Abstract
Ovarian carcinomas with mutations in the tumor suppressor BRCA2 are particularly sensitive to platinum compounds 1. However, such carcinomas ultimately develop cisplatin resistance. The mechanism of that resistance is largely unknown 2. Here we show that acquired resistance to cisplatin can be mediated by secondary intragenic mutations in BRCA2 that restore the wild-type BRCA2 reading frame. First, in a cisplatin-resistant BRCA2-mutated breast cancer cell line, HCC1428, a secondary genetic change in BRCA2 rescued BRCA2 function. Second, cisplatin selection of a BRCA2-mutated pancreatic cancer cell line, Capan-1 3,4, led to 5 different secondary mutations that restored the wild-type BRCA2 reading frame. All clones with secondary mutations were resistant both to cisplatin and to a poly(ADP-ribose) polymerase (PARP) inhibitor (AG14361). Finally, we evaluated recurrent cancers from patients whose primary BRCA2-mutated ovarian carcinomas were treated with cisplatin. The recurrent tumor that acquired cisplatin resistance had undergone reversion of its BRCA2 mutation. Our results suggest that secondary mutations that restore the wild-type BRCA2 reading frame may be a major clinical mediator of acquired resistance to platinum-based chemotherapy.
Women with ovarian cancer generally have a high initial response to platinum-based chemotherapy, but over time the majority of ovarian carcinomas become refractory, and most patients die with progressive chemoresistant disease 2. The molecular basis of initial platinum sensitivity and acquired resistance remains largely unknown. Individuals with germline mutations in BRCA1 or BRCA2 (BRCA1/2) have an increased risk of developing cancer in the breast and ovary 5 as well as in some other organs including the pancreas and prostate 6. BRCA2 is identical to the Fanconi anemia (FA) gene FANCD1 7. The BRCA2 protein directly binds to and regulates RAD51, an essential protein for DNA repair through homologous recombination (HR) 8. Tumors from BRCA1/2 mutation carriers usually have deletion of the wild-type allele at the BRCA1/2 locus and are BRCA1/2-deficient 9–11. BRCA1/2-deficient cancer cells are hypersensitive to DNA-crosslinking agents including cisplatin 1, 12. Therefore, cisplatin or its derivative, carboplatin, is a logical choice for the treatment of BRCA1/2-mutated tumors 13 and women with BRCA1/2-mutated ovarian carcinoma have a better prognosis than those without BRCA1/2 mutation if they receive platinum-based therapy 14, 15. However, even BRCA1/2-mutated tumors eventually develop platinum resistance. In several genetic disorders such as FA, spontaneous genetic alterations compensating for inherited disease-causing mutations have been described 16. These alterations in FA include secondary genetic changes of one of the mutated FA alleles, such as back-mutations to wild-type, compensatory mutations in cis, intragenic crossovers, and gene conversion 16–18. We speculated that secondary mutations in BRCA2-mutated alleles might also occur in cancer cells during chemotherapy.
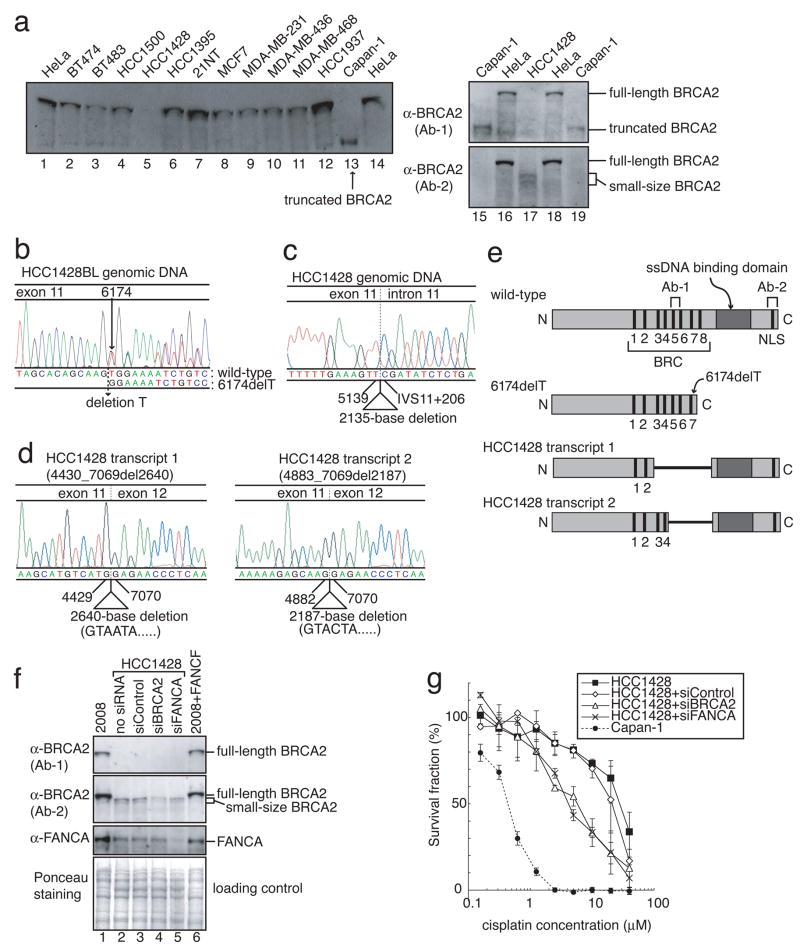
To study the mechanism of acquired cisplatin resistance of BRCA2-mutated cancer, first we screened 12 human breast cancer cell lines for alterations in BRCA2 protein expression (Fig. 1a). A pancreatic cancer cell line, Capan-1 3, 4, with a truncated BRCA2 protein, was used as a control.
