Abstract
Fungal activity is a major driver in the global nitrogen cycle, and mounting evidence suggests that fungal denitrification activity contributes significantly to soil emissions of the greenhouse gas nitrous oxide (N2O). The metabolic pathway and oxygen requirement for fungal denitrification are different from those for bacterial denitrification. We hypothesized that the soil N2O emission from fungi is formate and O2 dependent and that land use and landforms could influence the proportion of N2O coming from fungi. Using substrate-induced respiration inhibition under anaerobic and aerobic conditions in combination with 15N gas analysis, we found that formate and hypoxia (versus anaerobiosis) were essential for the fungal reduction of 15N-labeled nitrate to 15N2O. As much as 65% of soil-emitted N2O was attributable to fungi; however, this was found only in soils from water-accumulating landforms. From these results, we hypothesize that plant root exudates could affect N2O production from fungi via the proposed formate-dependent pathway.
The importance of fungal denitrification to the emission of nitrous oxide (N2O; an important greenhouse gas) from soils has been demonstrated for a number of systems. Up to 89% of soil N2O emissions could be attributed to fungal activity (25). Given that fungal biomass dominates in many ecosystems, its potential activity may be the dominant soil N2O-emitting process. Fungal denitrifiers are ecologically significant because most fungal isolates capable of denitrification appear to lack nitrous oxide reductase, the enzyme that reduces N2O to N2 (32, 58). Therefore, unlike the end product of bacterial denitrification, which is mostly N2, the end product of fungal denitrification is N2O.
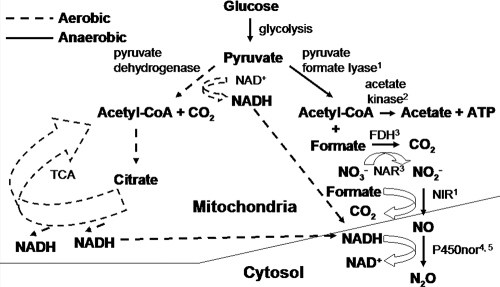
Accumulated evidence from work with fungal isolates indicates that the fungal pathway for the respiratory reduction of nitrogen oxides to nitric oxide (NO) and N2O is different from that of bacteria. Fungal nitric oxide reductase (P450nor) is a cytochrome p450-containing enzyme that receives electrons directly from NADH for the reduction of NO to N2O (32, 58). Consequently, small amounts of O2 (hypoxia) are required to generate NADH from the oxidation of citrate in the tricarboxylic acid cycle (Fig. 1). This is in contrast to bacterial denitrification, where successive enzymes in the pathway are increasingly sensitive to O2 inhibition (44, 59). The O2 requirement for fungal denitrification, however, has not been tested explicitly in soil.
FIG. 1.
Proposed O2- and formate-dependent fungal denitrification pathway developed from the cited works. 1, Kuwazaki et al., 2003 (24); 2, Zhou et al., 2002 (57); 3, Uchimura et al., 2002 (47); 4, Nakahara et al., 1993 (32); 5, Zhou et al., 2001 (58). TCA, tricarboxylic acid cycle; FDH, formate dehydrogenase; NAR, nitrate reductase; NIR, nitrite reductase; Acetyl-CoA, acetyl coenzyme A.
An interesting feature in some fungal denitrification pathways is the coupling of nitrate or nitrite reduction with formate (HCOO−) oxidation (Fig. 1) (24, 47). Low-molecular-weight organic acids, such as formate, are important root exudates (22) and are intermediates and by-products of anaerobic carbon metabolism (10). Formate (together with acetate) is the end product of the fermentation of citrate, oxaloacetate, and pyruvate. It is also produced from H2 and CO2 by a variety of anaerobic microorganisms (e.g., acetagens, sulfate reducers, and methanogens) (21). The amount of formate in aerobic soils is reported to range from 6 to 26% of the total extractable low-molecular-weight organic acids (51). Reported rhizosphere formate concentrations range from below the detection limit for clover (Trifolium repens) (9) to 117 μM for Norway spruce (Picea abies) (49) and 563 μM for quack grass (Elytrigia repens) (5). In addition to having external sources, fungi can produce formate. Under O2-limited conditions, formate is produced from the fermentation of pyruvate (57). Because of its various sources and relative ubiquity in soils, formate-dependent respiratory reduction of nitrogen oxides to N2O by fungi may be an important contributor to net N2O emissions from soils.
We hypothesized that soil N2O production by fungi is formate and O2 dependent and that land use and landform influence the proportion of N2O attributable to fungi. Land use factors, such as tillage, have an influence on fungi by physically disturbing the soil (17), while fertilizer applications can have an inhibitory effect on fungi (4, 16). Land use and landform also affect water distribution (18, 30) and the quantity and quality of soil organic matter (48), which, in turn, can affect soil fungi. Using substrate-induced respiration inhibition (SIRIN) under anaerobic and aerobic conditions, in combination with 15N gas analysis, we evaluated the importance of formate and O2 to N2O production by fungi in cultivated and uncultivated soils at the St. Denis National Wildlife Area in Saskatchewan, Canada.
MATERIALS AND METHODS
Study site.
