Abstract
NK cells identify infected, neoplastic, or MHC-disparate target cells via several different receptors. The NK cell receptor KLRE1 lacks known signaling motifs but has nevertheless been shown to regulate NK cell-mediated cytotoxicity. Here we demonstrate that KLRE1 forms functional heterodimers with either KLRI1 or KLRI2. Cotransfection with KLRE1 was necessary for surface expression of the NK cell receptor chains KLRI1 and KLRI2 in 293T cells. Moreover, KLRE1 can be coimmunoprecipitated with KLRI1 or KLRI2 from transfected NK cell lines. By flow cytometry, KLRE1 and KLRI1 showed colinear expression on NK cells, suggesting surface expression as heterodimers. Unlike other killer cell lectin-like receptors, KLRE1/KLRI1 and KLRE1/KLRI2 heterodimers predominantly migrated as single chains in SDS-PAGE, indicating noncovalent association. KLRI1 was coimmunoprecipitated with the tyrosine phosphatase Src homology region 2 domain-containing phosphatase 1. In accordance with an inhibitory function, anti-HA Ab induced reduced killing of FcR-bearing targets by KLRI1-HA-transfected NK cell lines in a redirected cytotoxicity assay. Reciprocally, KLRI2-HA transfectants displayed increased killing in this assay. Finally, Ab to KLRE1 induced inhibition in KLRI1-transfected cells but increased cytotoxicity in KLRI2 transfectants, demonstrating that KLRE/I1 is a functional inhibitory heterodimer in NK cells, whereas KLRE/I2 is an activating heterodimeric receptor.
NK cells recognize and kill certain tumor cells, infected cells, and MHC class I-disparate normal hemopoietic cells (1-3). The effector functions of NK cells are controlled by a balance of inhibitory and activating signals generated by a variety of receptors (4). Inhibitory receptors carry ITIM motifs in their cytoplasmic regions, which upon tyrosine phosphorylation recruit and activate the protein tyrosine phosphatase Src homology region 2 domain-containing phosphatase 1 (SHP-1).3 The activating NK receptors typically associate via charged amino acid residues in their transmembrane regions to adaptor molecules that contain either ITAM necessary for recruitment of Syk or ZAP70 tyrosine kinases (DAP12, FcεRIγ, CD3ζ; Ref. 4) or another tyrosine-based motif (YxNM) that recruits the p85 subunit of PI3K and/or Grb2-Vav1 (DAP10; Refs. 4 and 5). One of the two main categories of NK cell receptors consists of type II transmembrane receptors with extracellular C-type lectin-like domains, all encoded by the NK cell gene complex, and collectively termed killer cell lectin-like receptors (KLR). Included in this group are the Ly-49 (KLRA) and NKR-P1 (KLRB) multigene families, in addition to NKG2D (KLRK1), MAFA-1 (KLRG1), and CD94/NKG2 (KLRD1/KLRC1, −2, or −3; Refs 4 and 6). The Ly-49 multigene family members bind to MHC class I- or class I-like ligands, and either inhibit or activate NK effector functions (4, 6). Similarly, CD94 forms disulfide-linked heterodimers with either the ITIM-bearing, inhibitory NKG2A chain or the activating NKG2C chain that associates with the ITAM-bearing DAP12 adaptor (7-10).
We have previously reported the identification of KLRE1, a receptor expressed by mouse and rat NK cells (11, 12). In rat NK cells, KLRE1 was demonstrated to induce redirected inhibition in cytotoxicity assays (11). In contrast, data in the mouse indicated an activating function for KLRE1 (13), and KLRE1-deficient mice have a severely compromised capacity for rejection of allogeneic bone marrow transplants (14). Both rat and mouse KLRE1 lack known signaling motifs, suggesting that KLRE1 associates with other receptor chains that contain signaling motifs (11). In pursuit of these chains, we identified two novel NK cell receptors, KLRI1 and KLRI2 (15). KLRI1 contains two ITIM in tandem orientation. KLRI2 lacks ITIM but contains a charged lysine residue in the transmembrane region (15). Rat KLRE1 was previously shown to exist predominantly as a single chain in lysates from NK cells (11). Similarly, we have also shown that both KLRI1 and KLRI2 migrate as single chains in SDS-PAGE (15). These three chains thus differ from other C-type lectin-like NK receptors, which are expressed as disulfide-linked dimers (4, 6, 7).
Here, we demonstrate that KLRE1 forms noncovalent heterodimers with KLRI1 and KLRI2. Heterodimers of KLRE1 and KLRI1 (KLRE/I1) inhibit NK cell cytotoxicity, whereas heterodimers of KLRE1 and KLRI2 (KLRE/I2) induce killing in redirected lysis assays.
Materials and Methods
Expression constructs
The rat KLRE1-HA, KLRI1-FLAG, and KLRI2-HA expression constructs were generated as previously described (11, 15). The rat and mouse KLRI1-HA constructs, carrying a C-terminal HA epitope tag, were generated by PCR amplification followed by insertion into pECHA (a bicistronic, stable expression vector with SRα promoter upstream of a cloning site followed by an HA tag sequence). The KLRI2-HA + KLRE1 double-expression construct used in transient and stable 293T cell transfections were generated by PCR inserting a KLRI2-HA fragment (with a C-terminal HA tag) into pBudCE4.1 (Invitrogen) under the CMV promoter, and a rat KLRE1 (untagged) fragment under the EF-1α promoter. Single-expression constructs carrying either KLRE1 or KLRI2-HA were also generated. Mutated versions of KLRI2-HA (replacing lysine residue 95 with isoleucine) were generated from the KLRI2-HA and KLRE1 + KLRI2-HA constructs by site-directed mutagenesis (QuikChange; Stratagene). All expression constructs used were verified by sequencing.
