 96% versus 98%, but the change in curve shape was much more dramatic for the 16S rRNA gene library (Fig. 1). When the mcrA libraries from the bog and digester were compared, there was no difference in diversity or dominance indices (Table 3). The diversity indices for the mcrA libraries were estimated for a genus-level grouping and were compared with the 16S rRNA gene libraries at a genus-level grouping of 96% similarity. There was no difference in diversity when the 16S rRNA gene library was compared to the mcrA libraries of the bog and digester, but the 16S rRNA gene library did show less diversity than the other three libraries. Ecological indicators often underestimate the true richness of a community at low sample sizes (30), and as the 16S rRNA genes libraries were much smaller than the mcrA libraries, this may have contributed to the lower richness values for the 16S rRNA gene digester library.
FIG. 1.
Coleman rarefaction curves of 16S rRNA and mcrA gene clone libraries obtained from the acidic peatland and anaerobic digester environments. The 16S rRNA gene sequences were grouped by 98 or 96% similarity and mcrA sequences were grouped by 89% similarity using the program DOTUR. Rarefaction curves were generated using the program EstimateS at 100 randomizations made with sample replacement.
TABLE 3.
Diversity indices for bog and digester 16S rRNA gene and mcrA libraries using species-level groupings
| Library (% similarity) | Mean diversity index
|
|||
|---|---|---|---|---|
| Chao1 (95% confidence interval) | ACE ± SD | Shannon-Wiener index (H′) ± SD | Simpson's diversity index (1/D) ± SD | |
| 16S bog (98) | 28.02 (20.16, 65.43) | 30.2 ± 8.96 | 2.51 ± 0.16 | 10.99 ± 3.15 |
| 16S bog (96) | 20.66 (15.32, 54.5) | 19.07 ± 4.58 | 2.28 ± 0.14 | 8.44 ± 1.9 |
| 16S digester (98) | 17.33 (12.72, 48.36) | 17.19 ± 4.78 | 2.05 ± 0.15 | 6.85 ± 1.18 |
| 16S digester (96) | 10.73 (9.3, 24.78) | 10.74 ± 1.86 | 1.84 ± 0.12 | 5.59 ± 0.81 |
| mcrA bog | 25.83 (21.69, 52.1) | 24.27 ± 3.07 | 2.65 ± 0.09 | 11.93 ± 1.71 |
| mcrA digester | 26.06 (21.17, 57.1) | 24.81 ± 4.3 | 2.5 ± 0.08 | 9.94 ± 0.93 |
Phylogeny of bog and digester communities.
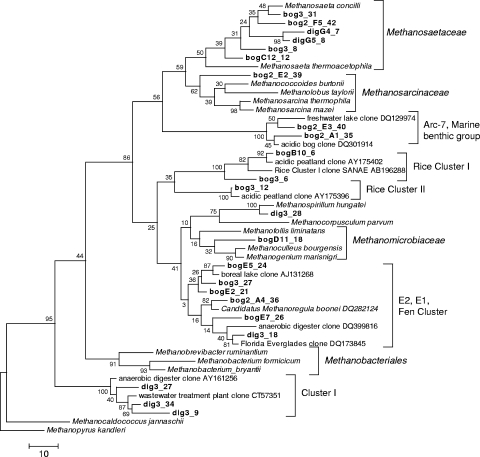
Sequences in the libraries were grouped at the family level using DOTUR, with minimum sequence similarity values of 93.5 and 79% for the 16S rRNA and mcrA genes, respectively. One representative sequence from each group was chosen for incorporation in the phylogenetic trees and was identified by BLAST analysis (see the supplemental material). The 16S rRNA gene sequences grouped into 15 clades for the bog and 7 clades for the digester, with no incidence of clones from both environments being present in the same clade (Fig. 2). For the 16S rRNA gene libraries, nearly all of the clones from Bear Meadows Bog closely grouped with clones from acidic freshwater environments, including two central New York State peatlands (4, 9) and a freshwater lake located downstream of an acidic Sphagnum bog (12). The 16S rRNA gene digester sequences were related to clones obtained from a variety of environments, including full-scale anaerobic digesters (GenBank accession numbers DQ386720, DQ399803, and AB266913; see Table S1 in the supplemental material), lotus field soil (GenBank accession number AB236107), and part of the Florida Everglades impacted by agricultural runoff (13).
