Abstract
We investigated the effect of codon optimization on the expression levels of heterologous proteins in Aspergillus oryzae, using the mite allergen Der f 7 as a model protein. A codon-optimized Der f 7 gene was synthesized according to the frequency of codon usage in A. oryzae by recursive PCR. Both native and optimized Der f 7 genes were expressed under the control of a high-level-expression promoter with their own signal peptides or in a fusion construct with A. oryzae glucoamylase (GlaA). Codon optimization markedly increased protein and mRNA production levels in both nonfused and GlaA-fused Der f 7 constructs. For constructs with native codons, analysis by 3′ rapid amplification of cDNA ends revealed that poly(A) tracts tended to be added within the coding region, producing aberrant mRNAs that lack a termination codon. Insertion of a termination codon between the carrier GlaA and native Der f 7 proteins in the GlaA fusion construct resulted in increases in mRNA and secreted-carrier-GlaA levels. These results suggested that mRNAs without a termination codon as a result of premature polyadenylation are degraded, possibly through the nonstop mRNA decay pathway. We suggest that codon optimization in A. oryzae results in elimination of cryptic polyadenylation signals in native Der f 7, thereby circumventing the production of truncated transcripts and resulting in an increase in steady-state mRNA levels.
The filamentous fungus Aspergillus oryzae has been used in the production of fermented foods, such as sake, soy sauce, and miso (soybean paste), in Japan for over a thousand years. In addition, A. oryzae has the ability to secrete large amounts of proteins and has recently become a favorable host for recombinant protein production (5). By use of a series of classical random mutagenesis and screening procedures, hypersecretion mutants of heterologous proteins in aspergilli have been obtained (9, 49). However, the secretion yields of heterologous proteins are low compared to those of homologous proteins or proteins from closely related fungal species and generally do not exceed tens of milligrams per liter (16). In order to improve the expression levels of heterologous proteins, several trials have been conducted, and some strategies have been reported to be effective in increasing the level of heterologous protein production (39). These results provide information on how to increase the expression levels of heterologous genes; however, there is little information on the mechanisms hampering heterologous gene expression. In this study, we investigate the effect of codon optimization on heterologous gene expression. Such optimization has been effective in improving secretion levels of heterologous proteins in several hosts (19). Studies of heterologous gene expression through codon optimization have reported improved heterologous protein production in filamentous fungal species, such as Aspergillus awamori (2, 15, 35), Aspergillus niger (27, 35), Neurospora crassa (13, 35), and Trichoderma reesei (47). Based on studies of bacteria, codon optimization is believed to improve translational efficiency, resulting in increased levels of protein production (4, 22, 25). Also, some studies of filamentous fungi have indicated that codon optimization increased mRNA levels, resulting in increased protein production (15, 27, 41, 47). Regardless, there is little information on the mechanism by which codon optimization increases the expression levels of heterologous genes in filamentous fungi. To elucidate such mechanisms in A. oryzae, we used Der f 7 as a model heterologous protein. Der f 7, a secretion protein of the house dust mite Dermatophagoides farinae, belongs to the group 7 mite allergens that react with immunoglobulin E in 50% of patients allergic to house dust (26, 43). House dust allergy is a serious problem among all allergic diseases, and specific allergens are required for diagnostics and immunotherapy. Allergens prepared from mites generally contain irrelevant proteins and can cause undesirable immune responses. Therefore, recombinant allergens present an attractive alternative to mite extracts. For clinical purposes, it is of great importance to prepare large quantities of specific recombinant allergens (3). Therefore, there have been many attempts to produce large quantities of mite allergen proteins by using Escherichia coli (54), Saccharomyces cerevisiae (6), Pichia pastoris (23, 53), and A. oryzae (44) as microbial hosts. With respect to these attempts, recombinant Der f 1 produced in A. oryzae has been reported to have immunoglobulin E-binding activity similar to that of native Der f 1, while recombinant Der f 1 produced in bacteria had weaker binding activity than native Der f 1 (44). These results suggested that A. oryzae is a suitable host for production of Der f 7 for immunotherapy.
MATERIALS AND METHODS
Strains, culture conditions, and DNA preparation.
A. oryzae NS4 (52) was used as a recipient strain for transformation. E. coli DH5α (supF44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 lacU169 Φ80lacZM15) was used for construction and propagation of plasmid DNAs. Czapek-Dox medium, containing 1% glucose, 0.3% NaNO3, 0.15% KCl, 0.1% KH2PO4, 0.05% MgSO4, and 0.1% methionine, was used as a selective medium for fungal transformation. For protein expression, YPM medium, containing 1% yeast extract, 2% peptone, and 2% maltose, was used. Transformation of A. oryzae was performed by the methods of Gomi et al. (12), and DNA manipulation and propagation were performed using standard DNA techniques (40).
Construction of a synthetic Der f 7 gene.
A codon-optimized Der f 7 gene was synthesized by PCR using eight mutually priming, overlapping oligonucleotides which were designed based on the A. oryzae codon usage database (http://www.kazusa.or.jp/codon/) (Table 1). Recursive PCR (1, 38) was performed using Ex-Taq polymerase (Takara Bio, Otsu, Japan) under the following reaction conditions: 30 cycles of denaturation at 94°C for 60 s, annealing at 60°C for 60 s, and elongation at 72°C for 60 s. The first PCR product was used as a template for the second PCR, which was performed under the same conditions, using the 3′ and 5′ terminal oligonucleotides as primers. The resulting product was cloned into a pCRII-TOPO cloning vector (Invitrogen, Tokyo, Japan) and subjected to DNA sequencing to select a clone containing the designed codon-optimized Der f 7 gene. The DNA cycle sequencing reaction was performed with a universal sequencing primer, using a Big-Dye Terminator 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) with an ABI PRISM 377 sequencer (Applied Biosystems). The plasmid clone pTDopt, containing a designed nucleotide sequence, was used in further study as the codon-optimized Der f 7 gene.
TABLE 1.
Oligonucleotides used for total synthesis of the codon-optimized Der f 7 gene
| Oligonucleotide no. | Sequence (5′→3′) |
|---|---|
| 1 | 5′-CCCGTCGACCATGATGAAGTTCTTGCTGATCGCTGCCGTCGCCTTCGTCGCCGTTTCGGCTGACCCCATTCACTACGACAAGATCACCGAGGAAATCAACAAGGCTATCG-3′a |
| 2 | 5′-ACGCTCGAACTTGTCGGCGTGGTCAGGGACCTTCATGGGATCGATGGTCTCGGACTGTTCGATAGCAGCAATGGCATCGTCGATAGCCTTGTTGATTT-3′ |
| 3 | 5′-CGACAAGTTCGAGCGTCACGTTGGTATCGTGGACTTCAAGGGTGAGTTGGCCATGCGCAACATCGAGGCTCGCGGCCTCAAGCAGATGAAGCGTCA-3′ |
| 4 | 5′-TCGACACGATATCATCGTGAACACCGATGAGCAAATGAGCCTTAACAATACCCTCTTCACCCTTGACATTAGCGTCACCCTGACGCTTCATCTGCTT-3′ |
| 5 | 5′-CGATGATATCGTGTCGATGGAGTACGATCTCGCCTACAAGCTGGGTGACCTTCATCCCACCACTCACGTCATCTCGGATATTCAGGACTTCGT-3′ |
| 6 | 5′-TGACAACATTAGCGAATTGGCGGACCTCGAAAGAGGTCATGGTGATGTTACCTTCGTCAGAAATCTCAAGGGAGAGGGCAACAACGAAGTCCTGAATATC-3′ |
| 7 | 5′-CAATTCGCTAATGTTGTCAACCACATCGGTGGCCTTTCCATCCTCGACCCCATTTTCGGCGTTCTCTCTGATGTCCTGACCGCTATCTTCCAAGACACCG-3′ |
| 8 | 5′-CCCTCTAGATTAATTCTTTTCCAGCTCACGCTTGAAGGCGGGGGCCAGGACCTTGGTCATTTCCTTACGGACGGTGTCTTGGAAGATAG-3′b |
The SalI site is underlined.
