Abstract
A novel whole-cell arsenite biosensor was developed using the photosynthetic bacterium Rhodopseudomonas palustris no. 7 and characterized. A sensor plasmid containing the operator-promoter region of the ars operon and arsR gene from Escherichia coli and the crtI gene from R. palustris no. 7 was introduced into a blue-green mutant with crtI deleted, R. palustris no. 711. The biosensor changed color in response to arsenite, and the change was obvious to the naked eye after 24 h without further manipulation. Real-time reverse transcription-PCR showed that the crtI mRNA was induced 3-fold at 3 h and 2.5-fold at 6 h after addition of 50 μg/liter arsenite compared with the no-arsenite control, and consistent with this, the relative levels of lycopene and rhodopin also increased compared with the control. Colorimetric analysis of the bacteria showed that the hue angle had clearly shifted from green-yellow toward red in an arsenic dose-dependent manner at 24 h after arsenite addition. This obvious shift occurred irrespective of the culture conditions before arsenite was added, indicating that the color change of the biosensor is stable in water samples containing various concentrations of dissolved oxygen. Finally, assays using samples prepared in various types of mineral water indicated that this biosensor could be used to screen groundwater samples for the presence of arsenite in a variety of locations, even where electricity is not available.
The contamination of groundwater with arsenic and its toxic effects on humans have been a serious problem in many parts of the world, including Bangladesh, India, and Vietnam (1, 3, 6, 15, 16). In these countries, millions of people are still exposed to considerable levels of arsenic in their drinking water (25). Several field kits have been developed to distinguish safe wells from unsafe wells. The advantages of field kits are their simplicity and portability; however, the levels of precision and reproducibility of the data that they generate tend to be low (8, 19). Atomic absorption spectroscopy has also been used to detect heavy metals present in the environment. The advantages of atomic absorption spectroscopy are its high levels of accuracy and reproducibility; however, this method requires expensive instrumentation and sometimes cannot be used outside the laboratory. Therefore, a sensitive and simple sensor system to evaluate arsenic in groundwater is needed.
Whole-cell biosensors have become a useful tool for monitoring environmental pollutants (2, 10, 35). A microbial sensor system is a simple, cost-effective, and sensitive method for measuring target molecules. Such systems indicate the presence of a target molecule by expressing a reporter protein, such as green fluorescent protein, luciferase, or β-galactosidase. Several arsenite biosensors harboring a reporter gene fused with the operator-promoter region of the ars operon have been constructed (4, 20, 23, 26, 28, 29). The ars operon of Escherichia coli contains two regulatory genes (arsR and arsD) and three structural genes (arsA, arsB, and arsC) and has been shown to confer resistance to arsenite in its host (5, 22). The ArsR repressor, the first gene product of the ars operon, controls transcription of the ars operon by binding to or dissociating from the operator-promoter region (Pars). In the absence of arsenite, ArsR binds to Pars and prevents the transcription of downstream genes (32). In the presence of arsenite, ArsR binds to the arsenite and dissociates from Pars (24). As a consequence of the ArsR dissociation, transcription of the downstream genes is initiated.
We recently reported that a carotenoid biosynthesis gene, crtA, can also be used as a novel reporter in whole-cell biosensors made from Rhodovulum sulfidophilum (9, 14). Two crtA-based biosensors have been established by disrupting the crtA gene and reintroducing it downstream of a DNA response element. The crtA-based arsenite biosensor indicates the presence of arsenite by a change in the bacterial color from yellow to red that is due to carotenoid conversion, and this change is obvious to the naked eye. The fact that no instrument is required to detect the reporter signal is a major advantage of this biosensor, enabling it to be used outside the laboratory. However, further improvements are needed to use the crtA-based biosensor with groundwater samples. Because R. sulfidophilum is a marine bacterium, an adequate concentration of salt needs to be added in order to analyze groundwater samples. Furthermore, because CrtA requires molecular oxygen to form 2-keto carotenoids (36), the bacterial color change is very sensitive to the amount of dissolved oxygen in the culture medium (14).
Therefore, to obtain a stable reporter signal, we selected the gene encoding phytoene dehydrogenase, crtI, as a reporter gene and the freshwater photosynthetic bacterium Rhodopseudomonas palustris as the host strain. R. palustris is a purple, nonsulfur photosynthetic bacterium that is ubiquitous in soil and freshwater; thus, this organism could be used for direct measurement of pollutants in groundwater samples. R. palustris synthesizes carotenoids through the spirilloxanthin pathway (27). In this pathway, CrtI catalyzes four desaturation steps and converts the colorless carotenoid phytoene into the red carotenoid lycopene without a requirement for molecular oxygen. This feature should also allow direct measurement of pollutants in groundwater samples irrespective of the concentration of dissolved oxygen. Furthermore, because a mutant strain of R. palustris with crtI deleted cannot synthesize any colored carotenoids and is blue-green (37), an obvious color change toward red would be obtained by reintroducing the crtI gene downstream of Pars. We made use of these advantages by generating a novel whole-cell arsenite biosensor and used this biosensor to detect arsenite in samples prepared with different types of mineral water (MW). This was the first evaluation of a carotenoid-based biosensor to determine its practical application to natural samples.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Throughout this study, the photosynthetic bacteria were grown in modified Okamoto medium (MOM) (13). MOM contains (in 1,000 ml deionized water at pH 7.0) 1.0 g yeast extract, 1.7 g disodium succinate, 1.0 g dl-malate, 1.0 g sodium pyruvate, 1.0 g sodium acetate, 1.0 g NaHCO3, 0.25 g MgSO4·7H2O, 0.2 g CaCl2·2H2O, 0.2 g NaCl, 0.02 g FeSO4, 41 mg KH2PO4, 495 mg K2HPO4, 5 ml of a 1 M NH4Cl solution, 1 ml trace metal mixture A5, 500 μg vitamin B1 hydrochloride, 500 μg nicotinic acid, 300 μg p-aminobenzoic acid, and 50 μg d-biotin. Trace metal mixture A5 contains (in 1,000 ml deionized water) 2.86 g H3BO4, 1.81 g MnCl2·4H2O, 0.22 g ZnSO4·7H2O, 0.08 g CuSO4·5H2O, 0.021 g Na2MoO4, 0.01 g CoCl2·6H2O, and 50 g Na2-EDTA. R. palustris no. 711 (37), which is a mutant of R. palustris no. 7 with crtI deleted, and the biosensor strain were incubated in MOM in glass tubes or rectangular glass bottles at 30°C under an incandescent lamp at a photon flux intensity of 40 to 45 μmol s−1 m−2. For anaerobic light conditions, the cultivation vessels were filled completely with MOM, covered with a screw cap or a double cap, and incubated without agitation. For aerobic dark conditions, 50-ml cultures were grown in 500-ml Erlenmeyer flasks covered with aluminum foil with strong agitation in the dark.
E. coli JM109 [recA1 endA1 gyrA96 thi hsdR17(rK− mK+) e14 (mcrA) supE44 relA1 Δ(lac-proAB)/F′ traD36 proAB+ lacIq lacZΔM15] was cultivated in Luria-Bertani medium. Antibiotics were added to the E. coli cultures at final concentrations of 50 μg/ml ampicillin, 30 μg/ml kanamycin, and 50 μg/ml chloramphenicol.