FIG. 2.
Phylogenetic tree for representative 16S rRNA gene sequences (shown in Table S1 in the supplemental material) obtained from an acidic peatland (clones labeled “bog”) and an anaerobic digester treating municipal wastewater sludge (clones labeled “dig”). The tree was constructed as a maximum parsimony tree using close-neighbor interchange level 1 and bootstrapped with 100 trials. All positions containing gaps and missing data were eliminated from the data set. The scale bar represents the number of changes over the whole sequence. The classification of clusters is based on data described previously by Vetriani et al. (62) for the marine benthic group, which was also named Arc-7 by Castro et al. (10); Großkopf et al. (25) for rice clusters I and II; Cadillo-Quiroz et al. (9) for E1 and E2, also named fen cluster by Juottonen et al. (32); and McHugh et al. for cluster I (45).
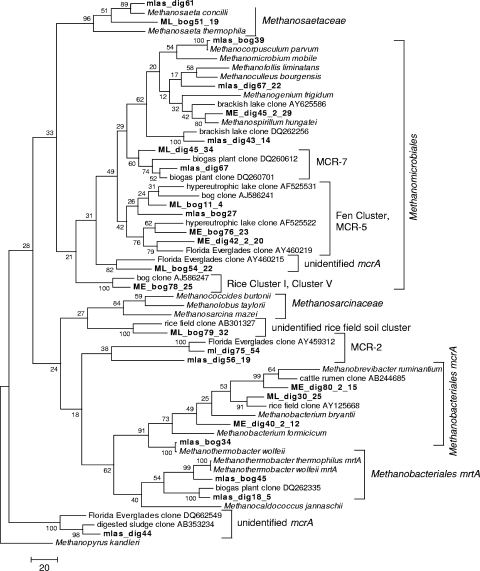
The mcrA sequences grouped into 12 clades for the bog and 14 clades for the digester, with two clades containing representative sequences from both the bog and digester libraries (Fig. 3). Trends are seen in the mcrA libraries that are similar to those seen with the 16S rRNA gene libraries, with most of the bog sequences being most closely related to clones from oligotrophic freshwater lakes and peatlands (17, 18, 19, 32) and the majority of the digester sequences being most closely related to clones from anaerobic digesters (GenBank accession numbers AY937276, AY937277, AY937285, DQ260701, DQ260681, DQ260578, and DQ262263; see Table S2 in the supplemental material) and pasture and rice paddy soil (22, 56).
FIG. 3.
Phylogenetic tree for representative mcrA sequences (shown in Table S2 in the supplemental material) obtained from an acidic peatland (bog) and an anaerobic digester treating municipal wastewater sludge (dig). The tree was constructed as a maximum parsimony tree using close-neighbor interchange level 1 and bootstrapped with 100 trials. All positions containing gaps and missing data were eliminated from the data set. The scale bar represents the number of changes over the whole sequence. The classification of clusters is based on data described previously by Castro et al. (10) for MCR-2, MCR-5, and MCR-7; Juottonen et al. (32) for the fen cluster; Großkopf et al. (27) for rice cluster I, also named cluster V by Galand et al. (20); and Lueders et al. (41) for the unidentified rice field soil cluster.
Both the bog and digester libraries were dominated by clones that were related to uncultured methanogen groups. The majority of clones from the bog environment were members of the fen cluster, a deeply branching clade of the Methanomicrobiales that was first identified by Galand et al. (20) but has also been labeled by other researchers as MCR-5 and E1 and E2 (4, 10). Three groups of uncultured methanogens were represented exclusively within the bog clone libraries. These include rice clusters I and II, first described in Großkopf et al. (25), and two sequences that are related (89% similarity) to a branch within cluster MCR-5 (Fig. 3). Members of this cluster are related to members of the Methanomicrobiales isolated from an oligotrophic area of the Florida Everglades (10). The majority of the digester 16S rRNA gene clones grouped with sequences from cluster I, isolated from a variety of bioreactors and described previously by McHugh et al. (45), and with sequences from a nutrient-impacted area of the Florida Everglades that are grouped in Methanomicrobiales clusters E1 and E2, as described previously by Cadillo-Quiroz et al. (9). Most of the mcrA digester clones group with cluster MCR-7, which was identified previously by Castro et al. (10) during a molecular survey of oligotrophic and nutrient-impacted areas of the Florida Everglades. A number of digester clones grouped into cluster MCR-2, a cluster also defined in the study described previously by Castro et al. (10). In that study, all sequences belonging to MCR-2 and MCR-7 were found only in the digester environment.