The St. Denis National Wildlife Area in central Saskatchewan, Canada (52°12′N, 106°5′W), is typical of the North American prairie pothole region. It contains 216 wetlands within an area of 3.84 km2 (19) in the Dark Brown soil zone. Soil types range from thin Typic Calciborolls and thick Typic Haploborolls for water-shedding landform elements to Albic Argiborolls and Argic Cryaquolls in water-accumulating elements (54). Soils (Weyburn Association) developed on loamy unsorted glacial till parent materials in hummocky terrain with slope classes ranging from 10 to 15% (31). Six ephemeral wetlands (three cultivated and three uncultivated) were selected for study. Ephemeral wetlands are those depressions in hummocky landscapes that contain standing water in the spring but typically dry out during the growing season (18).
A digital elevation model was used to subdivide the cultivated wetlands (CW) into convex (CX; topographically high positions with a positive profile curvature that sheds water) and cultivated depression center (CD; level positions, roughly circular in shape, which temporarily collect rain or snowmelt water) landform elements (55). These two landforms represent the extremes in terms of N2O emission, soil moisture conditions, and biological productivity within the cultivated landscape (19, 54, 55). Uncultivated wetlands (UW) were found in nonagricultural portions of the site. These were divided into two landform elements roughly equivalent to those in the cultivated wetlands. Basin centers (BC) are level areas covered by 99 nongrass plant species, collect rain and snowmelt water, and are analogous to the CD elements. Uncultivated wetlands also included nonlevel fringe areas covered with Bromus inermis, termed riparian grass (RG) areas. The RG elements represent the driest areas within uncultivated wetlands and, in this sense, are analogous to CX elements in the cultivated wetlands.
Soil sampling and soil carbon and nitrogen determination.
Three cultivated and three uncultivated wetlands (each constituting a land use replicate [6]) were sampled on 12 September 2006. Five soil cores (0 to 15 cm; 15-cm internal diameter [i.d.]) were collected from the individual landform elements in each of the six wetlands. The cores from each individual landform element were bulked together to form a composite sample for that location, yielding a total of 12 composite samples (2 land uses × 2 landforms × 3 replicates). Samples were transported on ice, and subsamples were collected for soil formate extraction and lyophilization (for phospholipid fatty acid [PLFA] extraction). The remaining soil was air dried just enough (<24 h) to pass through a 2-mm sieve without smearing and stored at −20°C. Inorganic nitrogen (2 M KCl extracts) (28), soil organic carbon (dry combustion) (37), and total nitrogen (Dumas method) (34) were determined by standard methods, and results are listed in Table 1.
TABLE 1.
Organic carbon and mineral and total nitrogen in soils of the St. Denis National Wildlife Area, Saskatchewan, Canadaa
| Land use | Landform | SOC (%) | NH4+ | NO3− | Total N (%) | C/N |
|---|---|---|---|---|---|---|
| Cultivated | CX | 2.3 (0.1) | 1.8 (0.1) | 1.7 (0.2) | 0.2 (0.0) | 9 (0) |
| CD | 3.2 (0.2) | 2.7 (0.1) | 2.9 (0.4) | 0.4 (0.0) | 9 (0) | |
| Uncultivated | RG | 2.4 (0.1) | 3.4 (0.4) | 1.6 (0.2) | 0.3(0.0) | 9 (0) |
| BC | 3.5 (0.1) | 4.7 (0.8) | 6.0 (1.6) | 0.4 (0.0) | 9 (0) |
Reported values are means (μg N g−1 soil) with standard errors in parentheses.
SIRIN incubations to determine the contribution of fungi to N2O emissions.
Substrate-induced respiration inhibition studies involved the following treatments: control soil (no added microbial inhibitor), cycloheximide-amended soil, and streptomycin-amended soil. Whereas cycloheximide was chosen for targeted suppression of fungal activities, streptomycin was chosen as a broad-spectrum bacterial activity inhibitor (3). Soils were removed from cold storage, thawed at room temperature, and packed into a 10-ml volume at the bottom of a 55-ml glass culture tube (22-mm i.d.) to yield bulk densities similar to those observed in the field. Soil water content was determined using standard procedures with an assumed particle density of 2.65 g cm−3 (46). Preliminary experiments determined that when repacked cores are initially wetted to 50% or 70% water-filled pore space (WFPS), a burst of N2O was observed at 24 h after wetting, declined to background levels at 48 h after wetting, and remained at background levels for several days thereafter. Thus, soil water content was first adjusted to 50% WFPS, and the culture tube was capped with Parafilm and stored at 4°C for 48 h prior to treatment.
Preliminary experiments also were conducted to determine the optimal cycloheximide and streptomycin concentrations for use with the CD and BC soils. Optimum concentrations of the inhibitors were determined using a modification of the method described by Laughlin and Stevens (25). The modifications included bringing the soils to 70% WFPS and adding potassium formate (2 mg formate-C g−1 soil; equivalent to about 2% of the maximum soil organic carbon found in the St. Denis soils) and potassium nitrate (80 μg NO3-N g−1 soil; this gave a ratio of soil organic carbon to total N [C/N ratio] of 24:1). The optimum concentration of both cycloheximide and streptomycin was found to be 4 mg inhibitor g−1 soil for the CD soils and 8 mg inhibitor g−1 soil for the BC soils. These levels of inhibitor also were applied to the respective upland soils. Because cycloheximide is soluble only in methanol, the cycloheximide stock solution consisted of 4 g of cycloheximide dissolved in 10 ml of methanol and dispersed in deionized water to a final volume of 50 ml. Streptomycin and control (no-biocide) solutions were made with a similar volume and methanol/water ratio. The addition of the inhibitors resulted in an increase in soil moisture content to approximately 60% WFPS. Thus, the tubes were recapped with Parafilm and incubated in the dark at 23°C for an additional 24 h prior to the addition of the formate and nitrate (11). After adjustment of the soils to a final moisture content of 70% WFPS, the tubes were sealed with butyl-rubber caps and incubated in the dark at 23°C for a 24-h incubation. After 24 h, a 20-ml gas sample was withdrawn from the headspace using a 20-ml disposable syringe equipped with a 25-gauge needle and injected into a preevacuated 12-ml Exetainer vial (Labco Ltd., United Kingdom). Headspace N2O concentrations were determined using a gas chromatograph equipped with an electron capture detector (55).