Construction of sensor plasmid.
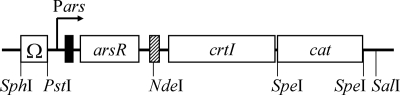
R. palustris-E. coli shuttle vector pMG103 (11) was used to construct the sensor plasmid. To prevent readthrough transcription from a promoter upstream of Pars, two oligonucleotides were synthesized (5′-CGCGCCGCGCGGGCTCCTCCCCGCGCGGCGCTTTTTTTCTGCA-3′ and 5′-GAAAAAAAGCGCCGCGCGGGGAGGAGCCCGCGCGGCGCGCATG-3′) and annealed to generate a terminator region derived from the pufC terminator in R. sulfidophilum (9), with the 5′ sticky end of a SphI site and the 3′ sticky end of a PstI site. The terminator region was inserted into the pGEM-T vector (Promega, United States) digested with SphI and PstI, generating pGEMΩ. Pars and the arsR gene (DDBJ accession number NC_00913) were amplified by PCR from the sensor plasmid pSENSE-AS (9) using primers parsS (5′-TTTCTGCAGGTCGACTCTAGAT-3′ [the PstI site is underlined]) and parsA (5′-CATATGATTACGGCTCGACTTTTAACTGCA-3′ [the NdeI site is in bold type, and a partial region of the ArsR-binding site is underlined]) (33, 34). The PCR fragment obtained by digestion with PstI and NdeI was inserted into pGEMΩ digested with PstI and NdeI, generating pGEMΩParsarsR. The ΩParsarsR region was amplified from pGEMΩParsarsR using primers M13 forward (5′-GTTTTCCCAGTCACGAC-3′) and M13 reverse (5′-CAGGAAACAGCTATGAC-3′) and digested with SphI and NdeI, generating a ΩParsarsR fragment. The open reading frame of the crtI gene containing an artificial Shine-Dalgarno sequence was amplified from R. palustris no. 7 genomic DNA using primers crtIS (5′-CCATATGCGTTTTTGGTTATGTAGGTACCC-3′ [the introduced NdeI site is in bold type, and a partial region of the ArsR-binding site is underlined]) and crtIA (5′-TCATGATGTCACCAGACTGTCGGCC-3′) and cloned into the pGEM-T vector, generating pGEMcrtI. pGEMcrtI was digested with NdeI and SalI, generating a crtI fragment. The ΩParsarsR fragment and the crtI fragment were then inserted into pMG103 digested with SphI and SalI, generating pMG103ΩParsarsRcrtI. As a result of ligation at the NdeI site, a second ArsR-binding site was generated downstream of the arsR gene. The introduction of a second ArsR-binding site has been shown to be effective for reducing the background expression of a reporter gene (26). The chloramphenicol resistance gene, which is a selectable marker for R. palustris transformation, was amplified by PCR from plasmid pLysS (Takara, Japan) using primers cprS (5′-CGTTGATCGGCACGTAACTAGTTCC-3′ [the introduced SpeI site is underlined]) and cprA (5′-CAGGCGTAGCACTAGTCGTTTAACG-3′ [the introduced SpeI site is underlined]). The chloramphenicol resistance gene fragment was obtained by digestion with SpeI and finally inserted into pMG103ΩParsarsRcrtI digested with SpeI, yielding pMG103ΩParsarsRcrtIcat. Figure 1 shows a schematic diagram of the structure of the sensor plasmid pMG103ΩParsarsRcrtIcat. All the DNA manipulations were carried out using standard protocols (21).
FIG. 1.
Schematic diagram of the arsenite sensor plasmid pMG103ΩParsarsRcrtIcat. The pufC terminator (Ω), arsR, crtI, and chloramphenicol acetyltransferase gene (cat) are indicated by open boxes. The operator-promoter region of the ars operon is indicated by a filled box, and an additional ArsR-binding site is indicated by a cross-hatched box.
Electroporation-mediated transfer of sensor plasmid and screening.
R. palustris no. 711 was semiaerobically grown in 50 ml of MOM until the optical density at 600 nm (OD600) was 0.5. The cells were collected by centrifugation and suspended in 1.5 ml of ice-cold sterile distilled water. The centrifugation and resuspension steps were repeated two times. The cell pellet was resuspended in 1 ml of 10% glycerol-water. One microgram of the sensor plasmid DNA was then added to 70 μl of the cell suspension, and the cell-DNA suspension was transferred to an ice-cold electroporation cuvette (gap width, 1 mm). Electroporation was carried out using a Gene Pulsar (Bio-Rad, United States) with the parameters set at 25 μF, 1.25 kV, and 250 Ω. The cells were immediately transferred to 1 ml of MOM and incubated on ice for 30 min. The cell suspension was then added to 4 ml of MOM in a glass tube and grown semiaerobically at 30°C overnight. The cells were diluted and spread onto MOM agar plates containing 200 μg/ml chloramphenicol. The chloramphenicol-resistant colonies were isolated after 5 to 6 days of incubation under light conditions. The presence of the sensor plasmid was confirmed by PCR using several sets of primers. A strain of R. palustris no. 711 transformed by the sensor plasmid was designated crtIBS.
Assay for detection of arsenite.
crtIBS cells were precultured under anaerobic light or aerobic dark conditions until the exponential growth phase (OD600, 0.5 to ∼0.7) and collected, and then the OD600 of each culture was adjusted to 0.5 with MOM. Each cell suspension was dispensed into 7.5-ml sterilized glass tubes (7.5 ml per tube), and the dispensed crtIBS cells were incubated with various concentrations of NaAsO2 [0, 0.5, 1.0, 5.0, 10, 50, 100, and 500 μg/liter as As(III)] for 24 h under light conditions. Color changes were detected by the naked eye or with a spectrophotometer.
To perform the arsenite detection assay with several types of MW, fivefold-concentrated MOM was prepared with deionized water and then diluted fivefold with MW. Anaerobically grown crtIBS cells were collected, and the OD600 was adjusted to 0.5 with MW-based MOM. The conditions used for the detection assay were the same as those described above. Water taken from a faucet was used as tap water. Three types of commercially available MW were selected and purchased from local stores in Japan. Table 1 shows the mineral composition of each type of MW.
TABLE 1.
Concentrations of minerals and pH in MWs used in this study
| Water | Concn (mg/liter)
|
Hardness (mg/liter) | pH | |||
|---|---|---|---|---|---|---|
| Na+ | K+ | Mg2+ | Ca2+ | |||
| Tap water | <20 | 1.8 | 11.7 | 37 | 7.3 | |
| Volvic | 11.6 | 6.2 | 8.0 | 11.5 | 60 | 7.0 |
| Evian | 7.0 | 26 | 80 | 304 | 7.2 | |
| Vittel | 7.3 | 4.9 | 19.9 | 91.0 | 307 | 7.3 |
Real-time RT-PCR assay.
