Abstract
The identification of activating mutations in NOTCH1 in the majority of T-cell acute lymphoblastic leukemias and lymphomas (T-ALL) has brought much interest in inhibiting NOTCH1 signaling as therapeutic target in this disease. Small molecule inhibitors of the γ-secretase complex, which mediates a critical proteolytic cleavage required for NOTCH1 activation, hold the promise of becoming an effective molecular therapy against relapsed and refractory T-ALL. Recent progress in the elucidation of the transcriptional regulatory networks downstream of oncogenic NOTCH1 has uncovered a central role of NOTCH1 signaling in promoting leukemic cell growth and revealed an intricate circuitry that connects NOTCH1 signaling with MYC and the PI3K-AKT signaling pathway. The identification of the downstream effector pathways controlled by NOTCH1 should pave the way for the rational design of anti-NOTCH1 therapies for the treatment of T-ALL.
Background
The NOTCH pathway is an evolutionary-conserved mechanism responsible for the direct transduction of developmental signals at the cell surface into changes in gene expression in the nucleus (1–3). The intrinsic elements of the NOTCH signaling pathway include: (i) the Delta and Serrate family of ligands (Delta-like 1, 3 and 4; and Jagged 1 and 2), (ii) the NOTCH receptors (NOTCH1–4), and (iii) the CSL (CBF1/Su(H)/LAG-1) transcription factor, a DNA binding protein that interacts with the activated forms of NOTCH receptors.
The mature NOTCH1 protein is an heterodimeric transmembrane receptor consisting of an extracellular subunit and a transmembrane and intracellular subunit, which are generated by furin protease cleavage from a precursor polypeptide during its maturation in the trans-Golgi network (4, 5). In resting conditions, the two NOTCH1 subunits remain associated forming a heterodimeric complex (2, 3). However, upon ligand-receptor interaction, the N-terminus fragment of NOTCH1 is pulled away from the complex. This triggers a double proteolytic processing of the transmembrane and intracellular portion of NOTCH1, first by an ADAM protease (6, 7) and subsequently by the γ-secretase complex (8–10). This final proteolytic cleavage releases the intracellular domains of NOTCH1 (ICN1) from the membrane, which then translocates to the nucleus, binds to the CSL DNA-binding protein (11), and recruit the MAML1 transcriptional coactivator to activate the expression of target genes (12). Transcriptional activation terminates NOTCH1 signaling through phosphorylation of the C-terminus PEST domain of ICN by CDK8, which results in FBXW7-CSF mediated degradation of the activated receptor in the proteasome (13). NOTCH1 reads and transduces extracellular signals in a quantic way, as one molecule of ligand activates the irreversible proteolytic cleavage and activation of one molecule of receptor, which in turn binds to one promoter to activate gene expression (1–3).
In the hematopoietic system, NOTCH1 plays a critical role in T-cell development and transformation (3, 14). During thymocyte development NOTCH1 signals are required for the commitment of multipotent hematopoietic progenitors to the T-cell lineage (15–18) and then along T-cell development for progression through the early DN1, DN2 and DN3 stages of thymocyte maturation (19). Aberrant NOTCH1 signaling was first identified in T-cell acute lymphoblastic leukemias (T-ALLs) harboring the t(7;9)(q34;q34.3), a recurrent chromosomal translocation which truncates the NOTCH1 gene and misplaces it next to the TCRB locus, leading to high levels of expression of a constitutively active intracellular form of NOTCH1 (20, 21). However, the most common mechanisms leading to aberrant NOTCH1 signaling are activating mutations in NOTCH1, which are present in over 50% of T-ALL cases (22). Activating NOTCH1 mutations located in the heterodimerization domain (HD alleles) and the juxtamembrane extracellular region (JME alleles) induce ligand independent activation of the receptor (23, 24), while truncating mutations deleting the PEST domain in the C-terminal region of the protein extend NOTCH1 signaling by impairing the proteasomal degradation of ICN1. In addition, mutations in FBXW7 involving three critical arginine residues that mediate the interaction of this F-box protein with the phosphodegron moiety in the NOTCH1 PEST domain, also extend NOTCH1 signaling by impairing the proteasomal degradation of ICN1 in 15% of TALL cases (25–28). Importantly, about 15–25% of these leukemias harbor two concurrent lesions activating NOTCH1; the first one inducing ligand independent activation of NOTCH1 –an HD or JME allele –and a second one leading to increased protein stability and extended duration of NOTCH1 signaling –a PEST truncation or FBXW7 mutation –(22, 25, 27, 28).
