Abstract
The Ssk1p response regulator of Candida albicans is required for oxidant adaptation, survival in human neutrophils, and virulence in a disseminated murine model of candidiasis. The activities of the Ssk1p are signaled through the HOG1 MAP kinase pathway (HOG=hyperosmotic glycerol). Further, we have previously shown that the amino acid residues D556 and D513 of the Ssk1p receiver domain are critical to the activities of the Ssk1p in oxidant stress adaptation and morphogenesis. Herein, we use transcriptional profiling to explain the oxidant sensitivity and defect in morphogenesis of two point mutants (D556N and D513K, respectively) compared to a WT strain. In the D556N mutant compared to WT cells during oxidative stress (5mM H2O2), a down regulation of genes associated with redox homeostasis and oxidative stress occurred that accounted for about 5% of all gene changes, including among others, SOD1 (superoxide dismutase), CAP1 (transcription factor required for some forms of oxidant adaptation), and three genes encoding glutathione biosynthesis proteins (GLR1, GSH1, and GSH2). Mutant SSK26 (D513K) was not sensitive to peroxide but was impaired in its yeast to hyphal transition. We noted down regulation of genes associated with morphogenesis and cell elongation. Virulence of each mutant was also evaluated in a rat vaginitis model of candidiasis. Clearance of the SSK1 null mutant (strain SSK21) and SSK25 from the vaginal canal was significantly greater than wild type (CAF2-1) or the D513K mutant (D513K), indicating that a specific change in a single amino acid of the Ssk1p may alter the ability of this strain to colonize the rat vaginal mucosa.
Keywords: response regulator, two-component proteins, point mutants
Introduction
Signal transduction pathways contribute to many cell functions including adaptation to external and internal stress conditions. Environmental stress such as high oxidants, either self-generated, from microbial competitors, or during phagocytosis by neutrophils or macrophages, requires perception of that signal by cells and relay via phosphotransfer to MAPK pathways. Subsequently, gene transcription occurs such that cells can adapt to oxidants. This paradigm has been suggested based upon mutant data in, for example, C. albicans, where strains lacking key phosphotransfer proteins are unable to adapt and consequently are more sensitive to specific oxidants or unable to undergo morphogenesis (yeast → hyphal transition). The role of anti-oxidant encoding genes in virulence has been established to further substantiate their importance. In Candida albicans, the HOG1 MAPK pathway and upstream two component signaling proteins such as Ssk1p are critical to oxidant adaptation and virulence (Alonso-Monge et al., 2003, 1999; Arana et al., 2005; Chauhan et al., 2006, 2003; Enjalbert et al., 2006; 1999; San Jose et al., 1996; Smith et al., 2004). The HOG1 pathway of Saccharomyces cerevisiae is critical to osmoadaptation (reviewed in Hohmann, 2002; Maeda et al., 1994; Posas et al., 1998, 1996).
The Ssk1p, a key phosphotransfer upstream protein in the HOG MAPK pathway of C. albicans, has previously been shown to provide an oxidant adaptation function (Chauhan, et al., 2003). A null mutant lacking Ssk1p was oxidant sensitive, more easily killed by human neutrophils than a parental, WT strain of C. albicans, and avirulent in a mouse model of hematogenously disseminated candidiasis (Chen et al., 2005; Calera et al., 2000; Calera et al., 1999). During peroxide stress in the ssk1 deletion mutant (strain SSK21), the downstream Hog1p MAPK was unphosphorylated in vitro (Chauhan et al., 2003). However, the same mutant exhibited a minor sensitivity to osmotic shock with NaCl and in this stress condition, Hog1p was phosphorylated, indicating that Ssk1p is required for down stream signaling to Hog1 in oxidant but is not critical to adaptation during osmotic stress. In addition to C. albicans, the Hog1 homologue (saKa) of Aspergillus fumigatus has also been shown to provide oxidant and nutrient adaptation functions (Chen et al., 2006; Xue et al., 2004). In Cryptococcus neoformans, processes such as stress adaptation and sexual reproduction require Hog1p, some of which is regulated by Ssk1p, although Skn7p can also provide stress adaptation in C. neoformans and C. albicans (Bahn et al., 2006; Singh et al., 2004; Wormley et al, 2005). Oxidant adaptation in fungi including human pathogenic fungi is described in Chauhan et al., 2006 and Moyer-Rowley, 2003.
Structure-function studies of the C. albicans Ssk1p response regulator protein have identified key residues within the receiver domain of this protein that are required for phosphotransfer from Ypd1p, the up stream histidine phosphotransfer (Hpt) intermediate protein (Menon et al., 2006; Calera et al., 1999). Ypd1p-Ssk1p phosphotransfer has been studied extensively in S. cerevisiae (Janiak-Spens et al., 1999; Porter et al., 2005, 2003; Stock and West, 2002; Thompson and Kay, 2000; West and Stock, 2001). In C. albicans, the D556 of the response regulator (receiver) domain, which is thought to be the site of phosphotransfer from the histidine phosphotransfer protein Ypd1p, corresponds to the D554 of Saccharomyces cerevisiae (Hohmann, 2002). In C. albicans, mutation of D556 (D556N) resulted in peroxide sensitivity and non-translocation of Hog1p to the nucleus even though the Hog1p is phosphorylated (Menon et al., 2006). In comparison, Hog1p phosphorylation and its nuclear translocation is unaffected in the D556N mutant (SSK25) when cells are stressed in 1.5 M NaCl.
The D513 residue of the response regulator domain is believed to be required for stabilization of the protein during phosphotransfer from Ypd1p. However, in a D513K point mutant (SSK26), peroxide resistance was similar to WT cells but this strain was unable to convert to hyphae in normally inducing conditions (Menon et al., 2006). To examine these mutants for transcript changes, we performed microarray analysis in YPD medium with or without hydrogen peroxide (WT and D556N) or in M199 medium (WT and D513K) at 37°C and compared their transcriptional profiles. Further, we evaluated the consequence of these point mutations in virulence by comparing strains in a rat model of vaginitis.