15N stable isotope incubation with N2O, ammonium, and nitrate analyses.
Modified SIRIN incubations were repeated for the CD and BC soils in which the N substrate was enriched with 15N. That is, the added KNO3 consisted of 78 μg KNO3-N g−1 soil and 2 μg K15NO3-15N g−1 soil (at 98% 15N enrichment). The 15N studies were conducted under both aerobic and anaerobic conditions. Anaerobic systems were prepared by replacing the headspace air in the culture tubes with ultrahigh-purity N2 (i.e., working in an anaerobic chamber under a N2 atmosphere). Gas samples were collected as described for SIRIN incubations, after which the soils were destructively sampled for ammonium and nitrate by extraction with 2 M KCl.
15N-labeled N2O was analyzed by isotope ratio mass spectrometry at the University of California—Davis Stable Isotope Facility (Davis, CA). The fractionation of 15N-labeled N2O was calculated as described by Arah (1) and Stevens et al. (41). The diffusion disk technique described by Stark and Hart (40) and modified by Bedard-Haughn et al. (7) was used to concentrate soil ammonium and nitrate for 15N analysis.
Soil N2O emissions in response to increasing concentrations of formate.
Soils were removed from cold storage, thawed at room temperature, and packed to a 10-ml volume at the bottom of a 55-ml glass culture tube to yield bulk densities similar to those observed in the field. The soil water content was adjusted to 50% WFPS, and the tubes were capped with Parafilm and stored at 4°C for 48 h. The soils were then amended with formate (at concentrations of 0.0, 0.1, 0.2, 0.4, 0.8, and 1.6 nmol formate-C g−1 soil) and adjusted to 70% WFPS; the tubes were then sealed with butyl-rubber caps and incubated in the dark at 23°C for 24 h. Gas sampling and analysis were carried out as described for the SIRIN incubations. The effect of formate additions on N2O emissions was modeled using the Enzyme Kinetics Module (single-substrate model) in SigmaPlot version 9.0 (Systat Software, Inc., San Jose, CA).
Soil formate extraction and analysis.
Soil formate was extracted using the centrifugation drainage technique described by van Hees et al. (50). Extracts were stored at −20°C prior to analysis and were analyzed using the capillary electrophoresis method for anion detection described by Swallow and Low (43). The analyses were carried out using a Waters Quanta 4000 capillary electrophoresis system (Waters Corporation, Millford, MA) equipped with a 60-cm by 75-μm (i.d.) fused silica column. The electrolyte buffer was 5 mM sodium chromate-0.4 mM OFM-BT (Waters Corporation) adjusted to pH 8 with lactic acid and filtered through a 0.45-μm Millipore membrane filter (Millipore, Billerica, MA). Anions were detected at 254 nm using indirect UV detection. Samples and standards were analyzed according to the following sequence: (i) 30-s hydrostatic injection, (ii) 5-min run time at 25 kV, (iii) 1.5-min rinse with 0.1 M NaOH, (iv) 4-min rinse with Nanopure deionized water, and (v) 5-min purge with electrolyte buffer.
PLFA extraction and analysis.
PLFAs were extracted from lyophilized soil samples (4.0 g) using the method described by White et al. (53). Briefly, lipids were extracted from soils in a mixture of methanol, chloroform, and phosphate buffer (2:1:0.8 [vol/vol/vol]). Lipids were separated on silica gel columns (Bond Elut, Varian, Inc., Mississauga, Canada) by sequential applications of chloroform, acetone, and methanol (with the methanol fraction containing the phospholipids) (20). Phospholipids were methylated and separated by using a Hewlett Packard 5890 Series II gas chromatograph equipped with a 25-m by 0.2-mm (i.d.) Ultra 2 column (J&W Scientific; MIDI, Inc., Newark, DE) and a flame ionization detector. Lipid peaks were identified by comparisons of retention times to those in the TSBA version 4.1 and CLIN version 4.0 lipid libraries (MIDI, Inc.). Methylnonadecanoate (19:0) fatty acid (1 μg) was added to each sample as the internal standard before the methylation step (20). The PLFA 18:2ω6,9 was used as the fungal biomass marker (2).
Statistical analysis.
The SIRIN and 15N gas data were imported into Minitab (v. 11.21; State College, PA) and analyzed using two-way analysis of variance (ANOVA) with a general linear model (α = 0.1) after verification that the data met the ANOVA assumptions (using the Anderson-Darling test for normality and Bartlett's and Levene's tests for homogeneity of variance). Classification variables were landform and treatment. Because there were significant differences in landform (P = 0.001) and landform versus treatment (P < 0.001), Tukey's pairwise comparison was used to assess the significance of treatment differences within a landform. PLFA data were compared using one-way ANOVA and Tukey's pairwise comparisons with landform as the classification variable.
