Abstract
Lectin-like transcript-1 (LLT1) (also named osteoclast inhibitory lectin or CLEC2D) is a ligand for the human NKR-P1A (CD161) receptor, present on NK cells and T cells. To further understand the physiological relevance of this interaction, we developed mAbs against LLT1, characterized the expression pattern of LLT1, and explored the functional consequence of LLT1 engagement of the NKR-P1A receptor on NK cells and T cells. LLT1 is expressed on TLR-activated plasmacytoid dendritic, TLR-activated monocyte-derived dendritic cells, and on B cells stimulated through TLR9, surface Ig, or CD40. Interactions between NKR-P1A on NK cells and LLT1 on target cells inhibit NK cell-mediated cytotoxicity and cytokine production and can inhibit TNF-α production by TCR-activated NKR-P1A+ CD8+ T cells. In contrast, NKR-P1A failed to inhibit or augment the TCR-dependent activation of NKR-P1A-bearing CD4+ T cells. Expression of LLT1 on activated dendritic cells and B cells suggests that it might regulate the cross-talk between NK cells and APCs.
Human NKR-P1A (CD161), encoded by the KLRB1 gene, represents the only human relative of the rodent NKR-P1 family, which includes NKR-P1C, the prototypic NK1.1 alloantigen defining mouse NK cells in C57BL/6 mice. It is a member of the C-type lectin superfamily, and the protein is a type II disulfide-linked homodimer (1). Human NKR-P1A is expressed on immature human NK cells, before acquisition of CD16 or CD56 (2), and expression of NKR-P1A can be up-regulated on human mature NK cells by IL-12 (3, 4). NKR-P1A is also expressed on human memory/effector CD4+ T cells, CD8+ T cells, γδ-TCR+ T cells, and ~60% of invariant NKT cells, as well as a subset of CD3+ thymocytes (1, 5, 6). Experiments examining the role of NKR-P1A on T cells have suggested a stimulatory role for this receptor; anti-NKR-P1A mAbs augmented the anti-CD3 mAb-induced proliferation of human CD1d-specific NK T cells (7) and induced the proliferation of immature thymocytes (8). In contrast, cross-linking with an anti-NKR-P1A mAb inhibits human NK cell-mediated cytotoxicity against FcR+ target cells (1).
We and others identified Lectin-like transcript-1 (LLT1),3 also known as osteoclast inhibitory lectin, as a physiologic ligand for NKR-P1A (9, 10). Interestingly, the gene encoding LLT1, CLEC2D, lies directly adjacent to the KLRB1 gene in the NK genomic complex, located on human chromosome 12 (11). Recently, the closest homologue to LLT1, activation-induced C-type lectin (AICL), and the closest homologue to NKR-P1A, NKp80, were shown to interact, providing another example of genomically linked C-type lectins recognizing one another (12). Functional studies indicated that the interaction of LLT1 on target cells with NKR-P1A on NK cells serves to inhibit NK cell-mediated cytotoxicity (9, 10). To further investigate the functional significance of the LLT1-NKR-P1A interaction, we developed mAbs and a polyclonal rabbit serum against LLT1. We demonstrate that LLT1 is expressed on many B cell lines and on activated primary B cells, and that expression of LLT1 on B cells inhibits NK cell degranulation and IFN-γ production. As CpG DNA stimulation gave the strongest LLT1 induction on B cells, we examined LLT1 expression following treatment with a panel of TLR ligands and found that TLR3, TLR4, TLR7, TLR8, and TLR9 stimulations all induced LLT1 on PBMC. TLR-activated plasmacytoid dendritic cells (pDC) and TLR-activated monocyte-derived dendritic (Mo-DC) cells both expressed LLT1. Lastly, we examined the function of NKR-P1A on T cells and found that whereas NKR-P1A inhibits degranulation and cytokine production in NK cells, NKR-P1A did not modulate TCR-induced cytokine production in CD4+ T cells or degranulation in CD8+ cells, but did partially inhibit TNF-α production in CD8+ T cells.
Materials and Methods
Constructs and transductions
The pMX-s-puro (13) retroviral plasmid was provided by Dr. T. Kitamura (University of Tokyo, Tokyo, Japan). AICL cDNA was provided by Dr. Jorg Hamann (University of Amsterdam, The Netherlands). AICL and LLT1 with a C-terminal V5 epitope tag were subcloned into pMX-s-puro and transfected with Lipofectamine 2000 (Invitrogen) into the Phoenix packaging cell lines (gifts from Dr. G. Nolan, Stanford University, Stanford, CA) (14) to produce retroviruses. Retroviruses in medium containing 8 μg/ml polybrene (Sigma-Aldrich) were used to infect IL-3+ Ba/F3 or 721.221 cells, as described (15, 16). Infected cells were selected in medium supplemented with 10% FCS, 2 mM L-glutamine, penicillin, streptomycin, and 1 μg/ml puromycin.