Materials and methods
Strains and culture conditions
Candida albicans strains used in this study are described in Table 1. Inocula for all experiments were prepared in YPD (1% yeast extract, 2% peptone and 2% glucose) at 30°C with shaking at 200 rpm, cells were washed, and adjusted to a specific cell number in YPD broth (see below) with/without 5mM H2O2 or M199 for microarray experiments. For the oxidative stress experiments, we used strains CAF2-1 (WT) and SSK25 (D556N) since the latter but not SSK26 was hypersensitive to peroxide stress. Wt and SSK25 were grown overnight in YPD broth at 30°C. Overnight cultures were than washed, and diluted to an OD600 of 0.1 in 500 ml of fresh YPD broth medium and grown at 30°C until cells reached mid-log phase (OD600 ~0.5). The mid-log phase cells of both CAF2-1(WT) & SSK25 (D556N) were then stressed with 5mM H2O2 for 10 min. At 0 and 10 min post-treatment, cultures were harvested by centrifugation at 3000 × g for 10 min, cells were washed, and the cell pellet was rapidly frozen prior to RNA extraction for microarray experiments. For the microarrays with strains CAF2-1 (WT) and the SSK26 (D513K), cells were grown overnight in YPD, washed, diluted to an appropriate inoculum, added to 500-ml of M199 (pH 7.0), and harvested immediately (0 time) or grown at 37°C for 3 hr in liquid Medium 199 (pH 7.0), a time in which germination approached 90%. After 3h, cultures were harvested by centrifugation at 3000 × g for 10 min, washed, and the cell pellet thus obtained was quick frozen and stored at −80°C for RNA extraction.
Table 1.
Strains of C .albicans used in this study
| Strains | Genotype | Reference |
|---|---|---|
| CAF2-1 | URA3/ura3Δ∷imm434 | Fonzi & Irwin, 1998 |
| SSK21 | ssk1:hisG-URA3-hisG/ssk1 ∷hisG ura3Δ∷imm434/ura3Δ∷imm434 | Calera et al., 2000 |
| SSK25 | ssk1∷hisG/SSK1 (D556N)-URA3 ura3Δ ∷imm434/ura3Δ∷imm434 | Menon et al., 2006 |
| SSK26 | ssk1∷hisG/SSK1 (D513K)-URA3 ura3Δ ∷imm434/ura3Δ∷imm434 | Menon et al., 2006 |
Microarray analysis
Total RNA for microarray analysis was extracted with the hot phenol method (Chauhan et al., 2003). Samples were stored at −80°C. DNA microarrays were fabricated at the University of Wisconsin Biotechnology Center by spotting C. albicans specific ~75 mer oligonucleotides on slides obtained from Qiagen (Valencia, CA) which correspond to roughly 6000 open reading frames (ORFs). Twenty micrograms of total RNA from each sample was used for microarray analysis. cDNA synthesis, and labeling of samples with Cy3 and Cy5 dyes was performed according to the SuperScript Indirect cDNA Labeling kit (Invitrogen). The Cy3 and the Cy5 labeled samples were hybridized to the array in 25% formamide, 5X SSC, and 0.1% SDS at 42°C overnight (~16 h). Following hybridization, the microarrays were washed at room temperature with the following (2 min each): 2X SSC + 0.1 % SDS, 1X SSC, and 0.2X SSC. The microarrays were then immediately dried and scanned with a GenePix 4000 microarray scanner (Molecular Devices). Two arrays per strain/RNA/condition were run to assess the consistency of data (i.e., technical replicates). For microarray data analysis, the TIGR microarray software suite (http://www.tm4.org/) was used. Experimental data from each hybridization was normalized using a LOWESS analysis and standard-deviation regulation, so that each gene in an array received a normalized expression value. Finally, the normalized data was imported into TIGR Mev (version 3.03), and it was further normalized using total intensity normalization and processed using low intensity cutoff filter, low percentage cutoff and variance data filters. This data was then analyzed by using Self Organizing Tree Algorithm (SOTA) to identify genes that are differentially expressed in mutant over WT strain. We used a >2-fold increase and a 0.5-fold decrease in both mutants compared to WT CAF2-1 strains of C. albicans samples to identify up- or down regulated genes.
RT-PCR
For verification of microarray data, RT-PCR was performed with strains CAF2-1, SSK25 (D556N), and SSK26 (D513K) (Table 1) using conditions described above, i.e., SSK25 with or without peroxide, and SSK26 (cultures were compared in YPD and M199). In brief, cultures were grown in 5-ml of YPD at 30°C overnight. These cultures were then used to inoculate 50-ml of YPD in order to obtain an initial optical density (OD600) between 0.1 and 0.15. All strains were then grown at 30°C until they reached mid log phase (OD600 ~0.5), cells were harvested by centrifugation, and total RNA was extracted using the hot phenol method (Chauhan et al., 2003). RT-PCR was performed using 1 µg of total RNA with primer sets for genes whose transcription was altered in each mutant (Table 2). Samples were processed by the one-step RT-PCR kit according to the manufacturer’s instructions (Qiagen, Valencia, CA.). 10 µl of each PCR amplification reaction was separated on a 1% agarose gel and stained with ethidum bromide. ACT1 served as an internal loading control.
Table 2.