Figure 1. HCC1428 is a cisplatin-resistant breast cancer cell line with a secondary BRCA2 mutation.
(Full-length blots are presented in Supplemental Figure 9) a, Cell lysates from HeLa, Capan-1 and indicated breast cancer cell lines were immunoblotted with BRCA2 antibodies. b, Genomic DNA sequence of BRCA2 in HCC1428BL lymphoblast. A heterozygous mutation (6174delT) was identified. c, Genomic DNA sequence of BRCA2 in HCC1428 breast cancer cell line. A homozygous 2135-base deletion from nt5140 (in exon 11) to IVS11+205 (in intron 11) was identified. d, Sequences of the two BRCA2 transcripts in HCC1428 breast cancer cell line. The two RT-PCR products shown in Fig. S1b were purified from the gel, and sequenced separately. HCC1428 transcripts 1 and 2 had a 2640-base deletion (nt4430 to 7069) and a 2187-base deletion (nt4883 to 7069), respectively, suggesting activation of the cryptic splice donor sites in exon11. e, Schematic presentation of BRCA2 proteins. The regions that the BRCA2 antibodies (Ab-1 and Ab-2) recognize are also depicted. HCC1428 transcripts 1 and 2 encode BRCA2 lacking 881 amino acids and 730 amino acids, respectively, both of which have ssDNA binding domains, nuclear localization signals (NLS) and some of the BRC repeats. f, Cell lysates from HCC1428 cells treated with indicated siRNA were immunoblotted with BRCA2 and FANCA antibodies. 2008 and 2008+FANCF were used as controls. g, Cisplatin sensitivity assessed by XTT assay. HCC1428 was resistant to cisplatin, and depletion of BRCA2 or FANCA sensitized HCC1428 to cisplatin (mean ± SEM, n=3).
From breast cancer cell line, HCC1428 19, BRCA2 protein was undetectable by western blotting using anti-BRCA2 (Ab-1), which recognizes the middle part of BRCA2 (Fig. 1a) but was detectable with anti-BRCA2 (Ab-2) that recognizes the BRCA2 C-terminus. The small size of HCC1428 BRCA2 identified by Ab-2 suggested that the protein might have an internal in-frame deletion. Constitutional DNA from a lymphoblast cell line (HCC1428BL) from the same patient had one wild-type BRCA2 allele and one mutant allele with a frameshift mutation, 6174delT, which is common in the Ashkenazi Jewish population (Figs. 1b and S1) 20.
Sequence of genomic DNA from HCC1428 cancer cells revealed no wild-type allele and a mutant allele with a 2135-basepair deletion surrounding the original 6174delT mutation and the exon 11/intron 11 junction (Figs. 1c and S1). Sequence of cDNA indicated that this deletion activated two cryptic splice donor sites in exon 11, resulting in expression of two transcripts with in-frame deletions (Figs. 1d, and S1). HCC1428 transcript 1 had a 2640-basepair deletion and encoded a protein lacking amino acids 1401 to 2281. Transcript 2 had a 2187-basepair deletion and encoded a protein lacking amino acids 1552 to 2281 (Figs. 1d, 1e and S1). Both HCC1428 BRCA2 proteins retain the single-strand DNA (ssDNA) binding domain and the C-terminus nuclear localization signals (NLS), whereas these domains are lost in BRCA2.6174delT (Fig. 1e). A recent report that only 1 BRC repeat plus ssDNA binding domain and NLS are sufficient for BRCA2 function 21 suggests that the novel BRCA2 proteins in HCC1428 might be functional. Indeed, HCC1428 cells were resistant to cisplatin (Fig. 1g). Furthermore, depletion of these novel BRCA2 proteins with siRNA restored sensitivity of HCC1428 cells to cisplatin (Figs. 1f and 1g).
HCC1428 was derived after chemotherapy from the pleural effusion of a 49 year-old woman with stage IV breast carcinoma who died 6 months later 19. We speculate that the patient’s chemotherapy selected in vivo for secondary mutations in BRCA2-mutated breast tumor cells.
To test this hypothesis, we selected in vitro for cisplatin-resistant clones from the cisplatin-sensitive BRCA2-mutated pancreatic cancer cell line Capan-1. Capan-1 has the mutant allele BRCA2.6174delT, but no wild-type allele (Fig. S5a) 3, 4. Fluorescence in situ hybridization (FISH) revealed that Capan-1 harbors at least two copies of the BRCA2 gene indicating duplication of the chromosome 13 with the mutant BRCA2 gene (Fig. S2).