RESULTS
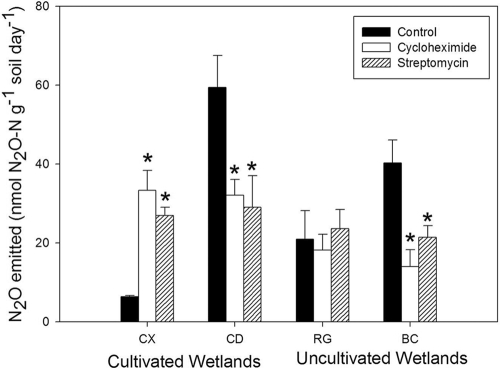
Nitrous oxide emissions attributable to fungal activity were found (P < 0.1) in both CD and BC soils (Fig. 2). Indeed, CD and BC soils incubated with cycloheximide produced 46% and 65% less N2O, respectively, than the control soil. The wide-spectrum bacterial inhibitor streptomycin also decreased N2O emissions in the CD (−51%) and BC (−47%) soils. The inhibitors had no significant effect on N2O emissions from the RG soils, but they produced increased emissions in soils from the CX landform elements. Either these water-shedding landforms, RG and CX, do not have significant fungal N2O emission, or the inhibitors were ineffective in these landforms. Thus, because our focus was on fungal denitrification, subsequent incubation studies designed to probe the regulation of fungal denitrification with 15N-labeled nitrate and inhibitors was restricted to the CD and BC soils.
FIG. 2.
Nitrous oxide emission from a SIRIN assay incubated at 70% WFPS for 24 h. Reported values are means with standard error bars (n = 3). An asterisk above the error bar denotes N2O emission from a treatment incubation with results that were significantly different from those of the control incubation (P < 0.1).
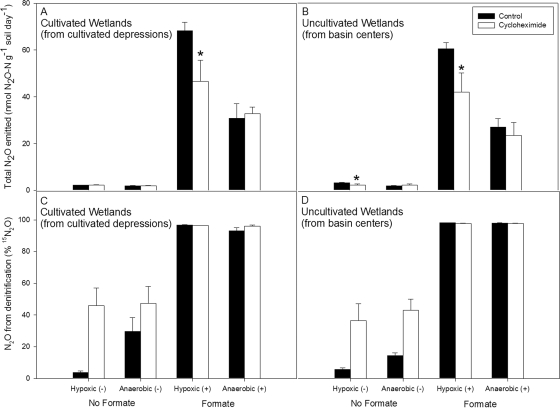
Relative to the appropriate controls, N2O emissions from the CD and BC soils exhibited a 30-fold increase when incubated with formate (Fig. 3A and B). Furthermore, the formate effect was most pronounced under hypoxic conditions (i.e., soils incubated at 70% WFPS with an aerobic headspace). In the presence of cycloheximide, N2O emissions from CD and BC soils decreased by about 30% when they were incubated in a low-oxygen environment. Conversely, there was no difference between incubations with or without the inhibitor when soils were incubated under anaerobic conditions (i.e., soils at 70% WFPS with a nitrogen atmosphere), suggesting a bacterial source for the N2O. In the presence of formate, nearly all of the N2O emitted was derived from the labeled 15NO3− pool (Fig. 3C and D). In the absence of formate, the majority of the N2O was derived from the unlabeled soil-N pool. Nitrogen-15 enrichment of the ammonium pool was not observed during the incubation period (data not shown).
FIG. 3.
SIRIN assay with 15N-labeled nitrate incubated at 70% WFPS for 24 h. Reported values are means with standard error bars (n = 3). (A and B) Total N2O emissions. An asterisk above the error bar denotes N2O emission from a treatment incubation with results that were significantly different from those of the control incubation (P < 0.1). (C and D) Percentage of emitted N2O attributable to denitrification or processes that oxidized 15NO3− to 15N2O.
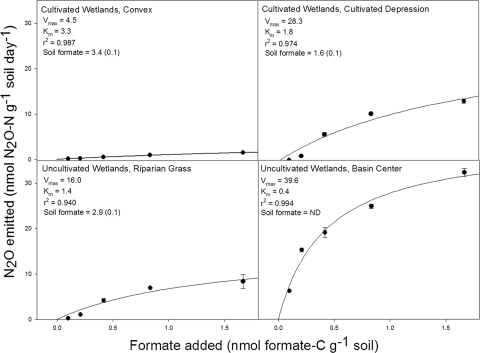
In general, N2O emissions increased as the amount of formate added to the soils increased (Fig. 4). Moreover, soil N2O emissions demonstrated a hyperbolic dependence on formate concentration. As such, Michaelis-Menten kinetics was used to describe the formate-dependent N2O emissions from all soils (r2 values ranged from 0.94 to 0.99). Soils from water-accumulating (CD and BC) landforms had greater affinity (lower Km values) for formate than soils from drier landforms (CX and RG). Not surprisingly, then, there was less extractable formate in soils from water-accumulating landforms than in soils from water-shedding or drier landforms.
FIG. 4.
N2O emitted as a function of added formate concentration. Closed circles indicate experimental means of N2O emitted with standard error bars (n = 3); the solid line is the modeled response. Units for Vmax are nanomoles N2O-N nanomoles formate-C−1 day−1. Km has the same units as the x axis. The r2 is the goodness of fit for the nonlinear regression line as determined by the Enzyme Kinetics Module for SigmaPlot 9.0 using the single-substrate (formate) model. Reported soil formate concentrations are means with the standard error in parentheses (nanomoles formate-C grams−1 soil). ND, not detected.