Primer sets for RT-PCR reactions.
| Gene | Sequence (5’ – 3’) |
|---|---|
| ECE1-F | GCTTTATCTTCTCAAGCTGCC |
| ECE1-R | GAAAGACTTTGTTAAACTCGTCG |
| SPT3-F | GTTGGTGTAGGAGCCG |
| SPT3-R | GCCATATCATATAGAG |
| RFG-F | CTTCCTAATAATAAATC |
| RFG-R | GATTATAGTCTCAATG |
| HAC1-F | CATGTTGAAAAACAAGAA |
| HAC1-R | GAATCATTTCAGTTGG |
| NRG1-F | AATTGACGGTGGTTGCACGTTGTC |
| NRG1-R | CATGTTGGCCATGGACATTGGTGT |
| EFG1-F | ACAACCAGGTCAACAGACTGGACA |
| EFG1-R | ACCAGACACATTACTGCCACCACT |
| CPH1-F | ACACAGAAGAGGATCAACGACGCA |
| CPH1-R | AGCAGTAGTAGCAACTGCACCAGA |
| TUP1-F | AGTGTTCAATGTCACCACCGGAGA |
| TUP1-R | CTTGCCTTCCAAGTGCCACAACTT |
| RBF1-F | GCGCAAGCTCAAAGAGAACACCAA |
| RBF1-R | TGCTGTTGCAGTTGTTGACTCTGG |
| HYR1-F | GGTGGAATTCAAGGTTTCCATG |
| HYR1-R | GTCGTAAAAACTCTGGTGCGTC |
| MIG-F | CAGCAACAATATTACCAAC |
| MIG-R | CAACAATCAACAACAACAG |
| RIM101-F | GGATACTACTCAAACAAC |
| RIM101-R | GCACAAGCTCCACAGC |
| HAP5-F | GACGACCAGCTTCCGCAG |
| HAP5-R | CATGCAGGAGGACATTAACTGC |
| MSB1-F | GCTGGTATTATGGACCCAGC |
| MSB1-R | CACCAACTATGTGTTTGTGTCC |
| GCN4-F | TATTGGAGTCGGGTTTCAGCACCA |
| GCN4-R | GCAACAACAACTTCACCACCAGCA |
| HWP1-F | ACCCACAACAACAACCACAAGAGC |
| HWP1-R | TGTTTCAGTGGCTGGAGCAGTAGA |
| FKH2-F | GCTCCAGAGGAGAAAACCAATG |
| FKH2-R | CATACGCATTACTAGTCGTTAGG |
| TSA1-F | GGCTCCAGTCGTTCAACAAC |
| TSA1-R | GATCATATTTAGTCCGAGCTTAG |
| TRR1-F | GCTGCTGGTGGTCAATTGAC |
| TRR1-R | CATCTTCGACAAGGACCTCAC |
| GSH2-F | GTTATGTACCCACCCAACTTTG |
| GSH2-R | GGAGATTATCGTTGGATCGTCG |
| SOD1-F | GGTTAAAGCTGTCGCTGTTG |
| SOD1R | CTACCGTTAGAACCGAGTAG |
| CAP1-F | GTTTCTACCGGTACACCTC |
| CAP1-R | GACAACTTTGATCTTGC |
| GSH1-F | GAGTTTATCAGCGGCAGCACC |
| GSH1-R | GAGAGGTTGGACCAAATCATC |
| ARH1-F | GATACGGGGTAGCACCAGATC |
| ARH1-R | GTTGAAACACCCCTCGATTTCC |
| GLR1-F | CCTCTGCAAGAAGAGCTGCCAAAT |
| GLR1-R | AATGTAACCAGCACCGACAATGGC |
| TPS3-F | ACCCGCAGCTGCTATTCCAGATAA |
| TPS3-R | TGTCGCGAACCATCTTGGGAACTA |
| HSP30-F | GCTTGGTTCTTGGGTTGGCCATTA |
| HSP30-R | AACACCAGTGTCTGGAACACCTGA |
Rat vaginitis model
The virulence of strains CAF2-1 (SSK1/SSK1), SSK21 (ssk1/ssk1), SSK25 (ssk1/SSK1 D556N), and SSK26 (ssk1/SSK1 D513K) in a rat model of experimental vaginal candidiasis was evaluated according to the protocol described by De Bernardis et al., 2002. Each C. albicans strain was grown in YPD medium at 28°C overnight. Cells were harvested by centrifugation, washed twice in calcium-and magnesium-free phosphate-buffered saline (PBS; Gibco-BRL) and suspended to a required density on the basis of hemocytometer counts prior to use. Ovariectomized, female, Wistar rats (80 to 100 g; Charles River, Wilmington, MA.) were injected subcutaneously with 0.5mg of estradiol benzoate (Benzatrone; Samil, Rome, Italy). Six days after the first estradiol treatment, groups of five rats were inoculated intravaginally with 107 yeast cells per 0.1 ml of each strain tested. The inoculum was dispensed into the vaginal cavity through a syringe equipped with a multipurpose calibrated tip (Combitip; PBI, Milan, Italy). Enumeration of C. albicans in the vaginal cavity was achieved by culturing 0.1-ml samples (using a calibrated plastic loop, Disponoi; PBI), obtained from each animal at designated times from 0 – 28 days, on Sabouraud agar containing chloramphenicol (50µg/ml). Cultures were incubated at 28°C for 48 to 72 h. For each time point per strain, a total of four Saboraud agar plates were inoculated and the average cfu/ml of vaginal fluid was obtained. Experiments were repeated at least one additional time.
Results
Microarray analysis of genes down regulated during oxidative stress in SSK25
For these experiments, strain SSK25 was compared to WT since we have previously reported its sensitivity to oxidants such as hydrogen peroxide (Menon et al., 2006). Cells of WT and SSK25 (D556N) were grown under identical conditions, i.e., YPD at 30°C with or without 5mM H2O2 for 10 min and their transcriptional profiles compared. The effect of oxidative stress on the transcription of WT and strain SSK25 resulted in a change of approximately 256 transcripts (both up and down regulated genes). The three largest groups of transcriptionally altered genes in peroxide-treated SSK25 (D556N) either encode proteins related to cell growth and maintenance (35%), energy metabolism/pathways (20%), or unknown functions (20%) (Supplementary data, S1).