Abs and immunofluorescent staining
Mouse anti-human NKR-P1A (CD161) mAbs DX1 and DX12 were produced in our laboratory (1), and B199.2 was purchased from GeneTex. PE-conjugated goat anti-mouse IgG was purchased from Jackson Immuno-Research Laboratories, streptavidin secondary reagents were from Becton Dickinson. FITC-conjugated anti-CD107a, PE-conjugated anti-CD56, PerCP-conjugated anti-CD4, PE-Cy7-conjugated CD8, allophycocyanin-conjugated anti-CD161 DX12, PE-conjugated anti-IL-2, allophycocyanin-conjugated anti-IFN-γ, Alexa700-conjugated anti-TNF-α, Abs against CD14, CD16, CD19, CD20, CD27, CD45RA, CD45RA, CD123, and CD236, and intracellular staining reagents were from BD Pharmingen. Anti-human LLT1 mAb 4C7 was purchased from Abnova.
To produce mouse anti-human LLT1 mAbs (R&D Systems; clones 402624 and 402659, both IgG1 isotype Abs), mice were immunized with Escherichia coli-expressed recombinant human LLT1 extracellular domain (aa 57–191; GenBank accession number NP_037401). Hybridoma supernatants were screened by ELISA for their ability to bind recombinant LLT1. Positive hybridomas were subsequently screened by using a cell-based ELISA for recognition of LLT1-transfected P815 cells, but not an irrelevant P815 transfectant. Hybridomas showing the desired reactivity were subcloned by limiting dilution and further screened on the LLT1-positive B cell lines 721.221 and Raji. Ab was purified from hybridoma supernatants by using Protein G affinity chromatography. We tested whether the anti-LLT1 mAbs (clones 4C7, 40262, or 402659) were capable of blocking the interaction with NKR-P1A by using our previously described NKR-P1A reporter cells (10) and found these mAbs lacked blocking activity (data not shown).
Anti-LLT1 antisera was made by immunizing rabbits with a fusion protein of GST and the intracellular domain of LLT1 (aa 1–33). Abs were affinity purified on a fusion protein of Mannose-binding protein and the intracellular domain of LLT1, covalently attached to CNBr-activated sepharose (Sigma-Aldrich).
Cells were incubated on ice for 30 min with mAb, washed, and subsequently stained with secondary Abs. Cells were fixed in 1% paraformal-dehyde in PBS and analyzed on a FACScan (BD Biosciences). The data were processed by using FlowJo software (TreeStar).
Biochemistry and Western blots
SDS-PAGE and Western blots were performed as described (15). Neuraminidase (Roche) and protein N-glycanase F (PNGase F; New England Biolabs) were used according to the manufacturer’s instructions. For immunoblots, anti-LLT1 affinity-purified Abs were used at 1 μg/ml and detected with HRP-conjugated goat-anti-rabbit IgG (Jackson ImmunoResearch Laboratories).
Primary cell cultures
Venous blood was obtained from healthy volunteers after obtaining informed consent, under procedures approved by the UCSF Committee on Human Research. Small, resting memory T cells were isolated as follows: PBMC were suspended in 30% Percoll in PBS with 10% FCS and layered over 40% Percoll in PBS with 10% FCS and centrifuged at 800 × g for 35 min. Low-buoyant density leukocyte were harvested from the interface and small, resting PBL were recovered from the pellet and then depleted of non-T cells and naive T cells with magnetic beads (Miltenyi Biotech) coated with mAbs against CD14, CD16, CD19, CD56, CD45RA, CD123, and CD236. Small, resting memory T cells were cultured in RPMI 1640 medium, 10% FCS, 2 mM glutamine, and penicillin and streptomycin (complete medium), supplemented with 200 U/ml human recombinant IL-2 (provided by the NCI BRB Preclinical Repository). For NK cell assays, the low-buoyant density cells from the Percoll interface layer were isolated and grown in complete medium with 200 U/ml IL-2. Experiments were performed on cells after at least 24 h culture in IL-2.
B cells were purified from human umbilical cord blood samples (collected from Royal Prince Alfred Hospital, Sydney, Australia) or spleens (provided by LifeLink, Australian Red Cross Blood Service, Sydney, Australia) by using a B cell negative isolation kit (Miltenyi Biotec). Splenic B cells were further separated into naive and memory subsets by sorting CD20+CD27− and CD20+CD27+ populations, respectively, using a FACSAria. The purity of the recovered B cells was typically >98%.
Mo-DC were generated as follows: CD14+ monocytes were positively selected from PBMC by using magnetic beads (Miltenyi Biotec) and cultured for 6 days in the presence of 25 ng/ml IL-4 and 50 ng/ml GM-CSF (R&D Systems) in complete medium. Cytokines were replenished on days 2 and 4. Cells were harvested on day 6 and verified to be CD14−, CD1α +, CD11c+, and HLA-DR+.
B cells were isolated from PBMC by using anti-CD19 mAb-conjugated magnetic beads (Miltenyi Biotec) and grown in Iscove’s DMEM supplemented with 10% FCS, 2 mM glutamine, penicillin and streptomycin, and 10 U/ml IL-2.
Human peripheral blood pDCs were positively selected from PBMC with anti-BDCA-4-coated microbeads and sorted by flow cytometry as CD3−CD4+CD8−CD11c−CD14−CD16−CD19−CD56− cells. pDCs were cultured at 1 × 106 cells/ml in complete media in the presence of various stimuli: 20 μg/ml IL-3 (R&D Systems), 5 μM CpG A (2216), 5μM CpG B (2006), 5 μM CpG C (C274) (Operon), heat-inactivated HSV (multiplicity of infection (MOI) 10), influenza virus (MOI 10), or 0.1 μg/ml R848 (Invivogen). After 20 h, total RNA was prepared and reverse-transcription was performed as described (17). For protein expression analysis, cells were activated for 48 h, and then harvested and lysed.