After incubation for 3 and 6 h with arsenite, the total RNA was extracted from crtIBS cells using an RNeasy mini kit (Qiagen, Germany) according to the manufacturer's instructions. First-strand cDNA was prepared using a PrimeScript reverse transcription (RT) reagent kit (Takara, Japan) according to the manufacturer's instructions. A real-time RT-PCR assay was performed with a 7500 real-time PCR system (Applied Biosystems, United States) using SYBR Premix Ex Taq II (Takara, Japan) and gene-specific primers rtcrtIS (5′-TCGCAACTTCCAAGATCACG-3′) and rtcrtIA (5′-TCATGATGTCACCAGACTGTCGGCC-3′). The amplification data were evaluated using the Sequence Detection System software (Applied Biosystems, United States). The expression of the crtI gene was normalized to that of rRNA. rRNA was amplified using primers rtrrnaS (5′-AGCCTACGATCTATAGCTGG-3′) and rtrrnaA (5′-GGAATTCCTCTCGGAAAACC-3′).
HPLC analysis.
After 24 h of incubation with arsenite, crtIBS cells were collected by centrifugation, stored at −80°C overnight, and freeze-dried. A mixture of acetone and methanol (7:2, vol/vol) was added to the lyophilized cells and vortex mixed for 2 min. The supernatants collected after centrifugation were analyzed using a high-performance liquid chromatography (HPLC) system equipped with a μBondapak C18 column (3.9 by 300 mm; pore size, 125-Å; Nihon Waters, Japan) eluted with methanol or acetonitrile-methanol (70:30, vol/vol). The elution patterns were monitored with a photodiode array detector (type 2996; Nihon Waters, Japan). All pigments were identified by using their absorption spectra and specific retention times.
Colorimetric analysis.
After 24 h of incubation with arsenite, crtIBS cells were colorimetrically analyzed by spectrophotometry (CM-3500d; Konica Minolta Sensing, Japan) as described previously (37). The colors based on the light spectra transmitted through the cultures were plotted in the CIE-L*a*b* color space (CIE is Commission Internationale de l'Eclairage). The CIE-L*a*b* color space is a general system for measuring the colors of objects that can also evaluate the differences in color between objects (30, 31). In the CIE-L*a*b* color space, the L* and a*b* coordinates are expressed as lightness and chroma values. An increase in the a* value from the center of the color space toward the plus direction indicates an increase in the red chroma, and an increase in the a* value from the center of the color space toward the minus direction indicates an increase in the green chroma. An increase in the b* value toward the plus direction indicates an increase in the yellow chroma, and an increase in the b* value toward the minus direction indicates an increase in the blue chroma. The hue angle (HA) indicates the change from the true red axis. An HA of 0° indicates pure red, while HAs of 90° and 180° indicate pure yellow and pure green, respectively. HA is defined by the following equation: HA = tan−1(b*/a*). ΔE*ab, which indicates the color difference between objects, was used to compare the colors of a sample exposed to arsenite and a sample containing no arsenite. ΔE*ab is defined by the following equation: ΔE*ab = [(ΔL*)2 +(Δa*)2 + (Δb*)2]1/2, where ΔL*, Δa*, and Δb* are differences in the L*, a*, and b* values between culture colors, respectively.
Evaluation of cross-reactivity with arsenate and antimonite.
crtIBS cells were precultured under anaerobic light conditions as described above and were incubated with various concentrations of Na2HAsO4·7H2O [0, 10, 50, 100, 200, 300, 400, 500, 1,000, and 5,000 μg/liter as As(V)] or SbCl3 [0, 0.5, 1.0, 5.0, 10, 50, 100, 250, 500, and 1,000 μg/liter as Sb(III)] for 24 h under light conditions. Color changes were detected by the naked eye or with a spectrophotometer.
Assay for detection of arsenite with iron.
Solutions of Fe(II) and Fe(III) were freshly prepared by dissolving FeSO4·7H2O and FeCl3 in deionized water. crtIBS was incubated with 50 μg/liter As(III) in the presence of various concentrations of iron (0, 0.5, 1.0, 5.0, 10, 50, and 100 mg/liter) for 24 h under light conditions. Then L*a*b* values were calculated by using the spectrophotometer, and effect of iron on the color change was analyzed.
Time-lapse study of the bacterial color change.
crtIBS cells were precultured under aerobic dark conditions and then were collected as described above. The cells were suspended in MOM supplemented with 0.5% tryptone and placed into two 190-ml cultivation vessels. The dispensed crtIBS cells were incubated in the presence or absence of 50 μg/liter As(III) under anaerobic light conditions, and time-lapse image capturing was started immediately. A series of pictures were taken with an Optio 750Z (Pentax, Japan) every 15 min for 24 h. The image stacks were processed for display using Microsoft Windows Movie Maker, version 2.1.
Double-blind test.
A double-blind test was carried out by using the following steps. (i) Researcher 1 incubated crtIBS cells with various concentrations of arsenite (0, 0.5, 1.0, 5.0, 10, 50, 100, and 500 μg/liter), as shown in Fig. 2. A sample containing 10 μg/liter As was also prepared as a standard for judging color. (ii) After 24 h, researcher 1 randomly labeled samples containing 0 to 500 μg/liter As with letters so that no one except researcher 1 would know the arsenite concentrations. (iii) Researcher 1 recruited 10 healthy volunteers who were not color blind. (iv) Researcher 2 asked the volunteers whether each of the labeled samples was redder than the standard (containing 10 μg/liter As) or not and divided the samples into two categories according to the judgments of the volunteers. Steps i to iv were repeated twice with other groups of volunteers, and a total of three independent experiments were carried out. To exclude deception and bias by researcher 1, step iv was conducted by researcher 2, who did not know the relationship between the arsenite concentrations and the letters.
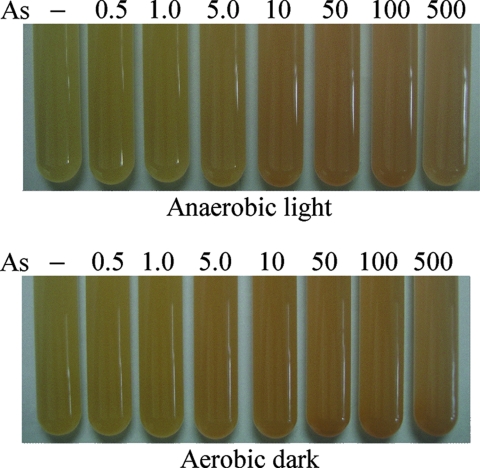
FIG. 2.
Bacterial color changes in crtIBS induced by arsenite. The preculture conditions are indicated below the panels. −, no arsenite; 0.5, 1.0, 5.0, 10, 50, 100, and 500, 0.5, 1.0, 5.0, 10, 50, 100, and 500 μg/liter As(III), respectively.
RESULTS
Detection of arsenite and evaluation of color change.
crtIBS cells were precultured under anaerobic light or aerobic dark conditions and then incubated with various concentrations of arsenite for 24 h under anaerobic light conditions. Figure 2 shows the color changes in crtIBS in response to the arsenite. Under both preculture conditions, the color of the biosensor changed toward red with increasing arsenite concentrations, and the changes could be detected by the naked eye.