Transfections
For transient 293T cell transfections, 6 μg DNA was preincubated with 20 μl of Lipofectamine (Invitrogen) for 15 min at room temperature in 0.5 ml of Opti-MEM (Invitrogen), mixed with 2 ml of Opti-MEM and added to a 60−80% confluent monolayer of 293T cells in a 25-cm2 flask. After 6 h of incubation at 37°C, 2.5 ml of RPMI 1640 containing 20% FCS (Invitrogen) was added to the cells; 40 h after transfection, cells were harvested using 0.5 mM EDTA in PBS. For stable transfections, 3 × 106 RNK-16 or RNKDA1 cells (11) were mixed with 20 μg of linearized plasmid at 4°C in complete medium (RPMI 1640 with 1 mM sodium pyruvate, 1% antibiotic/antimycotic solution, 10% FCS (all Invitrogen), and 5 × 10−5 M 2-ME) and electroporated in a 2-mm cuvette at 120 V, 960 μF (GenePulser; Bio-Rad Laboratories). Stably transfected cells were selected by cultivation in complete medium supplemented with 2 mg/ml geneticin (G-418 disulfate; Duchefa Biochemie) and tested for gene expression by flow cytometry. Rat rIL-2 (16) equivalent to 1000 IU/ml of human IL-2 was added to the RNKDA1 cell medium at all times.
Flow cytometry
For single-color analysis, 293T cells (107 cells/ml) were incubated with anti-HA (10 μg/ml HA.11; Covance Research Products), anti-FLAG (2 μg/ml M2; Sigma-Aldrich), or anti-rat KLRE1 (2 μg/ml WEN27) (11) primary mAb in a total volume of 50 μl at 4°C for 30 min, washed three times, and incubated with FITC-conjugated F(ab′)2 fragments of sheep anti-mouse IgG (Sigma-Aldrich). For two-color analysis, cells were labeled in five steps with 2 μg/ml anti-rat KLRE1 mAb (WEN27) or isotype-matched (IgG1) irrelevant mAb (W3/25; anti-rat CD4; Ref. 17) FITC-conjugated F(ab′)2 fragments of sheep anti-mouse IgG (Sigma-Aldrich); mouse IgG (Sigma-Aldrich); then either biotinylated anti-rat NKp46 (WEN23; Ref. 18), biotinylated anti-HA (HA.11; Covance Research Products) mAb; or isotype-matched (IgG1) irrelevant mAb (biotinylated AKS1; anti-bovine NKp46; Ref. 19) and finally streptavidin-Alexa 647 conjugate (Molecular Probes). Labeled cells were analyzed on a flow cytometer (FACSCalibur, BD Biosciences).
Biochemistry
Transiently transfected 293T cells were lysed in 1% Igepal CA-630 (Sigma-Aldrich), 20 mM Tris-HCl, 100 mM NaCl, 10 mM sodium pyrophosphate, 1 mM Na3VO4, 1 mM PMSF, 5 mM EDTA, and a protease inhibitor mixture (Sigma-Aldrich) at 4°C for 30 min. Protein samples were subjected to SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes (Immobilon P; Millipore). Membranes were probed with either 1 μg/ml rabbit anti-FLAG Ab (Sigma-Aldrich) or 2 μg/ml anti-HA mAb (Covance Research Products) followed by incubation with HRP conjugates of either goat anti-rabbit Ig or sheep anti-mouse Ig (Jackson ImmunoResearch Laboratories). For coimmunoprecipitation of SHP-1, RNKDA1 cells were stimulated with pervanadate in PBS (pH 7.4), 1 mM Na3VO4 at 37°C for 5 min, followed by lysis in 1% digitonin (Calbiochem), 25 mM Tris-HCl, 150 mM NaCl (pH 7.5), 1 mM NaF, 1 mM PMSF, 1 mM Na3VO4 10 μg/ml leupeptin, and 10 μg/ml aprotinin as previously described (11). Lysates were subjected to immunoprecipitation using anti-HA affinity matrix (Roche) at 4°C for 2 h. Subsequently, PVDF membranes were probed with either 0.2 μg/ml anti-SHP-1 rabbit Ab or 0.4 μg/ml rabbit anti-HA Ab (Santa Cruz Biotechnology), followed by incubation with HRP-conjugated goat anti-rabbit IgG (Jackson Immuno Research Laboratories). The anti-rat KLRE1 mAb WEN27 has low sensitivity in Western blotting. To this end, an affinity-purified, KLRE1-specific polyclonal rabbit Ab was generated by immunization with a synthetic peptide derived from the cytoplasmic region of rat KLRE1. For KLRE1 coimmunoprecipitations, RNKDA1 cells were lysed in 50 mM n-octylglucoside (Sigma-Aldrich), 0.5% Igepal CA-630 (Sigma-Aldrich), 20 mM Tris-HCl, 100 mM NaCl (pH 7.5), 1 mM NaF, 1 mM PMSF, 1 mM Na3VO4, 10 μg/ml leupeptin, and 10 μg/ml aprotinin.
Lysates were subjected to immunoprecipitation using anti-HA (Roche) or anti-FLAG (EZview M2 affinity gel; Sigma-Aldrich) affinity matrix at 4°C for 2 h. Subsequently, PVDF membranes were probed with 2 μg/ml anti-HA mAb (Covance Research Products), 1 μg/ml rabbit anti-FLAG Ab (Sigma-Aldrich), or 1.3 μg/ml rabbit anti-rat KLRE1 Ab, followed by incubation with HRP-conjugated goat anti-mouse or goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories) and chemiluminescent detection (Super Signal Substrate System from Pierce Bio-technology; BioMax MR film from Eastman Kodak).