The XbaI site is underlined.
Construction of expression vectors.
The Der f 7 gene with native codons and an original signal sequence was amplified by PCR on pGEX-Der f 7 (a gift from S. Kawamoto, Hiroshima University), using primers 5′-CCCGTCGACATGATGAAATTTTTGTTGAT-3′ (the SalI restriction site is underlined) and 5′-CCCTCTAGATTAATTTTTTTCCAATTCAC-3′ (the XbaI restriction site is underlined). An amplified fragment was inserted between the improved glaA promoter and the agdA terminator of an expression vector, pNGA142 (46), which contains the niaD gene as a selectable marker, yielding pDer/ntv. The Der f 7 gene fragment with optimized codons, obtained by digestion of pTDopt with XbaI and SalI, was also cloned into XbaI/SalI-digested pNGA142, yielding pDer/opt.
pGlaDer/ntv and pGlaDer/opt were constructed based on pNGL, which contains a glucoamylase A (GlaA) catalytic domain region in the NotI and PmaCI sites of pNGA142. A DNA fragment encoding the GlaA catalytic domain was prepared by PCR with A. oryzae genomic DNA by using primers 5′-CCCGCGGCCGCATGGTGTCTTTCTCCTCTTG-3′ (the NotI restriction site is underlined) and 5′-CCCCACGTGAGTCGTAGAGCAAGCTGACG-3′ (the PmaCI restriction site is underlined) and was inserted into NotI/PmaCI-digested pNGA142, resulting in pNGL. The Der f 7 gene without a signal sequence was amplified by PCR using the following primers: 5′-CCCCACGTGAAAAGAGATCCAATTCACTATGATAA-3′ (the PmaCI restriction site is underlined) and 5′-CCCATATGTTGGTTTTTTTCCAATTCACG-3′ (the NdeI restriction site is underlined) for the native-codon construct and 5′-CCCCACGTGAAACGCGACCCCATTCACTACGACAA-3′ (the PmaCI restriction site is underlined) and 5′-CCCCATATGTTAATTCTTTTCCAGCTCAC-3′ (the NdeI restriction site is underlined) for the codon-optimized construct. Both sense primers contained hexanucleotides encoding a cleavage site (shown in italics) for a Kexin-like protease (KexB) (34), Lys-Arg. PCR-amplified fragments were inserted in the PmaCI and NdeI sites of pNGL to be fused in-frame at the 3′ terminus of the nucleotide sequence encoding the GlaA catalytic domain through the KexB cleavage sequence Lys-Arg.
pGlaDer/ntv-stop was constructed by deletion of two nucleotides (5′-CG-3′) located immediately upstream of the KexB cleavage site of the GlaA::Der f 7 fusion. That created a frameshift and a termination codon occurring 8 codons after the deletion site.
SDS-PAGE and Western blot analysis.
Proteins secreted by transformants cultured in YPM medium for 24 h were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (30) on 12.5% polyacrylamide gels and strained with Coomassie brilliant blue (CBB) R-250. Proteins in the medium samples were concentrated by precipitation with 10% trichloroacetic acid (TCA). For Western analysis, 20 μl of culture medium was electrophoresed and transferred onto a polyvinylidene difluoride membrane (Nippon Genetics, Tokyo, Japan) by using a semidry blotting system (Bio-Rad, Hercules, CA) with 3-(cyclohexylamino)-1-propanesulfonic acid buffer. Membranes were incubated with rabbit anti-Der f 7 polyclonal antibody and then detected with an immunostaining kit (Konica, Tokyo, Japan), using peroxidase-conjugated goat-rabbit antibodies (Promega, Madison, WI).
Quantitative RT-PCR analysis.
Total RNAs were extracted from mycelia of transformants cultured for 20 h at 30°C using Isogen (Nippon Gene, Tokyo, Japan) according to the manufacturer's instructions. Five micrograms of total RNA was treated with DNase I (Nippon Gene), and first-strand cDNAs were synthesized using murine leukemia virus reverse transcriptase (Invitrogen) with an oligo(dT) primer. Synthesized cDNA was treated with RNase H, and quantitative reverse transcription-PCR (RT-PCR) was performed with DyNAmo (Finnzymes, Espoo, Finland), using the following primers: for the native-codon Der f 7 gene, 5′-CGTGGCTTCTTCTACACACCC-3′ (forward) and 5′-AGCTCCAGTTGTGCCACTTGT-3′ (reverse); for the codon-optimized Der f 7 gene, 5′-GACGATGCCATTGCTGCTAT-3′ (forward) and 5′-ACACCGATGAGCAAATGAGC-3′ (reverse); and for the histone gene, 5′-CAAGCGTATCTCTGCCATGA-3′ (forward) and 5′-CACCGAAACCGTAGAGGGTA-3′ (reverse). Reactions and subsequent analyses were performed with the DNA Engine Opticon system (MJ Research, San Francisco, CA). The relative mRNA levels were normalized to that of the histone H4 gene, used as a reference gene (33).
Northern blot analysis.
A digoxigenin (DIG)-labeled double-strand DNA probe was synthesized on pDer/ntv-opt as a template by using a PCR DIG probe kit (Roche, Indianapolis, IN). Five micrograms of total RNA was electrophoresed on a 1.5% formaldehyde-agarose gel. RNA quality was evaluated by visualization of rRNA stained with ethidium bromide. Transfer of the total RNAs onto a Hybond N+ nylon membrane (Amersham Biosciences, Amersham, United Kingdom), prehybridization, and hybridization were performed according to the DIG luminescence kit protocol (Roche). Signal detection was performed with CSPD (Roche) as a substrate, according to the manufacturer's instructions, before exposure to X-ray film (Fuji Film, Tokyo, Japan).
3′-RACE analysis and poly(A) addition site mapping.