Next, to evaluate the difference between the color of cells exposed to arsenite (0.5 to 500 μg/liter As) and the color of cells cultured without arsenite (As− cells), the L*, a*, and b* values of the colors of the cultures were calculated and plotted in the CIE-L*a*b* color space using the spectrophotometer (Table 2). The a* values indicated green chroma for As− cells (−0.26 under anaerobic light conditions and −0.07 under aerobic dark conditions), and there was a shift to red chroma in response to arsenite. With increasing arsenite concentrations, the a* and b* values tended to increase and decrease, respectively, in a dose-dependent manner for crtIBS cells precultured under both anaerobic light and aerobic dark conditions. The HA calculated using the a* and b* values for As− cells incubated under anaerobic light conditions indicated that the cells were in the green-yellow group (92.80°). In contrast, with increasing arsenite concentrations, the HA tended to shift toward the red group (0°) in a dose-dependent manner. The same tendency was observed for crtIBS cells incubated under aerobic dark conditions; the HA for As− cells indicated the yellow-green group (90.71°), and the HA tended to shift toward the red group in a dose-dependent manner. The HA difference (ΔHA) between cells exposed to arsenite and As− cells ranged from −4.50° to −28.01°, decreased markedly from an As concentration of 1.0 μg/liter to an As concentration of 10 μg/liter, and reached a plateau at an As concentration of 50 μg/liter. These observations corresponded to how easy it was to distinguish color differences by the naked eye at As concentrations above 10 μg/liter (from −23.62° to −28.01° under anaerobic light conditions and from −19.05° to −21.76° under aerobic dark conditions) (Fig. 2). The WHO's guideline value for arsenic is 10 μg/liter. The L* value also decreased in the cultures exposed to arsenite. The ΔE*ab calculated using the L*, a*, and b* values tended to increase in a dose-dependent manner (up to 2.52 under anaerobic light conditions and up to 2.22 under aerobic dark conditions). These data indicate that the change in color was due to the increase in the a* value and the decreases in the L* and b* values in cells incubated with arsenite.
TABLE 2.
CIE-L*a*b* color values for the bacterial cultures after incubation with arsenite in deionized water
| As concn (μg/liter) | L*a | a*a | b*a | HA | ΔHA | ΔL* | Δa* | Δb* | ΔE*ab |
|---|---|---|---|---|---|---|---|---|---|
| Anaerobic light conditions | |||||||||
| 0 | 92.81 | −0.26 | 5.32 | 92.80 | |||||
| 0.5 | 92.68 | 0.16 | 5.39 | 88.30 | −4.50 | −0.12 | 0.42 | 0.08 | 0.45 |
| 1.0 | 92.66 | 0.33 | 5.29 | 86.43 | −6.37 | −0.15 | 0.60 | −0.03 | 0.62 |
| 5.0 | 92.74 | 0.92 | 4.81 | 79.17 | −13.63 | −0.06 | 1.18 | −0.51 | 1.29 |
| 10 | 92.67 | 1.65 | 4.34 | 69.18 | −23.62 | −0.14 | 1.91 | −0.97 | 2.15 |
| 50 | 92.62 | 1.89 | 4.13 | 65.41 | −27.39 | −0.19 | 2.15 | −1.18 | 2.46 |
| 100 | 92.52 | 1.91 | 4.23 | 65.70 | −27.10 | −0.29 | 2.17 | −1.09 | 2.44 |
| 500 | 92.54 | 1.94 | 4.12 | 64.79 | −28.01 | −0.27 | 2.21 | −1.20 | 2.52 |
| Aerobic dark conditions | |||||||||
| 0 | 92.77 | −0.07 | 5.65 | 90.71 | |||||
| 0.5 | 92.77 | 0.39 | 5.54 | 85.97 | −4.74 | 0.00 | 0.46 | −0.12 | 0.48 |
| 1.0 | 92.58 | 0.69 | 5.55 | 82.91 | −7.80 | −0.19 | 0.76 | −0.10 | 0.79 |
| 5.0 | 92.19 | 1.47 | 5.41 | 74.80 | −15.91 | −0.58 | 1.54 | −0.24 | 1.67 |
| 10 | 91.96 | 1.79 | 5.40 | 71.66 | −19.05 | −0.81 | 1.87 | −0.25 | 2.05 |
| 50 | 92.00 | 1.96 | 5.19 | 69.31 | −21.40 | −0.77 | 2.04 | −0.47 | 2.22 |
| 100 | 92.02 | 1.92 | 5.17 | 69.63 | −21.08 | −0.75 | 2.00 | −0.49 | 2.19 |
| 500 | 92.13 | 1.94 | 5.04 | 68.95 | −21.76 | −0.64 | 2.01 | −0.61 | 2.20 |
The values are the means of three measurements.
Carotenoid analysis by HPLC after detection of arsenite.
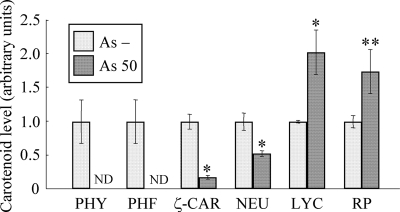
To confirm that the color change toward red was due to accumulation of red carotenoids, a pigment analysis was performed by using HPLC. Figure 3 shows a comparison of the carotenoid levels in As− cells and cells exposed to 50 μg/liter As. The level of lycopene, a red carotenoid which is the end product of a four-step desaturation reaction catalyzed by CrtI, was about twofold greater in cells exposed to 50 μg/liter As than in As− cells. The concentration of rhodopin, which is the downstream product of lycopene, was about 1.7-fold greater in cells exposed to 50 μg/liter As. In contrast, phytoene, which is the colorless carotenoid precursor in the reaction catalyzed by CrtI, was not detected in cells exposed to 50 μg/liter As. Similarly, phytofluene, which is the second product of a four-step desaturation reaction catalyzed by CrtI, was not detected. The concentrations of ζ-carotene and neurosporene, which are yellow carotenoids that are intermediate metabolites of CrtI, were decreased in cells exposed to 50 μg/liter As and were one-fifth and one-half the concentrations in As− cells, respectively. These data indicate that the color changes in bacterial cells exposed to 50 μg/liter As were mainly due to accumulation of the red carotenoids lycopene and rhodopin and reductions in the levels of phytoene, phytofluene, ζ-carotene, and neurosporene.
FIG. 3.
Comparison of the carotenoid contents of As− cells (As −) and cells exposed to 50 μg/liter As (As 50). The relative values were calculated from the peak areas in an HPLC chromatogram, and the values for As− cells were defined as 1.0. The data are the means ± standard deviations of three independent experiments. The asterisks indicate statistically significant differences between As− cells and cells exposed to 50 μg/liter As (*, P < 0.01; **, P < 0.05, Student's t test). ND, not detected; PHY, phytoene; PHF, phytofluene; ζ-CAR, ζ-carotene; NEU, neurosporene; LYC, lycopene; RP, rhodopin.
Induction of crtI expression by arsenite.