For microarray experiments, human peripheral B cells were positively selected from PBMC with anti-CD19-coated microbeads and sorted by flow cytometry as CD3−CD4−CD8−CD11c−CD14−CD16−CD20+ CD56−BDCA2− cells. B cells were cultured at 1 × 106 cells/ml for 20 h in complete medium in the presence of 5 μM CpG B (2006) or IL-2 (50 U/ml) and IL-10 (100 U/ml). Memory B cells were positively selected from PBMC with anti-CD19-coated microbeads and sorted by flow cytometry as CD3−CD4−CD8−CD11c−CD14−CD16−CD20+CD27+CD56−BDCA2− cells.
Microarray data
A gene expression database, which included the major human immune cell types in peripheral blood, was established as described (17). The Positional Dependent Nearest Neighbor model (18) was used to estimate the gene expression values from the probe intensity values. The final expression output was normalized with the numerical value of one representing the estimated threshold of basal expression.
Quantitative RT-PCR
Sense ATTACACCATCTGAATTGCCTGC and antisense GCGCCAAAT TAAGGTAGCTTTAATA primers were used with probe ACCCAGGTT GTCTGCATTCAAAAGAGCA to detect LLT1 transcripts by using an ABI 7300 Real Time PCR System. LLT1 transcript levels are shown relative to controls; GAPDH for pDC and PBMC Donor A and B cells, or S18 for PBMC Donor B.
Cellular assays
For mAb cross-linking studies, B cells were cultured for 48 h in 24-well tissue culture plates coated with N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-tri-methylammonium methylsulfate (DOTAP) and mAbs, as described (16). In other B cell experiments, human splenic and cord blood B cells were cultured (2 × 105/ml) in the absence or presence of recombinant human CD40L, F(ab) ′2 fragments of goat anti-human Ig (H and L chains; Jackson ImmunoResearch Laboratories; 2 μg/ml), CpG-2006 (1 μg/ml, Proligo) or combinations of these for 48 h. Expression of LLT1 was then determined by flow cytometry or Western blotting as detailed above.
For other stimulations, low buoyant density lymphocytes, Mo-DC, and B cells were treated for 48 h with 1 μg/ml E. coli LPS (Sigma-Aldrich), 5μM CpG-B-2006 (Operon) for B cells, 2 μM each CpG-A, B, C (Operon) for cells other than B cells, 50 μg/ml poly I:C (Amersham Pharmacia), 1–5 μg/ml R837 (Invivogen), 5 μg/ml CL-075 (Invivogen), 10 μg/ml zymosan (Sigma-Aldrich), peptidoglycan 10 μg/ml (Invivogen), or 1 ng/ml IL-1μ (R&D Systems).
T cells (1.5 × 105/well) in 200 μl of complete medium were cocultured in 96-well round-bottom plates with irradiated (3000 rad) mouse P815 cells (2 × 104/well) and stimulating mAbs for 6 h to induce the production of cytokines, in the presence of Golgiplug and Golgistop (BD Pharmingen). For P815 stimulations, 0.5 μg/ml anti-CD3 (clone Leu 4) and 0.5 μg/ml anti-CD28 (clone L293) were used for suboptimal stimulation and anti-NKR-P1A or an isotype-matched control IgG1 was added at 0.25 μg/well. For T cell degranulation assays, cells were cocultured with human CD80-transfected P815 cells (P815-B7.1) (19) and 0.5 μg/ml anti-CD3 for 2 h, stained with FITC-conjugated anti-CD107a and Abs against CD4, CD8, and CD161, and then analyzed on an LSRII flow cytometer (BD Pharmingen). For cytokine assays, cells were cultured for 6 h, as described above, stained for surface proteins CD4, CD8, and CD161, and then fixed and permeabilized, and stained for intracellular cytokines according to the manufacturer’s instructions. Stimulations with Raji cells were as follows: 2.5 ng/ml Staphylococcus enterotoxin B (SEB, Toxin Technologies) was used to stimulate T cells (1.5 × 106/well) cocultured with Raji cells (0.25 × 106/well) in the presence of Golgiplug with or without blocking Abs (0.5 μg/well) for 6 h, and then stained as above.
For NK cell assays, low-buoyant density cells (1.5 × 105/well) in 200 μl of complete medium were cocultured in 96-well round-bottom plates with irradiated (3000 rad) mouse P815 cells (1.5 × 104/well). For P815 experiments, cells were stimulated with 2.5 μg/ml anti-2B4 (clone C1.7) and 2.5 μg/ml either isotype-matched control Ig or anti-NKR-P1A mAb for 2.5 h in the presence of Golgiplug and Golgistop (BD Pharmingen), and then stained with mAbs against FITC-conjugated anti-CD107a, PE-conjugated anti-CD56 (BD Pharmingen), and PE-Cy5-conjugated anti-CD3, and then analyzed by flow cytometry. For Raji experiments, low-buoyant density cells (1.5 × 105/well) were cocultured with Raji cells (2 × 104/well) for 6 h with Golgiplug and Golgistop alone or in the presence of, isotype-matched control F(ab′)2 or blocking anti-NKR-P1A F(ab′)2, were stained with CD107a, CD56, and CD3 mAbs, and then stained for cytokines as described above.