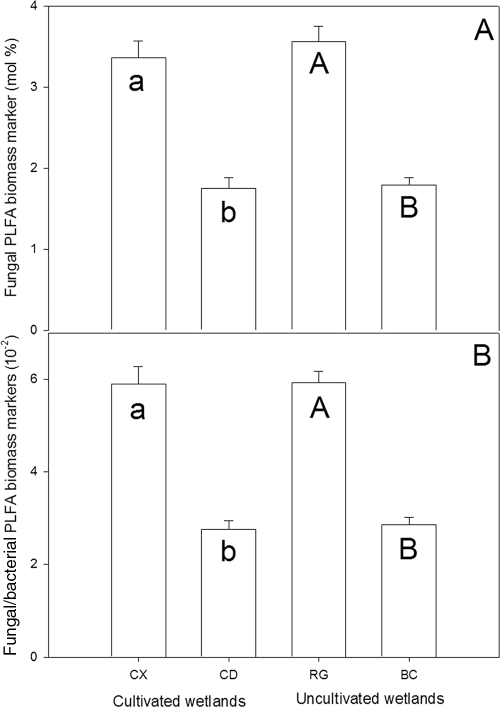
Nitrous oxide emissions attributable to fungal activity were greatest in the water-accumulating landforms (Fig. 2). However, the water-accumulating landforms yielded the least extractible fungal PLFA (Fig. 5A) and exhibited the lowest fungus/bacterium PLFA ratios (Fig. 5B).
FIG. 5.
Fungal biomass in soil as determined by PLFA extraction. Shown are the mol% of fungal PFLA biomass marker (A) and fungi/bacteria PLFA biomass marker (B). Reported values are means with standard error bars (n = 3). Different letters below error bars denote significant differences (α = 0.1); lowercase letters are used for comparisons between soils of cultivated wetlands, and uppercase letters are used for comparisons between soils of uncultivated wetlands.
DISCUSSION
The O2 and formate dependence of N2O emissions associated with fungi can be explained by the proposed fungal denitrification pathway (Fig. 1). To date, fungal nitric oxide reductase activity and the P450nor genotype have been reported for only a few fungi isolated from soil and wastewater reactors (52, 56, 58). In all cases, the expression of the P450nor gene was essential for the proposed fungal denitrification pathway. Thus, it was not surprising to find that fungal denitrification was not directly linked to fungal biomass. Rather, it is likely linked to the presence and expression of a specific functional genotype, P450nor.
The hyperbolic dependence of N2O emission on formate concentration, as modeled by Michaelis-Menten kinetics, supports our contention, outlined in Fig. 1, that a formate dehydrogenase/nitrate reductase couple and/or a formate-dependent nitrite reductase is directly linked to N2O emissions. The enzymatic characteristics of the formate-dependent N2O emissions differed between soils of water-accumulating landforms (CD and BC) and the soils from the corresponding water-shedding or drier landforms (CX and RG). For example, the higher affinity (lower Km) for formate in CD and BC soils would impart a competitive advantage to those fungi in soils with low soil formate concentrations. Also, the potential maximum formate-dependent N2O emission per unit formate-C (Vmax) was 2.5 to 6 times greater in soils from water-accumulating than from upland landforms. The reported enzymatic parameters and magnitude of the formate-dependent N2O emissions may be underestimated, because experimental additions of formate can encourage nitrogen immobilization by increasing the C/N ratio (29).
Formate stimulated nonfungal N2O emissions. In the bacterial pathway, formate mediates the reduction of nitrate to ammonia (nitrate ammonification) (8), with N2O being released as a result of the nonspecific action of dissimilatory nitrate reductase. Although energetically less favorable than denitrification (42), nitrate ammonification is reported to be the more important process in respiratory nitrate reduction under anaerobic conditions and when nitrate is limiting (44, 45). However, this did not appear to be the case; i.e., we did not observe any 15N enrichment of the ammonium pool as would have been expected. Alternatively, bacteria capable of mixotrophy (e.g., Paracoccus denitrificans) can denitrify nitrate using formate/H2 as an electron donor (38, 39).
Under hypoxic conditions, heterotrophic nitrification may also contribute to N2O release. Evidence suggests this is an important N transformation process for organic N and NH4+ in a variety of soil systems (11, 26, 35). Heterotrophic nitrification derives energy from organic C oxidation rather than NH3 oxidation (23), with N2O being a product of the incomplete oxidation of hydroxylamine. Castaldi and Smith (11) demonstrated that fungal heterotrophic nitrification can be the dominant N2O-emitting process in woodland and arable soils. In addition, heterotrophic nitrifiers produce N2O from denitrification of nitrification products (i.e., fungal nitrifier denitrification) (12). Fungal heterotrophic nitrification would produce primarily 14N2O. Under hypoxic conditions with formate, 15N2O dominated, whereas without formate, only 10% of the N2O was 15N2O. Thus, under hypoxic conditions, there may be two fungal pathways contributing to N2O release: fungal denitrification via P450nor and fungal heterotrophic nitrification. Our results suggest that the relative importance of these two processes is linked to soil formate concentrations.
Nitrous oxide emissions associated with fungi were landform, but not land use, dependent. It is often reported that fungal biomass is greater in native (uncultivated) soils than in adjacent cultivated soils and that fungal biomass increases with decreasing land use intensity (e.g., in no-tillage versus conventional-tillage soils) (13, 14, 17). Such was not the case in the present study; instead, differences in the fungal community appeared to be linked to landscape position (i.e., landform) through its influence on water redistribution. For example, fungal biomass (mycorrhizal and saprotrophic) has been shown to decline in soils that experience periods of flooding or high soil water content (30). This presumably reflects decreased mycorrhizal associations (33) or decreased saprotrophic activity under conditions of decreased O2 availability imposed during these periods.