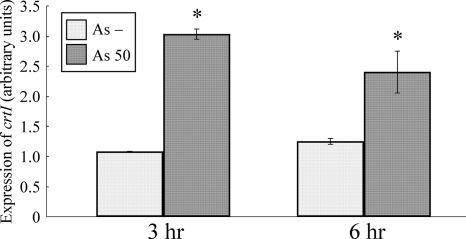
To confirm that the accumulation of red carotenoids was due to the expression of crtI on the sensor plasmid in response to arsenite, real-time RT-PCR assays were carried out using total RNA extracted from crtIBS cultured in the absence or presence of 50 μg/liter As(III). The crtI mRNA was induced 3-fold at 3 h and 2.5-fold at 6 h by addition of arsenite compared with the control (Fig. 4). The induction of crtI expression by arsenite indicates that the Pars derived from E. coli regulated the downstream gene expression in R. palustris. These results support the idea that the red carotenoids that accumulated in cells exposed to 50 μg/liter As were synthesized by CrtI translated from the mRNA transcribed under the Pars driver in the sensor plasmid.
FIG. 4.
Induction of crtI mRNA in response to arsenite. The expression level in As− cells (As −) was defined as 1.0. The data are the means ± standard deviations of three independent experiments. The asterisks indicate statistically significant differences between As− cells and cells exposed to 50 μg/liter As (As 50) (P < 0.05, Student's t test).
Detection of arsenite in various types of MW.
It is assumed that groundwater contains potential inhibitors of the arsenite-sensing system of biosensors. Commercially available MWs are derived from pure groundwater bottled without filtering or sterilization. Therefore, to test the potential usefulness of the carotenoid-based whole-cell biosensor, we created artificial arsenic-contaminated waters based on various MWs and performed an arsenite detection analysis using MW-based medium. The L*, a*, and b* values for the culture colors were calculated 24 h after the addition of 10 or 50 μg/liter As(III).
Obvious differences between the color of cultures incubated in the absence of As and the color of cultures incubated in presence of 10 or 50 μg/liter As were observed with all the MWs and could be recognized by the naked eye. With increasing arsenite concentrations, the a* value increased in a dose-dependent manner and the b* value decreased in a dose-dependent manner in all the MWs. The same tendencies shown in Table 2 were observed for the L*, a*, and b* values for crtIBS cells incubated with tap water-based MOM (Table 3). The a* value indicated green chroma for As− cells (−0.13) and shifted to red chroma for cells exposed to 10 and 50 μg/liter As (1.09 and 1.40, respectively). In contrast, the a* values for As− cells incubated with Volvic-, Evian-, and Vittel-based MOM indicated red chroma (0.09 in Volvic-based MOM, 1.33 in Evian-based MOM, and 1.58 in Vittel-based MOM). However, red chroma obviously increased for cells exposed to 10 and 50 μg/liter As (1.47 and 1.56 in Volvic-based MOM, 2.73 and 3.12 in Evian-based MOM, 2.86 and 3.06 in Vittel-based MOM, respectively).
TABLE 3.
CIE-L*a*b* color values for the bacterial cultures after incubation with arsenite in several types of water
| As concn (μg/liter) | L*a | a*a | b*a | HA | ΔHA | ΔL* | Δa* | Δb* | ΔE*ab |
|---|---|---|---|---|---|---|---|---|---|
| Tap water | |||||||||
| 0 | 93.62 | −0.13 | 4.81 | 91.55 | |||||
| 10 | 93.26 | 1.09 | 4.54 | 76.50 | −15.05 | −0.36 | 1.22 | −0.27 | 1.30 |
| 50 | 93.30 | 1.40 | 4.10 | 71.15 | −20.40 | −0.32 | 1.53 | −0.71 | 1.72 |
| Volvic | |||||||||
| 0 | 93.65 | 0.09 | 4.50 | 88.85 | |||||
| 10 | 93.23 | 1.47 | 4.01 | 69.87 | −18.98 | −0.43 | 1.38 | −0.49 | 1.52 |
| 50 | 93.22 | 1.56 | 3.87 | 68.05 | −20.80 | −0.44 | 1.47 | −0.62 | 1.65 |
| Evian | |||||||||
| 0 | 90.85 | 1.33 | 6.67 | 78.72 | |||||
| 10 | 90.49 | 2.73 | 5.89 | 65.13 | −13.59 | −0.36 | 1.40 | −0.77 | 1.64 |
| 50 | 90.45 | 3.12 | 5.67 | 61.18 | −17.54 | −0.40 | 1.79 | −0.99 | 2.08 |
| Vittel | |||||||||
| 0 | 90.55 | 1.58 | 6.69 | 76.71 | |||||
| 10 | 90.01 | 2.86 | 6.36 | 65.79 | −10.92 | −0.55 | 1.27 | −0.33 | 1.43 |
| 50 | 90.33 | 3.06 | 5.72 | 61.86 | −14.85 | −0.22 | 1.47 | −0.97 | 1.78 |
The values are the means of three measurements.
As a consequence of the changes in a* and b* values, the HA shift toward red was observed for all samples of cells exposed to 10 and 50 μg/liter As (91.55° to 76.50° to 71.15° in tap water-based MOM, 88.85° to 69.87° to 68.05° in Volvic-based MOM, 78.72° to 65.13° to 61.18° in Evian-based MOM, and 76.71° to 65.79° to 61.86° in Vittel-based MOM). The ΔHAs observed for cells incubated with 10 μg/liter As (−15.05 in tap water-based MOM, −18.98 in Volvic-based MOM, −13.59 in Evian-based MOM, and −10.92 in Vittel-based MOM) and with 50 μg/liter As (−20.40 in tap water-based MOM, −20.80 in Volvic-based MOM, −17.54 in Evian-based MOM, and −14.85 in Vittel-based MOM) were lower than the ΔHAs observed with deionized water, irrespective of the preculture conditions (Table 2). However, the ΔHAs observed for the MWs were still evident to the naked eye. The L* value decreased for all the samples of cells exposed to 10 and 50 μg/liter As. The ΔE*ab calculated using the L*, a*, and b* values increased in a dose-dependent manner for all samples of cells exposed to 10 and 50 μg/liter As. These results suggested that in natural water crtIBS is able to sense arsenite and that the color change is elicited via the same mechanism that is used in deionized water-based MOM.
Cross-reactivities with arsenate and antimonite.
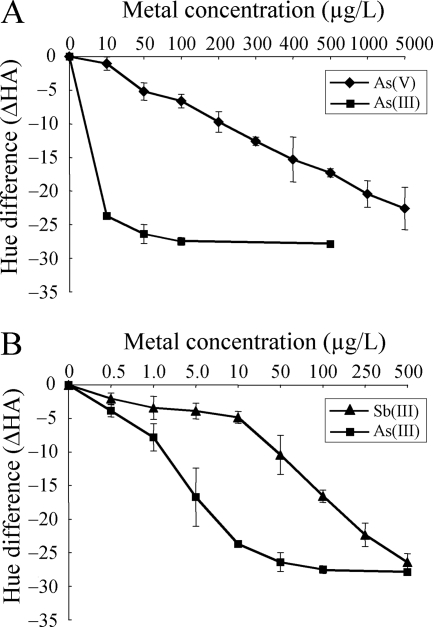
crtIBS changed color in response to both arsenate and antimonite with sensitivity lower than that observed for arsenite. The sensitivities to arsenate and antimonite were evaluated by using ΔHA calculated from a* and b* values. The ΔHA in response to 10 μg/liter As was 24° (Fig. 5A). However, more than 5,000 μg/liter As(V) was needed to obtain this ΔHA (Fig. 5A). These results indicated that the sensitivity of arsenate detection by crtIBS is 500-fold lower than the sensitivity of arsenite detection. In contrast, 250 μg/liter Sb(III) was needed to obtain the same ΔHA (Fig. 5B), which indicated that the sensitivity of antimonite detection by crtIBS is 25-fold lower the sensitivity of arsenite detection.