Cytotoxicity assay
Standard 4-h redirected cytotoxicity assays were set up as follows: 5 × 106 P388D1, P815, or YAC-1 target cells were labeled with 100 μCi of Na251CrO4 (Amersham Biosciences) for 1 h at 37°C, washed three times in PBS, resuspended to 105 cells/ml in complete medium, and added to 96-well plates at 100 μl/well. Untransfected or stably transfected effector RNK-16 cell lines were preincubated in complete medium with either 2 μg/ml anti-HA (HA.11; Covance), 5 μg/ml anti-rat KLRE1 (WEN27; Ref. 11), 2 μg/ml anti-rat NKp46 (WEN23; Ref. 18), 2 μg/ml anti-rat CD43 (WEN1; Ref. 20), all mouse IgG1 mAbs; or no Ab for 20 min at room temperature and mixed with target cells in different ratios as indicated. After a 4-h incubation at 37°C, the supernatants were harvested and counted in a gamma counter (Minaxi Auto Gamma 5000 Series; Packard). Spontaneous release was between 5 and 10% for all experiments. Specific lysis was calculated from median values as previously described (21).
Results
KLRE1 is necessary for surface expression of KLRI1
As we have previously demonstrated, KLRE1 was expressed on the cell surface of transfected 293T cells as a disulfide-linked homodimer. In NK cell lysates, however, KLRE1 appeared to be expressed mostly in a noncovalently linked form (11). Similarly, rat KLRI1 and KLRI2 are expressed predominantly as noncovalently linked molecules in transfected NK cells (15). To investigate whether KLRE1 and KLRI1 could dimerize, an expression construct encoding rat KLRI1-FLAG and expression constructs encoding rat KLRI1-FLAG or KLRE1-HA (both with extracellular C-terminal epitope tags) were generated. Whereas transfected NK cells readily express KLRI1 (15), rat KLRI1-FLAG was not detected on the cell surface of transiently transfected 293T cells in flow cytometry analysis using an anti-FLAG mAb. Cotransfection with a rat KLRE1-HA expression construct, however, induced cell surface expression of KLRI1-FLAG (Fig. 1A). Using an anti-HA mAb, rat KLRE1-HA was identified on the cell surface by flow cytometry both in single rat KLRE1-HA transfections and in cotransfections with rat KLRI1-FLAG. The same observations were made when cotransfecting with untagged rat KLRE1 and detection with the anti-KLRE1 mAb WEN27 (data not shown). Efficient translation into KLRI1 protein in single transfected cells was verified by Western blotting (Fig. 1B). In similar transfections with mouse KLRE1 and mouse KLRI1-HA expression constructs (using the anti-KLRE1 mAb DX6; Ref. 11), KLRI1-HA was detected on the cell surface only when cotransfected with KLRE1 (data not shown). These data demonstrate that, in both species, KLRI1 is dependent on KLRE1 for cell surface expression in 293T cells. The transfection experiments were also performed in Chinese hamster ovary cells, with similar observations (data not shown).
FIGURE 1.
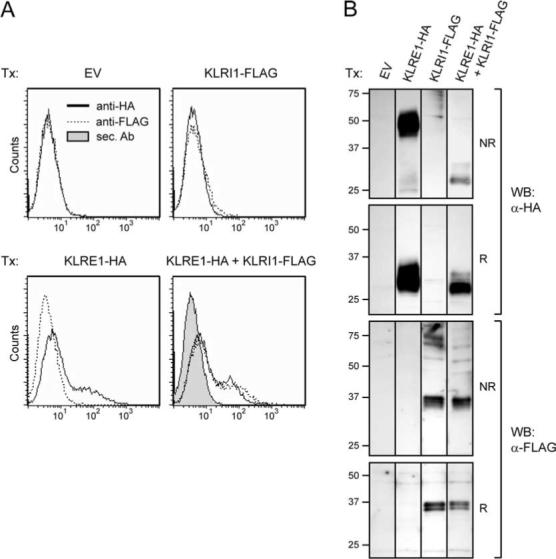
Surface expression of KLRI1 depends on coexpression of KLRE1. A, Flow cytometry analysis of 293T cells transiently transfected (Tx) with KLRI1-FLAG, KLRE1-HA, KLRI1-FLAG + KLRE1-HA, or empty vector (EV). Surface expression was detected using anti (α)-HA  or anti-FLAG
or anti-FLAG 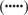 mAbs (both IgG1); or secondary (sec.) Ab alone
mAbs (both IgG1); or secondary (sec.) Ab alone  . B, Western blot (WB) analysis of whole-cell lysates of transfected 293T cells, under reducing (R) or nonreducing (NR) conditions, using anti-HA or anti-FLAG Abs as indicated. Relative molecular masses are indicated.
. B, Western blot (WB) analysis of whole-cell lysates of transfected 293T cells, under reducing (R) or nonreducing (NR) conditions, using anti-HA or anti-FLAG Abs as indicated. Relative molecular masses are indicated.
In parallel to flow cytometry, whole-cell lysates of transfected 293T cells were subjected to Western blot analysis using anti-HA and anti-FLAG Abs. As in transfected NK cells (15), rat KLRI1-FLAG protein was predominantly detected as two bands of ∼35 and ∼37 kDa under both reducing and nonreducing conditions (Fig. 1B). Importantly, under nonreducing conditions, rat KLRE1 migrated as a disulfide-linked homodimer in lysates from single transfections, but predominantly as a monomer in lysates from cotransfected cells (Fig. 1B). These results suggest that KLRE1 preferentially associates noncovalently with KLRI1 instead of forming disulfide-linked homodimers. The existence of KLRE1/KLRI1 heterodimers finds precedence in another KLR heterodimeric receptor. CD94 forms covalent heterodimers with either the ITIM-bearing NKG2A or the DAP12-binding NKG2C chains (7-10, 22). We have previously demonstrated that rat KLRE1 and NKG2A do not dimerize in cotransfected 293T cells (11). To investigate a possible association between CD94 and KLRI1, 293T cells were transiently transfected with rat CD94-HA and KLRI1-FLAG expression constructs. Flow cytometry analysis of cotransfected cells showed no surface expression of rat KLRI1-FLAG, indicating that KLRI1 does not dimerize with CD94 (data not shown).