The importance of mutations activating the NOTCH1 pathway is highlighted by the potential role of NOTCH1 as a therapeutic target in T-ALL. Given the strict requirement of γ-secretase cleavage for NOTCH1 activation it was recognized early on that inhibition of this proteolytic step could be exploited to effectively abrogate the function of oncogenic NOTCH1 in T-ALL cells. Importantly, the presenilin γ-secretase complex has been the focus of extensive research over the last decade because of its role in the generation of amyloid deposits in the brains of patients with Alzheimer’s disease; and small molecule γ-secretase inhibitors (GSIs), originally designed for the treatment of neurodegenerative disorders, effectively block NOTCH1 signaling. In vitro studies showed that GSI treatment of T-ALL cell lines harboring activating mutations in NOTCH1 induced cell cycle arrest in G1 (22, 29, 30) prompting the investigation of these agents for the treatment of relapsed and refractory T-ALL (31). However, the target genes and effector pathways that mediate the oncogenic activity of NOTCH1 remained largely unknown precluding a fully rational design of anti-NOTCH therapies. This gap has started to be filled as result of microarray gene expression profiling studies of T-ALL cells treated with GSIs. These analyses have demonstrated a major role of NOTCH1 in the regulation of cell growth and metabolism (29, 32). Thus, NOTCH1 inhibition in T-ALL cells results in the transcriptional downregulation of numerous genes involved in biosynthesis pathways such as ribosome biosynthesis, protein translation and amino acid and nucleotide biosynthesis (29, 32, 33) (Figure 1). Indeed, many of these anabolic genes were shown to be direct targets of NOTCH1 by ChIP-on-chip analysis, thus highlighting the role of NOTCH1 as direct regulator of cell growth (29). Importantly, MYC is also a direct trascriptional target downstream of oncogenic NOTCH1 (29, 32, 33) and MYC regulates the expression of anabolic genes and pathways downstream of NOTCH1 (34). This NOTCH1-MYC transcriptional regulatory loop places the control of cell growth anabolic pathways at the core of the mechanisms mediating T-cell transformation by oncogenic NOTCH1 (Figure 1).
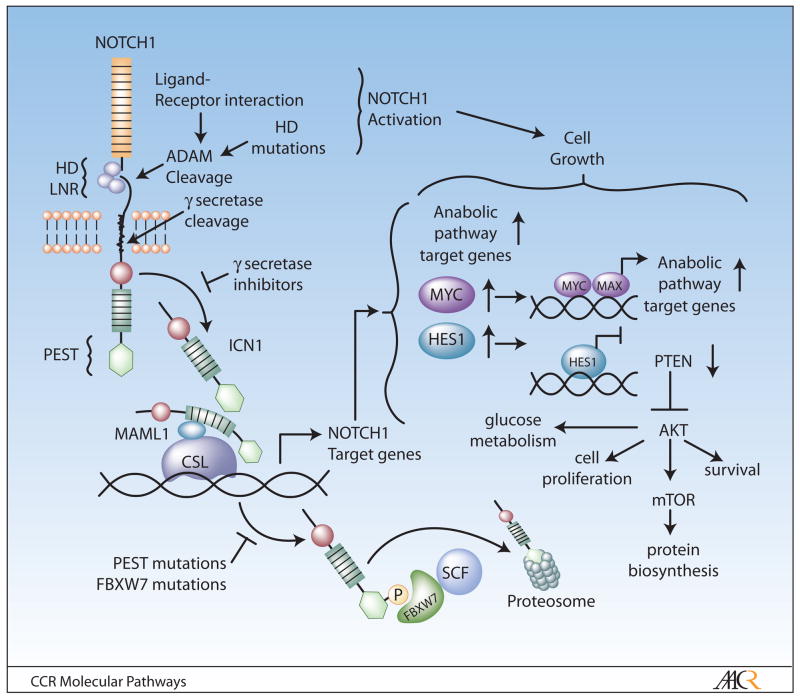
Figure 1. Schematic representation of the NOTCH1 signaling pathway and transcriptional networks promoting leukemic cell growth downstream of oncogenic NOTCH1.