Results
Human LLT1 is a disulfide-bonded, glycosylated homodimer
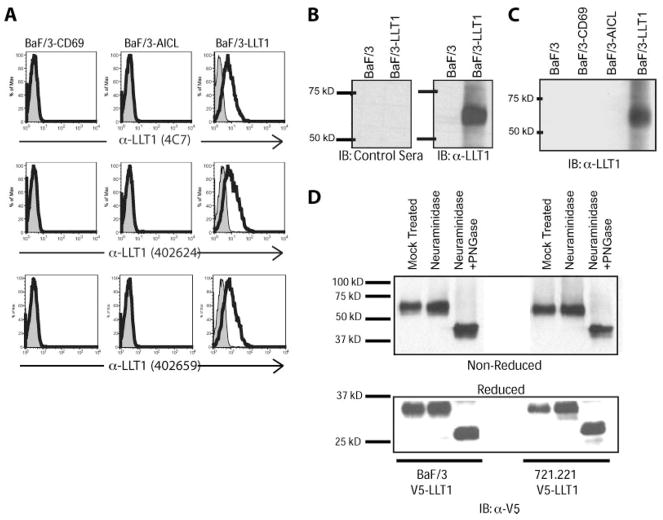
To understand the functional role of an NKR-P1A and LLT1 interaction, we first sought to determine under what conditions and in which cell types LLT1 is expressed. To accomplish this, we created mAbs (R&D Systems clones 402624 and 402659) against the extracellular domain of LLT1. Mice were immunized with the extracellular domain of LLT1 in soluble recombinant form, and then boosted with an injection of mouse Ba/F3 cells expressing human LLT1. Hybridomas were screened for reactivity against the soluble recombinant protein and for reactivity against mouse Ba/F3 cells expressing LLT1. We also characterized and used a recently commercially available Ab, clone 4C7, against LLT1. These mAbs specifically recognized human LLT1-transfected Ba/F3 cells, but did not recognize parental untransfected Ba/F3 or Ba/F3 expressing human AICL, the closest homologue of LLT1 (Fig. 1A).
FIGURE 1.
LLT1 is a glycosylated disulfide-bonded homodimer. A, mAbs against LLT1 specifically recognize LLT1-transfected cells but not untransfected cells or cells transfected with related protein, AICL. B, Anti-LLT1 antiserum, but not control preimmune sera, specifically detects LLT1. C, Anti-LLT1 antiserum specifically recognizes NKR-P1A, but not the related C-type lectins ACIL or CD69. D, Protein N glycanase F (PNGase) treatment of LLT1 transfectants reveals that LLT1 is an N-glycosylated dimer.
We also created an affinity-purified rabbit antiserum against the intracellular domain of LLT1 for immunoblots. This anti-LLT1 antiserum, but not preimmune control serum, specifically recognized LLT1-transfected Ba/F3 cells, but not untransfected Ba/F3 or AICL-transfected Ba/F3 (Fig. 1, B and C). SDS-PAGE analysis revealed that LLT1 migrated at ~65 kD in its nonreduced form. When lysates were treated with neuraminidase and PNGase, a significant shift in migration was observed, indicating the presence of N-linked glycans on LLT1 (Fig. 1D). Reduced monomers of LLT1 migrated at ~35 kD or at 25 kD when treated with neuraminidase and PNGase, which is close to the predicted 22 kD m.w. of the LLT1 core polypeptide.