KLRI2 surface expression in transfected cells depends on KLRE1
Like KLRI1, single transfections with rat KLRI2 constructs did not induce surface expression in 293T cells. Cotransfections with KLRE1, however, induced surface expression of KLRI2-HA, suggesting that KLRE1 and KLRI2 form heterodimers (Fig. 2). Whereas transfected NK cell lines expressed high levels of KLRI2 (15), the level of surface expression of KLRI2 was consistently found to be low in several types of transfection experiments in non-NK cells. Mouse and rat KLRI2 contain a positively charged residue (lysine) in the transmembrane region, similar to activating receptors that associate with ITAM-bearing adaptor proteins. Lack of the appropriate ITAM-bearing adaptor protein in 293T cells thus provides a possible explanation for the observed low surface expression of KLRI2 in these cells. To this end, mutated constructs were generated with the transmembrane lysine replaced by an isoleucine residue (KLRI2-HA K95I). The mutated constructs consistently induced high levels of surface expression in transient as well as stably transfected 293T cells (Fig. 2).
FIGURE 2.
KLRI2 depends on KLRE1 for cell surface expression in 293T cells. A, Flow cytometry analysis of 293T cells transiently transfected (Tx) with wild-type KLRI2-HA or KLRI2-HA carrying a mutated transmembrane region (KLRI2-HA K95I) either alone or in combination with KLRE1. B, Flow cytometry analysis of 293T cells untransfected or stably transfected with wild-type or mutated KLRI2-HA. EV, Empty vector. Surface expression was detected by anti-rat KLRE1 mAb WEN27  , anti-HA (−), or secondary (sec.) Ab alone
, anti-HA (−), or secondary (sec.) Ab alone  .
.
KLRE1 coimmunoprecipitates with KLRI1 and KLRI2
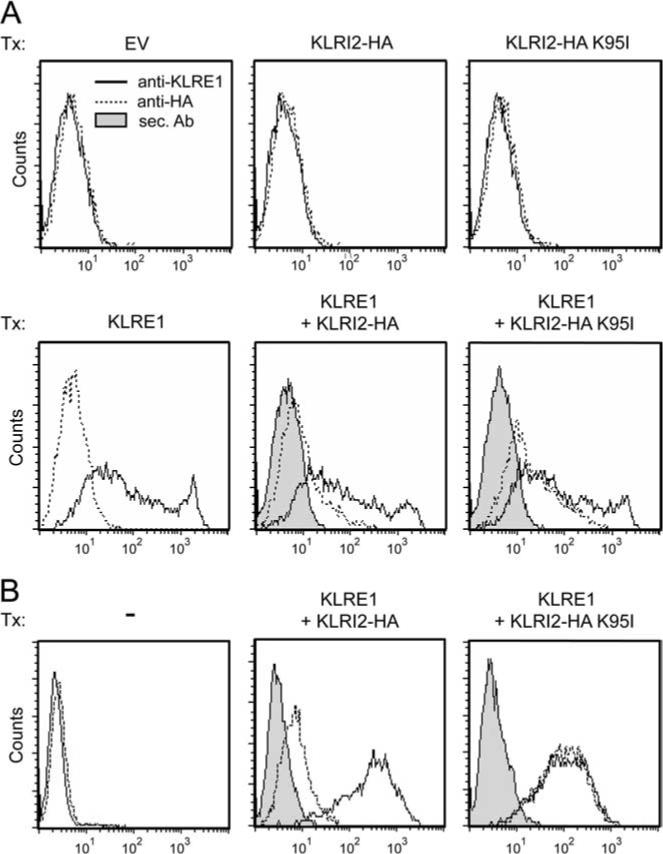
The rat NK cell line RNKDA1 was stably transfected with KLRI1-HA or KLRI2-FLAG expression constructs, and surface expression of endogenous KLRE1 as well as the epitope-tagged KLRI1 or KLRI2 chains was verified by flow cytometry. The transfectants were lysed, immunoprecipitated with anti-HA or anti-FLAG affinity matrix, and subjected to Western blot analysis under nonreducing conditions. In the KLRI1-HA transfectants, anti-HA immunoprecipitation yielded a strong ∼24 kDa band that reacted with a polyclonal Ab toward KLRE1, representing a KLRE1 single chain. The KLRE1 band observed in whole cell lysates had the same molecular mass. KLRI2-FLAG transfectants, similarly, yielded a strong ∼24-kDa KLRE1 single-chain band upon FLAG immunoprecipitation (Fig. 3). These observations indicate that KLRE1 forms noncovalently linked homodimers with either KLRI1 or KLRI2 in NK cells. Although not readily detectable in whole-cell lysates, a KLRE1 band of ∼70 kDa was also consistently observed in both KLRI1-HA and KLRI2-FLAG-transfected cells. This band was markedly diminished under reducing conditions, suggesting that it represented a disulfide-linked complex (Fig. 3). Anti-HA and anti-FLAG Abs also reacted with a band of similar mobility, suggesting that the ∼70-kDa band represented a complex containing both KLRE1 and KLRI1-HA or KLRE1 and KLRI2-FLAG (data not shown). Because this band was not clearly visible in whole-cell lysates, and thus unlikely to represent more than a very small fraction of KLRE/KLRI receptors, it may represent a minor fraction of covalently bonded heterodimers selectively enriched by immunoprecipitation.