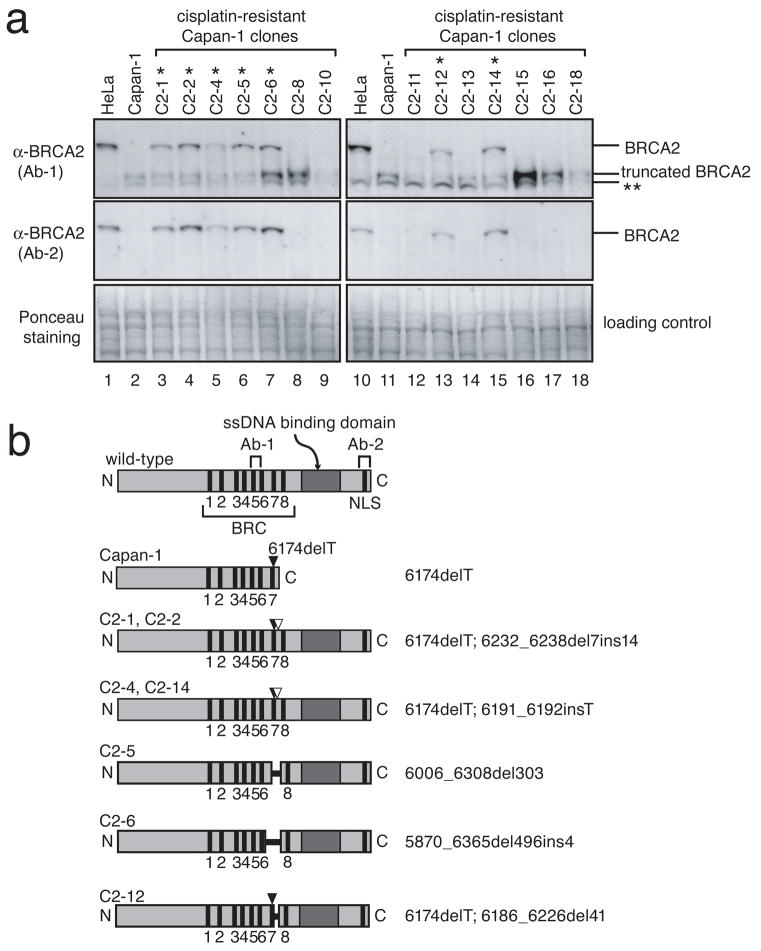
After selection in cisplatin, we obtained 14 cisplatin-resistant Capan-1 clones out of 12 million cells (Fig. S3 and Table S1). Importantly, in 7 of these 14 clones, BRCA2 expression, at close to the length of the wild-type protein, was now detectable (Fig. 2). The other 7 cisplatin-resistant clones still lacked BRCA2 protein expression.
Figure 2. Secondary genetic changes in mutated BRCA2 in cisplatin-resistant clones of a pancreatic cancer cell line, Capan-1.
(Full-length blots are presented in Supplemental Figure 9) a, Seven Capan-1 clones indicated with a single asterisk (*) restored apparently full-length BRCA2 protein expression. Cell lysates from indicated cells were immunoblotted with BRCA2 antibodies (Ab-1 and Ab-2). A double asterisk (**) indicates a band presumed to be nonspecific. b, Schematic presentation of BRCA2 proteins encoded by transcripts in Capan-1 clones. In all of the BRCA2-restored Capan-1 clones, additional genetic changes in exon 11 of BRCA2 were detected (Table S1, Figs. S4 and S5). All of these secondary genetic changes (shown as white arrow heads or black horizontal bars) cancel the frameshift caused by the original mutation (6174delT, black arrow heads), and the encoded BRCA2 proteins have intact C-terminal regions containing a single strand DNA (ssDNA) binding domain and nuclear localization signals (NLS).
In all 7 cisplatin-resistant clones with restored, nearly full-length BRCA2 protein expression, we identified additional BRCA2 mutations that corrected the frameshift caused by the 6174delT mutation (Figs. 2b, S4, and S5, and Table S1). These secondary genetic changes included a small deletion, insertion, and deletion/insertion at sites close to the original mutation, and in-frame deletions surrounding the original mutation site. Interestingly, in each clone we observed both the original mutant BRCA2.6174delT sequence and the 6174delT sequence with additional mutations (Figs. S4 and S5) indicating that the secondary mutations occurred on only one of the duplicated mutant BRCA2 copies (Fig. S5g). None of the 7 cisplatin-resistant clones lacking BRCA2 protein expression harbored additional mutations in BRCA2.
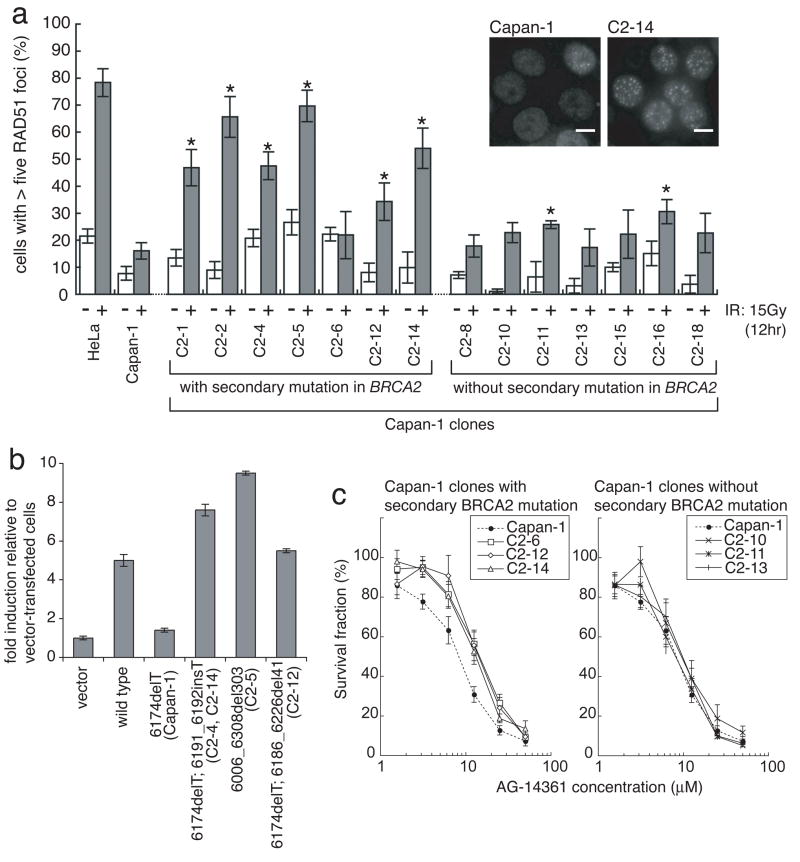
Next, we assessed the function of the restored BRCA2 proteins in the 14 cisplatin-resistant Capan-1 clones by evaluating RAD51 foci formation after exposure to ionizing radiation (IR) (Fig. 3a). IR-induced RAD51 foci formation was impaired in parental Capan-1 1. In contrast, in 6 of the 7 cisplatin-resistant Capan-1 clones with restored BRCA2 expression, IR-induced RAD51 foci formation was significantly improved, suggesting that these novel BRCA2 isoforms are functional.