LLT1 is expressed on activated B cells
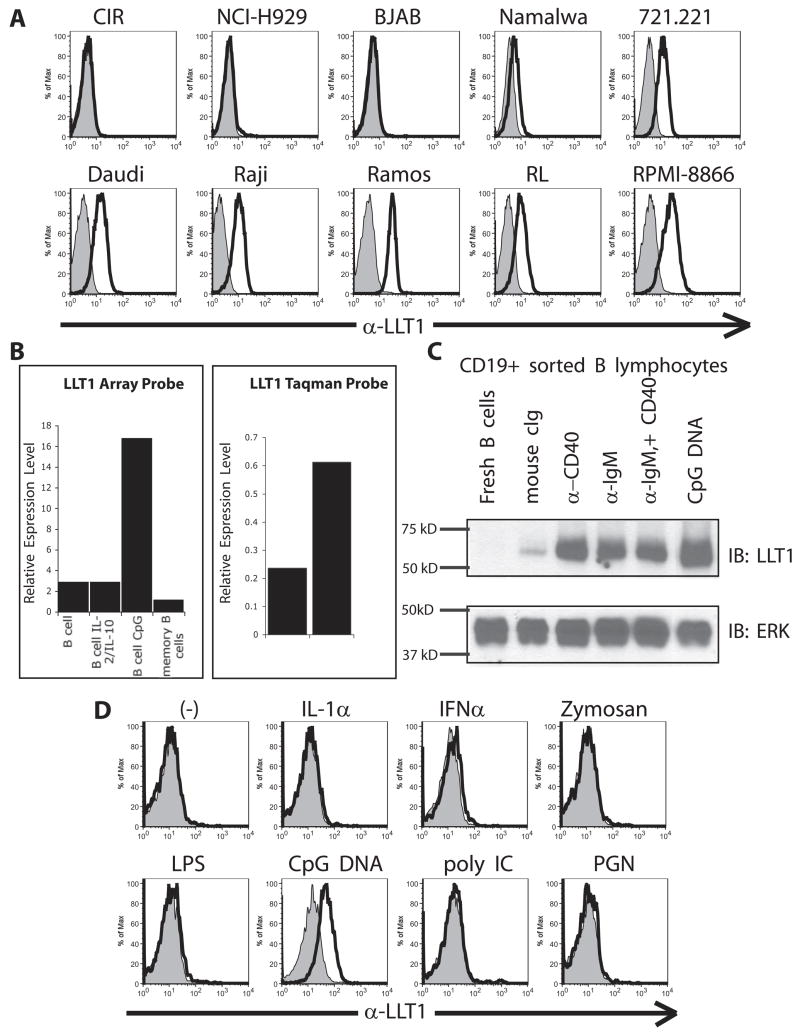
We had previously detected LLT1 on the EBV-transformed B cell line 721.221 and reported that LLT1 on this target cell inhibited NK cell-mediated cytotoxicity through interactions with NKR-P1A on NK cells (10). We examined a panel of human B cell lines for LLT1 expression and detected LLT1 protein on a number of B cell lines, including Daudi, Raji, Ramos, RL, RPMI-8866, and Namalwa (Fig. 2A). These cell lines all expressed LLT1 mRNA by RT-PCR and demonstrated LLT1 protein by Western blot analysis (data not shown). Interestingly, out of the five Burkitt’s lymphoma cell lines analyzed (i.e., BJAB, Namalwa, Daudi, Raji, and Ramos), four expressed LLT1. There was no correlation, however, between LLT1 expression and EBV transformation status.
FIGURE 2.
Activated B cells express LLT1. A, Many immortalized B cell lines express LLT1. B, Microarray expression data reveal that B cells treated with CpG, but not resting or memory B cells, express LLT1. Quantitative RT-PCR confirms that CpG induces LLT1 expression in B cells. C, Western blot analysis of purified B cells demonstrates that activation through CD40, IgM, or TLR9 is sufficient to induce LLT1 protein in sorted B cells. D, CpG DNA, but not other TLR stimuli, induces LLT1 protein on the surface of B cells.
In examining whether primary B cells express LLT1, we found that freshly isolated cord blood, peripheral blood, and tonsillar B cells, as well as splenic naive, memory, and plasma cells, were negative for LLT1 or expressed only low amounts of LLT1, depending on the donor. However, CpG-activated blood B cells expressed significantly higher levels of LLT1 (Fig. 2, B–D). We also detected increased LLT1 mRNA transcripts in CpG-activated B cells by quantitative RT-PCR (Fig. 2B). To confirm this was a direct effect of CpG on B cells, to exclude the requirement of other cell types, and to determine whether other methods of activating B cells can induce LLT1, we examined LLT1 expression in purified B cells. B cells isolated from PBMC were treated with plate-bound Abs against CD40 or IgM or were stimulated with CpG for 48 h and then lysed for examination by Western blot. Whereas resting peripheral blood B cells expressed very low amounts of LLT1, stimulation via CD40 or IgM cross-linking induced LLT1 protein, as did CpG DNA treatment (Fig. 2C). Treatment with CpG DNA consistently resulted in the highest induction of LLT1 expression. These observations were also made when expression of LLT1 was examined on naive splenic and cord blood B cells that had been stimulated with anti-Ig, CD40L, or CpG (data not shown). These data indicated that IgM, CD40, or CpG stimulation of B cells alone is sufficient to induce LLT1 expression.
To examine the specificity of CpG DNA stimulation, we treated PBMC with a panel of TLR stimuli and examined B cell expression of LLT1. We detected LLT1 on CpG-treated B cells, but not on B cells treated with other TLR stimuli (Fig. 2D). These results are consistent with the expression of TLRs by B cells because they express abundant levels of TLR9 (and TLR10), but low or negligible levels of TLR1, TLR2, TLR4, and TLR7 (20 –22).