Of all transcriptional changes, we chose to focus upon the family of redox homeostasis & oxidative stress response genes, since the SSK25 mutant was peroxide sensitive (Menon et al, 2006). Of those, we observed 11 genes assigned an anti-oxidant function that were down-regulated to the greatest extent (see methods) in SSK25 compared to the WT cells (about 4.3% of the total changes) In Table 3, microarray data are expressed as a differential expression ratio (SSK25/WT) in the presence of peroxide. Adaptation to oxidative stress requires the induction of genes that play a role in detoxification of the reactive oxygen intermediates (ROI), like superoxide, peroxides, and hydroxide radicals. To verify the change in transcription of oxidative stress genes listed in Table 3, we performed RT-PCR of those genes. Each of the down regulated genes listed in Table 3 as determined by micorarray also was down regulated in strain SSK25 as determined by RT-PCR compared to wt cells (Figure 1). In this regard, the down regulation was either greater for peroxide stress (GSH1, ARH1, GSH2, TSA1 and CAP1) or occurred about equally in strain SSK25 with or without peroxide stress, for example FKH2, GLR1, TRR1, and SOD1. Two significantly up regulated genes were also identified by microarray analysis (Table 3) (HSP30, TPS3), but their regulation was verified by RT-PCR reactions only for HSP30 (Figure 1). The decreased transcription of the genes that are involved in oxidative stress adaptation in strain SSK25 may explain in part the oxidant sensitive phenotype of the mutant compared to the strain CAF2-1.
Table 3.
Genes that encode redox potential or anti-oxidant proteins that are differentially expressed in strain D556N Ssk1p response regulator by microarray analysis are listed.
| Gene name | Description | Differential expression ratio1 |
|---|---|---|
| GSH1 | Gamma-glutamylcysteine | 0.18 |
| GLR1 | Glutathione reductase, Cap1-induced | 0.16 |
| GSH2 | Glutathione synthase | 0.30 |
| TSA1 | Alkyl hydroperoxide peroxidase | 0.24 |
| TRR1 | Thioredoxin reductase | 0.23 |
| SOD1 | Cu,ZN-superoxide dismutase | 0.28 |
| ARH1 | Adrenodoxin reductase and ferredoxin-NADP+ reductase | 0.18 |
| FKH2 | Forhkead transcription factor | 0.21 |
| CAP1 | Transcription factor, oxidative stress | 0.50 |
| HSP30 | Heat shock protein, response to stress | 5.10 |
| TPS3 | trehalose-phosphate synthase, regulatory subunit | 2.90 |
Differential expression is calculated as the following ratio: D556N/WT under conditions of peroxide stress (YPD + 5mM H2O2).
Figure 1.
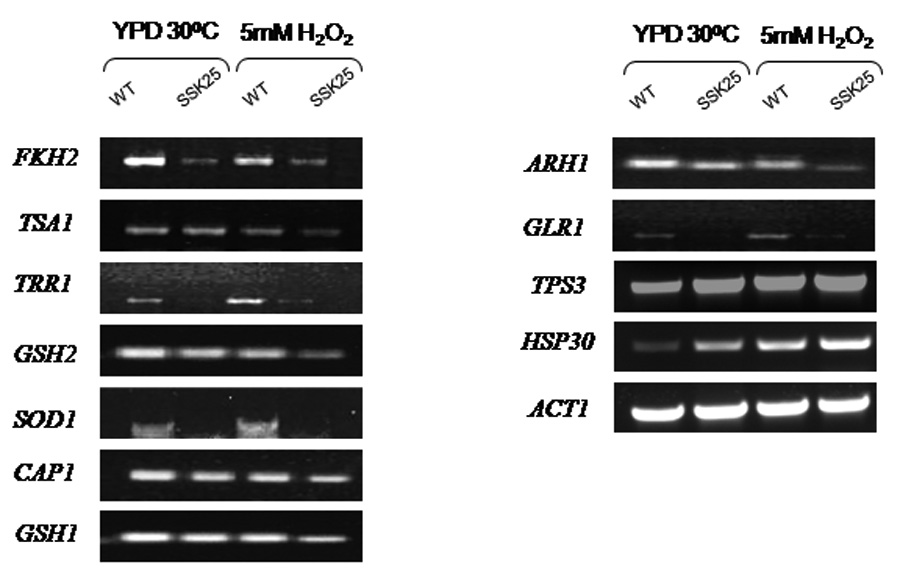
RT-PCR of SSK25 genes that are down regulated during peroxide stress (YPD + 5mM H2O2) compared to WT cells. The genes represented were chosen for analysis based upon microarray data of only those associated with redox homeostasis and oxidative stress response. CAF2-1 and D556N were grown in YPD + peroxide as described in the text. RT-PCR reactions are shown for both strains with and without H2O2. ACT1 was used as a loading control
Other oxidative stress profiling studies in C. albicans have revealed induction of genes that function in the adaptation to reactive oxygen species (Smith et al., 2004; Enjalbert et al., 2006). In these studies, most of the core functions required for tolerance to peroxide stress are involved in vacuolar protein sorting, cell wall biosynthesis and maintenance, transcription and protein synthesis. Transcription of genes encoding proteins involved in mitochondrial function, ubiquitin processing, and the pentose phosphate pathway were also up regulated in response to oxidative stress.