The polyadenylation sites of Der f 7 mRNAs from Der/ntv and Der/opt were mapped using analysis by 3′ rapid amplification of cDNA ends (3′-RACE). Approximately 5 μg of total RNA, prepared for Northern blot analysis as described above, was used for cDNA synthesis using a GeneRacer kit (Invitrogen). PCR amplification was performed using the sense primers (5′-CCCCACGTGAAAAGAGATCCAATTCACTATGATAA-3′ for native Der f 7 and 5′-CCCCACGTGAAACGCGACCCCATTCACTACGACAA-3′ for codon-optimized Der f 7) used for amplification of the Der f 7 gene for the fusion construct and the antisense primer supplied with the kit. Amplified products were inserted into a pCRII-TOPO cloning vector (Invitrogen), and the resulting plasmids were digested with EcoRI, followed by agarose electrophoresis to compare the inserted fragment lengths of the obtained clones. The DNA cycle sequencing reaction was performed as described above.
Nucleotide sequence accession number.
The nucleotide sequence of the codon-optimized Der f 7 gene constructed in this study has been deposited in the DDBJ/GenBank/EMBL databases under accession number AB441028.
RESULTS
Design and construction of codon-optimized Der f 7.
Comparison of the codon usage of Der f 7 with that of A. oryzae by use of a codon usage database (http://www.kazusa.or.jp/codon/) showed that the frequencies of individual codons are different from those in A. oryzae. The GC content of the coding region of native Der f 7 was 37.8%, while that of A. oryzae genes is ≈55% (http://www.kazusa.or.jp/codon/). Based on codon usage, a codon-optimized Der f 7 gene was designed. Codons infrequently used in A. oryzae were replaced by more-frequently used codons, and portions of other codons were altered to reflect the usage of individual codons (Table 2). Consequently, 43.7% of the codons in native Der f 7 were altered. Reconstitution of the gene by codon optimization resulted in a GC content of 52.8%.
TABLE 2.
Comparison of codon usage in A. oryzae genes with that in native and codon-optimized Der f 7 genes
| Amino acid | Codon | % Frequencya
|
||
|---|---|---|---|---|
| A. oryzae | Der f 7
|
|||
| Native | Optimized | |||
| Ala | GCT | 27 | 50 | 50 |
| GCC | 31 | 30 | 50 | |
| GCA | 23 | 20 | 0 | |
| GCG | 20 | 0 | 0 | |
| Arg | CGT | 18 | 58 | 58 |
| CGC | 23 | 0 | 42 | |
| CGA | 17 | 42 | 0 | |
| CGG | 18 | 0 | 0 | |
| AGA | 13 | 0 | 0 | |
| AGG | 12 | 0 | 0 | |
| Asn | AAT | 45 | 58 | 58 |
| AAC | 55 | 42 | 42 | |
| Asp | GAT | 53 | 94 | 36 |
| GAC | 47 | 6 | 64 | |
| Cys | TGT | 46 | 0 | 0 |
| TGC | 54 | 0 | 0 | |
| Gln | CAA | 43 | 100 | 32 |
| CAG | 57 | 0 | 68 | |
| Glu | GAA | 44 | 88 | 38 |
| GAG | 56 | 12 | 62 | |
| Gly | GGT | 28 | 82 | 74 |
| GGC | 31 | 9 | 26 | |
| GGA | 24 | 9 | 0 | |
| GGG | 17 | 0 | 0 | |
| His | CAT | 47 | 76 | 24 |
| CAC | 53 | 24 | 76 | |
| Ile | ATT | 36 | 67 | 29 |
| ATC | 50 | 24 | 71 | |
| ATA | 14 | 9 | 0 | |
| Leu | TTA | 7 | 17 | 0 |
| TTG | 18 | 65 | 17 | |
| CTT | 19 | 17 | 17 | |
| CTC | 23 | 0 | 36 | |
| CTA | 11 | 0 | 0 | |
| CTG | 22 | 0 | 29 | |
| Lys | AAA | 36 | 93 | 0 |
| AAG | 64 | 7 | 100 | |
| Met | ATG | 100 | 100 | 100 |
| Phe | TTT | 38 | 60 | 0 |
| TTC | 62 | 40 | 100 | |
| Pro | CCT | 27 | 18 | 18 |
| CCC | 26 | 0 | 82 | |
| CCA | 25 | 82 | 0 | |
| CCG | 22 | 0 | 0 | |
| Ser | AGT | 13 | 0 | 0 |
| AGC | 18 | 0 | 0 | |
| TCT | 18 | 33 | 33 | |
| TCC | 20 | 22 | 33 | |
| TCA | 14 | 12 | 0 | |
| TCG | 16 | 33 | 33 | |
| Thr | ACT | 24 | 12 | 12 |
| ACC | 32 | 55 | 88 | |
| ACA | 24 | 33 | 0 | |
| ACG | 20 | 0 | 0 | |
| Trp | TGG | 100 | 100 | 100 |
| Tyr | TAT | 47 | 69 | 0 |
| TAC | 53 | 31 | 100 | |
| Val | GTT | 27 | 40 | 40 |
| GTC | 33 | 25 | 50 | |
| GTA | 13 | 25 | 0 | |
| GTG | 27 | 10 | 10 | |
Percent frequencies of individual codons are shown for each corresponding amino acid.
The codon-optimized Der f 7 gene was synthesized in two rounds of recursive PCR (1, 38). The resulting PCR products were cloned, and 1 clone, whose sequence corresponded to the designed codon-optimized Der f 7 gene, was obtained out of 15 clones. Using the native Der f 7 gene and the synthesized, codon-optimized Der f 7 gene, we constructed Der f 7 expression vectors pDer/ntv and pDer/opt. These contained the Der f 7 gene that was inserted downstream of the improved glaA promoter (46). In addition, we constructed the expression vectors pGlaDer/ntv (for native Der f 7) and pGlaDer/opt (for codon-optimized Der f 7) to evaluate the effect of codon optimization in the fusion construct. Der f 7 without a signal sequence was fused to the catalytic domain of A. oryzae GlaA at the N terminus through a KexB cleavage site. Both types of expression vectors were constructed based on a fungal high-level-expression vector, pNGA142 (46).
A. oryzae NS4 was transformed using the above-mentioned vectors. Transformants containing one copy of an expression vector at the resident niaD locus as determined by Southern blot analysis (data not shown) were selected. Two independent transformants were selected for further analysis in individual transformation experiments.
Codon optimization improves Der f 7 production in nonfused and fused constructs.
To investigate the effect of codon optimization on protein production levels, the transformants and the parental strain were cultured, and the culture supernatants were subjected to SDS-PAGE analysis. CBB staining showed that α-amylase signals of around 55 kDa were similar in all samples, indicating no difference in protein production ability among the transformants. Western blot analysis showed that two signals, of ≈25 and 30 kDa, were detected in the culture supernatants of transformants Der/opt-1 and -2 (Fig. 1B). In contrast, no signals were detected in those of transformants Der/ntv-1 and -2. This result indicates that codon optimization increased the amount of secreted Der f 7 from an undetectable level to a detectable level in the nonfused construct.
FIG. 1.