The two subunits of the mature NOTCH1 receptor are generated by cleavage by a furin protease in the trans-Golgi network soon after translation of NOTCH1 precursor protein. The extracellular subunit responsible for ligand-receptor interaction and the transmembrane subunit responsible for triggering transcription activation interact by their respective heterodimerization (HD) domains. Upon binding to its ligands –Delta-like 1, 3 and 4; Jagged 1 and 2– the transmembrane portion of NOTCH1 is sequentially cleaved by ADAM proteases and then by the γ-secretase complex. This final proteolytic cleavage liberates the active intracellular fragment of NOTCH1 (ICN1), which translocates to the nucleus and activates the expression of target genes by forming a ternary complex with the CSL DNA binding protein and the MAML1 transcriptional coactivator. Small molecule inhibitors of the γ-secretase block NOTCH1 activation by retaining the receptor at the membrane. NOTCH1 promotes cell growth by transcriptional upregulation of genes involved in anabolic pathways and also by transcriptional upregulation of MYC. In addition NOTCH1 induces the expression of HES1, a transcriptional repressor that promotes the upregulation of the PI3K-AKT signaling pathway by transcriptional downregulation of PTEN. Termination of NOTCH1 signaling is mediated by phosphorylated-coupled degradation of the activated receptor in the proteasome via FBXW7-SCF.
A prominent role of NOTCH1 as a regulator of cell growth in immature T-cells was further supported by several lines of evidence indicating that activation of the PI3K-AKT-mTOR pathway is also an important outcome downstream of NOTCH1 signaling. In early work, Sade and coworkers showed that the Src family protein tyrosine kinase p56lck is required for NOTCH1-mediated activation of AKT in T-cells (35). Furthermore phosphoproteomic analysis of T-ALL cells treated with NOTCH1 inhibitors showed a marked decrease in the phosphorylation of mTOR targets (36). In addition, elegant work by Ciofani and coworkers showed that Notch signals regulate the trophic state (cell size, glucose uptake and glycolysis) of T-cell precursors and promote cell survival through maintenance of cellular metabolism (37). Importantly, these critical effects of Notch signaling in the growth, proliferation and survival of T-cell progenitors were mediated by activation of the PI3K-Akt signaling pathway (37).
The PI3K-AKT signal transduction pathway mediates multiple cellular responses triggered by the engagement of growth factor receptors, including increased cell growth, proliferation and survival (38–43). Activation of PI3K by receptor tyrosine kinases and G protein-coupled receptors triggers the phosphorylation of phosphatidylinositol 4,5 biphosphate (PIP2) to generate phosphatidylinositol triphosphate (PIP3). Accumulation of PIP3 at the plasma membrane recruits AKT and induces its phosphorylation by PDK1 and the mTOR-Rictor complex at Thr308 and Ser473, respectively (44, 45). In turn, AKT activates the phosphorylation of multiple substrates that promote cell growth, increased glucose uptake and oxidation, cell cycle progression and cell survival by direct and indirect mechanisms (38–41). Signaling by the PI3K-AKT pathway is terminated by the PTEN phosphatase, which dephosphorylates and thereby inactivates PIP3 (38–41).
In this context, activation of the PI3K-AKT pathway downstream of NOTCH1 signaling promotes cell growth at multiple levels and plays an important role in T-cell transformation. However, the specific mechanisms connecting these two pathways remained unclear until a combination of genomic analysis in T-ALL cells and a forward genetic screen in Drosophila identified the PTEN downregulation as a major component of the oncogenic program downstream of NOTCH1 (Figure 1) (34). These studies showed that PTEN expression in T-ALL is controlled by a dual input transcriptional regulatory circuit operated by HES1, a transcriptional repressor directly controlled by NOTCH1, and MYC, also a direct NOTCH1 target (34). Both HES1 and MYC bind and regulate the PTEN promoter, with HES1 working as a strong transcriptional repressor and MYC as a weaker transcriptional activator, so that the overall output downstream of NOTCH1 activation is a controlled downregulation of PTEN transcripts (Figure 1) (34).