Microarray analysis of genes down regulated in SSK26 (D513K) grown in the hyphal inducing medium M199
We also compared the transcriptional profiles of strains SSK26 and CAF2-1. In these experiments, strains were grown in Medium 199 (37°C) because of the constitutive yeast morphology of strain SSK26 compared to the filamentous growth of CAF2-1. Profiling of wild type and SSK26 (D513K) under non-inducing and hyphal-inducing conditions revealed that a total of 283 genes were affected, a majority of which were up regulated in the wild type strain under these hyphal inducing conditions (data not shown). In this case, those genes that encode functions related to cellular metabolism and cell maintenance displayed the largest number of transcriptional changes (40%) (Supplementary data, S2). Since strain SSK26 is defective in its conversion of yeast to hyphae, we focused on the gene group associated with among other things, cell elongation, and cytoskeleton functions (5% of total change). The following genes were down regulated in SSK26 compared to WT cells: HYR1, HWP1, ECE1, MIG1, GCN4, RFG1 (ROX1), MSB1 (cell polarity), RBF1, RIM101, HAC1, HAP5, TUP1, NRG1, EFG1, and CPH1 (Table 4). Most of these genes either are required for or up regulated during hyphal growth of the organism (Kumamoto & Vinces, 2005). Again, RT-PCR was done to verify microarray data (Figure 2). As with strain SSK25, transcriptional changes in SSK26 either were specific for conditions (in this experiment) inducing hypha formation (M199 pH 7.0) or occurred regardless of whether cells were induced (M-199 medium) or not induced to form hyphae (YPD medium). Thus genes NRG1, EGF1, CPH1, TUP1, RBF1, HYR1, MIG1, GCN4, HAC1, RIM101, and HWP1 were all down regulated in both growth conditions (Figure 2). Genes down regulated in M-199 medium more than YPD included, ECE1, SPT3, HAP5, and MSB1 (Figure 2). Surprisingly, genes encoding yeast transcription factors or suppressors of hyphal growth (TUP1, MIG1, RBF1, and NRG1) were also down regulated in strain SSK26 even though SSK26 constitutively produces yeast growth (Table 4, and Figure 2).
Table 4.
Microarray analysis of genes that encode morphogenesis functions that are differentially expressed in the Ssk1p response regulator D513K point mutant, strain SSK26.
| Gene name | Description | Differential expression ratio |
|---|---|---|
| HYR1 | Hyphal regulated protein | 0.331 |
| HWP1 | Hyphal cell wall protein | 0.37 |
| ECE1 | Cell elongation protein | 0.31 |
| MSB1 | Cell polarity-related protein | 0.28 |
| MIG1 | Transcriptional hyphal suppressor | 0.15 |
| HAC1 | Transcription factor | 0.5 |
| TUP1 | Repressor of hyphal formation | 0.14 |
| RFG1 | Hyphal regulator | 0.4 |
| GCN4 | Transcriptional activator, filamentous growth | 0.22 |
| RIM101 | Alkaline induction of filamentation | 0.27 |
| HAP5 | Transcription, morphogenesis | 0.29 |
| RBF1 | Transcription factor | 0.18 |
| NRG1 | Transcriptional repressor, hyphal growth | 0.17 |
| EFG1 | Transcription factor, hyphal growth | 0.19 |
| CPH1 | Transcription factor, hyphal growth | 0.2 |
As in Table 3, data are expressed as D513K/WTt in Medium199, 37°C.
Figure 2.
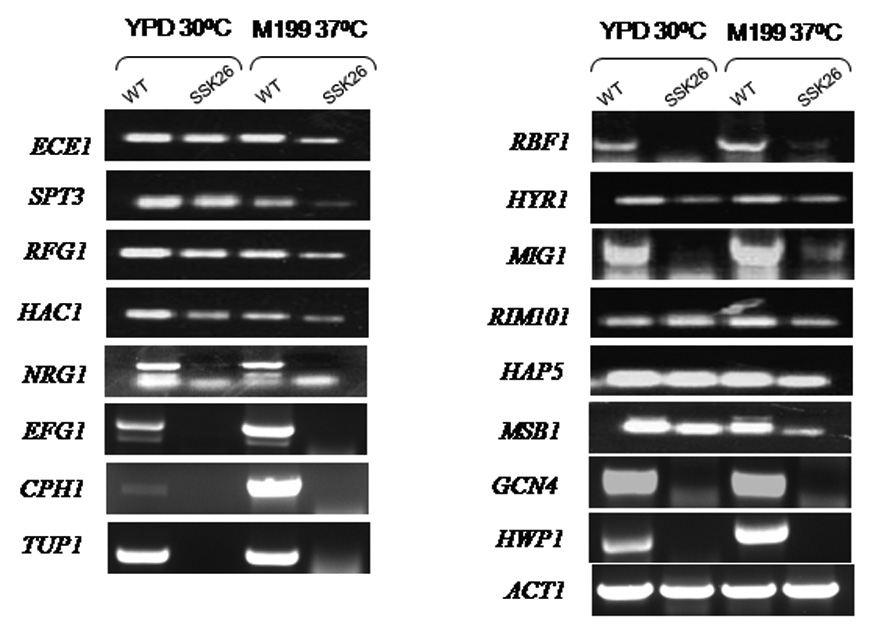
RT-PCR of genes from SSK26 (D513K) that are down regulated compared to WT during growth in YPD or M-199 medium. The genes represented were chosen for analysis based upon microarray data of only those associated with cell cycle, DNA replication and cell elongation and include reactions for both strains grown in YPD or M199 medium. ACT1 was used as a loading control
Virulence in a rat model of candida vaginitis
The virulence of each point mutant as well as strain SSK21 (ssk1/ssk1) and the wild type strain CAF2-1 was evaluated in a rat model of candida vaginitis (Table 5). This model measures persistence on the vaginal mucosa as a disease index. Our data indicate that Ssk1p is required for optimal colonization of the rat vaginal mucosa and that maximal virulence also requires the D556 residue in the receiver domain. It would appear that mutation to D513 (D513K), even though a non-conservative change, has little effect on virulence. The reduced vaginal colonization by strain SSK21 may be related to the down regulation of the host recognition protein Als1p (agglutinin-like sequences) in this strain (Chauhan et al., 2003; Hoyer, 2001). Strain SSK21 (ssk1/ssk1) is also less adherent to human esophageal tissue in vitro and that phenotype may be due to the same transcriptional change just described (Li et al., 2002; Bernhardt, et al., 2001).