Stimulation through TLR3, TLR4, TLR7, TLR7, or TLR9 induces LLT1
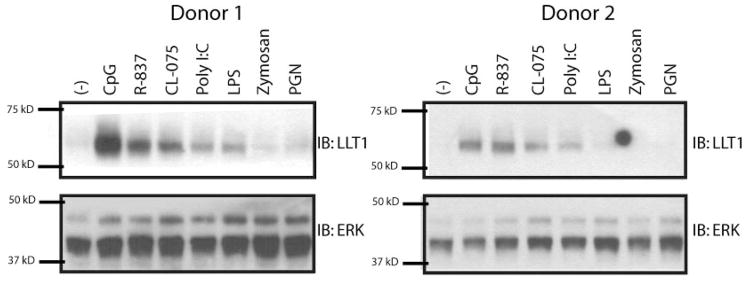
Previous work suggested that LLT1 mRNA is expressed in osteo-blast cell lines and that LLT1 transcript levels increased with IL-1α treatment (23). A prior report also demonstrated that PMA-stimulated PBMC express LLT1 mRNA (24). After observing that LLT1 was induced on CpG-treated B cells, we examined whether stimulation by other TLR agonists might induce LLT1 expression on peripheral blood leukocytes. To accomplish this, we treated low buoyant density PBMC, enriched for monocytes, NK cells, B cells, and dendritic cells (DC), with various TLR stimuli for 48 h and then tested for LLT1 protein by Western blot analysis. The TLR9 ligand CpG DNA induced LLT1 expression, as did TLR7 ligand R-837 and TLR8 ligand CL-075, and to a lesser extent TLR3 ligand poly I:C and TLR4 ligand LPS (Fig. 3). TLR2 ligands, peptidoglycan, and zymosan did not induce LLT1 (Fig. 3).
FIGURE 3.
TLR stimulations of peripheral blood low buoyant density mononuclear cells (LBD-PBMC). Immunoblots of TLR-stimulated LBD-PBMC lysates demonstrate that LLT1 protein is induced by ligands of TLR3, TLR4, TLR7, TLR8, or TLR9. All findings were reproduced in at least three independent experiments.
Although other groups have reported LLT1 expression on monocytes and NK cells (25), we examined resting monocytes, resting and IL-2 activated NK cells, and resting and IL-2 activated T cells and found no evidence for LLT1 protein by flow cytometry and Western blot analysis (data not shown). We did, however, see LLT1 expression on NK cells lines NKL and YT2C2, suggesting it may be possible for NK cells to express this protein under some circumstances; interestingly NKL and YT2C2 do not express NKR-P1A (data not shown).
Activated plasmacytoid DCs and monocyte-derived DCs express LLT1 in a TLR-specific fashion
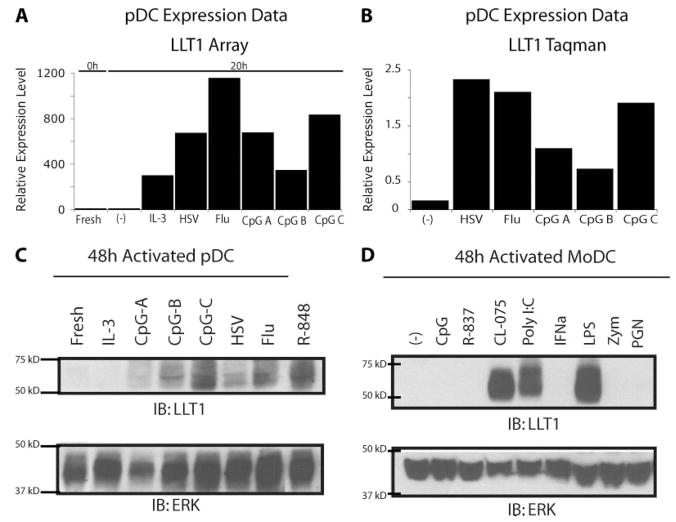
Because treatment of B cells with TLR9 ligand CpG DNA potently induced LLT1, we examined LLT1 expression in pDCs, which also express TLR9, and are responsive to CpG DNA (20 –22). pDCs, also known as IFN-producing cells, respond to viral infections by rapidly producing large amounts of type I IFN. To determine whether LLT1 is expressed by pDCs we examined microarray data from isolated pDCs treated with a panel of stimuli. Whereas freshly isolated pDC expressed little LLT1 mRNA, the amount of LLT1 was increased significantly after treatment with inactivated influenza virus, inactivated HSV-1, or CpG DNA of various compositions (Fig. 4A). To confirm these data, we performed quantitative RT-PCR on pDC cDNA and again observed more LLT1 transcripts following pDC activation with inactivated viruses or CpG DNA (Fig. 4B). Western blot analysis confirmed that LLT1 protein is induced in activated pDCs (Fig. 4C). LLT1 protein was induced strongest after stimulation with TLR9 ligand CpG-C DNA, TLR7/8 ligand R-848, and the TLR7 ligand inactivated influenza virus.
FIGURE 4.
TLR-activated pDC and activated Mo-DC express LLT1. A, Microarray expression data from purified pDC demonstrate that pDCs treated with inactivated viruses or CpG DNAs increased LLT1 mRNA. B, Quantitative RT-PCR of purified pDC samples confirmed increased levels of LLT1 transcripts in activated pDCs. C, Activated pDCs express LLT1 protein. D, Mo-DC express LLT1 after TLR3, TLR4, or TLR8 stimulation, but not after TLR2 activation. All findings were reproduced in at least two independent experiments.
We then examined whether DC other than pDC can express LLT1. We generated Mo-DC by culturing CD14+ sorted monocytes with IL-4 and GM-CSF for 6 days. Mo-DC were treated with various stimuli for 48 h and assayed for LLT1 protein. Mo-DCs expressed LLT1 after stimulation with TLR3 ligand poly I:C, TLR4 ligand LPS, and TLR8 ligand CL-075 (Fig. 4D). This induction of LLT1 in Mo-DC apparently is TLR-specific because the TLR2 ligands zymosan and peptidoglycan did not induce LLT1 protein.