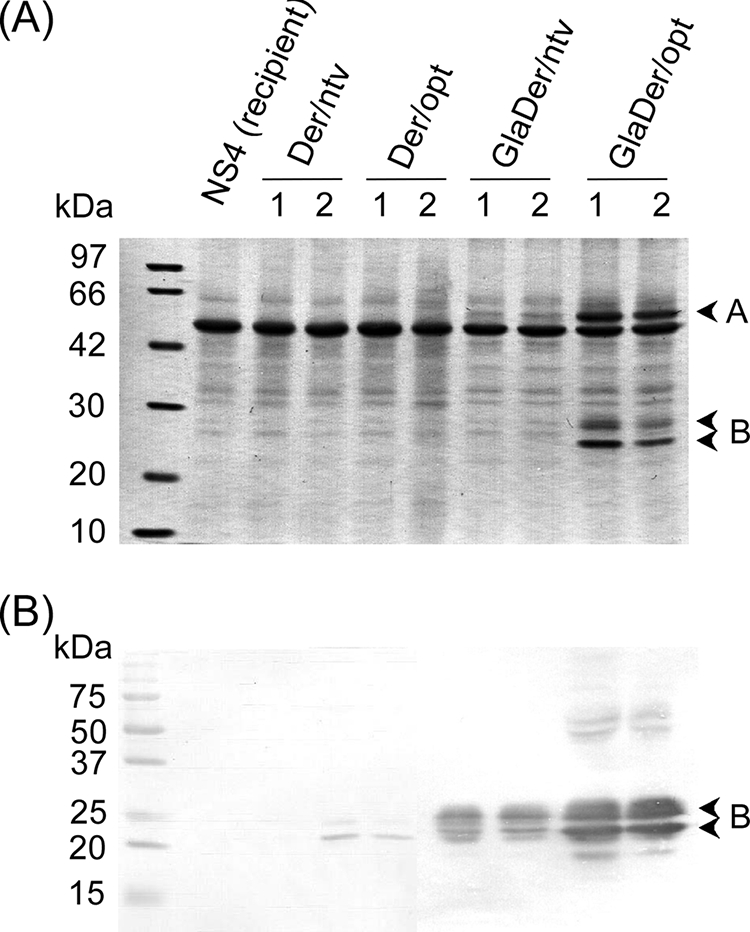
SDS-PAGE analysis of the transformants. (A) CBB staining. (B) Western blotting. Two independent transformants harboring a single copy of each expression cassette were grown in YPM medium for 24 h at 30°C. The culture supernatant (100 μl) concentrated fivefold by precipitation with TCA was loaded on a 12.5% SDS-polyacrylamide gel. Anti-Der f 7 antibodies were used for Western blotting. Arrowheads designated A and B indicate the carrier GlaA protein and Der f 7, respectively.
In the fusion constructs, two distinct signals, of ≈25 and 30 kDa, were detected by Western blot analysis in the culture supernatants of transformants GlaDer/opt-1 and -2, whereas weaker signals were detected in those of transformants GlaDer/ntv-1 and -2 (Fig. 1B). Signal intensity quantification of the bands by use of NIH image software (National Institutes of Health, Bethesda, MD) indicated that the signal intensities of the GlaDer/opt band were three- to fivefold higher than those of GlaDer/ntv. In addition, protein bands corresponding to the signals detected by Western blot analysis were observed only in the culture supernatants of transformants GlaDer/opt-1 and -2 by CBB staining (Fig. 1A and B). Furthermore, bands of ≈60 kDa, corresponding to the size of the catalytic domain of GlaA, were detected in both GlaDer/ntv and GlaDer/opt (Fig. 1A), although the signal intensity of the band in GlaDer/ntv was noticeably weak. Compared with the result for the nonfused construct, the fusion strategy markedly increased secretion yields of Der f 7 in both the native and the codon-optimized constructs. However, secretion yield in GlaDer/opt was much higher than that in GlaDer/ntv, indicating that codon optimization might synergistically improve production of Der f 7 when a fusion strategy is employed.
In this study, the nucleotide sequence around the start codon did not follow Kozak's rule in the fusion constructs (the NotI site of the primer contains a “C” at position −3 relative to the AUG codon). This structure might be expected to negatively affect translation efficiency (29). However, because both pGlaDer/ntv and pGlaDer/opt had the same sequence at position −3, the translation efficiencies in the two constructs were not expected to differ from each other. Additionally, the amount of recombinant Der f 7 produced in GlaDer/opt was much higher (>5-fold) than that in Der/opt, which has a “G” at position −3 (Fig. 1). The ratio of steady-state mRNA levels between GlaDer/opt and Der/opt was ≈5-fold (Fig. 2) (see below). These observations were associated with the positive effect of GlaA, the carrier protein fused to Der f 7, but these results suggested that the sequence around the start codon did not significantly affect the translation efficiency.
FIG. 2.
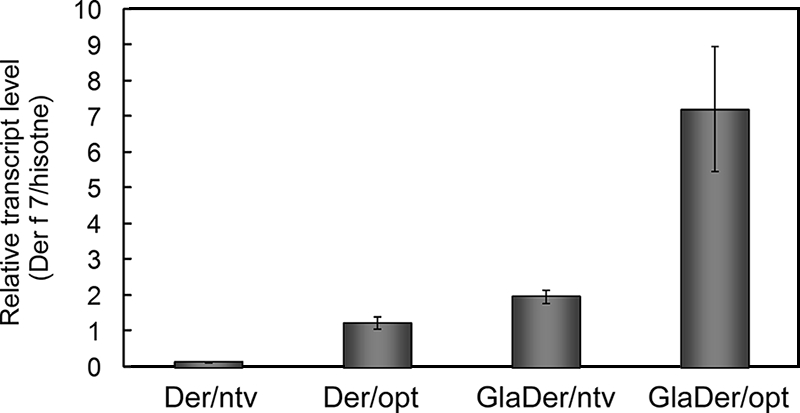
Quantitative RT-PCR analysis of Der f 7 mRNAs in the transformants. Total RNAs were extracted from transformant mycelia grown in YPM medium for 24 h. cDNAs were subjected to real-time PCR analysis using specific primers as described in Materials and Methods. The relative expression level for each gene was normalized to that for the histone H4 gene. The values are means of results from three independent experiments, and the error bars denote standard errors.
SDS-PAGE analysis of mite Der f 7 indicated that two bands, of 30 and 31 kDa, were glycosylated forms of a 25-kDa protein (43). The medium sample of the transformants was treated with endoglycosidase H, confirming that the 30-kDa band was a glycosylated form of the 25-kDa protein (data not shown).
Comparison of the Der f 7 mRNA levels.
To investigate whether differences in secretion yields of Der f 7 between transformants containing codon-optimized Der f 7 and those containing native Der f 7 could be explained at the transcriptional level, Der f 7 mRNA levels in each transformant were compared by quantitative RT-PCR analysis. Total RNAs were extracted from the transformants cultivated in YPM medium for 20 h. Transformants containing the codon-optimized Der f 7 gene (Der/opt or GlaDer/opt) had higher Der f 7 mRNA levels than transformants containing the native-codon Der f 7 gene (Der/ntv or GlaDer/ntv) (Fig. 2). The Der f 7 mRNA level of transformant Der/opt was ≈10-fold higher than that of transformant Der/ntv. Similarly, that of transformant GlaDer/opt was ≈3- to 5-fold higher than that of GlaDer/ntv. These data suggested that codon optimization increased the steady-state mRNA level, resulting in significant improvement of the secretion yield of Der f 7.