Although the majority of clones obtained in this study were closely related to other uncultured clone sequences, a number of clone sequences from both libraries were closely related to cultured methanogen species. The bog environment included clones related to Methanocorpusculum parvum (GenBank accession no. AF414045) and mrtA of Methanothermobacter thermophilus (accession no. AY289753), and several clones from the digester environment were closely related to Methanospirillum hungatei strain JF-1 (accession no. NC007796) and Methanobrevibacter ruminantium mcrA (accession no. AF414406).
DISCUSSION
The methanogens of the bog and digester environments were found to be very different from one another, and although this result was expected, the degree of difference in comparing sequences from both environments was surprising. There was nearly no overlap in either 16S rRNA or mcrA gene sequences even at a level of similarity representing the average similarity within a methanogen family. Few studies have compared methanogens in natural wetlands to those found in constructed environments, and the present study is the first to directly compare methanogen communities in a naturally occurring wetland and a constructed anaerobic reactor. In addition to containing very different methanogen sequences, both the bog and digester libraries were dominated by clones belonging to uncultured environmental groups. The majority of clones from both the 16S rRNA gene and mcrA libraries of the bog environment were related to the fen cluster, a deeply branching clade within the Methanomicrobiales. The fen cluster may no longer be considered an uncultured group, as Brauer et al. (7) recently isolated and described a member of this group, “Candidatus Methanoregula boonei,” which was found to be acid tolerant and hydrogenotrophic. The Fen cluster has been identified in a number of other peatland environments, many of which are acidic, oligotrophic bogs. The majority of clones from the 16S rRNA gene library of the digester environment grouped with cluster I, members of which have been found in a variety of anaerobic digesters (14, 45, 50). The majority of clones in the mcrA library from the digester grouped with clade MCR-7, which has been found in a wide range of environments including oligotrophic and nutrient-impacted areas of the Florida Everglades (10), biogas plants (GenBank accession no. DQ260701, DQ260681, DQ260578, and DQ262263; see Table S2 in the supplemental material), and brackish lake sediment (2). Cluster I is related to the Methanobacteriales, and MCR-7 is found within the Methanomicrobiales, so it is likely that members of cluster I and MCR-7 are hydrogenotrophic. This suggests that both environments are dominated by hydrogenotrophic methanogens, although acetotrophic methanogen sequences closely related to Methanosaeta were also found in both environments. Members of the Methanosaeta, which are strictly acetotrophic, appear to be ubiquitous and have been isolated from a number of different environments including freshwater marshes and lakes (10-12), acidic peatlands (4, 18, 32), rice field soil (41), and anaerobic digesters treating municipal solid waste and sewage sludge (24, 46), dairy wastewater (40), or industrial wastewater (15). Despite the abundance of clones representing hydrogenotrophic methanogens in all libraries, it should be noted that sequence abundance in a clone library does not necessarily relate to the abundance of the organism in the environment.
The analyses of sequences from accepted methanogen taxonomy performed in this study illustrate the difficulty in gene fragment-based surveys of environmental microorganisms. Springer et al. (60) found that the average mcrA sequence distance between species was approximately three times greater than the distance of the 16S rRNA gene sequences. This correlates well to the difference between the average sequence distances found for the 16S rRNA gene and mcrA in this study. Despite this fact, the range of similarity within a methanogen genus varied from 88.2 to 99.9% for the 16S rRNA genes and from 69.3 to 100.0% for mcrA. Currently, the definition of species applied to methanogen 16S rRNA gene clone libraries varies from researcher to researcher, and there is no species definition applied to mcrA libraries. Ecological richness estimators can be used for microbial communities only if strict OTU definitions are consistently applied (30). The consistent application of an OTU definition also makes it possible to compare diversity measurements among studies. The goal for determining average sequence similarity within genera and families of methanogens in this study was primarily to calculate richness estimators and characterize differences between the two methanogen communities that we studied, but these data were also used to relate mcrA sequence disparity to phylogenetic distance.