We did not observe fungus-linked N2O emissions in the CX and RG landforms on the basis of our cycloheximide inhibition experiment. The cycloheximide concentrations were optimized for the CD and BC landforms and then applied to the CX and RG landforms. We hypothesized that since soils of CD and BC landforms have the highest levels of organic matter (Table 1), the levels of cycloheximide that inhibit fungi in these soils would be effective in the soils of water-shedding landforms, CX and RG. If this was incorrect, we may have underestimated the importance of fungus-linked N2O emissions in the water-shedding landform elements.
Technical limitations and questions associated with the specificity of the inhibitors and substrate (formate) dictate that future investigations will require novel ideas to validate the findings of this work. To validate the specific activity of the inhibitors, quantification of 18S and 16S rRNA gene fragments or transcripts (36) could shed light on how the 18S or 16S pool is affected by the different inhibitor treatments. Isolation and characterization of the P450nor and formate dehydrogenase enzymes by mass spectrometry (32, 47) from soil would provide credence to our hypothesis as shown in Fig. 1. Inoculation with and activity monitoring of fungi with wild-type and mutant genotypes of these two enzymes into soil may also support our arguments.
For the first time, soil N2O emissions related to fungi are linked to soil formate concentration and O2 availability. This finding poses interesting management considerations. First, the selection of crop varieties that exude smaller amounts of formate (9) could mitigate N2O production from fungi in areas where fungal denitrification is prevalent. Second, the benefits of improved soil fertility and plant growth through H2 release from nodules of legumes (15, 27) could be countered by increased N2O production from soil fungi because of formate production from H2 and CO2 (21).
Acknowledgments
We thank the Natural Science and Engineering Research Council of Canada, BIOCAP Canada Foundation, and Green Crop Network for funding this work.
Footnotes
Published ahead of print on 12 September 2008.
REFERENCES
- 1.Arah, J. R. M. 1997. Apportioning nitrous oxide fluxes between nitrification and denitrification using gas-phase mass spectrometry. Soil Biol. Biochem. 29:1295-1299. [Google Scholar]
- 2.Bååth, E. 2003. The use of neutral lipid fatty acids to indicate the physiological conditions of soil fungi. Microb. Ecol. 45:373-383. [DOI] [PubMed] [Google Scholar]
- 3.Badalucco, L., F. Pomare, S. Grego, L. Landi, and P. Nannipieri. 1994. Activity and degradation of streptomycin and cycloheximide in soil. Biol. Fertil. Soils 18:334-340. [Google Scholar]
- 4.Bardgett, R. D., and E. McAlister. 1999. The measurement of soil fungal:bacterial biomass ratios as an indicator of ecosystem self-regulation in temperate meadow grasslands. Biol. Fertil. Soils 29:282-290. [Google Scholar]
- 5.Baziramakenga, R., R. R. Simard, and G. D. Leroux. 1995. Determination of organic acids in soil extracts by ion chromatography. Soil Biol. Biochem. 27:349-356. [Google Scholar]
- 6.Bedard-Haughn, A., A. L. Matson, and D. J. Pennock. 2006. Land use effects on gross nitrogen mineralization, nitrification, and N2O emissions in ephemeral wetlands. Soil Biol. Biochem. 38:3398-3406. [Google Scholar]
- 7.Bedard-Haughn, A., K. W. Tate, and C. van Kessel. 2004. Using nitrogen-15 to quantify vegetative buffer effectiveness for sequestering nitrogen in runoff. J. Environ. Qual. 33:2252-2262. [DOI] [PubMed] [Google Scholar]
- 8.Berks, B. C., S. J. Ferguson, J. W. B. Moir, and D. J. Richardson. 1995. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta Bioenerg. 1232:97-173. [DOI] [PubMed] [Google Scholar]
- 9.Bolan, N. S., R. Naidu, S. Mahimairaja, and S. Baskaran. 1994. Influence of low-molecular-weight organic acids on the solubilization of phosphates. Biol. Fertil. Soils 18:311-319. [Google Scholar]
- 10.Bott, M. 1997. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch. Microbiol. 167:78-88. [PubMed] [Google Scholar]
- 11.Castaldi, S., and K. A. Smith. 1998. Effect of cycloheximide and N2O and NO3− production in a forest and an agricultural soil. Biol. Fertil. Soils 27:27-34. [Google Scholar]
- 12.Crenshaw, C. L., C. Lauber, R. L. Sinsabaugh, and L. K. Stavely. 2008. Fungal control of nitrous oxide production in semiarid grassland. Biogeochemistry 87:17-27. [Google Scholar]
- 13.de Vries, F. T., J. Bloem, N. van Eekeren, L. Brusaard, and E. Hoffland. 2007. Fungal biomass in pastures increases with age and reduced N input. Soil Biol. Biochem. 39:1620-1630. [Google Scholar]
- 14.de Vries, F. T., E. Hoffland, N. van Eekeren, L. Brussaard, and J. Bloem. 2006. Fungal/bacterial ratios in grasslands with contrasting nitrogen management. Soil Biol. Biochem. 38:2092-2103. [Google Scholar]
- 15.Dong, Z., L. Wu, B. Kettlewell, C. D. Caldwell, and D. B. Layzell. 2003. Hydrogen fertilization of soils: is this a benefit of legumes in rotation? Plant Cell Environ. 26:1875-1879. [Google Scholar]
- 16.Donnison, L. M., G. S. Griffith, and R. D. Bardgett. 2000. Determinants of fungal growth and activity in botanically diverse haymeadows: effects of litter type and fertilizer additions. Soil Biol. Biochem. 32:289-294. [Google Scholar]
- 17.Frey, S. D., E. T. Elliott, and K. Paustian. 1999. Bacterial and fungal abundance and biomass in conventional and no-tillage agroecosystems along two climatic gradients. Soil Biol. Biochem. 31:573-585. [Google Scholar]
- 18.Hayashi, M., G. Van der Kamp, and D. L. Rudolph. 1998. Water and solute transfer between a prairie wetland and adjacent uplands. 1. Water balance. J. Hydrol. 207:42-55. [Google Scholar]
- 19.Hogan, J. M., and F. M. Conly. 2002. St. Denis National Wildlife Area land cover classification: 1997. Technical Report Series, no. 384. Canadian Wildlife Service, Prairie and Northern Region, Saskatoon, Canada.