3′-RACE analysis and polyadenylation site mapping.
As described above, codon optimization of Der f 7 could be effective for improving mRNA transcript yield and could result in an increase in protein production level. However, the precise mechanism by which codon optimization increased the expression level of the heterologous gene was unclear. Because truncation of mRNA has been reported to occur in heterologous gene expression in filamentous fungi (14, 15, 42), we examined the lengths of Der f 7 transcripts by RT-PCR and then sequenced the amplified products in transformants Der/ntv and Der/opt.
RT-PCR was performed using the total RNAs used for Northern analysis as described above. RT-PCR products were cloned with the TA-cloning system, and clones were arbitrarily selected to be digested with EcoRI prior to agarose gel electrophoresis. The Der f 7 cDNA fragments in Der/opt were mostly uniform in length, whereas those in Der/ntv were shorter and varied considerably (Fig. 3). This suggested that expression of the Der f 7 construct with native codons results in formation of truncated mRNAs and that codon optimization prevents truncation of transcripts. Further DNA sequencing of amplified fragments revealed that multiple polyadenylation sites were located within the coding region but not in the 3′ untranslated region in the construct pDer/ntv. Yet, polyadenylation sites occurred within the 3′ untranslated region in all cDNA clones examined in the construct pDer/opt (Fig. 4). In addition, premature termination of transcription occurred in the fusion construct pGlaDer/ntv (see Fig. S1 in the supplemental material). These results suggest that formation of aberrant mRNA without a termination codon is correlated with mRNA levels.
FIG. 3.
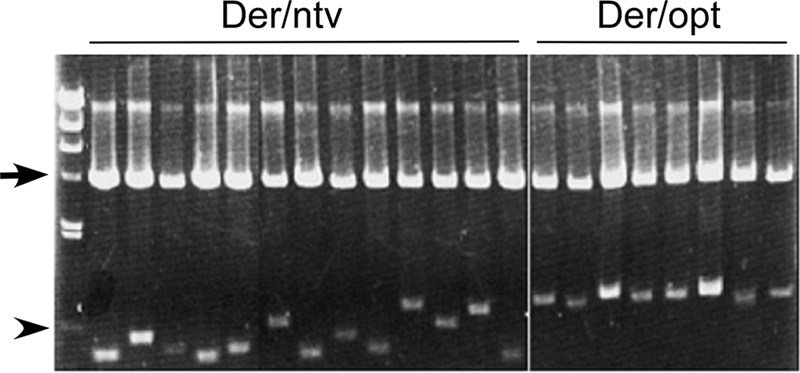
Analysis of the inserted fragments of two 3′-RACE clones. First-strand cDNA was synthesized using an oligo(dT) anchor primer. Then, PCR was performed using the anchor primer and the Der f 7-specific primer, and amplified PCR fragments were cloned by insertion into the pCRII TA-cloning vector. The resulting plasmid DNAs were isolated and digested with EcoRI, followed by agarose gel electrophoresis to compare the lengths of the inserted fragments. The arrow indicates the bands of the TA-cloning vector used for cloning of the RT-PCR products, and the arrowhead indicates a 500-bp molecular marker.
FIG. 4.
Polyadenylation sites of Der f 7 mRNA in the transformants Der/ntv and Der/opt. Arrows indicate the polyadenylation sites of the two sequenced 3′-RACE clones. The nucleotide sequence of the coding region is indicated in uppercase letters, and the terminator sequence is indicated in lowercase letters. Numbers beside the arrow indicate the number of clones containing the poly(A) tract at the indicated position.
Translation termination codon insertion upstream of Der f 7.
As described above, we showed that expression of the heterologous gene with high-level AT content resulted in truncation of the transcripts and that codon optimization could prevent formation of such aberrant mRNAs, possibly by eliminating AT-rich sequences. Premature polyadenylation within the coding region forms truncated mRNA without a translational termination codon, the so-called nonstop mRNA. Recently, the nonstop mRNA decay pathway was proposed as the mechanism underlying the correlation between premature polyadenylation and decreased mRNA levels (10, 48). This pathway is thought to play a role in the quality control of mRNAs. mRNAs that lack a translational termination codon are degraded through the pathway to prevent the synthesis of potentially deleterious proteins from aberrant mRNAs. Der f 7 mRNAs containing a premature poly(A) tract within the coding region have no in-frame translation termination codon. We therefore presumed that Der f 7 mRNAs are rapidly degraded through the nonstop mRNA decay pathway in the construct pDer/ntv.
It has been reported that transcript degradation was avoided by the addition of a translation termination codon upstream of the poly(A) tract in nonstop gene constructs (10). If Der f 7 mRNAs were degraded through such a nonstop mRNA decay pathway, insertion of a termination codon upstream of the polyadenylation site would increase the Der f 7 mRNA level. Premature polyadenylation occurred in the fusion construct pGlaDer/ntv as well as in the direct construct pDer/ntv. Therefore, we constructed a frameshift mutant of the fusion construct (pGlaDer/ntv-stop) in which the translation termination codon was located downstream of the GlaA catalytic domain. Schematic diagrams of the Der f 7 genes of pGlaDer/ntv and pGlaDer/ntv-stop and their sequence alignment are shown in Fig. 5. These two constructs were introduced into A. oryzae, and the mRNA levels were determined by Northern blot analysis. The mRNA level of the fusion gene increased by ≈2- to 3-fold by occurrence of a translational termination codon between glaA and Der f 7 (Fig. 5B), suggesting that the native Der f 7 mRNA is degraded by the nonstop mRNA decay pathway. The fusion constructs pGlaDer/ntv and pGlaDer/ntv-stop produced a protein of ≈60 kDa, corresponding to the GlaA catalytic domain generated by cleavage of GlaA::Der f 7 with a Kexin-like protease or by termination of translation downstream of the catalytic domain. Densitometric analyses following SDS-PAGE of the medium samples showed that the transformant GlaDer/ntv-stop produced the GlaA catalytic domain at a level ≈5-fold higher than that expressed by the transformant GlaDer/ntv (Fig. 5C).
FIG. 5.
Expression analysis of GlaA::Der f 7 fusion genes with or without a premature termination codon. (A) Nucleotide and amino acid sequences around the fused region of pGlaDer/ntv and pGlaDer/ntv-stop. Two deleted nucleotides are underlined. (B) Northern blot analysis. Total RNAs were extracted from transformant mycelia cultured in YPM medium for 24 h using the native Der f 7 gene as a probe. (C) CBB staining of SDS-polyacrylamide gel. Culture supernatants of 24-h-grown transformants were concentrated fivefold by TCA precipitation, followed by SDS-PAGE. Arrowheads indicate the protein band corresponding to the GlaA catalytic domain.