LLT1 on Raji cells inhibits NK cell activation, but does not affect TCR-dependent stimulation of NKR-P1A+ CD4+ T cells
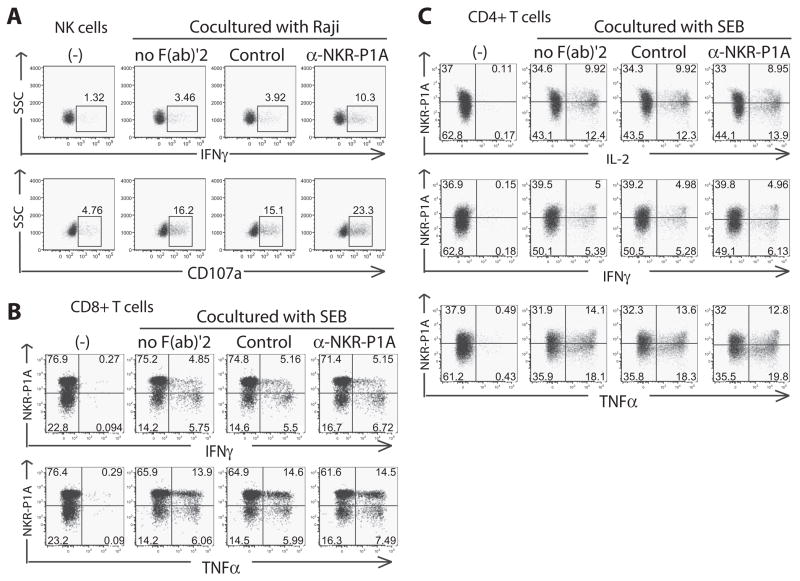
In addition to being on NK cells, NKR-P1A is expressed on ~30 –50% of CD4+ CD45RO+ T cells and 30 – 60% of CD8+ CD45RO+ effector/memory T cell subsets in adult human peripheral blood (1, 6). Although an inhibitory role for NKR-P1A has been established for NK cells, no function has been previously reported for NKR-P1A on conventional effector or memory T cells. First, to confirm that LLT1 was a functional ligand for NKRP-1A on Raji cells, we cocultured Raji cells with IL-2-activated peripheral blood NK cells and examined NK cell activation in the presence or absence of an anti-NKR-P1A F(ab′)2. Because these anti-NKR-P1A F(ab′)2 fragments lack Fc domains, they cannot bind to Fc receptors and cross-link NKR-P1A, and thus are a useful reagent to block the interaction with LLT1, as previously reported (10). We examined NK cell degranulation by using CD107a (LAMP-1) surface staining, as well as IFN-γ production by using intracellular cytokine staining. We observed a marked increase in both degranulation and cytokine production by NK cells cocultured with Raji cells in the presence of blocking anti-NKR-P1A F(ab′)2 (Fig. 5A). This demonstrated that Raji cells express functional LLT1, which serves to inhibit degranulation and cytokine production in NK cells through interactions with NKR-P1A on NK cells.
FIGURE 5.
LLT1 on Raji cells inhibits peripheral blood NKR-P1A+ NK cells, but does not affect peripheral blood NKR-P1A+ T cells. A, Blocking NKR-P1A with F(ab′)2 demonstrates that LLT1 on Raji cells specifically interacts with NKR-P1A on primary human NK cells to functionally inhibit degranulation and cytokine production. B, Blocking NKR-P1A with F(ab′)2 demonstrates that LLT1 on Raji cells does not affect NKR-P1A+ CD8+ IFN-γ or TNF-α production. C, Blocking NKR-P1A with F(ab′)22 reveals that LLT1 on Raji cells does not induce any specific effects on NKR-P1A+ CD4+ IL-2, IFN-γ or TNF-α production. Data are representative of at least four independent experiments.
Having validated that LLT1 on Raji cells functionally suppresses NK cell activation, we used these LLT1+ Raji cells as APCs to stimulate NKR-P1A+ T cells in the presence or absence of neutralizing anti-NKR-P1A F(ab′)2. To enrich for resting effector/memory T cells, we first isolated small, resting high buoyant-density peripheral blood leukocytes from PBMC by Percoll density centrifugation, and then depleted cells positive for CD14, CD19, CD56, CD45RA, CD123, or CD236. T cells were cultured overnight, and then incubated with LLT1+ Raji cells in the presence of the superantigen, SEB, at a suboptimal dose (based on prior titration) with or without blocking anti-NKR-P1A F(ab′)2. Cells were stimulated for 6 h and analyzed for cytokine production by intracellular staining. We observed that although anti-NKR-P1A F(ab′)2 enhanced NK cell activation, anti-NKR-P1A F(ab′)2did not affect SEB-induced production of IFN-γ or TNF-α by CD8+ T cells (Fig. 5B) or IL-2, IFN-γ, or TNF-α by CD4+ T cells (Fig. 5C). Experiments were performed at varying concentrations of SEB in the presence of saturating amounts of NKR-P1A F(ab′)2, and no specific increase or decrease of cytokine production was noted (data not shown). Additional experiments performed in the presence of CTLA-4-IgG Fc to block B7-CD28 interactions also showed no additional effect of blocking NKR-P1A, compared with blocking with CTLA-4-Ig Fc (data not shown).