The clone libraries for ME, ML, and mlas as the forward primers were compared to detect any difference in mcrA sequence recovery among the three forward primers. In general, a clone group sharing at least 80% similarity was detected by all three primers. Clone groups with only one or two clones were not detected by all three primers, but this is likely a limitation of the number of clones sampled in the different libraries. With both primers ML and mlas, clone groups with more than one clone were obtained solely with that primer. This was most dramatic for forward primer mlas, which had four groups of unique clones. These results suggest that forward primers ML and mlas may capture gene sequences not captured by forward primer ME. The ME primer set has been reported to not detect sequences from Methanosaeta sp. or sequences of the isoenzyme mrtA (2), a finding mimicked in the current study.
A few authors looked at primer bias in obtaining mcrA sequences from the natural environment. Lueders et al. (44) found that a cluster of mcrA sequences, which was named “unidentified rice field soil mcrA” by those authors, could be detected with the MCR, but not the ME, primer pair. Nercessian et al. (47) amplified mcrA sequences from deep-sea hydrothermal vents with ML and ME primer sets and recovered different sequences in both libraries, with only some overlap in clones (47). Juottonen et al. (33) examined potential bias in the three mcrA primer sets in obtaining clones from drained peatland sediment. Similar clone sequences were obtained with all sets, but four minor OTUs were not detected by all sets, although those authors could not rule out the number of clones sequenced as being responsible for these differences in coverage. The dominant OTUs obtained with each primer set differed, which in turn affected ecological diversity measurements of the clone libraries, namely, the Shannon-Wiener and Simpson's dominance indices (33). Results from these studies suggest that more comprehensive libraries of mcrA diversity may be obtained by using two or more primer sets. A limitation in comparing the results of these studies is that different PCR parameters, notably the annealing temperature, were used in each study. Lueders and Friedrich previously demonstrated that the numbers and types of clones obtained in an mcrA library generated with the ML primer set was determined by the choice of annealing temperature (42), so both the choice of primer set and PCR conditions will affect the clones obtained in a library.
Supplementary Material
Acknowledgments
We thank Shandra D. Justicia León and Allison Speers for their assistance in primer development and clone library construction.
This study was supported by the Penn State Biogeochemical Research Initiative for Education (NSF IGERT grant DGE-9972759).
Footnotes
Published ahead of print on 5 September 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arbeli, Z., A. Brenner, and A. Abeliovich. 2006. Treatment of high-strength dairy wastewater in an anaerobic deep reservoir: analysis of the methanogenic fermentation pathway and the rate-limiting step. Water Res. 40:3653-3659. [DOI] [PubMed] [Google Scholar]
- 2.Banning, N., F. Brock, J. C. Fry, R. J. Parkes, E. R. C. Hornibrook, and A. J. Weightman. 2005. Investigation of the methanogen population structure and activity in a brackish lake sediment. Environ. Microbiol. 7:947-960. [DOI] [PubMed] [Google Scholar]
- 3.Bapteste, E., C. Brochier, and Y. Boucher. 2005. Higher-level classification of the archaea: evolution of methanogenesis and methanogens. Archaea 1:353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basiliko, N., J. B. Yavitt, P. M. Dees, and S. M. Merkel. 2003. Methane biogeochemistry and methanogen communities in two northern peatland ecosystems, New York state. Geomicrobiol. J. 20:563-577. [Google Scholar]
- 5.Bodik, I., B. Herdova, and M. Drtil. 2000. Anaerobic treatment of the municipal wastewater under psychrophilic conditions. Bioprocess Eng. 22:385-390. [Google Scholar]
- 6.Boone, D. R., W. B. Whitman, and P. Rouviere. 1993. Diversity and taxonomy of methanogens, p. 35-80. In J. G. Ferry (ed.), Methanogens. Chapman and Hall, Inc., New York, NY.