DISCUSSION
Codon optimization has been considered an effective strategy for improving the expression levels of heterologous genes that contain codons rarely used in the host organism (19). In the present study, we demonstrated that reconstituting the mite allergen gene Der f 7 altered to fit Aspergillus codon usage could substantially improve the levels of gene expression and protein production for Der f 7 in A. oryzae. It has been suggested that codon optimization does not affect transcript levels but can alleviate the translation inefficiency often caused by ribosomal pauses at rare codons interrupting translation elongation (21, 24, 45, 51). In contrast, it has been reported that in bacteria (50), yeast (37), plants (7, 8), mammalian cells (28), and filamentous fungi (15, 27, 41), native heterologous gene transcripts with rare codons were hardly detectable or were small. By codon optimization, however, heterologous gene transcripts were stabilized or became full-length mRNAs. We observed that codon optimization of the Der f 7 gene resulted in significantly increased levels of mRNA in this study, consistent with these observations. However, it could not be ruled out that translational efficiency plays an important part in the increase in Der f 7 production level caused by codon optimization. Truncated transcripts could not be clearly detected in the pDer/ntv or pGlaDer/ntv construct by Northern blot analyses. However, partial codon optimization of Der f 7, in which chimeric genes were constructed by substituting native fragments (≈100 nucleotides [nt]) for the corresponding codon-optimized fragments, resulted in small amounts of transcripts that were shorter than expected (data not shown). This indicates that truncation of the transcripts occurs in the expression of the Der f 7 gene with rare codons for A. oryzae. 3′-RACE analysis clearly demonstrated that premature polyadenylation within the coding region of the gene occurred only in the constructs containing native codons. This suggested that cryptic polyadenylation signals exist within the coding region of the native Der f 7 gene.
There are no known experimental data on the 3′-end processing of mRNAs, including for sequences that function as polyadenylation signals in filamentous fungi. However, the regulatory sequences involved in mRNA 3′-end processing have been extensively studied for mammalian (20), yeast (17, 18), and plant (31) cells. Among these, the hexanucleotide AAUAAA or its related sequences (AUUAAA and AAAAAA) situated between 10 and 30 nt upstream of the 3′-end processing site are known to act as a polyadenylation signal. In addition, a near-upstream element (NUE) generally located within 10 nt upstream of the 3′-end processing site has been characterized as U rich (17). According to these observations, consensus sequences related to AAUAAA could also act as polyadenylation signals in Aspergillus spp., although other sequences or combinations of those sequences might also act as polyadenylation signals. To identify the canonical sequences that might function as a polyadenylation signal in native Der f 7 mRNA, we examined those sequences upstream of multiple polyadenylation sites in Der f 7 mRNAs. There was an AAAAAA sequence immediately upstream of the termination codon but no sequences that perfectly matched AAUAAA or AUUAAA. However, because the native Der f 7 gene is a highly AT-biased gene (GC content of 37.8%), several AU-rich sequences are found upstream of multiple polyadenylation sites. These AU-rich sequences might be involved in premature polyadenylation. Furthermore, sequences related to U-rich motifs are found near the poly(A) sites and likely function as a NUE. In contrast, in the codon-optimized Der f 7 gene, AU-rich and U-rich sequences are absent in the coding region. Taken together, the results suggest that the AU-rich and U-rich sequences present in the coding region of native Der f 7 pre-mRNA are involved in incorrect 3′-end processing. It is also suggested that codon optimization eliminates the AU-rich and U-rich sequences and results in the prevention of premature polyadenylation. Further studies of 3′-end processing mechanisms in filamentous fungal groups, including Aspergillus spp., is needed to obtain more information on the cis-regulatory sequences, such as polyadenylation signals and NUEs.
We observed a correlation between the decreased Der f 7 mRNA level and premature polyadenylation in the pDer/ntv construct. Similar observations have been reported for heterologous gene expression in filamentous fungal species, such as A. awamori and Schizophyllum commune (15, 41, 42). These results have indicated that premature polyadenylation within a coding region results in the production of unstable, aberrant transcripts that can be degraded rapidly. However, the mechanisms underlying these findings have not been reported. Recently, a novel mRNA surveillance mechanism in eukaryotes that allows recognition and degrading of aberrant transcripts that lack a termination codon was identified (10, 48). This nonstop mRNA decay pathway is reported to remove the aberrant mRNAs without a termination codon that could encode a truncated protein. Therefore, we examined whether the nonstop mRNA decay pathway is involved in the degradation of aberrant Der f 7 mRNA lacking a termination codon. For this purpose, pGlaDer/ntv-stop, a fusion construct with an artificial translational termination codon upstream of the sequence encoding Der f 7, was used. The addition of a termination codon between the GlaA catalytic domain and Der f 7 resulted in increased mRNA levels and a concomitant increase in the amount of GlaA catalytic domain produced. This result indicated that the nonstop mRNA decay pathway could be relevant to the degradation of aberrant transcripts lacking a termination codon in A. oryzae. Taken together, these observations suggest a model by which codon optimization increases the mRNA level of the heterologous Der f 7 gene. That is, a portion of the heterologous gene transcripts containing a cryptic polyadenylation signal(s) is prematurely polyadenylated, and the resulting aberrant mRNAs are degraded by the nonstop mRNA decay system. Codon optimization can eliminate the cryptic polyadenylation signals in the coding region and thus result in the production of full-length mRNA. Although it is plausible that native full-length mRNA with rare codons is unstable and degrades rapidly due to its secondary structure and/or translational pausing, we suggest that aberrant transcripts resulting from premature polyadenylation are most likely degraded through the nonstop mRNA decay pathway. The mRNA decay pathway is thought to be conserved among eukaryotes (10), and we conclude that filamentous fungi also have this pathway. However, in the genome sequence databases of A. oryzae (32), Aspergillus nidulans (11), and Aspergillus fumigatus (36), we could not find any gene homologous to SKI7 that is reported to play an important role in the nonstop mRNA decay pathway in S. cerevisiae (48). Therefore, identification of genes encoding the Ski7p homologue or other associate proteins is necessary to clarify the involvement of this pathway in the degradation of aberrant mRNA in A. oryzae.
We report that the transcript of the heterologous Der f 7 gene that contains the AT-biased codon is polyadenylated prematurely within the coding region. Multiple cryptic sequence elements in the Der f 7-coding region might be recognized by fungal cells as polyadenylation signals, although precise sequences have not been elucidated. Recognition of these signals appears to be one factor contributing to the low accumulation of full-length Der f 7 transcripts in A. oryzae. Altering the codon usage to represent a fungal-preferred codon bias could result in a decrease in the A and T contents of the heterologous genes and eliminate the cryptic polyadenylation signals within the coding region. In addition, we have proposed that the nonstop mRNA decay pathway, involving aberrant mRNAs that lack a termination codon due to gene AT bias, is involved in degrading heterologous gene transcripts in A. oryzae.
Supplementary Material
Acknowledgments
We thank Seiji Kawamoto for providing the anti-Der f 7 antibody and the plasmid pGEX-Der f 7.