FIGURE 3.
KLRI1 and KLRI2 coimmunoprecipitate with KLRE1. RNKDA1 cells stably transfected with KLRI1-HA (A) or KLRI2-FLAG (B) expression constructs were lysed in n-octylglucoside + Igepal buffer, immunoprecipitated with anti-HA or anti-FLAG affinity matrix, separated by SDS-PAGE under nonreducing (NR) or reducing (R) conditions, and subjected to Western blot analysis with a polyclonal Ab toward rat KLRE1. Relative molecular masses are indicated. RNKDA1 cells express both KLRE1 and KLRI1 endogenously, providing an explanation as to why a KLRE1 homodimeric band is not observed in this case, as was the case with 293T cells single-transfected (Tx) with KLRE1 (Fig. 1B). WCL, Whole-cell lysates.
Colinear surface expression of KLRE1 and KLRI1 on NK cells
To investigate whether the formation of noncovalent KLRE1/KLRI1 heterodimers in 293T cells was enforced by a lack of appropriate partner chains, the rat NK cell line RNK-16 was stably transfected with a rat KLRI1-HA expression construct. RNK-16 cells express endogenous KLRE1 on the cell surface identified by flow cytometry. No KLRI1 mRNA was detectable by RT-PCR from this cell line (15). Two-color flow cytometry demonstrated a colinear distribution of KLRE1 and KLRI1-HA surface expression in transfected RNK-16 cells. As a control, cells stained with anti-HA and anti-rat NKp46 mAbs displayed independent expression levels. These data indicate that KLRI1 associates with KLRE1 and not with other KLR chains expressed by NK cells (including CD94, which is expressed in RNK-16 cells; Ref. 23) and suggest a model in which KLRI1 and KLRE1 bind each other exclusively (Fig. 4).
FIGURE 4.
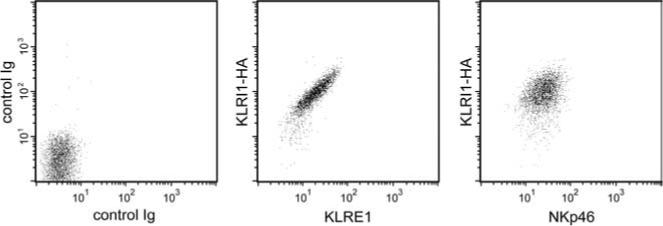
Colinear surface expression of KLRE1 and KLRI1 in NK cells. Two-color flow cytometry analysis of RNK-16 cells stably transfected with KLRI1-HA, using biotinylated anti-HA mAb and either anti-rat KLRE1 (center) or anti-rat NKp46 mAbs (right). Left, Control staining with isotype-matched nonrelevant mAbs.
KLRI1 associates with the protein tyrosine phosphatase SHP-1
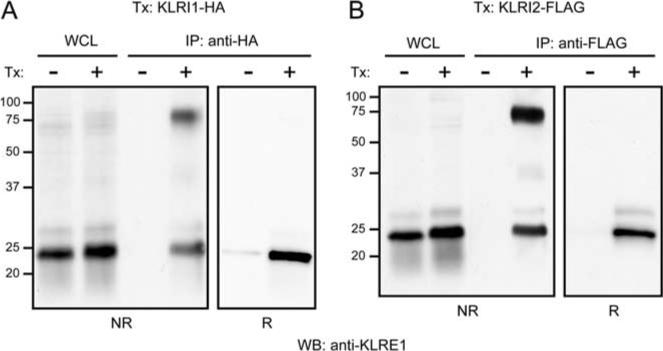
Mouse and rat KLRI1 have two tandem-orientated ITIM in the cytoplasmic domain, suggesting an inhibitory function. We have previously demonstrated in untransfected NK cells that KLRE1 can be coimmunoprecipitated with the tyrosine phosphatase SHP-1, despite the absence of ITIM motifs, and proposed that this association was indirect and involved a partner chain with ITIM motifs (11). To assess the ability of KLRI1 to recruit SHP-1, the IL-2-dependent rat NK cell line RNKDA1 was stably transfected with a rat KLRI1-HA expression construct. Untransfected and rat KLRI1-HA transfected cells were incubated in PBS with or without sodium pervanadate, lysed in digitonin buffer, and subjected to immunoprecipitations with anti-HA affinity matrix. By Western blotting, SHP-1-specific bands were identified in anti-HA precipitates from transfected but not untransfected cells (Fig. 5). The amount of coprecipitated SHP-1 was higher from pervanadate-stimulated cells, indicating that association was likely dependent on ITIM phosphorylation. Nevertheless, a substantial SHP-1 band was reproducibly observed with unstimulated cells, suggesting that one or both KLRI1 ITIMs to some extent might be constitutively phosphorylated. These results suggest that KLRI1 recruits the phosphatase SHP-1, similar to KLR receptors that have an inhibitory function in NK cells.
FIGURE 5.