Figure 3. Functional analyses of the restored BRCA2 proteins.
a, Ionizing radiation (IR)-induced RAD51 foci formation is restored in most of BRCA2-restored Capan-1 clones. Indicated cells were irradiated (15 Gy) and fixed 12 hours after IR. Cells were immunostained with RAD51 antibody. Representative pictures of immunostained cells after IR are shown, together with quantification of the cells with at least five RAD51 foci before (−, white bars) and 12 hours after IR (+, grey bars) (mean values of at least three independent experiments ± SEM). Asterisks (*) indicate significant difference with irradiated parental Capan-1 cells (p<0.05, unpaired t test). Scale bar = 40 μm. b, Quantitation of HR induced by I-SceI in VC8-DR-GFP cells transiently transfected with wild-type and mutant forms of FLAG-tagged human BRCA2 cDNA. The proportion of GFP-positive cells for each construct relative to vector control is shown (mean ± SEM, n=3). c, BRCA2-restored Capan-1 clones are resistant to a PARP inhibitor. Capan-1, its clones with secondary BRCA2 mutation (C2-6, C2-12 and C2-14) and clones without secondary BRCA2 mutation (C2-10, C2-11 and C2-13) were treated with a PARP inhibitor (AG14361) at the indicated concentrations for 6 days, and survival fraction was measured by XTT assay (mean ± SEM, n=6).
In most of the cisplatin-resistant Capan-1 clones without secondary mutations, IR-induced RAD51 foci formation remained impaired (Fig. 3a), suggesting that these clones acquired cisplatin resistance through mechanisms other than the restoration of the BRCA2-RAD51 pathway.
Next, we analyzed the homologous recombination (HR)-based DNA double-strand-break (DSB) repair function of some of the novel BRCA2 proteins using an I-SceI-dependent DR-GFP reporter assay in BRCA2-deficient VC-8 Chinese hamster cells 22. The proportions of GFP-positive cells arising through the repair of I-SceI-induced DSB by HR after transfection of various mutant BRCA2 constructs were compared (Figs. 3b and S6, Table S2). Transfection of a wild-type BRCA2 construct resulted in about 5-fold more GFP-positive cells compared to transfection of vector control or the BRCA2.6174delT mutant. Transfection of constructs with any of the secondary BRCA2 mutations resulted in GFP-positive frequencies equal to or greater than that of the construct with wild-type BRCA2. These results indicate that the novel BRCA2 proteins efficiently promote HR.
Poly(ADP-ribose) polymerase (PARP) inhibitors selectively kill BRCA1/2-deficient tumor cells 23, 24 and are expected to become a therapeutic option for patients with BRCA1/2-mutated cancers 13. However, it remains unclear whether BRCA1/2-mutated tumors with acquired cisplatin resistance are sensitive to PARP inhibitors. We tested the sensitivity of cisplatin-resistant Capan-1 clones to a PARP inhibitor (AG14361) (Fig. 3c). Both parental Capan-1 and cisplatin resistant clones without secondary mutations were sensitive to the PARP inhibitor. In contrast, Capan-1 clones with secondary BRCA2 mutations were resistant, consistent with the restoration of functional BRCA2 in these clones, although the differences of the sensitivity between parental Capan-1 and these clones were relatively small.
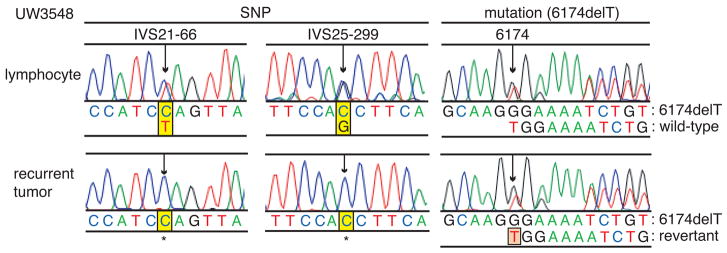
Finally, we analyzed tissues of 5 patients with BRCA2 mutations and recurrent ovarian carcinomas previously treated with platinum. Three recurrent tumors were clinically refractory to platinum and two were sensitive (Table S3). Platinum-refractory tumor UW3548 revealed genetic reversion of BRCA2.6174delT (Fig. 4). Evidence for genetic reversion is as follows. Constitutional DNA of this patient was heterozygous for BRCA2.6174delT and at two single nucleotide polymorphisms (SNPs). As expected, the microdissected specimen of the recurrent tumor was hemizygous at each SNP, reflecting loss of heterozygosity of BRCA2 in the tumor. However, in the same tumor sample, both sequences of BRCA2.6174delT and wild-type BRCA2 were detected, suggesting that the recurrent tumor had acquired wild-type BRCA2 by genetic reversion, or back mutation to wild-type. We speculate that the appearance of both mutant and wild-type sequences resulted from duplication of the mutant BRCA2 followed by genetic reversion of one of the duplicates, similar to the situation occurring in Capan-1 clones with secondary mutations (Figs. S4 and S5). A larger clinical study is warranted to determine the prevalence and clinical significance of in vivo secondary mutations in BRCA2 in human tumors.