- 20.Högberg, M. N. 2006. Discrepancies between ergosterol and the phospholipid fatty acid 18:2ω6,9 as biomarkers for fungi in boreal forest soils. Soil Biol. Biochem. 38:3431-3435. [Google Scholar]
- 21.Horn, M. A., C. Matthies, K. Kusel, A. Schramm, and H. L. Drake. 2003. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 69:74-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, D. L. 1998. Organic acids in the rhizosphere: a critical review. Plant Soil 205:25-44. [Google Scholar]
- 23.Kuenen, J. G., and L. A. Robertson. 1994. Combined nitrification-denitrification processes. FEMS Microbiol. Rev. 15:109-117. [Google Scholar]
- 24.Kuwazaki, S., N. Takaya, A. Nakamura, and H. Shoun. 2003. Formate-forming fungal catabolic pathway to supply electrons to nitrate respiration. Biosci. Biotechnol. Biochem. 67:937-939. [DOI] [PubMed] [Google Scholar]
- 25.Laughlin, R. J., and R. J. Stevens. 2002. Evidence for fungal dominance of denitrification and codenitrification in a grassland soil. Soil Sci. Soc. Am. J. 66:1540-1548. [Google Scholar]
- 26.Laughlin, R. J., R. J. Stevens, C. Muller, and C. J. Watson. 2008. Evidence that fungi can oxidize NH4+ to NO3− in a grassland soil. Eur. J. Soil Sci. 59:285-291. [Google Scholar]
- 27.Maimaiti, J., Y. Zhang, J. Yang, Y. P. Cen, D. B. Layzell, M. Peoples, and Z. M. Dong. 2007. Isolation and characterization of hydrogen-oxidizing bacteria induced following exposure of soil to hydrogen gas and their impact on plant growth. Environ. Microbiol. 9:435-444. [DOI] [PubMed] [Google Scholar]
- 28.Maynard, D. G., Y. P. Kalra, and J. A. Crumbaugh. 2007. Nitrate and exchangeable ammonium nitrogen, p. 71-80. In M. R. Carter and E. G. Gregorich (ed.), Soil sampling and methods of analysis, 2nd ed. CRC Press, Boca Raton, FL.
- 29.McLain, J. E. T., and D. A. Martens. 2006. N2O production by heterotrophic N transformations in a semiarid soil. Appl. Soil. Ecol. 32:253-263. [Google Scholar]
- 30.Mentzer, J. L., R. M. Goodman, and T. C. Balser. 2006. Microbial response over time to hydrologic and fertilization treatments in a simulated wet prairie. Plant Soil 284:85-100. [Google Scholar]
- 31.Miller, J. J., D. F. Acton, and R. J. St. Arnaud. 1985. The effect of groundwater on soil formation in a morainal landscape in Saskatchewan. Can. J. Soil Sci. 65:293-307. [Google Scholar]
- 32.Nakahara, K., T. Tanimoto, K. Hatano, K. Usuda, and H. Shoun. 1993. Cytochrome P450-55a1 (P450dnir) acts as nitric oxide reductase employing NADH as the direct electron donor. J. Biol. Chem. 268:8350-8355. [PubMed] [Google Scholar]
- 33.Rickerl, D. H., F. O. Sancho, and S. Ananth. 1994. Vesicular-arbuscular endomycorrhizal colonization of wetland plants. J. Environ. Qual. 23:913-916. [DOI] [PubMed] [Google Scholar]
- 34.Rutherford, P. M., W. B. McGill, J. M. Arocena, and C. T. Figueiredo. 2007. Total nitrogen, p. 239-250. In M. R. Carter and E. G. Gregorich (ed.), Soil sampling and methods of analysis, 2nd ed. CRC Press, Boca Raton, FL.
- 35.Schimel, J. P., M. K. Firestone, and K. S. Killham. 1984. Identification of heterotrophic nitrification in a Sierran Forest soil. Appl. Environ. Microbiol. 48:802-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma, S., Z. Szele, R. Schilling, J. C. Munch, and M. Schloter. 2006. Influence of freeze-thaw stress on the structure and function of microbial communities and denitrifying populations in soil. Appl. Environ. Microbiol. 72:2148-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skjemstad, J. O., and J. A. Baldock. 2007. Total and organic carbon, p. 225-237. In M. R. Carter and E. G. Gregorich (ed.), Soil sampling and methods of analysis, 2nd ed. CRC Press, Boca Raton, FL.