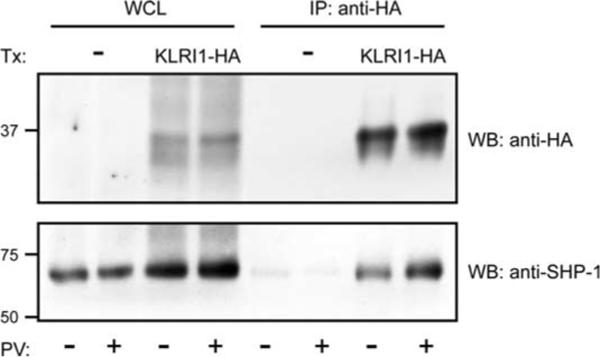
KLRI1 associates with the protein tyrosine phosphatase SHP-1. Western blots (WB) of whole-cell lysate (WCL) and anti-HA immunoprecipitates (IP) under nonreducing conditions. Stably transfected (Tx: KLRI1-HA) or nontransfected (Tx: —) RNKDA1 cells were either untreated (PV: —) or stimulated (PV: +) with pervanadate. Relative molecular masses are indicated.
KLRE1 inhibits or activates NK cell-mediated cytotoxicity dependent on association with KLRI1 or KLRI2
To assess signaling functions of KLRI1 and KLRI2, the rat NK cell line RNK-16 was stably transfected with rat KLRI1-HA, rat KLRI2-HA, or mouse KLRI1-HA expression constructs (Fig. 6A). KLRI-HA+ transfectants were used as effector cells in redirected lysis assays using the FcR+ target cell lines P388D1 and P815 and the FcR− target cell line YAC-1. When rat KLRI1-HA- or mouse KLRI1-HA-transfected cells were used as effector cells, preincubation with anti-HA mAb induced redirected inhibition toward P388D1 (Fig. 6B). In contrast, rat KLRI2-HA transfected cells preincubated with anti-HA mAb killed P388D1 and P815 more efficiently compared with preincubation with no Ab or control Ab (Fig. 6, B and C), demonstrating that KLRI1 functions as an inhibitory receptor, whereas KLRI2 activates NK cell-mediated cytotoxicity. None of the Abs had any effect on the FcR− control target cells YAC-1 (Fig. 6D).
FIGURE 6.
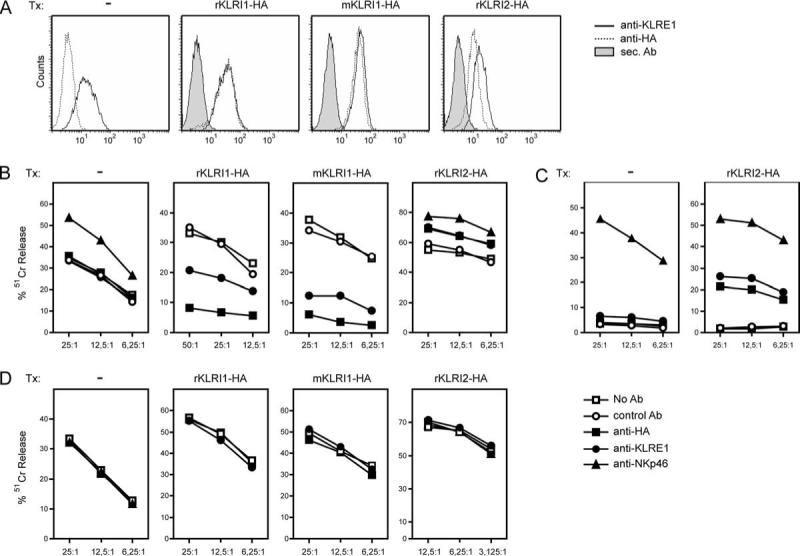
KLRE1/KLRI1 heterodimers inhibit whereas KLRE1/KLRI2 heterodimers activate NK cell-mediated cytotoxicity. A, Flow cytometry analysis of RNK-16 cells untransfected (Tx—) or stably transfected (Tx) with rat KLRI1-HA, rat KLRI2-HA, or mouse KLRI1-HA expression constructs. Surface expression was detected by anti-HA  or anti-KLRE1
or anti-KLRE1  mAbs; or secondary (sec.) Ab alone
mAbs; or secondary (sec.) Ab alone  . RNK-16 cells endogenously express KLRE1 and small amounts of KLRI2, but no KLRI1, as detected by RT-PCR (15) B–D, Redirected cytotoxicity assay using transfected or untransfected RNK-16 cells as effector cells and the tumor lines P388D1, P815, or YAC-1 as target cells. The effector cells were preincubated with anti-HA (■) or anti-rat KLRE1 (•) mAb before incubation with P388D1 (FcR+; B) or P815 (FcR+; C) target cells. Preincubation with anti-rat NKp46 (▲) or anti-rat CD43 (○) mAb served as positive and negative controls, respectively. The FcR− cell line YAC-1 was used as control target cells (D). Ordinate, Percentage specific lysis; abscissa, E:T ratios. Median values from one representative experiment (of three) are shown. Similar observations were made with two additional independent rat KLRI1-HA-, KLRI2-HA-, and mouse KLRI1-HA-stable transfectants.
. RNK-16 cells endogenously express KLRE1 and small amounts of KLRI2, but no KLRI1, as detected by RT-PCR (15) B–D, Redirected cytotoxicity assay using transfected or untransfected RNK-16 cells as effector cells and the tumor lines P388D1, P815, or YAC-1 as target cells. The effector cells were preincubated with anti-HA (■) or anti-rat KLRE1 (•) mAb before incubation with P388D1 (FcR+; B) or P815 (FcR+; C) target cells. Preincubation with anti-rat NKp46 (▲) or anti-rat CD43 (○) mAb served as positive and negative controls, respectively. The FcR− cell line YAC-1 was used as control target cells (D). Ordinate, Percentage specific lysis; abscissa, E:T ratios. Median values from one representative experiment (of three) are shown. Similar observations were made with two additional independent rat KLRI1-HA-, KLRI2-HA-, and mouse KLRI1-HA-stable transfectants.