Table 5.
Vaginal colonization by strains of C. albicans (CAF2-1, wt; SSK21,ssk1/ssk1; SSK25, D556N; and SSK26, D513K). Rats were infected intravaginally with 107 yeast cells. Vaginal samples were taken at the times indicated, aliquots plated on Sabouraud agar medium containing chloramphenicol, and cfu per ml determined.
| CFU/ml VAGINAL FLUID × 103 | ||||
|---|---|---|---|---|
| DAYS | CAF2-1 | SSK21 | SSK25 | SSK26 |
| 0 | >100 | >100 | >100 | >100 |
| 1 | >100 | 60±2 | 50.5 ± 1.8 | 75.8 ± 1.8 |
| 2 | 95 ± 3.1 | 58±2.4 | 43.2 ± 2.6 | 67.6 ±1.4 |
| 5 | 77 ± 2.5 | 46±2 | 41.4 ± 2.5 | 60.2 ± 1.3 |
| 7 | 60.7 ± 1.1 | 40±1.5 | 35.6 ± 1.8 | 53.6 ± 1.2 |
| 14 | 38.5 ± 1.1 | 19±2.2 | 11.2 ± 0.8 | 28.8 ± 2.8 |
| 21 | 19.6 ± 1.2 | 12±1.7 | 4.8 ± 1.4 | 12.8 ±1.2 |
| 28 | 0 | 0 | 0 | 1.2 ± 0.6 |
Discussion
The Hpt (histidine phosphotransfer protein) Ypd1p of fungi such as C. albicans (Calera et al., 2000) and S. cerevisiae (Hohmann, 2000) is phosphorylated by the Sln1p membrane-associated histidine kinase. The Ssk1p response regulator protein in turn catalyzes and is the substrate for phosphate transfer from Ypd1p during unstressed conditions such that phosphorylated Ssk1p is unable to activate the Ssk2/22 MAPKKK of the HOG1 MAPK pathway. In oxidatively- or osmotically-stressed cells, phosphotransfer does not occur among these proteins and unphosphorylated Ssk1p in turn now activates the HOG1 MAPK via the Ssk2/22 (MAPKKK) and Pbs2p (MAPKK) proteins resulting in cell adaptation to stress (Arana et al., 2005; Walia and Calderone, 2008).
Phosphotransfer from Ypd1p requires the D556 amino acid residue of the Ssk1p receiver domain. Paradoxically, since the Ssk1p of the D556N mutant is not phosphorylated, then that mutant should behave similarly to peroxide resistant wild type cells at comparable concentrations of the inhibitor. However, the D556N mutant is oxidant sensitive (Menon et al, 2006). Our explanation of this observation relates to the fact that for adaptation to oxidant stress to occur, phosphorylated Hog1p (via Ssk1p activation of the HOG1 MAPK pathway) must translocate to the nucleus for transcriptional regulation of stress genes. We have previously shown that during peroxide stress, in fact while Hog1p is phosphorylated, it is not translocated to the nucleus; hence, the D556N mutant is oxidant sensitive. Also, it would appear that the D556 residue, in addition to its phosphorylation role, also is either directly or indirectly required for Hog1p translocation. Interestingly, the D556N mutant is not osmosensitive, but Hog1p is both phosphorylated and translocated to the nucleus (Menon et al, 2006). In this case, we have previously shown in C. albicans that the Ssk1p-driven Hog1 MAPK pathway is not critical to osmoadaptation (Chauhan, et al, 2003). Based upon this observation, we speculate that the Sho1p–Ypd1p-Pbs1p-Hog1p driven pathway (or another) appears to be more important to provide osmoadaptation.
In E. coli, the D13 residue of the CheY RR protein is critical to the phosphorylation of D57 (D556 in C. albicans) and forms part of a divalent metal binding site that is required for flagella rotation and chemotaxis (Stock et al, 1985). Because of the critical role this residue plays in phosphotransfer among RR proteins, we also examined its function in the Ssk1p of C. albicans (Menon et al, 2006). We found that the D513K mutant like wt cells was oxidant- and osmoresistant, and that Hog1p was phosphorylated and able to translocate to the nucleus under both stress conditions. However, a major difference in the D513K mutant versus the D556N mutant was that in the D513K mutant, Hog1p was constitutively phosphorylated in the absence of stress and in hyphal-inducing media (Menon et al., 2006). We hypothesized that constitutive activation of Hog1p is at least partially responsible for the morphogenesis defect of D513K (failure to undergo hyphal conversion), analogous to the sustained phosphorylation of Hog1p in S. cerevisiae which results in at least a partial activation of the pheromone response pathway in cells that are osmotically stressed (Yale and Bohnert, 2001).
In order to investigate the transcriptional profiling of the D556N and D513K mutants, we used microarray analysis. In these experiments, entire arrays were done of both mutants, but data presented above focused upon gene families that might reflect the oxidant sensitive phenotype in the D556N, while in the D513K mutant, gene families associated with morphogenesis functions were highlighted. Since the morphogenesis phenotype of D556N and the oxidant phenotype of D513K each was like wt cells, our mutants were only compared to wt cells. However, the entire microarray analysis of each mutant is available as supplementary data, S1 and S2.