Figure 4. Genetic reversion of BRCA2 in platinum-resistant recurrent BRCA2-mutated ovarian cancer.
DNA sequences of BRCA2 in peripheral blood lymphocytes and a microdissected specimen of the recurrent tumor from a patient (UW3548) with BRCA2-mutated ovarian cancer previously treated with cisplatin. In the lymphocytes, heterozygous single nucleotide polymorphisms (SNPs) of the BRCA2 locus (IVS21-66C/T and IVS25-299C/G) were detected, in addition to a heterozygous mutation (6174delT). In the microdissected recurrent tumor specimen, loss of heterozygosity (LOH) of the SNPs was confirmed, but mixed sequences of 6174delT and wild-type BRCA2 were detected.
The second platinum-refractory relapsed tumor (CS2) presented a complex profile. This patient carried germline mutations in both BRCA1 and BRCA2 (Fig. S7). The primary ovarian tumor was sensitive to platinum and showed loss of the wild-type BRCA1 allele and heterozygosity of BRCA2, indicating that carcinogenesis was primarily driven by the BRCA1 mutation. Importantly, the recurrent platinum-refractory tumor had lost the mutant BRCA2 allele. We speculate that loss of the mutant BRCA2 allele was the result of selection by chemotherapy and contributed to platinum resistance.
The tumor of a third patient (UW174) recurred 5 months after her primary chemotherapy, so her tumor is defined as clinically refractory to platinum. No secondary mutation in BRCA2 was detectable in the tumor specimen of this patient. Recurrent tumors of two other patients (CS9 and CS15) were sensitive to cisplatin. No secondary genetic alterations in BRCA2 were identified in either of these tumors.
Taken together, our data demonstrate that secondary mutations that correct the frameshift caused by mutated BRCA2 alleles are a mechanism of acquired resistance to cisplatin (Fig. S8). Testing for secondary mutations in platinum-treated BRCA2-mutated cancers may be clinically important, because tumors with secondary mutations are likely to be resistant to both cisplatin and PARP inhibitors. Theoretically, it might be possible to re-sensitize these tumors to cisplatin and to PARP inhibitors by treatment with drugs such as proteasome inhibitors that inhibit RAD51 recruitment to sites of DNA repair 25. In contrast, platinum-resistant BRCA2-mutated tumors without secondary BRCA2 mutations may remain sensitive to PARP inhibitors.
Treatment with cisplatin could facilitate secondary mutations by increasing the mutation rate via DNA damaging effects. Alternatively, since BRCA2 is involved in error-free DNA repair, HR, it is possible that BRCA2 deficiency itself promotes secondary mutations through compensatory utilization of more error-prone DNA repair processes 26.
Secondary mutations are relevant as a mechanism of resistance only in those tumors that contain frameshift mutations in BRCA2, which occurs in only 5% of ovarian carcinomas. However, BRCA2 mRNA expression is undetectable in 13% of ovarian carcinoma 27. We speculate that other mechanisms yet to be identified could restore BRCA2 in acquired cisplatin resistance in these sporadic cases.
Our findings may be applicable to cancers other than ovary. Secondary mutations in BRCA2 occurred in a mitomycin C-resistant human acute myeloid leukemia cell line with biallelic BRCA2 mutations derived from an FA (D1 subtype) patient 28 and BRCA2-mutated Chinese hamster fibroblast lines 29, suggesting that secondary mutations in BRCA2 may have a general role in resistance to DNA-crosslinking agents in cells derived from various organs.
Secondary mutation of mutated BRCA2 is reminiscent of mechanisms of acquired resistance of Philadelphia-chromosome-positive leukemia to imatinib 30, such as BCR-ABL point mutations that prevent imatinib binding. In both cases, the disease-causing mutations increase tumor sensitivity to specific agents, but development of further mutations in the disease-causing genes during targeted drug treatment leads to drug resistance. It will be important to explore mechanisms for restoring drug sensitivity.
METHODS SUMMARY
HCC1428, Capan-1, and a lymphoblast cell line (HCC1428BL) were purchased from the American Type Culture Collections. Cisplatin-resistant Capan-1 clones were generated by long-term culture in cisplatin (2 μM)-containing medium. PCR-amplified BRCA2 gene fragments were analyzed for sequence alterations with BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). All nucleotide numbers refer to the wild-type cDNA human sequence of BRCA2 (accession no. U43746; version U43746.1 GI: 1161383) and BRCA1 (U14680.1).
For western blot analysis, anti-BRCA2 Ab-1 (1:100; OP95, EMD Biosciences), anti-BRCA2 Ab-2 (1:100; PC146, EMD Biosciences), and anti-FANCA (1:1000, (#7488) (a gift from Dr. M Hoatlin)) were used. For immunofluorescence microscopy, cells were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) for 10 min, followed by permeabilization with 0.25% Triton-X-100 in PBS (10 min). A primary antibody (anti-RAD51 (Ab-1) (1:1000, PC130, EMD Biosciences)) and a secondary antibody (Alexa Fluor 488 goat anti-rabbit IgG (H+L) antibody (A-11034, Invitrogen)) were used.