In cytotoxicity assays, preincubation with anti-KLRE1 mAb induced redirected inhibition by rat KLRI1-HA- and mouse KLRI1-HA-transfected RNK-16 cells. In contrast, anti-KLRE1 mAb induced enhanced killing by rat KLRI2-HA-transfected RNK-16 cells (Fig. 6), demonstrating that KLRE1 associates with KLRI1 or KLRI2 to form functional heterodimers that inhibit or activate NK cell-mediated cytotoxicity, respectively. Preincubation of untransfected RNK-16 cells with anti-KLRE1 mAb had no significant effect on any target cell line tested, as expected because this cell line does not express endogenous KLRI1 and transcribes only very low endogenous amounts of KLRI2 (15).
Discussion
In this study, we have demonstrated that the NK cell receptor KLRE1 is expressed as a heterodimer with either KLRI1 or KLRI2. We previously cloned the rat and mouse KLRI1 and −2 receptors (15) as a direct result of searching the rat genomic trace sequence database for a partner chain for rat KLRE1, proposed to exist based on lack of known signaling motifs in the cytoplasmic region of KLRE1 (11).
Similar to other NK cell receptor pairs or families, the KLRE/I NK cell receptor pair consists of one inhibitory (KLRE/I1, short for KLRE1/KLRI1) and one activating (KLRE/I2, short for KLRE1/KLRI2) heterodimer. These results provide a possible explanation for conflicting reports. KLRE1 has been demonstrated to mediate inhibition of NK cell cytotoxicity in different rat strains but appeared to activate killing in mouse NK cell alloreactivity assays (11, 13, 14). Like most other KLRs, expression of KLRE/I1 and KLRE/I2 heterodimers is expected to be individually regulated on NK cells. Accordingly, different strains of mice and rats may generate subpopulations of NK cells that express either receptor alone or both receptors, thus leading to different outcomes when anti-KLRE1 mAb are used to cross-link these receptors. IL-2-activated NK cells from a panel of inbred rat strains were analyzed for KLRI1 and KLRI2 expression by Northern blot analysis. Indeed, the ratios of KLRI1 to KLRI2 transcripts varied between strains. However, IL-2-activated NK cells from the two extreme strains, DA (high KLRI1, low KLRI2) and LEW (low KLRI1, high KLRI2), both yielded redirected inhibition with an anti-KLRE1 mAb (data not shown). To further investigate this, surface expression levels of KLRI1 and KLRI2 on individual NK cells from different mouse and rat strains need to be investigated, allowing functional characterization of KLRE/I1 and -I2 subsets. These experiments await specific mAbs, currently being generated in our laboratory.
The activating signaling properties of KLRI2 are likely to be mediated through noncovalent association with an adaptor protein with a negatively charged transmembrane residue, as is the case for similar activating KLR chains as well as Ig-like NK cell receptors encoded by the leukocyte receptor gene complex. Thus, the observed low surface expression of wild-type KLRI2 in 293T cells is most likely due to a lack of expression of the appropriate adaptor protein in this cell type. In line with this, replacement of the positively charged lysine residue in the KLRI2 transmembrane region with a hydrophobic residue allowed efficient surface expression. This suggests that KLRI2 chains that fail to associate with the appropriate adaptor (missing in 293T, a human embryonic kidney-derived cell line) through a salt bridge involving the transmembrane lysine residue (85K) are retained intracellularly. 293T cells stably transfected with KLRE1 and KLRI2 were cotransfected with adaptor proteins known to associate (4) with KLR members or other activating NK cell receptors (DAP10, DAP12, FcεRIγ, or CD3ζ). None of these significantly enhanced surface expression of KLRI2 (data not shown), suggesting that another adaptor may associate with KLRI2 in NK cells. Efforts to identify the KLRI2-associated adaptor are ongoing.
Sequence comparisons and phylogram analysis of mouse and rat KLRI1 and −2 demonstrated intraspecies homogenization of their extracellular domains, suggesting that KLRE/I1 and KLRE/I2 bind structurally related or possibly identical ligands (15). Although no ligand has presently been identified, KLRE1-deficient mice have a reduced capacity for in vivo NK-mediated rejection of allogeneic bone marrow transplants (14), suggesting MHC-encoded ligands. No orthologs of rodent Klre1, Klri1, or Klri2 have been detected in the human genome sequence. It remains to be investigated to what extent the KLRE/I receptors in rat and mouse may serve as functional homologs of specific human NK cell receptors, in the same way that rodent Ly-49 receptors appear to be functional homologs of human killer cell Ig-like receptors.
The KLRE/I1 and KLRE/I2 pair of receptors further extends the receptor repertoire of rodent NK cells. Like CD94/NKG2 receptors, the KLRE/I pair of heterodimeric receptors harbors opposite signaling capacities. Further studies are warranted to explain the apparently prominent role of KLRE/I receptors in allogeneic bone marrow transplantation (13, 14), as well as in regulation of other NK cell effector functions.
Acknowledgments
We thank Wendi Jensen and Marianne Lauritzen for technical assistance.
Footnotes
This work was supported by the Research Council of Norway, the Norwegian Cancer Society, Bergljot and Sigurd Skaugen's fund, Anders Jahre's fund, and National Institutes of Health Grant AI068129. L.L.L. is an American Cancer Society Research Professor.
Abbreviations used in this paper: SHP-1, Src homology region 2 domain-containing phosphatase 1; KLR, killer cell lectin-like receptor; PVDF, polyvinylidene difluoride.
Disclosures The authors have no financial conflict of interest.