- 7.Brauer, S. L., H. Cadillo-Quiroz, E. Yashiro, J. B. Yavitt, and S. H. Zinder. 2006. Isolation of a novel acidiphilic methanogen from an acidic peat bog. Nature 442:192-194. [DOI] [PubMed] [Google Scholar]
- 8.Burggraf, S., T. Mayer, R. Amann, S. Schadhauser, C. R. Woese, and K. O. Stetter. 1994. Identifying members of the domain Archaea with rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 60:3112-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cadillo-Quiroz, H., S. Brauer, E. Yashiro, C. Sun, J. Yavitt, and S. Zinder. 2006. Vertical profiles of methanogenesis and methanogens in two contrasting acidic peatlands in central New York state, USA. Environ. Microbiol. 8:1428-1440. [DOI] [PubMed] [Google Scholar]
- 10.Castro, H., A. Ogram, and K. R. Reddy. 2004. Phylogenetic characterization of methanogenic assemblages in eutrophic and oligotrophic areas of the Florida Everglades. Appl. Environ. Microbiol. 70:6559-6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan, O. C., P. Claus, P. Casper, A. Ulrich, T. Lueders, and R. Conrad. 2005. Vertical distribution of structure and function of the methanogenic archaeal community in Lake Dagow sediment. Environ. Microbiol. 7:1139-1149. [DOI] [PubMed] [Google Scholar]
- 12.Chan, O. C., M. Wolf, D. Hepperle, and P. Casper. 2002. Methanogenic archaeal community in the sediment of an artificially partitioned acidic bog lake. FEMS Microbiol. Ecol. 42:119-129. [DOI] [PubMed] [Google Scholar]
- 13.Chauhan, A., and A. Ogram. 2006. Fatty acid-oxidizing consortia along a nutrient gradient in the Florida Everglades. Appl. Environ. Microbiol. 72:2400-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chouari, R., D. Le Paslier, P. Daegelen, P. Ginestet, J. Weissenbach, and A. Sghir. 2005. Novel predominant archaeal and bacterial groups revealed by molecular analysis of an anaerobic sludge digester. Environ. Microbiol. 7:1104-1115. [DOI] [PubMed] [Google Scholar]
- 15.Collins, G., A. Woods, S. McHugh, M. W. Carton, and V. O'Flaherty. 2003. Microbial community structure and methanogenic activity during start-up of psychrophilic anaerobic digesters treating synthetic industrial wastewaters. FEMS Microbiol. Ecol. 46:159-170. [DOI] [PubMed] [Google Scholar]
- 16.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Earl, J., G. Hall, R. W. Pickup, D. A. Ritchie, and C. Edwards. 2003. Analysis of methanogen diversity in a hypereutrophic lake using PCR-RFLP analysis of mcr sequences. Microb. Ecol. 46:270-278. [DOI] [PubMed] [Google Scholar]
- 18.Galand, P. E., H. Fritze, R. Conrad, and K. Yrjala. 2005. Pathways for methanogenesis and diversity of methanogenic archaea in three boreal peatland ecosystems. Appl. Environ. Microbiol. 71:2195-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galand, P. E., H. Juottonen, H. Fritze, and K. Yrjala. 2005. Methanogen communities in a drained bog: effect of ash fertilization. Microb. Ecol. 49:209-217. [DOI] [PubMed] [Google Scholar]
- 20.Galand, P. E., S. Saarnio, H. Fritze, and K. Yrjala. 2002. Depth related diversity of methanogen archaea in Finnish oligotrophic fen. FEMS Microbiol. Ecol. 42:441-449. [DOI] [PubMed] [Google Scholar]
- 21.Garcia, J. L., B. K. C. Patel, and B. Ollivier. 2000. Taxonomic phylogenetic and ecological diversity of methanogenic archaea. Anaerobe 6:205-226. [DOI] [PubMed] [Google Scholar]
- 22.Gattinger, A., M. G. Hoefle, M. Schloter, A. Embacher, F. Bohme, J. C. Munch, and M. Labrenz. 2007. Traditional cattle manure application determines abundance, diversity and activity of methanogenic archaea in arable European soil. Environ. Microbiol. 9:612-624. [DOI] [PubMed] [Google Scholar]
- 23.Girguis, P. R., V. J. Orphan, S. J. Hallam, and E. F. DeLong. 2003. Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl. Environ. Microbiol. 69:5472-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin, M. E., K. D. McMahon, R. I. Mackie, and L. Raskin. 1998. Methanogenic population dynamics during start-up of anaerobic digesters treating municipal solid waste and biosolids. Biotechnol. Bioeng. 57:342-355. [DOI] [PubMed] [Google Scholar]
- 25.Großkopf, R., S. Stubner, and W. Liesack. 1998. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl. Environ. Microbiol. 64:4983-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hales, B. A., C. Edwards, D. A. Ritchie, G. Hall, R. W. Pickup, and J. R. Saunders. 1996. Isolation and identification of methanogen-specific DNA from blanket bog feat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hallam, S. J., P. R. Girguis, C. M. Preston, P. M. Richardson, and E. F. DeLong. 2003. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 69:5483-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoj, L., R. A. Olsen, and V. L. Torsvik. 2005. Archaeal communities in high arctic wetlands at Spitsbergen, Norway (78 degrees N) as characterized by 16S rRNA gene fingerprinting. FEMS Microbiol. Ecol. 53:89-101. [DOI] [PubMed] [Google Scholar]
- 29.Hori, T., S. Haruta, Y. Ueno, M. Ishii, and Y. Igarashi. 2006. Dynamic transition of a methanogenic population in response to the concentration of volatile fatty acids in a thermophilic anaerobic digester. Appl. Environ. Microbiol. 72:1623-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Innis, M., D. Gelfand, and J. J. Sninsky. 1999. PCR applications: protocols for functional genomics. Academic Press, San Diego, CA.