Cisplatin sensitivity and PARP inhibitor sensitivity of cells were determined either by crystal violet or XTT (3′-1-phenylaminocarbonyl-3,4-tetrazolium-bis(4-methoxy-6-nitrobenzenesulfonic acid)) assays. A PARP inhibitor, AG14631 is a gift of Pfizer. Expression of targeted genes was knocked down by transient transfection of siRNA directed against FANCA (5′-AAGGGTCAAGAGGGAAAAATA-3′), BRCA2 (5′-AACAACAATTACGAACCAAAC-3′), and negative control (5′-AATTCTCCGAACGTGTCACGT-3′). HR assay was done as previously described 22. A BAC clone, RP11-37E23, was used as a probe for FISH on metaphase chromosome preparations of Capan-1 cells. Three DNA samples from BRCA2-mutated ovarian cancer patients were obtained from Cedars-Sinai Medical Center (Los Angeles, CA) through the Pacific Ovarian Cancer Research Consortium, and two were from the tissue bank of University of Washington (Seattle, WA). The study was approved by Institutional Review Boards of Fred Hutchinson Cancer Research Center and University of Washington.
METHODS
Cell lines
Capan-1, HCC1428BL and breast cancer cell lines (BT-474, BT-483, HCC38, HCC1500, HCC1428, HCC1395, MCF7, MDA-MB-231, MDA-MB-436, MDA-MB-468) were purchased from American Type Culture Collections (Manassas, VA). 21NT was a gift from Dr. K Polyak (Dana Farber Cancer Institute). HCC1937, 2008 and 2008+FANCF were described 31, 32. HCC1428BL were grown in RPMI with 10% fetal calf serum (FCS) and other cell lines were grown in DMEM with 10% FCS. Gamma irradiation was delivered using a linear accelerator. 1.2 ×107 Capan-1 cells (106 cells/15 cm tissue culture plate × 12 plates) were selected in cisplatin (2 μM)-containing medium. Resistant colonies, appearing after 5–6 weeks, were picked, and expanded in normal medium.
RNA extraction
Total RNAs were extracted using Trizol (Invitrogen; Carlsbad, CA), and 2 μg of total RNA was placed in 20 μL reaction and converted to first-strand cDNA using Superscript III RNase H(-) Reverse Transcriptase and oligo(dT)15 (Invitrogen).
Genomic DNA extraction, PCR and Sequencing of BRCA2 and BRCA1
Genomic DNA was extracted from cell lines using QIAamp DNA Blood Mini Kit (Qiagen; Valencia, CA). PCR was performed using Expand High Fidelity PCR System (Roche). Sequences of PCR products were analyzed with BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems; Foster City, CA). Primers are listed in Tables S4 and S5. All nucleotide numbers refer to the wild-type cDNA human sequence of BRCA2 (accession no. U43746; version U43746.1 GI: 1161383) and BRCA1 (U14680.1) (GenBank database).
Western blotting
Cells were lysed with 1x sample buffer (50 mM Tris-HCl pH6.8, 86 mM 2-mercaptoethanol, 2% SDS), boiled for 5 min, and subjected to electrophoresis using NuPAGE 3–8% Tris-Acetate Gel (Invitrogen). Proteins were transferred to nitrocellulose membranes. After blocking with 5% non-fat dried milk in TBS-T (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% Tween 20), the membrane was incubated with the primary antibody (BRCA2 Ab-1 (a middle part (a.a. 1651–1821) of BRCA2) (1:100; OP95, EMD Biosciences; San Diego, CA), BRCA2 Ab-2 (C-terminus (a.a.3245–3418) of BRCA2) (1:100; PC146, EMD Biosciences), FANCA (1:1000, #7488, gift from Dr. M Hoatlin, Oregon Health and Science University) diluted in TBS-T, and incubated with the appropriate horseradish peroxidase-linked secondary antibody (Amersham). Chemiluminescence was used for detection.
Immunofluorescence microscopy
Cells were fixed with 4% paraformaldehyde in PBS for 10 min and permeabilized with 0.25% Triton-X-100 in PBS. After blocking in blocking buffer (3% bovine serum albumin/0.05% Triton X-100/0.04% sodium azide/PBS), a primary antibody (RAD51 (1:1000, PC130, EMD Biosciences)) diluted in blocking buffer was added and incubated for 1–2 hours. A secondary antibody (Alexa Fluor 488 goat anti-rabbit IgG (H+L) antibody (A-11034, Invitrogen)) (1:1000) was added and incubated for 1 hour. The nuclei were counterstained with 4′, 6-diamidino-2-phenylindole dihydrochloride. Images were captured on a microscope (TE2000, Nikon, Tokyo) through a CCD camera and processed using MetaVue (Universal Imaging, Downington, PA) and Photoshop (Adobe). Cells with five or more RAD51 foci were scored as positive. At least 100 cells per experimental point were scored.