Transcriptional profiling of the D556N point mutant correlates with its peroxide sensitivity, as 9 genes associated with oxidative stress adaptation were down regulated as determined by microarray and that result was verified by RT-PCR during peroxide stress. As for the D513K mutant, 15 genes (by microarray) with functions associated with morphogenesis in C. albicans were altered transcriptionally compared to wt cells. By RT-PCR, of these 15 genes, transcriptional changes fell into 2 categories; four genes were down regulated in M199 only (M199 medium induces hyphal formation), while nine genes were down regulated in both YPD and M199. Thus, the phenotype of each mutant correlated with transcriptional changes as verified by RT-PCR from microarray data.
To investigate the requirement of the D556 and D513 amino acids in virulence, we used the rat model of vaginitis which is distinguished from the human disease in at least two important ways. First, cellular infiltrates including neutrophils are not observed and second, the rat vaginal canal does not harbor a commensal population of C. albicans (De Bernardis et al., 2002). Nevertheless, the model has been useful in evaluating a number of C. albicans mutants for virulence. We show that the D556N mutation but not D513K affects colonization. The reasons for this difference in virulence are not understood. In summary, we have identified D556 as an important residue in at least the oxidative stress response and virulence, probably through its activation of the HOG1 MAPK pathway. As candidiasis continues to be an important disease of patients (Wisplinghoff, 2004), Ssk1p may be useful target in drug discovery. In fact, recent data indicate that ssk1 deletion mutants are hypersensitive to fluconazole and voriconazole both of which are cidal to the mutant (Chauhan et al, 2007).
Supplementary Material
The following supplementary material is available for this article:
Table S1. Expression profile of all genes in strain SSK26 following growth in M199 medium at 370C for 3 hrs.
Table S2. Expression profile of all genes in strain SSK25 following treatment with 5 mM H2O2.
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.XXXX.XXXX.2007.00XXX.x
(This link will take you to the article abstract.)
Acknowledgements
The research in this paper was supported by a National Institutes of Health grant (NIH-NIAID-43465) to R.C. NC is a recipient of an American Heart Association (AHA) National Scientist Development grant, 0635108N and an NIH grant (NIH-NIAID RAI076084A).
Footnotes
Publisher's Disclaimer: Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alonso-Monge R, Navarro-Garcia M, Molero G, Diez-Orejas R, Gustin M, Pla J, Sanchez M, Nombela C. Role of mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J Bacteriol. 1999;181:3058–3068. doi: 10.1128/jb.181.10.3058-3068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Monge R, Navarro-Garcia F, Roman E, Negredo A, Eximan B, Nombela C, Pla J. The Hog1 MAP kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eucaryotic Cell. 2003;2:351–361. doi: 10.1128/EC.2.2.351-361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana DM, Nombela C, Alonso-Monge R, Pla J. The Pbs2 MAP kinase kinase is essential for the oxidative stress response in the fungal pathogen Candida albicans. Microbiology. 2005;151:1033–1049. doi: 10.1099/mic.0.27723-0. [DOI] [PubMed] [Google Scholar]
- Bahn YS, Kojima K, Cox GM, Heitman J. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development and virulence of Cryptococcus neoformans. Mol Biol Cell. 2006;17:3122–3135. doi: 10.1091/mbc.E06-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt J, Herman D, Sheridan M, Calderone R. Adherence and invasion studies of Candida albicans strains utilizing in vitro models of esophageal candidiasis. J Infect Dis. 2001;184:1170–1175. doi: 10.1086/323807. [DOI] [PubMed] [Google Scholar]
- Calera JA, Calderone R. Identification of a putative response regulator, two-component phosphorelay gene (CaSSK1) from Candida albicans. Yeast. 1999;15:1243–1254. doi: 10.1002/(SICI)1097-0061(19990915)15:12<1243::AID-YEA449>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Calera JA, Zhao X-J, Calderone R. Defective hyphal formation and avirulence caused by a deletion of the CSSK1 response regulator gene in Candida albicans. Infect Immun. 2000;68:518–525. doi: 10.1128/iai.68.2.518-525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calera JA, Herman D, Calderone R. Identification of YPD1, a gene of Candida albicans which encodes a two-component phospho-histidine intermediate protein. Yeast. 2000;16:1053–1059. doi: 10.1002/1097-0061(200008)16:11<1053::AID-YEA598>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Chauhan N, Kruppa M, Calderone R. The Ssk1p response regulator and Chk1p histidine kinase mutants are hypersensitive to fluconazole and voriconazole. Antimicrob Agents Chemother. 2007;51:3747–3751. doi: 10.1128/AAC.00929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan N, Latge J-P, Calderone R. Signaling and oxidant adaptation in Aspergillus fumigatus and Candida albicans. Nature Microbiol Rev. 2006;4:435–444. doi: 10.1038/nrmicro1426. [DOI] [PubMed] [Google Scholar]
- Chauhan N, Inglis D, Roman E, Pla J, Li D, Calera J, Calderone R. The SSK1 of Candida albicans is associated with oxidative stress adaptation and cell wall biosynthesis. Eucaryotic Cell. 2003;2:1018–1024. doi: 10.1128/EC.2.5.1018-1024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Calderone R, Richert J, Li D. Deletion of the SSK1 response regulator gene in Candida albicans contributes to enhanced killing by human polymorphonuclear neutrophils. Infect Immun. 2005;73:865–871. doi: 10.1128/IAI.73.2.865-871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Sarfati J, Latge J-P, Calderone R. The sakA (Hog1) and tscB (Sln1) two-component proteins in oxidant adaptation in Aspergillus fumigatus. Med Mycol. 2006;44:211–218. doi: 10.1080/13693780500338886. [DOI] [PubMed] [Google Scholar]