Clinical-translational advances
The induction of cell cycle arrest and a decrease in cell size in T-ALL cell lines treated with GSIs is in agreement with the role of NOTCH1 as a direct regulator of cell growth. However, the most striking direct implication of the elucidation of the transcriptional circuitry controlling cell growth downstream of NOTCH1 is that it sets the conceptual framework to understand the mechanisms that mediate primary resistance to GSI therapy in T-ALL. It was recognized early on that only a fraction of T-ALL cell lines with activating mutations in NOTCH1 show a decreased cell size and cell cycle arrest response when treated with GSIs (22). However, analysis of GSI sensitive and GSI resistant T-ALL cell lines showed no differences in the effect of GSI treatment on inhibition of NOTCH1 processing, the rate and kinetics of ICN1 clearance or the transcriptional response triggered by NOTCH1 inhibition in these cells (34). These results demonstrated that GSI resistance was not the result of increased drug metabolism, increased drug export or decreased ability of this GSI to interact with and inhibit the γ-secretase complex; and supported that activation of an alternative oncogenic pathway was taking over the role of NOTCH1 as central regulator of growth and metabolism in GSI resistant T-ALL cells. This hypothesis was confirmed upon discovering that mutational loss of PTEN and consequent constitutive activation of the PI3K-AKT pathway was present in GSI resistant T-ALL lines, but not in GSI-sensitive leukemias (34). Further analysis established a mechanistic link between loss of PTEN, activation of AKT and increased metabolism as a mechanism of resistance to NOTCH1 inhibition in T-ALL cell lines (34). Importantly, loss of PTEN was also found in 17% of primary T-ALL samples, suggesting that primary resistance to GSI therapy may be readily present in a significant fraction of T-ALL patients (34).
Treatments that target oncogenic signaling pathways controlling the growth and survival of malignant tumor cells are based on the concept of oncogene addiction, which proposes that adaptation to oncogenic signals require an irreversible rewiring of the cellular machinery which renders tumor cells dependent on continuous oncogene activity for proliferation and survival (46). Following on this premise we predicted that constitutive AKT activation could circumvent the requirement of NOTCH1 signaling for cell growth and proliferation, at the expense of inducing an oncogene addiction switch that would render PTEN null/GSI-resistant cells addicted to AKT signaling. Treatment of PTEN-positive/GSI-sensitive and PTEN-null/GSI-resistant TALL cells with an AKT inhibitor confirmed this prediction showing a 10 fold higher sensitivity to this drug in cells resistant to NOTCH-inhibition (34). Thus, small molecule inhibitors of PI3K-AKT signaling currently under clinical development (47) may constitute an effective treatment in T-ALL tumors with mutational loss of PTEN.
Future developments
Despite strong rationale and much enthusiasm towards GSIs as a targeted therapy against T-ALL tumors harboring activating mutations in NOTCH1, the results of the first clinical trial testing the MK-0752 GSI, in relapsed and refractory T-ALL have shown no significant clinical responses and a high incidence of dose limiting gastrointestinal toxicity (31). Thus, new approaches aiming to increase the therapeutic window of GSIs are required for the successful implementation of anti-NOTCH based therapies in T-ALL. Combination therapies exploiting the interaction of NOTCH inhibitors with drugs targeting the PI3K-AK-mTOR pathway such as rapamycin and perifosin may increase the intrinsic activity of GSIs and facilitate dose schedules that avoid the development of gastrointestinal toxicity.
Recent studies have shown that, in addition to its direct and indirect effects on cell growth and metabolism, NOTCH1 signaling may regulate the activity of p53 (48) and the NFKB pathway (49). Although the specific mechanisms mediating these interactions have not been elucidated yet, these results suggest that combinations of GSIs with DNA damaging agents inducing p53 activation, or with drugs interfering with the activity of the NFKB pathway such as bortezomib, may have a synergistic effect in the treatment of T-ALL.
Acknowledgments
This work was supported by the National Institutes of Health (R01CA120196 and R01CA129382 to A.F.), the WOLF Foundation and the Leukemia and Lymphoma Society (grants 1287–08 and 6237–08 to A.F). Adolfo Ferrando is a Leukemia & Lymphoma Society Scholar.