This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas, Applied Genomics (no. 17019001), from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 12 September 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Brocca, S., C. Schmidt-Dannert, M. Lotti, L. Alberghina, and R. Schmid. 1998. Design, total synthesis, and functional overexpression of the Candida rugosa lip1 gene coding for a major industrial lipase. Protein Sci. 7:1415-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardoza, R. E., S. Gutierrez, N. Ortega, A. Colina, J. Casqueiro, and J. F. Martin. 2003. Expression of a synthetic copy of the bovine chymosin gene in Aspergillus awamori from constitutive and pH-regulated promoters and secretion using two different pre-pro sequences. Biotechnol. Bioeng. 83:249-259. [DOI] [PubMed] [Google Scholar]
- 3.Chapman, M. D., A. M. Smith, L. D. Vailes, and L. K. Arruda. 1997. Defined epitopes: In vivo and in vitro studies using recombinant allergens. Int. Arch. Allergy Immunol. 125:102-104. [DOI] [PubMed] [Google Scholar]
- 4.Chen, G. T., and M. Inouye. 1994. Role of the AGA/AGG codons, the rarest codons in global gene expression in Escherichia coli. Genes Dev. 8:2641-2652. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, T., H. Woeldike, F. Boel, S. B. Mortensen, K. Hjortshoej, L. Thim, and M. Hansen. 1988. High level expression of recombinant genes in Aspergillus oryzae. Bio/Technology 6:1419-1422. [Google Scholar]
- 6.Chua, K. Y., P. K. Kehal, W. R. Thomas, P. R. Vaughan, and I. G. Macreadie. 1992. High-frequency binding of IgE to the Der p allergen expressed in yeast. J. Allergy Clin. Immunol. 89:95-102. [DOI] [PubMed] [Google Scholar]
- 7.De Rocher, E. J., T. C. Vargo-Gogola, S. H. Diehn, and P. J. Green. 1998. Direct evidence for rapid degradation of Bacillus thuringiensis toxin mRNA as a cause of poor expression in plants. Plant Physiol. 117:1445-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diehn, S. H., W.-L. Chiu, E. J. De Rocher, and P. J. Green. 1998. Premature polyadenylation at multiple sites within a Bacillus thuringiensis toxin gene-coding region. Plant Physiol. 117:1433-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn-Coleman, N. S., P. Bloebaum, R. M. Berka, E. Bodie, N. Robinson, G. Armstrong, M. Ward, M. Przetak, G. L. Carter, and R. LaCost. 1991. Commercial levels of chymosin production by Aspergillus. Bio/Technology 9:976-981. [DOI] [PubMed] [Google Scholar]
- 10.Frischmeyer, P. A., A. van Hoof, K. O'Donnell, A. L. Guerrerio, R. Parker, and H. C. Dietz. 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295:2258-2261. [DOI] [PubMed] [Google Scholar]
- 11.Galagan, J. E., S. E. Calvo C. Cuomo, L.-J. Ma, J. Wortman, S. Batzoglou, S.-I. Lee, M. Baştürkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. Á. Peñalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 12.Gomi, K., Y. Iimura, and S. Hara. 1987. Integrative transformation of Aspergillus oryzae with a plasmid containing the Aspergillus nidulans argB gene. Agric. Biol. Chem. 51:2549-2555. [Google Scholar]
- 13.Gooch, V. D., A. Mehra, L. F. Larrondo, J. Fox, M. Touroutoutoudis, J. J. Loros, and J. C. Dunlap. 2008. Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot. Cell 7:28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouka, R. J., P. J. Punt, J. G. M. Hessing, and C. A. M. J. J. van den Hondel. 1996. Analysis of heterologous protein production in defined recombinant Aspergillus awamori strains. Appl. Environ. Microbiol. 62:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gouka, R. J., P. J. Punt, and C. A. M. J. J. van den Hondel. 1997. Glucoamylase gene fusions alleviate limitations for protein production in Aspergillus awamori at the transcriptional and (post)translational levels. Appl. Environ. Microbiol. 63:488-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouka, R. J., P. J. Punt, and C. A. M. J. J. van den Hondel. 1997. Efficient production of secreted proteins by Aspergillus: progress, limitations and prospects. Appl. Microbiol. Biotechnol. 47:1-11. [DOI] [PubMed] [Google Scholar]
- 17.Graber, J. H., C. R. Cantor, S. C. Mohr, and T. F. Smith. 1999. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. USA 96:14055-14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graber, J. H., G. D. McAllister, and T. F. Smith. 2002. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 30:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gustafsson, C., S. Govindrajan, and J. Minshull. 2004. Codon bias and heterologous protein expression. Trends Biotechnol. 22:346-353. [DOI] [PubMed] [Google Scholar]
- 20.Hu, J., C. S. Lutz, J. Wilusz, and B. Tian. 2005. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA 11:1485-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, S., L. Li, J. Qiao, Y. Guo, L. Cheng, and J. Liu. 2006. Codon optimization, expression, and characterization of an internalizing anti-ErbB2 single-chain antibody in Pichia pastoris. Protein Expr. Purif. 47:249-257. [DOI] [PubMed] [Google Scholar]
- 22.Ikemura, T. 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151:389-409. [DOI] [PubMed] [Google Scholar]
- 23.Jacquet, A., M. Magi, H. Petry, and A. Bollen. 2002. High-level expression of recombinant house dust mite allergen Der p 1 in Pichia pastoris. Clin. Exp. Allergy 32:1048-1053. [DOI] [PubMed] [Google Scholar]
- 24.Kane, J. F. 1995. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr. Opin. Biotechnol. 6:494-500. [DOI] [PubMed] [Google Scholar]
- 25.Kane, J. F., B. N. Violand, D. F. Curan, N. R. State, K. L. Duffin, and G. Bogosian. 1992. Novel in-frame two codon translational hop during synthesis of bovine placental lactogen in a recombinant strain of Escherichia coli. Nucleic Acids Res. 20:6707-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawamoto, S., T. Aki, M. Yamashita, A. Tategaki, T. Fujimura, S. Tsuboi, T. Katsutani, O. Suzuki, S. Shigeta, Y. Murooka, and K. Ono. 2002. Toward elucidating the full spectrum of mite allergens—state of the art. J. Biosci. Bioeng. 94:285-298. [DOI] [PubMed] [Google Scholar]
- 27.Koda, A., T. Bogaki, T. Minetoki, and M. Hirotsune. 2005. High expression of a synthetic gene encoding potato alpha-glucan phosphorylase in Aspergillus niger. J. Biosci. Bioeng. 100:531-537. [DOI] [PubMed] [Google Scholar]
- 28.Kofman, A., M. Graf, L. Deml, H. Wolf, and R. Wagner. 2003. Codon usage-mediated inhibition of HIV-1 gag expression in mammalian cells occurs independently of translation. Tsitologiia 45:94-100. [PubMed] [Google Scholar]