- De Bernardis F, Boccanera M, Adriani D, Girlamo A, Cassone A. Intravaginal and intransal immunizations are equally effective in inducing vaginal antibodies and conferring protection against vaginal candidiasis. Infect Immun. 2002;70:2725–2729. doi: 10.1128/IAI.70.5.2725-2729.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B, Smith D, Alam I, Nicholls S, Brown AJ, Quinn J. Role of Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell. 2006;17:1018–1032. doi: 10.1091/mbc.E05-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer L. The ALS gene family of Candida albicans. Trends Microbiol. 2001;9:176–180. doi: 10.1016/s0966-842x(01)01984-9. [DOI] [PubMed] [Google Scholar]
- Janiak-Spens F, Sparling JM, Gurfinkel M, West AH. Differential stabilities of phosphorylated response regulator domains reflect functional roles of the yeast osmoregulatory SLN1 and SSK1 proteins. J Bacteriol. 1999;181:411–417. doi: 10.1128/jb.181.2.411-417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA, Vinces MD. Contribution of hyphae and hyphal coregulated genes to Candida albicans virulence. Cellular Microbiol. 2005;7:1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- Li D, Bernhardt J, Calderone R. Temporal expression of the Candida albicans genes CHK1 and CSSK1, adherence and morphogenesis in a model of reconstituted human esophageal epithelial candidiasis. Infect Immun. 2002;70:1558–1565. doi: 10.1128/IAI.70.3.1558-1565.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Menon V, Li D, Chauhan N, Rajenadrum R, Dubrovska A, West AH, Calderone R. Functional studies of the Ssk1p response regulator protein of Candida albicans as determined by the phenotypic analysis of receiver domain point mutants. Mol Microbiol. 2006;62:997–1013. doi: 10.1111/j.1365-2958.2006.05438.x. [DOI] [PubMed] [Google Scholar]
- Moye-Rowley S. Regulation of the transcriptional response to oxidative stress in fungi: similarities and differences. Eukaryotic Cell. 2003;2:381–389. doi: 10.1128/EC.2.3.381-389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SW, Xu Q, West AH. Ssk1p response regulator binding surface on histidine containing phosphotransfer protein Ypd1p. Eukaryotic Cell. 2003;2:27–33. doi: 10.1128/EC.2.1.27-33.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SW, West AH. A common docking site for response regulators on the yeast phosphorelay protein YPD1. Biochim. Biophys. Acta. 2005;1748:138–145. doi: 10.1016/j.bbapap.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 "two-component" osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Posas F, Saito H. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. The EMBO J. 1998;17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Jose C, Monge R, Perez-Diaz R, Pla J, Nombela C. The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus, Candida albicans. J Bacteriol. 1996;178:5850–5852. doi: 10.1128/jb.178.19.5850-5852.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Chauhan N, Ghosh A, Dixon F, Calderone R. SKN7 of Candida albicans: mutant construction and phenotype analysis. Infect Immun. 2004;72:2390–2394. doi: 10.1128/IAI.72.4.2390-2394.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DA, Nicholls S, Morgan B, Brown AJ, Quinn J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell. 2004;14:1460–1467. doi: 10.1091/mbc.E04-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, West AH. histidine kinases in signal transduction. Elsevier Science (USA); 2002. Response regulators and their interactions with histidine protein kinases, cp. 12; pp. 237–271. [Google Scholar]
- Stock A, Koshland DE, Stock J. Homologies between the Salmonella typhimurium CheY protein and proteins involved in the regulation of chemotaxis, membrane protein synthesis, and sporulation. Proc Natl Acad Sci USA. 1985;82:7989–7993. doi: 10.1073/pnas.82.23.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason P, Kay R. Eukaryotic signal transduction via histidine-aspartate phosphorelay. J Cell Sci. 2000;113:3141–3150. doi: 10.1242/jcs.113.18.3141. [DOI] [PubMed] [Google Scholar]
- Walia A, Calderone R. The SSK2 MAPKKK of Candida albicans is required for oxidant adaptation in vitro. FEMS Yeast Res. 2008;8:287–299. doi: 10.1111/j.1567-1364.2007.00329.x. [DOI] [PubMed] [Google Scholar]
- West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- Wisplinghoff H, Bischoff T, Tallent S, Seifert H, Wenzel R, Edmond M. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- Wormley FL, Heinrich G, Miller JL, Perfect JR, Cox GM. Identification and characterization of an SKN7 homologue from Cryptococcus neoformans. Infect Immun. 2005;73:5022–5030. doi: 10.1128/IAI.73.8.5022-5030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T, Nguyen C, Romans A, Kontoyiannis D, May GS. A mitogen-activated protein kinase that senses nitrogen regulates conidial germination and growth in Aspergillus fumigatus. Eukaryotic Cell. 2004;3:557–560. doi: 10.1128/EC.3.2.557-560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yale J, Bohnert HJ. Transcript expression in Saccharomyces cerevisiae at high salinity. J.Biol Chem. 2001;276:15996–16007. doi: 10.1074/jbc.M008209200. [DOI] [PubMed] [Google Scholar]
- Yamada-Okabe T, Mio T, Ono N, Kashima Y, Matsui M, Arisawa A, Yamada-Okabe H. Roles of three histidine kinase genes in hyphal development and virulence of the pathogenic fungus, Candida albicans. J Bacteriol. 1999;181:7243–7247. doi: 10.1128/jb.181.23.7243-7247.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following supplementary material is available for this article:
Table S1. Expression profile of all genes in strain SSK26 following growth in M199 medium at 370C for 3 hrs.
Table S2. Expression profile of all genes in strain SSK25 following treatment with 5 mM H2O2.
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.XXXX.XXXX.2007.00XXX.x
(This link will take you to the article abstract.)