Cytotoxicity assays
Drug sensitivity was determined by crystal violet or XTT (3′-1-phenylaminocarbonyl-3,4-tetrazolium-bis(4-methoxy-6-nitrobenzenesulfonic acid)) assays as previously described 33–35. Cells were seeded onto 12-well plates at 6 × 103 cells/well in DMEM-10% FCS (1.5 ml). After cells attached, the medium was replaced with DMEM-10% FCS containing cisplatin or a PARP inhibitor, AG14631 (a gift of Pfizer, La Jolla, CA) 36, at various concentrations. After incubation for 10 days, monolayers were fixed for 5 to 10 min in 10% methanol and 10% acetic acid. Adherent cells were stained with 1% crystal violet in methanol (0.5 ml/well). The adsorbed dye was resolubilized with methanol containing 0.1% SDS (0.3 ml/well). Dye solution (200 μl) was transferred to 96-well plates and diluted (1:2) in methanol. Crystal violet concentrations were measured photometrically (595 nm) in a microplate reader. For XTT assays, cells were seeded in 96-well plates at 2 × 103 cells/well (for HCC1428 and Capan-1 in Figure 1g) or 8 × 102 cells/well (for Capan-1 and its clones in Figure 3c), and cisplatin or AG14631 was added at varying concentrations. After 5 or 6 days, the number of viable cells was measured by the XTT assay as previously described 37. For HCC1428 cells, we only used XTT assays, because HCC1428 do not grow in the condition for the crystal violet assays.
siRNA transfection
HCC1428 cells (3×105 cells) were plated in 6-well plates at the time of transfection in 2 ml of complete media. siRNA oligos (FANCA (5′-AAGGGTCAAGAGGGAAAAATA-3′) 38, BRCA2 (5′-AACAACAATTACGAACCAAAC-3′) 38, and negative control (5′-AATTCTCCGAACGTGTCACGT-3′)) were diluted in 400 μL DMEM and were mixed with 12 μL of HiPerFect reagent (Qiagen). Complex formation was allowed to proceed for 10 min at room temperature prior to addition to cells. Final concentration of siRNA was 50 nM. Two days after transfection, cells were seeded in 96-well plates for cisplatin sensitivity assay.
Homologous recombination (HR) assay
Nucleotide changes were incorporated into BRCA2 cDNA pcDNA3.1 subclones using the QuikChange kit (Stratagene; La Jolla, CA). Mutant partial cDNA fragments were subcloned into a FLAG-tagged full-length BRCA2 cDNA pcDNA3.1 plasmid. VC8-DR-GFP cells 22 were transfected with either pcDNA3.1 vector, pcBAsce1 vector containing the I-SceI restriction endonuclease gene, or pcBAsce1 plus various FLAG tagged BRCA2-pcDNA3.1 constructs. The percentage of GFP positive cells was quantitated by flow cytometric analysis 72 hours after transfection as described 22. In parallel, the percentage of VC8-DR-GFP cells expressing the various constructs were determined by immunofluorescence with anti-FLAG antibody and were used to normalize the HR data for VC8 transfection efficiency.
Fluorescence In Situ Hybridization (FISH)
A BAC clone, RP11-37E23, was used as a probe for FISH on metaphase chromosome preparations of Capan-1 cells using procedures detailed previously 39.
Clinical specimens
Three DNA samples from BRCA2-mutated ovarian cancer patients were obtained from Cedars-Sinai Medical Center through the Pacific Ovarian Cancer Research Consortium, and two DNA samples were from the tissue bank of University of Washington. DNA from UW3548 was obtained after laser capture microdissection. The study was approved by Institutional Review Boards of Fred Hutchinson Cancer Research Center and University of Washington.
Supplementary Material
Supplementary Information is available online.
Acknowledgments
We thank Drs. MC King and CW Drescher for discussions, Dr. B Trask for overseeing the FISH analyses in her lab and comments on the manuscript, JW Huang for comments on the manuscript, and Drs. M Hoatlin and K Polyak for reagents. We thank Pfizer for AG14361. We thank the Pacific Ovarian Cancer Research Consortium (P50 CA83636, PI: N Urban) for clinical specimens. This work was supported by grants from the National Institutes of Health/National Cancer Institute (R01CA125636 to T.T.) (K08CA96610-01 to E.M.S.), Searle Scholars Program (to T.T.), V Foundation (to T.T.), and Hartwell Innovation Fund (to T.T.), and start-up funds from the Fred Hutchinson Cancer Research Center (to T.T.) and a gift from the Yvonne Betson Trust (to E.M.S.).
Footnotes
Author Contributions:
W.S. performed most of the experiments. E.M.S., B.Y.K., and N.U. provided clinical samples and expertise on ovarian cancer. E.M.S. performed laser capture micro dissection and DNA extractions. M.K.A., D.J.F. and F.J.C. performed homologous recombination assays. J.H. sequenced BRCA2 in HCC1428. C.F. performed FISH analysis. E.V. performed siRNA experiments shown in Figs 1f and 1g. C.J. performed PARP inhibitor sensitivity assays in Fig 3c. T.T., W.S., and E.M.S. wrote the manuscript.
Author Information:
The authors declare that they have no competing financial interests. Correspondence and requests for materials should be addressed to T.T. (ttaniguc@fhcrc.org).