References
- 1.Wu J, Lanier LL. Natural killer cells and cancer. Adv. Cancer Res. 2003;90:127–156. doi: 10.1016/s0065-230x(03)90004-2. [DOI] [PubMed] [Google Scholar]
- 2.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu. Rev. Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 3.Rolstad B, Vaage JT, Naper C, Lambracht D, Wonigeit K, Joly E, Butcher GW. Positive and negative MHC class I recognition by rat NK cells. Immunol. Rev. 1997;155:91–104. doi: 10.1111/j.1600-065x.1997.tb00942.x. [DOI] [PubMed] [Google Scholar]
- 4.Lanier LL. NK cell recognition. Annu. Rev. Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 5.Upshaw JL, Arneson LN, Schoon RA, Dick CJ, Billadeau DD, Leibson PJ. NKG2D-mediated signaling requires a DAP10-bound Grb2-Vav1 intermediate and phosphatidylinositol-3-kinase in human natural killer cells. Nat. Immunol. 2006;7:524–532. doi: 10.1038/ni1325. [DOI] [PubMed] [Google Scholar]
- 6.Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat. Rev. Immunol. 2003;3:304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 7.Lazetic S, Chang C, Houchins JP, Lanier LL, Phillips JH. Human natural killer cell receptors involved in MHC class I recognition are disulfide-linked heterodimers of CD94 and NKG2 subunits. J. Immunol. 1996;157:4741–4745. [PubMed] [Google Scholar]
- 8.Braud VM, Allan DS, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 9.Vance RE, Kraft JR, Altman JD, Jensen PE, Raulet DH. Mouse CD94/NKG2A is a natural killer cell receptor for the nonclassical major histocompatibility complex (MHC) class I molecule Qa-1b. J. Exp. Med. 1998;188:1841–1848. doi: 10.1084/jem.188.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vance RE, Jamieson AM, Raulet DH. Recognition of the class Ib molecule Qa-1b by putative activating receptors CD94/NKG2C and CD94/ NKG2E on mouse natural killer cells. J. Exp. Med. 1999;190:1801–1812. doi: 10.1084/jem.190.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westgaard IH, Dissen E, Torgersen KM, Lazetic S, Lanier LL, Phillips JH, Fossum S. The lectin-like receptor KLRE1 inhibits natural killer cell cytotoxicity. J. Exp. Med. 2003;197:1551–1561. doi: 10.1084/jem.20021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilhelm BT, Mager DL. Identification of a new murine lectin-like gene in close proximity to CD94. Immunogenetics. 2003;55:53–56. doi: 10.1007/s00251-003-0540-6. [DOI] [PubMed] [Google Scholar]
- 13.Koike J, Wakao H, Ishizuka Y, Sato TA, Hamaoki M, Seino K, Koseki H, Nakayama T, Taniguchi M. Bone marrow allograft rejection mediated by a novel murine NK receptor, NKG2I. J. Exp. Med. 2004;199:137–144. doi: 10.1084/jem.20030851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimizu E, Koike J, Wakao H, Seino K, Koseki H, Kakiuchi T, Nakayama T, Taniguchi M. Role of a NK receptor, KLRE-1, in bone marrow allograft rejection: analysis with KLRE-1-deficient mice. Blood. 2004;104:781–783. doi: 10.1182/blood-2003-10-3468. [DOI] [PubMed] [Google Scholar]
- 15.Saether PC, Westgaard IH, Flornes LM, Hoelsbrekken SE, Ryan JC, Fossum S, Dissen E. Molecular cloning of KLRI1 and KLRI2, a novel pair of lectin-like natural killer-cell receptors with opposing signaling motifs. Immunogenetics. 2005;56:833–839. doi: 10.1007/s00251-004-0759-x. [DOI] [PubMed] [Google Scholar]
- 16.McKnight AJ, Classon BJ. Biochemical and immunological properties of rat recombinant interleukin- 2 and interleukin-4. Immunology. 1992;75:286–292. [PMC free article] [PubMed] [Google Scholar]
- 17.Brideau RJ, Carter PB, McMaster WR, Mason DW, Williams AF. Two subsets of rat T lymphocytes defined with monoclonal antibodies. Eur. J. Immunol. 1980;10:609–615. doi: 10.1002/eji.1830100807. [DOI] [PubMed] [Google Scholar]
- 18.Westgaard IH, Berg SF, Vaage JT, Wang LL, Yokoyama WM, Dissen E, Fossum S. Rat NKp46 activates natural killer cell cytotoxicity and is associated with FcεRIγ and CD3ζ. J. Leukocyte Biol. 2004;76:1200–1206. doi: 10.1189/jlb.0903428. [DOI] [PubMed] [Google Scholar]
- 19.Storset AK, Kulberg S, Berg I, Boysen P, Hope JC, Dissen E. NKp46 defines a subset of bovine leukocytes with natural killer cell characteristics. Eur. J. Immunol. 2004;34:669–676. doi: 10.1002/eji.200324504. [DOI] [PubMed] [Google Scholar]
- 20.Hoysaeter S, Jensen W, Fossum S. Expression of CD43 epitopes on NK and T cells. Scand. J. Immunol. 1994;40:437–442. doi: 10.1111/j.1365-3083.1994.tb03486.x. [DOI] [PubMed] [Google Scholar]
- 21.Rolstad B, Fossum S. Allogeneic lymphocyte cytotoxicity (ALC) in rats: establishment of an in vitro assay, and direct evidence that cells with natural killer (NK) activity are involved in ALC. Immunology. 1987;60:151–157. [PMC free article] [PubMed] [Google Scholar]
- 22.Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998;8:693–701. doi: 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 23.Dissen E, Berg SF, Westgaard IH, Fossum S. Molecular characterization of a gene in the rat homologous to human CD94. Eur. J. Immunol. 1997;27:2080–2086. doi: 10.1002/eji.1830270836. [DOI] [PubMed] [Google Scholar]