FIG. 5.
Cross-reactivities with arsenate and antimonite. The ΔHA was calculated by subtracting the HA value for a sample not exposed to metal from the HA for a sample exposed to metal. (A) Comparison of the dose-response decreases in ΔHA for arsenite (▪) and arsenate (⧫). (B) Comparison of the dose-response decreases in ΔHA for arsenite (▪) and antimonite (▴). The data are means ± standard deviations of two independent experiments.
Effect of iron on the bacterial color change induced by arsenite.
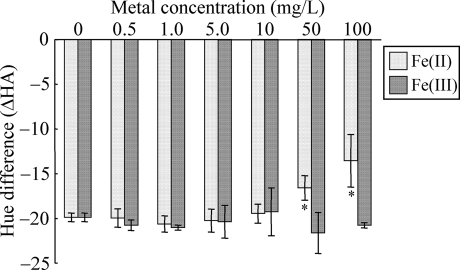
It has been reported previously that arsenic-contaminated groundwater often contains a significant amount of iron (3, 29). Furthermore, iron in the groundwater has been shown to affect light emission by a luciferase-based arsenite biosensor (29). The effect of iron on the color change induced by arsenite was evaluated by using ΔHA calculated from a* and b* values. The hue shift toward red induced by 50 μg/liter As(III) significantly decreased when crtIBS cells were incubated with 50 or 100 mg/liter Fe(II) (Fig. 6). In contrast, the hue shift was not affected by 0.5 to 10 mg/liter Fe(II) or by 0.5 to 100 mg/liter Fe(III). The bacterial color changes in the presence of 50 or 100 mg/liter Fe(II), however, were still obvious to the naked eye (data not shown). These results indicate that the effect of Fe(II) on crtIBS is not so serious that it should affect the use of this strain in analyses.
FIG. 6.
Effect of iron on the hue shift induced by arsenite. The ΔHA was calculated by subtracting the HA value for a sample not exposed to iron or arsenite from the HA for a sample exposed to iron or arsenite. For exposure to arsenite, 50 μg/liter As(III) was used. The data are means ± standard deviations of three independent experiments. The asterisks indicate statistically significant differences between the samples not exposed to iron and the samples exposed to iron (P < 0.05, Student's t test).
Bacterial color change within 24 h.
The real-time RT-PCR assay showed that the crtI mRNA was induced at 3 h (Fig. 4). To examine whether a bacterial color change could be recognized by the naked eye within 24 h, a time-lapse study was carried out. Development of color could be seen as early as 12 h, and a difference between the colors of As− cells and cells exposed to 50 μg/liter As could be observed after 12 h (see Movie S1 in the supplemental material). These results indicate that 12 h may be enough time to recognize a bacterial color change, but more than 12 h may be necessary for reliable recognition by the naked eye.
Evaluation of bacterial color change by double-blind test.
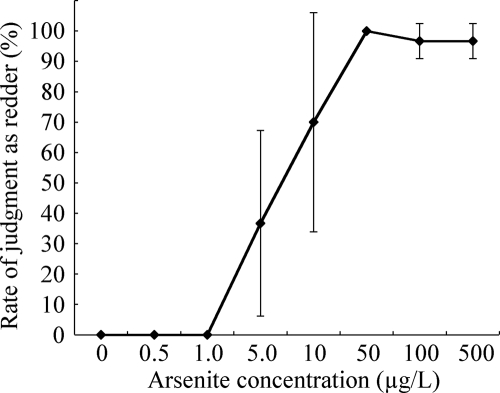
A color difference recognized by the naked eye is a very subjective parameter, so we carried out a double-blind test to evaluate whether a bacterial color change could be easily distinguished by untrained individuals. Importantly, all 30 subjects (100%) concluded that cells exposed to 0, 0.5, and 1.0 μg/liter As were not redder than the standard (cells exposed to 10 μg/liter As) (Fig. 7). In contrast, 11 of the 30 subjects (36.7%) concluded that samples exposed to 5.0 μg/liter As were redder than the standard, although the arsenite concentration of the standard was higher than the arsenite concentration of the samples exposed to 5.0 μg/liter As. All 30 subjects (100%) concluded that the samples exposed to 50 μg/liter As were redder than the standard, and a majority of the subjects (96.7%) concluded that the samples exposed to 100 and 500 μg/liter As were redder than the standard. These data indicate that the bacterial color changes induced by As(III) concentrations less than 1.0 μg/liter and the bacterial color changes induced by As(III) concentrations ranging from 50 to 500 μg/liter can be easily distinguished by untrained individuals when the color of cells exposed to 10 μg/liter As is employed as the standard. In other words, they imply that the bacterial color changes in response to unacceptable levels of As(III) (more than 10 μg/liter) can be easily distinguished when the color of cells exposed to 0 to 1.0 μg/liter As is employed as a standard.
FIG. 7.
Double-blind evaluation of the bacterial color change induced by arsenite. The rate of judgment as redder was calculated by dividing the number of subjects who judged a sample redder than the standard exposed to 10 μg/liter As by the total number of subjects (n = 10). The data are means ± standard deviations of three independent experiments.
DISCUSSION
In this study, we developed a novel arsenite biosensor that exhibits a clear color change after a reporter event. Because a blue-green mutant would have the greatest potential for an enhanced color change, we selected a mutant with crtI deleted, R. palustris no. 711 (37), as a host strain for the biosensor. The color of crtIBS expectedly changed from a cool background color to a warm color in deionized water- and tap water-based MOM. However, in the absence of As crtIBS cells were yellow-green, and this color was different from that of blue-green R. palustris no. 711. Pigment analysis by HPLC revealed that a certain level of colored carotenoids accumulated in the As− cells. A basal level of transcription is necessary to repress the operon in the absence of target molecules in negative-feedback loop systems (12). It is assumed that in crtIBS, the arsR gene transcribed from the sensor plasmid represses the transcription from Pars. Since the arsR gene and crtI gene are thought to be transcribed from the sensor plasmid as a polycistronic mRNA, a basal level of crtI gene transcript is probably also produced. The slight yellow color in the absence of arsenite may therefore be due to the carotenogenesis of phytoene catalyzed by CrtI protein translated from this basal crtI gene transcript. In addition, it has been reported that considerable light is emitted from uninduced E. coli- and Bacillus subtilis-based luminescent biosensors harboring a sensor plasmid containing Pars and arsR derived from Staphylococcus aureus (28). It is possible that E. coli ArsR may not completely repress the transcription from Pars in R. palustris. Recently, an ars operon was reported to be present in the R. palustris genome (18). Therefore, stricter regulation might be obtained if the Pars derived from R. palustris were used in combination with a second ArsR-binding site (26) in R. palustris-based biosensors. The bacterial color change in crtIBS was stable under both anaerobic and aerobic preculture conditions. In contrast, the bacterial color change in crtA-based biosensors is very sensitive to dissolved oxygen in the culture medium (14). Therefore, our findings demonstrate that crtIBS could be used for water samples containing various oxygen concentrations. The stable bacterial color change in crtIBS was also observed in MW-based media. However, crtIBS cells were yellow even in the absence of arsenite in Volvic-, Evian-, and Vittel-based MOM. The ΔHA and ΔE*ab obtained using MW-based media were lower than those obtained using deionized water-based medium. It is possible that ions or other unknown compounds in the MWs influence the arsenite-sensing ability of crtIBS. Despite the decrease in ΔHA and ΔE*ab, however, the bacterial color changes in response to 10 and 50 μg/liter As(III) were still sufficient for recognition by the naked eye. These data suggest that crtIBS is likely to be sensitive enough to assess whether groundwater should be considered safe to drink.