- 32.Juottonen, H., P. E. Galand, E. S. Tuittila, J. Laine, H. Fritze, and K. Yrjala. 2005. Methanogen communities and bacteria along an ecohydrological gradient in a northern raised bog complex. Environ. Microbiol. 7:1547-1557. [DOI] [PubMed] [Google Scholar]
- 33.Juottonen, H., P. E. Galand, and K. Yrjala. 2006. Detection of methanogenic archaea in peat: comparison of PCR primers targeting the mcrA gene. Res. Microbiol. 157:914-921. [DOI] [PubMed] [Google Scholar]
- 34.Kashyap, D. R., K. S. Dadhich, and S. K. Sharma. 2003. Biomethanation under psychrophilic conditions: a review. Bioresource Technol. 87:147-153. [DOI] [PubMed] [Google Scholar]
- 35.Krumholz, L. R., J. L. Hollenback, S. J. Roskes, and D. B. Ringelberg. 1995. Methanogenesis and methanotrophy within a sphagnum peatland. FEMS Microbiol. Ecol. 18:215-224. [Google Scholar]
- 36.Kumar, S., K. Tamura, and M. Nei. 2004. Mega3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 37.Reference deleted.
- 38.Lettinga, G., S. Rebac, S. Parshina, A. Nozhevnikova, J. B. van Lier, and A. J. M. Stams. 1999. High-rate anaerobic treatment of wastewater at low temperatures. Appl. Environ. Microbiol. 65:1696-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lettinga, G., S. Rebac, and G. Zeeman. 2001. Challenge of psychrophilic anaerobic wastewater treatment. Trends Biotechnol. 19:363-370. [DOI] [PubMed] [Google Scholar]
- 40.Liu, W. T., O. C. Chan, and H. H. P. Fang. 2002. Microbial community dynamics during start-up of acidogenic anaerobic reactors. Water Res. 36:3203-3210. [DOI] [PubMed] [Google Scholar]
- 41.Lueders, T., K. J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analyses of methyl-coenzyme M reductase alpha-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 42.Lueders, T., and M. W. Friedrich. 2003. Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl. Environ. Microbiol. 69:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luton, P. E., J. M. Wayne, R. J. Sharp, and P. W. Riley. 2002. The mcrA gene as an alternative to 16s rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521-3530. [DOI] [PubMed] [Google Scholar]
- 44.Marchesi, J. R., A. J. Weightman, B. A. Cragg, R. J. Parkes, and J. C. Fry. 2001. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol. Ecol. 34:221-228. [DOI] [PubMed] [Google Scholar]
- 45.McHugh, S., M. Carton, T. Mahony, and V. O'Flaherty. 2003. Methanogenic population structure in a variety of anaerobic bioreactors. FEMS Microbiol. Lett. 219:297-304. [DOI] [PubMed] [Google Scholar]
- 46.McMahon, K. D., D. D. Zheng, A. J. M. Stams, R. I. Mackie, and L. Raskin. 2004. Microbial population dynamics during start-up and overload conditions of anaerobic digesters treating municipal solid waste and sewage sludge. Biotechnol. Bioeng. 87:823-834. [DOI] [PubMed] [Google Scholar]
- 47.Nercessian, O., N. Bienvenu, D. Moreira, D. Prieur, and C. Jeanthon. 2005. Diversity of functional genes of methanogens, methanotrophs and sulfate reducers in deep-sea hydrothermal environments. Environ. Microbiol. 7:118-132. [DOI] [PubMed] [Google Scholar]
- 48.Nusslein, B., K. J. Chin, W. Eckert, and R. Conrad. 2001. Evidence for anaerobic syntrophic acetate oxidation during methane production in the profundal sediment of subtropical Lake Kinneret (Israel). Environ. Microbiol. 3:460-470. [DOI] [PubMed] [Google Scholar]