References
- 1.Yuan SS, et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 2.Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer. 2003;3:502–516. doi: 10.1038/nrc1123. [DOI] [PubMed] [Google Scholar]
- 3.Goggins M, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- 4.Abbott DW, Freeman ML, Holt JT. Double-strand break repair deficiency and radiation sensitivity in BRCA2 mutant cancer cells. J Natl Cancer Inst. 1998;90:978–985. doi: 10.1093/jnci/90.13.978. [DOI] [PubMed] [Google Scholar]
- 5.Li AJ, Karlan BY. Genetic factors in ovarian carcinoma. Curr Oncol Rep. 2001;3:27–32. doi: 10.1007/s11912-001-0039-y. [DOI] [PubMed] [Google Scholar]
- 6.van Asperen CJ, et al. Cancer risks in BRCA2 families: estimates for sites other than breast and ovary. J Med Genet. 2005;42:711–719. doi: 10.1136/jmg.2004.028829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howlett NG, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 8.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 9.Neuhausen SL, Marshall CJ. Loss of heterozygosity in familial tumors from three BRCA1-linked kindreds. Cancer Res. 1994;54:6069–6072. [PubMed] [Google Scholar]
- 10.Collins N, et al. Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12-13. Oncogene. 1995;10:1673–1675. [PubMed] [Google Scholar]
- 11.Gudmundsson J, et al. Different tumor types from BRCA2 carriers show wild-type chromosome deletions on 13q12-q13. Cancer Res. 1995;55:4830–4832. [PubMed] [Google Scholar]
- 12.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 13.Tutt AN, et al. Exploiting the DNA repair defect in BRCA mutant cells in the design of new therapeutic strategies for cancer. Cold Spring Harb Symp Quant Biol. 2005;70:139–148. doi: 10.1101/sqb.2005.70.012. [DOI] [PubMed] [Google Scholar]
- 14.Foulkes WD. BRCA1 and BRCA2: Chemosensitivity, Treatment Outcomes and Prognosis. Fam Cancer. 2006;5:135–142. doi: 10.1007/s10689-005-2832-5. [DOI] [PubMed] [Google Scholar]
- 15.Boyd J, et al. Clinicopathologic features of BRCA-linked and sporadic ovarian cancer. JAMA. 2000;283:2260–2265. doi: 10.1001/jama.283.17.2260. [DOI] [PubMed] [Google Scholar]
- 16.Hirschhorn R. In vivo reversion to normal of inherited mutations in humans. J Med Genet. 2003;40:721–728. doi: 10.1136/jmg.40.10.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamanoue S, et al. Myeloid lineage-selective growth of revertant cells in Fanconi anaemia. Br J Haematol. 2006;132:630–635. doi: 10.1111/j.1365-2141.2005.05916.x. [DOI] [PubMed] [Google Scholar]
- 18.Xia B, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 19.Gazdar AF, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer. 1998;78:766–774. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 20.Neuhausen S, et al. Recurrent BRCA2 6174delT mutations in Ashkenazi Jewish women affected by breast cancer. Nat Genet. 1996;13:126–128. doi: 10.1038/ng0596-126. [DOI] [PubMed] [Google Scholar]
- 21.Saeki H, et al. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc Natl Acad Sci U S A. 2006;103:8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu K, et al. Functional evaluation and cancer risk assessment of BRCA2 unclassified variants. Cancer Res. 2005;65:417–426. [PubMed] [Google Scholar]
- 23.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 24.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 25.Jacquemont C, Taniguchi T. Proteasome function is required for DNA damage response and fanconi anemia pathway activation. Cancer Res. 2007;67:7395–7405. doi: 10.1158/0008-5472.CAN-07-1015. [DOI] [PubMed] [Google Scholar]
- 26.Tutt A, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilton JL, et al. Inactivation of BRCA1 and BRCA2 in ovarian cancer. J Natl Cancer Inst. 2002;94:1396–1406. doi: 10.1093/jnci/94.18.1396. [DOI] [PubMed] [Google Scholar]
- 28.Ikeda H, et al. Genetic Reversion in an Acute Myelogenous Leukemia Cell Line from a Fanconi Anemia Patient with Biallelic Mutations in BRCA2. Cancer Res. 2003;63:2688–2694. [PubMed] [Google Scholar]
- 29.Wiegant WW, Overmeer RM, Godthelp BC, van Buul PP, Zdzienicka MZ. Chinese hamster cell mutant, V-C8, a model for analysis of Brca2 function. Mutat Res. 2006;600:79–88. doi: 10.1016/j.mrfmmm.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 31.Taniguchi T, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 32.Garcia-Higuera I, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi T, et al. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- 34.Chirnomas D, et al. Chemosensitization to cisplatin by inhibitors of the Fanconi anemia/BRCA pathway. Mol Cancer Ther. 2006;5:952–961. doi: 10.1158/1535-7163.MCT-05-0493. [DOI] [PubMed] [Google Scholar]
- 35.Naf D, Kupfer GM, Suliman A, Lambert K, D’Andrea AD. Functional activity of the fanconi anemia protein FAA requires FAC binding and nuclear localization. Mol Cell Biol. 1998;18:5952–5960. doi: 10.1128/mcb.18.10.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skalitzky DJ, et al. Tricyclic benzimidazoles as potent poly(ADP-ribose) polymerase-1 inhibitors. J Med Chem. 2003;46:210–213. doi: 10.1021/jm0255769. [DOI] [PubMed] [Google Scholar]
- 37.Yamashita T, Barber DL, Zhu Y, Wu N, D’Andrea AD. The Fanconi anemia polypeptide FACC is localized to the cytoplasm. Proc Natl Acad Sci U S A. 1994;91:6712–6716. doi: 10.1073/pnas.91.14.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruun D, et al. siRNA depletion of BRCA1, but not BRCA2, causes increased genome instability in Fanconi anemia cells. DNA Repair (Amst) 2003;2:1007–1013. doi: 10.1016/s1568-7864(03)00112-5. [DOI] [PubMed] [Google Scholar]
- 39.Trask BJ. Fluorescence in situ hybridization. In: Birren B, et al., editors. Genome Analysis: A Laboratory Manual. Vol. 4. Cold Spring Harbor Laboratory Press; 1999. pp. 303–413. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is available online.