Iron is known to influence the solubility of inorganic arsenic by adsorption under oxic conditions. This may lead to underestimation of the arsenic content by biosensor cells. In our study, the bacterial color change was affected by Fe(II) at concentrations ranging from 50 to 100 mg/liter, but it was not affected by 0.5 to 10 mg/liter Fe(II) or 0.5 to 100 mg/liter Fe(III). It is possible that coprecipitation of arsenite with iron might be inhibited by the anaerobic sensing conditions, leading to maintenance of the bioavailability of arsenite. In addition, the EDTA in the culture medium used in this study (MOM) might efficiently chelate iron, leading to inhibition of coprecipitation. Further improvements for practical use may be needed to obtain more a stable bacterial color change even in the presence of high concentrations of iron.
crtIBS exhibited >500-fold-lower sensitivity to arsenate. Despite the fact that we used Pars and arsR from E. coli, the sensitivity of crtIBS to arsenate was significantly lower than the sensitivities of E. coli-based luciferase biosensors (23, 26). It has been reported that arsenate is reduced to arsenite by the reductase in E. coli, and arsenite induces light emission. Therefore, it is possible that R. palustris has low reductase activity for production of arsenite under sensing conditions. Further improvements are needed to detect arsenate for practical use of crtIBS. crtIBS reacted to antimonite with slightly lower sensitivity (25-fold lower than the sensitivity to arsenite). It has been reported that in Vietnam the groundwater often contains only 1 to 4 μg/liter antimonite, so the bioreporter responses may not be substantially affected by antimony contamination.
It took 24 h to recognize the color change in crtIBS by the naked eye. In contrast, E. coli-based arsenite biosensors indicate the presence of arsenite within 1 h when instruments are used to measure it (4, 26). Because the naked eye is not as sensitive as these instruments, the difference in detection time between the E. coli-based biosensors and crtIBS is thought to be partially due to the detection methods. It has been reported that the E. coli ars operon is induced within 5 min in response to arsenite (17). The induction of the E. coli ars operon in R. palustris in the short term has not been determined. However, our real-time RT-PCR analysis revealed that crtI mRNA was induced 3 h after the addition of arsenite. The time-lapse study also revealed that bacterial color development could be seen as soon as 12 h after addition. It is possible that a short-term bioassay could be developed with crtIBS if a spectrophotometer were used to measure the color change. Furthermore, a portable spectrophotometer that can measure small differences in color is available (7), and arsenite detection using such a device could be performed outside the laboratory. Using crtIBS, results obtained with a portable spectrophotometer could be confirmed later using the naked eye.
To date, a luciferase-based arsenite biosensor using E. coli has been established (29). This biosensor can detect arsenic quantitatively at concentrations ranging from 0 to 75 μg/liter. A total of 194 groundwater samples were analyzed, and the correlation with atomic absorption spectroscopy data was assessed. Use of this method resulted in more than 90% correct measurements. At arsenic concentrations lower than 10 μg/liter, 8.0% of the samples were considered false negative. At arsenic concentrations higher than 10 μg/liter, 2.4% of the samples were considered false positive. These results are far more accurate than the results obtained with a chemical field kit. They imply that arsenic levels above and below the WHO's guideline value are distinguished quite reliably. Our sensor is not suitable for quantitative analysis when detection is carried out by using the naked eye. However, the double-blind test showed that the upper acceptable limits of arsenic concentrations (10 and 50 μg/liter) were easily distinguished from 0 to 1.0 μg/liter arsenic. Thus, crtIBS could be used as a first screening tool to distinguish whether groundwater contains unacceptable levels of arsenite, and this tool could be available to many people.
Supplementary Material
Acknowledgments
This work was supported by grant-in-aid 05A22703a from Industrial Technology Research Program in 2005 of the New Energy and Industrial Technology Development Organization (NEDO) of Japan to I.M. This work was also supported by a Grant-in-Aid for Specially Promoted Research from Utsunomiya University.
We thank the Center for Bioscience Research and Education at Utsunomiya University for technical assistance.
Footnotes
Published ahead of print on 5 September 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bagla, P., and J. Kaiser. 1996. India's spreading health crisis draws global arsenic experts. Science 274:174-175. [DOI] [PubMed] [Google Scholar]
- 2.Belkin, S. 2003. Microbial whole-cell sensing systems of environmental pollutants. Curr. Opin. Microbiol. 6:206-212. [DOI] [PubMed] [Google Scholar]
- 3.Berg, M., H. C. Tran, T. C. Nguyen, H. V. Pham, R. Schertenleib, and W. Giger. 2001. Arsenic contamination of groundwater and drinking water in Vietnam: a human health threat. Environ. Sci. Technol. 35:2621-2626. [DOI] [PubMed] [Google Scholar]
- 4.Cai, J., and M. S. DuBow. 1997. Use of a luminescent bacterial biosensor for biomonitoring and characterization of arsenic toxicity of chromated copper arsenite (CCA). Biodegradation 8:105-111. [DOI] [PubMed] [Google Scholar]
- 5.Carlin, A., W. Shi, S. Dey, and B. P. Rosen. 1995. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J. Bacteriol. 177:981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowdhury, T. R., G. K. Basu, B. K. Mandal, B. K. Biswas, G. Samanta, U. K. Chowdhury, C. R. Chanda, D. Lodh, S. L. Roy, K. C. Saha, S. Roy, S. Kabir, Q. Quamruzzaman, and D. Chakraborti. 1999. Arsenic poisoning in the Ganges delta. Nature 401:545-547. [DOI] [PubMed] [Google Scholar]
- 7.Dale Marcy, A., and P. L. Drake. 2007. Development of a field method for measuring manganese in welding fume. J. Environ. Monit. 9:1199-1204. [DOI] [PubMed] [Google Scholar]
- 8.Erickson, B. E. 2003. Field kits fail to provide accurate measure of arsenic in groundwater. Environ. Sci. Technol. 37:35A-38A. [DOI] [PubMed] [Google Scholar]