References
- 1.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–89. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 2.Aster JC, Pear WS, Blacklow SC. Notch Signaling in Leukemia. Annu Rev Pathol. 2007 doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aster JC, Pear WS, Blacklow SC. Notch Signaling in Leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–91. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 5.Logeat F, Bessia C, Brou C, et al. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci U S A. 1998;95:8108–12. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brou C, Logeat F, Gupta N, et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–16. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 7.Mumm JS, Schroeter EH, Saxena MT, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 8.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–5. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 9.De Strooper B, Annaert W, Cupers P, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–22. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 10.Struhl G, Greenwald I. Presenilin-mediated transmembrane cleavage is required for Notch signal transduction in Drosophila. Proc Natl Acad Sci U S A. 2001;98:229–34. doi: 10.1073/pnas.011530298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–82. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 12.Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26:484–9. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 13.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell. 2004;16:509–20. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 14.Radtke F, Wilson A, Mancini SJ, MacDonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. 2004;5:247–53. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 15.Jaleco AC, Neves H, Hooijberg E, et al. Differential effects of Notch ligands Delta-1 and Jagged-1 in human lymphoid differentiation. J Exp Med. 2001;194:991–1002. doi: 10.1084/jem.194.7.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pear WS, Radtke F. Notch signaling in lymphopoiesis. Semin Immunol. 2003;15:69–79. doi: 10.1016/s1044-5323(03)00003-4. [DOI] [PubMed] [Google Scholar]
- 17.Pui JC, Allman D, Xu L, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 18.Radtke F, Wilson A, Stark G, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–58. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt TM, Ciofani M, Petrie HT, Zuniga-Pflucker JC. Maintenance of T cell specification and differentiation requires recurrent notch receptor-ligand interactions. J Exp Med. 2004;200:469–79. doi: 10.1084/jem.20040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellisen LW, Bird J, West DC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–61. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 21.Palomero T, Barnes KC, Real PJ, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006;20:1279–87. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- 22.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 23.Malecki MJ, Sanchez-Irizarry C, Mitchell JL, et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26:4642–51. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sulis ML, Williams O, Palomero T, et al. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008 doi: 10.1182/blood-2007-12-130096. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson BJ, Buonamici S, Sulis ML, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–35. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Neil JN, Mouton PR, Tizabi Y, et al. Catecholaminergic neuronal loss in locus coeruleus of aged female dtg APP/PS1 mice. J Chem Neuroanat. 2007;34:102–7. doi: 10.1016/j.jchemneu.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malyukova A, Dohda T, von der Lehr N, et al. The Tumor Suppressor Gene hCDC4 Is Frequently Mutated in Human T-Cell Acute Lymphoblastic Leukemia with Functional Consequences for Notch Signaling. Cancer Res. 2007;67:5611–6. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- 28.Akhoondi S, Sun D, von der Lehr N, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–12. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 29.Palomero T, Lim WK, Odom DT, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–6. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis HD, Leveridge M, Strack PR, et al. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chem Biol. 2007;14:209–19. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 31.Deangelo D, Stone R, Silverman L, et al. A phase I clinical trial of the notch inhibitor MK-0752 in patients with T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) and other leukemias. Journal of Clinical Oncology, 2006 ASCO Annual Meeting Proceedings Part I. 2006;24:6585. [Google Scholar]
- 32.Weng AP, Millholland JM, Yashiro-Ohtani Y, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma VM, Calvo JA, Draheim KM, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26:8022–31. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palomero T, Sulis ML, Cortina M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007 doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sade H, Krishna S, Sarin A. The anti-apoptotic effect of Notch-1 requires p56lck-dependent, Akt/PKB-mediated signaling in T cells. J Biol Chem. 2004;279:2937–44. doi: 10.1074/jbc.M309924200. [DOI] [PubMed] [Google Scholar]
- 36.Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ. Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood. 2007;110:278–86. doi: 10.1182/blood-2006-08-039883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–8. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 38.Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–75. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 39.Sulis ML, Parsons R. PTEN: from pathology to biology. Trends Cell Biol. 2003;13:478–83. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 40.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–92. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 41.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–9. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 42.Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Curr Opin Oncol. 2006;18:77–82. doi: 10.1097/01.cco.0000198021.99347.b9. [DOI] [PubMed] [Google Scholar]
- 43.Hay N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell. 2005;8:179–83. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 44.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 45.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 46.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–4. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 47.Stephens L, Williams R, Hawkins P. Phosphoinositide 3-kinases as drug targets in cancer. Curr Opin Pharmacol. 2005;5:357–65. doi: 10.1016/j.coph.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 48.Beverly LJ, Felsher DW, Capobianco AJ. Suppression of p53 by Notch in lymphomagenesis: implications for initiation and regression. Cancer Res. 2005;65:7159–68. doi: 10.1158/0008-5472.CAN-05-1664. [DOI] [PubMed] [Google Scholar]
- 49.Vilimas T, Mascarenhas J, Palomero T, et al. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nat Med. 2007;13:70–7. doi: 10.1038/nm1524. [DOI] [PubMed] [Google Scholar]