- 49.Panikov, N. S., and S. N. Dedysh. 2000. Cold season CH4 and CO2 emission from boreal peat bogs (West Siberia): winter fluxes and thaw activation dynamics. Global Biogeochem. Cycles 14:1071-1080. [Google Scholar]
- 50.Pender, S., M. Toomey, M. Carton, D. Eardly, J. W. Patching, E. Colleran, and V. O'Flaherty. 2004. Long-term effects of operating temperature and sulphate addition on the methanogenic community structure of anaerobic hybrid reactors. Water Res. 38:619-630. [DOI] [PubMed] [Google Scholar]
- 51.Purdy, K. J., D. B. Nedwell, and T. M. Embley. 2003. Analysis of the sulfate-reducing bacterial and methanogenic archaeal populations in contrasting Antarctic sediments. Appl. Environ. Microbiol. 69:3181-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rocheleau, S., C. W. Greer, J. R. Lawrence, C. Cantin, L. Laramee, and S. R. Guiot. 1999. Differentiation of Methanosaeta concilii and Methanosarcina barkeri in anaerobic mesophilic granular sludge by fluorescent in situ hybridization and confocal scanning laser microscopy. Appl. Environ. Microbiol. 65:2222-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schulz, S., and R. Conrad. 1996. Influence of temperature on pathways to methane production in the permanently cold profundal sediment of Lake Constance. FEMS Microbiol. Ecol. 20:1-14. [Google Scholar]
- 56.Sheppard, S. K., A. J. McCarthy, J. P. Loughnane, N. D. Gray, I. M. Head, and D. Lloyd. 2005. The impact of sludge amendment on methanogen community structure in an upland soil. Appl. Soil Ecol. 28:147-162. [Google Scholar]
- 57.Sizova, M. V., N. S. Panikov, T. P. Tourova, and P. W. Flanagan. 2003. Isolation and characterization of oligotrophic acido-tolerant methanogenic consortia from a sphagnum peat bog. FEMS Microbiol. Ecol. 45:301-315. [DOI] [PubMed] [Google Scholar]
- 58.Smith, K. S., and C. Ingram-Smith. 2007. Methanosaeta, the forgotten methanogen? Trends Microbiol. 15:150-155. [DOI] [PubMed] [Google Scholar]
- 59.Sorensen, A. H., V. L. Torsvik, T. Torsvik, L. K. Poulsen, and B. K. Ahring. 1997. Whole-cell hybridization of Methanosarcina cells with two new oligonucleotide probes. Appl. Environ. Microbiol. 63:3043-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Springer, E., M. S. Sachs, C. R. Woese, and D. R. Boone. 1995. Partial gene-sequences for the A-subunit of methyl-coenzyme-M reductase (mcrI) as a phylogenetic tool for the family Methanosarcinaceae. Int. J. Syst. Bacteriol. 45:554-559. [DOI] [PubMed] [Google Scholar]
- 61.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence-analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 62.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A. L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagner, D., A. Lipski, A. Embacher, and A. Gattinger. 2005. Methane fluxes in permafrost habitats of the Lena Delta: effects of microbial community structure and organic matter quality. Environ. Microbiol. 7:1582-1592. [DOI] [PubMed] [Google Scholar]
- 64.Whitman, W. B., T. L. Bowen, and D. R. Boone. 2006. The methanogenic bacteria, p. 165-207. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 3. Springer-Verlag, New York, NY. [Google Scholar]
- 65.Zheng, D., and L. Raskin. 2000. Quantification of Methanosaeta species in anaerobic bioreactors using genus- and species-specific hybridization probes. Microb. Ecol. 39:246-262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.