- 9.Fujimoto, H., M. Wakabayashi, H. Yamashiro, I. Maeda, K. Isoda, M. Kondoh, M. Kawase, H. Miyasaka, and K. Yagi. 2006. Whole-cell arsenite biosensor using photosynthetic bacterium Rhodovulum sulfidophilum. Rhodovulum sulfidophilum as an arsenite biosensor. Appl. Microbiol. Biotechnol. 73:332-338. [DOI] [PubMed] [Google Scholar]
- 10.Harms, H., M. C. Wells, and J. R. van der Meer. 2006. Whole-cell living biosensors—are they ready for environmental application? Appl. Microbiol. Biotechnol. 70:273-280. [DOI] [PubMed] [Google Scholar]
- 11.Inui, M., J. H. Roh, K. Zahn, and H. Yukawa. 2000. Sequence analysis of the cryptic plasmid pMG101 from Rhodopseudomonas palustris and construction of stable cloning vectors. Appl. Environ. Microbiol. 66:54-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacob, F., and J. Monod. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3:318-356. [DOI] [PubMed] [Google Scholar]
- 13.Maeda, I., H. Miyasaka, F. Umeda, M. Kawase, and K. Yagi. 2003. Maximization of hydrogen production ability in high-density suspension of Rhodovulum sulfidophilum cells using intracellular poly(3-hydroxybutyrate) as sole substrate. Biotechnol. Bioeng. 81:474-481. [DOI] [PubMed] [Google Scholar]
- 14.Maeda, I., H. Yamashiro, D. Yoshioka, M. Onodera, S. Ueda, M. Kawase, H. Miyasaka, and K. Yagi. 2006. Colorimetric dimethyl sulfide sensor using Rhodovulum sulfidophilum cells based on intrinsic pigment conversion by CrtA. Appl. Microbiol. Biotechnol. 70:397-402. [DOI] [PubMed] [Google Scholar]
- 15.Nickson, R., J. McArthur, W. Burgess, K. M. Ahmed, P. Ravenscroft, and M. Rahman. 1998. Arsenic poisoning of Bangladesh groundwater. Nature 395:338. [DOI] [PubMed] [Google Scholar]
- 16.Nordstrom, D. K. 2002. Public health. Worldwide occurrences of arsenic in ground water. Science 296:2143-2145. [DOI] [PubMed] [Google Scholar]
- 17.Owolabi, J. B., and B. P. Rosen. 1990. Differential mRNA stability controls relative gene expression within the plasmid-encoded arsenical resistance operon. J. Bacteriol. 172:2367-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin, J., B. P. Rosen, Y. Zhang, G. Wang, S. Franke, and C. Rensing. 2006. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. USA 103:2075-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahman, M. M., D. Mukherjee, M. K. Sengupta, U. K. Chowdhury, D. C. Lodh, S. Roy, M. Selim, Q. Quamruzzaman, A. H. Milton, S. M. Shahidullah, M. T. Rahman, and D. Chakraborti. 2002. Effectiveness and reliability of arsenic field testing kits: are the million dollar screening projects effective or not? Environ. Sci. Technol. 36:5385-5394. [DOI] [PubMed] [Google Scholar]
- 20.Ramanathan, S., W. Shi, B. P. Rosen, and S. Daunert. 1997. Sensing antimonite and arsenite at the subattomole level with genetically engineered bioluminescent bacteria. Anal. Chem. 69:3380-3384. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.San Francisco, M. J., C. L. Hope, J. B. Owolabi, L. S. Tisa, and B. P. Rosen. 1990. Identification of the metalloregulatory element of the plasmid-encoded arsenical resistance operon. Nucleic Acids Res. 18:619-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott, D. L., S. Ramanathan, W. Shi, B. P. Rosen, and S. Daunert. 1997. Genetically engineered bacteria: electrochemical sensing systems for antimonite and arsenite. Anal. Chem. 69:16-20. [DOI] [PubMed] [Google Scholar]
- 24.Shi, W., J. Dong, R. A. Scott, M. Y. Ksenzenko, and B. P. Rosen. 1996. The role of arsenic-thiol interactions in metalloregulationof the ars operon. J. Biol. Chem. 271:9291-9297. [DOI] [PubMed] [Google Scholar]
- 25.Smedley, P. L., and D. G. Kinniburgh. 2002. A review of the source, behaviour and distribution of arsenic in natural waters. Appl. Geochem. 17:517-568. [Google Scholar]
- 26.Stocker, J., D. Balluch, M. Gsell, H. Harms, J. Feliciano, S. Daunert, K. A. Malik, and J. R. van der Meer. 2003. Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in portable water. Environ. Sci. Technol. 37:4743-4750. [DOI] [PubMed] [Google Scholar]
- 27.Takaichi, S. 1999. Carotenoids and carotenogenesis in anoxygenic photosynthetic bacteria, p 39-69. In H. A. Frank, A. J. Young, G. Britton, and R. J. Cogdell (ed.), The photochemistry of carotenoids. Kluwer, Dordrecht, The Netherlands.
- 28.Tauriainen, S., M. Karp, W. Chang, and M. Virta. 1997. Recombinant luminescent bacteria for measuring bioavailable arsenite and antimonite. Appl. Environ. Microbiol. 63:4456-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trang, P. T., M. Berg, P. H. Viet, N. Van Mui, and J. R. van der Meer. 2005. Bacterial bioassay for rapid and accurate analysis of arsenic in highly variable groundwater samples. Environ. Sci. Technol. 39:7625-7630. [DOI] [PubMed] [Google Scholar]
- 30.Tsubura, S., and R. Yamaguchi. 2005. Clinical evaluation of a new bleaching product “Polanight” in a Japanese population. Odontology 93:52-55. [DOI] [PubMed] [Google Scholar]
- 31.Wiegand, A., D. Vollmer, M. Foitzik, R. Attin, and T. Attin. 2005. Efficacy of different whitening modalities on bovine enamel and dentin. Clin. Oral Investig. 9:91-97. [DOI] [PubMed] [Google Scholar]
- 32.Wu, J., and B. P. Rosen. 1991. The ArsR protein is a trans-acting regulatory protein. Mol. Microbiol. 5:1331-1336. [DOI] [PubMed] [Google Scholar]
- 33.Wu, J., and B. P. Rosen. 1993. Metalloregulated expression of the ars operon. J. Biol. Chem. 268:52-58. [PubMed] [Google Scholar]
- 34.Xu, C., W. Shi, and B. P. Rosen. 1996. The chromosomal arsR gene of Escherichia coli encodes a trans-acting metalloregulatory protein. J. Biol. Chem. 271:2427-2432. [DOI] [PubMed] [Google Scholar]
- 35.Yagi, K. 2007. Applications of whole-cell bacterial sensors in biotechnology and environmental science. Appl. Microbiol. Biotechnol. 73:1251-1258. [DOI] [PubMed] [Google Scholar]
- 36.Yeliseev, A. A., J. M. Eraso, and S. Kaplan. 1996. Differential carotenoid composition of the B875 and B800-850 photosynthetic antenna complexes in Rhodobacter sphaeroides 2.4.1: involvement of spheroidene and spheroidenone in adaptation to changes in light intensity and oxygen availability. J. Bacteriol. 178:5877-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida, K., D. Yoshioka, K. Inoue, S. Takaichi, and I. Maeda. 2007. Evaluation of colors in green mutants isolated from purple bacteria as a host for colorimetric whole-cell biosensors. Appl. Microbiol. Biotechnol. 76:1043-1050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.