- 29.Kozak, M. 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361:13-37. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavaged structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Loke, J. C., E. A. Stahlberg, D. G. Strenski, B. J. Haas, P. C. Wood, and Q. Q. Li. 2005. Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol. 138:1457-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machida, M., K. Asai, M. Sano, T. Tanaka, T. Kumagai, G. Terai, K. Kusumoto, T. Arima, O. Akita, Y. Kashiwagi, K. Abe, K. Gomi, H. Horiuchi, K. Kitamoto, T. Kobayashi, M. Takeuchi, D. W. Denning, J. E. Galagan, W. C. Nierman, J. Yu, D. B. Archer, J. W. Bennett, D. Bhatnagar, T. E. Cleveland, N. D. Fedorova, O. Gotoh, H. Horikawa, A. Hosoyama, M. Ichinomiya, R. Igarashi, K. Iwashita, P. R. Juvvadi, M. Kato, Y. Kato, T. Kin, A. Kokubun, H. Maeda, N. Maeyama, J. Maruyama, H. Nagasaki, T. Nakajima, K. Oda, K. Okada, I. Paulsen, K. Sakamoto, T. Sawano, M. Takahashi, K. Takase, Y. Terabayashi, J. R. Wortman, O. Yamada, Y. Yamagata, H. Anazawa, Y. Hata, Y. Koide, T. Komori, Y. Koyama, T. Minetoki, S. Suharnan, A. Tanaka, K. Isono, S. Kuhara, N. Ogasawara, and H. Kikuchi. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157-1161. [DOI] [PubMed] [Google Scholar]
- 33.Maeda, H., M. Sano, Y. Maruyama, T. Tanno, T. Akao, Y. Totsuka, M. Endo, R. Sakurada, Y. Yamagata, M. Machida, O. Akita, F. Hasegawa, K. Abe, K. Gomi, T. Nakajima, and Y. Iguchi. 2004. Transcriptional analysis of genes for energy catabolism and hydrolytic enzymes in the filamentous fungus Aspergillus oryzae using cDNA microarrays and expressed sequence tags. Appl. Microbiol. Biotechnol. 65:74-83. [DOI] [PubMed] [Google Scholar]
- 34.Mizutani, O., A. Nojima, M. Yamamoto, K. Furukawa, T. Fujioka, Y. Yamagata, K. Abe, and T. Nakajima. 2004. Disordered cell integrity signaling caused by disruption of the kexB gene in Aspergillus oryzae. Eukaryot. Cell 3:1036-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson, G., O. Kozlova-Zwinderman, A. J. Collis, M. R. Knight, J. R. S. Fincham, C. P. Stanger, A. Renwick, J. G. M. Hessing, P. J. Punt, C. A. M. J. J. van den Hondel, and N. D. Read. 2004. Calcium measurement in living filamentous fungi expressing codon-optimized aequorin. Mol. Microbiol. 52:1437-1450. [DOI] [PubMed] [Google Scholar]
- 36.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafon, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151-1156. [DOI] [PubMed] [Google Scholar]
- 37.Outchkourov, S. N., W. J. Stiekema, and M. A. Jongsma. 2002. Optimization of the expression of equistatin in Pichia pastoris. Protein Expr. Purif. 24:18-24. [DOI] [PubMed] [Google Scholar]
- 38.Prodromou, C., and L. H. Pearl. 1992. Recursive PCR: a novel technique for total gene synthesis. Protein Eng. 5:827-829. [DOI] [PubMed] [Google Scholar]
- 39.Punt, P. J., N. van Biezen, A. Conesa, A. Alvers, J. Mangnus, and C. A. M. J. J. van den Hondel. 2002. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 20:200-2006. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsh, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Scholtmeijer, K., H. A. B. Wösten, J. Springer, and J. G. T. Wessels. 2001. Effect of introns and AT-rich sequences on expression of the bacterial hygromycin B resistance gene in the basidiomycete Schizophyllum commune. Appl. Environ. Microbiol. 67:481-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuren, F. H. J., and J. G. T. Wessels. 1998. Expression of heterologous genes in Schizophyllum commune is often hampered by the formation of truncated transcripts. Curr. Genet. 3:151-156. [DOI] [PubMed] [Google Scholar]
- 43.Shen, H. D., K. Y. Chua, W. L. Lin, K. H. Hsieh, and W. R. Thomas. 1995. Molecular cloning and immunological characterization of the house dust mite allergen Der f 7. Clin. Exp. Allergy 25:1000-1006. [DOI] [PubMed] [Google Scholar]
- 44.Shoji, H., H. Horiuchi, and M. Takagi. 1999. Production of recombinant Der fI (a major mite allergen) by Aspergillus oryzae. Biosci. Biotechnol. Biochem. 63:703-709. [DOI] [PubMed] [Google Scholar]
- 45.Sinclair, G., and F. Y. Choy. 2002. Synonymous codon usage bias and the expression of human glucocerebrosidase in the methylotrophic yeast, Pichia pastoris. Protein Expr. Purif. 26:96-105. [DOI] [PubMed] [Google Scholar]
- 46.Tamalampudi, S., M. M. T. Rahman, S. Hama, Y. Suzuki, A. Kondo, and H. Fukuda. 2007. Development of recombinant Aspergillus oryzae whole-cell biocatalyst expressing lipase-encoding gene from Candida antarctica. Appl. Microbiol. Biotechnol. 75:387-395. [DOI] [PubMed] [Google Scholar]
- 47.Te'o, V. S. J., A. E. Cziferszky, P. L. Bergquist, and K. M. H. Nevallainen. 2000. Codon optimization of xylanase gene xynB from the thermophilic bacterium Ditoglomus thermophilum for expression in the filamentous fungus Trichoderma reesei. FEMS Microbiol. Lett. 190:13-19. [DOI] [PubMed] [Google Scholar]
- 48.van Hoof, A., P. A. Frischmeyer, H. C. Diets, and R. Parker. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295:2262-2264. [DOI] [PubMed] [Google Scholar]
- 49.Ward, P. P., C. S. Piddington, G. A. Cunningham, X. Zhou, R. D. Wyatt, and O. M. Conneely. 1995. A system for production of commercial quantities of human lactoferrin: a broad spectrum natural antibiotic. Bio/Technology 13:498-503. [DOI] [PubMed] [Google Scholar]
- 50.Wu, X., H. Jornvall, D. B. Kurt, and U. Opperman. 2004. Codon optimization reveals critical factors for high level expression of two rare codon genes in Escherichia coli: RNA stability and secondary structure but not tRNA abundance. Biochem. Biophys. Res. Commun. 313:89-96. [DOI] [PubMed] [Google Scholar]
- 51.Xia, X. 1998. How optimized is the translational machinery in Escherichia coli, Salmonella typhimurium and Saccharomyces cerevisiae? Genetics 149:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamada, O., B. R. Lee, and K. Gomi. 1997. Transformation system for Aspergillus oryzae with double auxortophic mutations, niaD and sC. Biosci. Biotechnol. Biochem. 61:1367-1369. [Google Scholar]
- 53.Yasuhara, T., T. Takai, T. Yuuki, H. Okudaira, and Y. Okumura. 2001. Biologically active recombinant forms of a major house dust mite group 1 allergen Der f 1 with full activities of both cysteine protease and IgE binding. Clin. Exp. Allergy 31:116-124. [DOI] [PubMed] [Google Scholar]
- 54.Yuuki, T., Y. Okumura, T. Ando, H. Yamakawa, M. Suko, M. Haida, and H. Okudaira. 1991. Cloning and expression of cDNA coding for the major house dust mite allergen Der f II in Escherichia coli. Agric. Biol. Chem. 55:1233-1238. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