- 38.Smith, R. L., M. L. Ceazan, and M. H. Brooks. 1994. Autotrophic, hydrogen-oxidizing, denitrifying bacteria in groundwater, potential agents for bioremediation of nitrate contamination. Appl. Environ. Microbiol. 60:1949-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, R. L., D. N. Miller, and M. H. Brooks. 2001. In situ stimulation of groundwater denitrification with formate to remediate nitrate contamination. Environ. Sci. Technol. 35:196-203. [DOI] [PubMed] [Google Scholar]
- 40.Stark, J. M., and S. C. Hart. 1996. Diffusion technique for preparing salt solutions, Kjeldahl digests, and persulfate digests for nitrogen-15 analysis. Soil Sci. Soc. Am. J. 60:1846-1855. [Google Scholar]
- 41.Stevens, R. J., R. J. Laughlin, L. C. Burns, J. R. M. Arah, and R. C. Hood. 1997. Measuring the contributions of nitrification and denitrification to the flux of nitrous oxide from soil. Soil Biol. Biochem. 29:139-151. [Google Scholar]
- 42.Strohm, T. O., B. Griffin, W. G. Zumft, and B. Schink. 2007. Growth yields in bacterial denitrification and nitrate ammonification. Appl. Environ. Microbiol. 73:1420-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swallow, K. W., and N. H. Low. 1994. Capillary zone electrophoretic analysis of the minor anions present in orange juice and orange pulpwash. J. Agric. Food Chem. 42:2808-2811. [Google Scholar]
- 44.Tiedje, J. M. 1994. Denitrifiers, p. 245-267. In R. W. Weaver, J. S. Angle, and P. J. Bottomley (ed.), Methods of soil analysis. Part 2. Microbiological and biochemical properties, vol. 5. Soil Science Society of America, Madison, WI. [Google Scholar]
- 45.Tiedje, J. M. 1988. Ecology of denitrification and dissimilatory nitrate reduction to ammonium, p. 179-244. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. John Wiley and Sons, Inc., Toronto, Canada.
- 46.Topp, G. C., and P. A. Ferré. 2002. Water content, p. 417-545. In J. Dane and C. Topp (ed.), Methods of soil analysis. Part 1. Physical and mineralogical properties. Soil Science Society of America, Madison, WI.
- 47.Uchimura, H., H. Enjoji, T. Seki, A. Taguchi, N. Takaya, and H. Shoun. 2002. Nitrate reductase-formate dehydrogenase couple involved in the fungal denitrification by Fusarium oxysporum. J. Biochem. 131:579-586. [DOI] [PubMed] [Google Scholar]
- 48.van der Wal, A., J. A. van Veen, W. Smant, H. T. S. Boschker, J. Bloem, P. Kardol, W. H. van der Putten, and W. de Boer. 2006. Fungal biomass development in a chronosequence of land abandonment. Soil Biol. Biochem. 38:51-60. [Google Scholar]
- 49.van Hees, P. A. W., A.-M. T. Andersson, and U. S. Lundstrom. 1996. Separation of organic low molecular weight aluminium complexes in soil solution by liquid chromatography. Chemosphere 33:1951-1966. [Google Scholar]
- 50.van Hees, P. A. W., D. L. Jones, and D. L. Godbold. 2002. Biodegradation of low molecular weight organic acids in coniferous forest podzolic soils. Soil Biol. Biochem. 34:1261-1272. [Google Scholar]
- 51.van Hees, P. A. W., D. L. Jones, G. Jentschke, and D. L. Godbold. 2005. Organic acid concentrations in soil solution: effects of young coniferous trees and ectomycorrhizal fungi. Soil Biol. Biochem. 37:771-776. [Google Scholar]
- 52.Watsuji, T., N. Takaya, A. Nakamura, and H. Shoun. 2003. Denitrification of nitrate by the fungus Cylindrocarpon tonkinense. Biosci. Biotechnol. Biochem. 67:1115-1120. [DOI] [PubMed] [Google Scholar]
- 53.White, D. C., W. M. Davis, J. S. Nickels, J. D. King, and R. J. Bobbie. 1979. Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia 40:51-62. [DOI] [PubMed] [Google Scholar]
- 54.Yates, T. T., B. C. Si, R. E. Farrell, and D. J. Pennock. 2006. Probability distribution and spatial dependence of nitrous oxide emission: temporal change in hummocky terrain. Soil Sci. Soc. Am. J. 70:753-762. [Google Scholar]
- 55.Yates, T. T., B. C. Si, R. E. Farrell, and D. J. Pennock. 2006. Wavelet spectra of nitrous oxide emission from hummocky terrain during spring snowmelt. Soil Sci. Soc. Am. J. 70:1110-1120. [Google Scholar]
- 56.Zhang, L., N. Takaya, T. Kitazume, T. Kondo, and H. Shoun. 2001. Purification and cDNA cloning of nitric oxide reductase cytochrome P450nor (CYP55A4) from Trichosporon cutaneum. Eur. J. Biochem. 268:3198-3204. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, Z. M., N. Takaya, A. Nakamura, M. Yamaguchi, K. Takeo, and H. Shoun. 2002. Ammonia fermentation, a novel anoxic metabolism of nitrate by fungi. J. Biol. Chem. 277:1892-1896. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, Z. M., N. Takaya, M. A. C. Sakairi, and H. Shoun. 2001. Oxygen requirement for denitrification by the fungus Fusarium oxysporum. Arch. Microbiol. 175:19-25. [DOI] [PubMed] [Google Scholar]
- 59.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]