Anti-NKR-P1A mAb inhibits anti-CD3 mAb-induced TNF-α production by CD8+ T cells
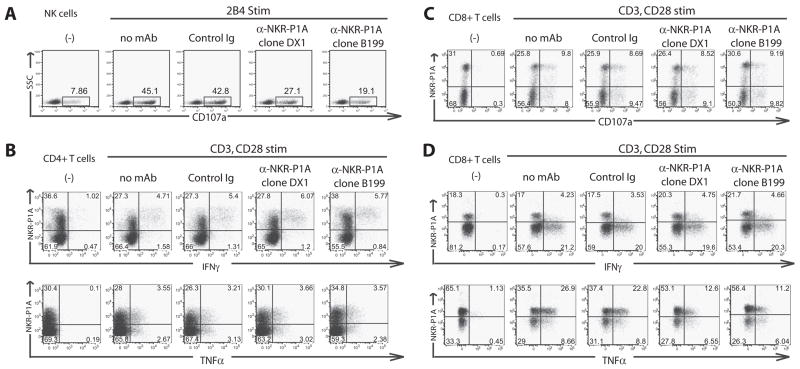
To further assess the role of NKR-P1A in T cells vs NK cells, we used mAb-redirected activation with Fc receptor-bearing mouse P815 stimulator cells, a system we have previously used to cross-link and study inhibitory receptors (e.g., inhibitory KIR) on NK cells (1, 26) and activating receptors (e.g., CD3) on T cells (19, 27). In this assay system, T cells or NK cells are cocultured with the Fc receptor-bearing P815 cell line in the presence or absence of intact IgG Abs against activating or inhibitory receptors on the T cell and NK cells. The anti-receptor Abs are cross-linked by binding to the Fc receptors on the P815 cells and function as receptor agonists. When cross-linked by the Fc receptors on P815, intact IgG mAbs against inhibitory receptors suppress cytotoxicity or cytokine production by the effector cells, whereas intact IgG mAbs against activating receptors enhance cytotoxicity or cytokine production. First, we demonstrated that anti-NKR-P1A mAb suppressed anti-CD244 (2B4) mAb-redirected degranulation of IL-2-activated peripheral blood NK cells. When the activating receptor CD244 was cross-linked with a specific anti-CD244 mAb by the Fc receptors on P815 target cells, we detected a significant increase in NK cell degranulation as measured by surface CD107a staining (Fig. 6A). However, when anti-NKR-P1A mAb, but not isotype-matched control Abs, was added, we observed a marked decrease in NK degranulation, confirming an inhibitory role for NKR-P1A on NK cell function.
FIGURE 6.
Cross-linking NKR-P1A with FcR+ P815 cells inhibits NK cell function and can inhibit NKR-P1A+ CD8+ T cell TNF-α production. A, Cross-linking NKR-P1A with P815 cells inhibits 2B4-induced degranulation in peripheral blood NK cells. B, Cross-linking NKR-P1A does not affect anti-CD3 induced IFN-γ or TNF-α production by NKR-P1A+ CD4+ T cells. C, Cross-linking NKR-P1A does not affected anti-CD3 induced degranulation on NKR-P1A+ CD8+ T cells. D, Cross-linking NKR-P1A does not affect IFN-γ production by NKR-P1A+ CD8+ T cells, but does inhibit TNF-α production in three of seven donors analyzed. Data are representative of at least three independent experiments.
To test the role of NKR-P1A in T cells using this system, we cocultured resting effector/memory T cells (isolated as described above) with P815 cells in the presence of suboptimal concentrations of anti-CD3 and anti-CD28, with or without anti-NKR-P1A. We also used CD80 (B7.1)-transfected P815 cells and suboptimal anti-CD3 mAb stimulation with similar results. We observed no specific differences in IFN-γ or TNF-α production by NKR-P1A+ CD4+ T cells when NKR-P1A was co-cross-linked with anti-NKR-P1A mAb by P815 cells (Fig. 6B). We did observe that a higher frequency of NKR-P1A+ CD4+ T cells produce IFN-γ or TNF-α than NKR-P1A− CD4+ cells in this memory-enriched T cell population, in agreement with prior findings (6). Because NKR-P1A inhibits NK cell degranulation (Fig. 6A), we tested the effects of NKR-P1A on T cell degranulation. We observed that whereas NKR-P1A+ CD8+ T cells degranulated after CD3 and CD28 stimulation, mAb-mediated cross-linking of NKR-P1A did not have any specific effects on degranulation in these cells (Fig. 6C). Lastly, we tested the effects of cross-linking NKR-P1A on cytokine production by NKR-P1A+ CD8+ T cells and found that whereas engaging NKR-P1A did not significantly effect IFN-γ production, cross-linking NKR-P1A did result in a decrease in TNF-α-producing cells in three of seven blood donors analyzed (Fig. 6D).
Discussion
Although LLT1 was identified recently as a ligand for human NKR-P1A, little is known about the expression or regulation of this molecule or the physiological relevance and context of this interaction. In this study, we sought to define the cellular sources of LLT1, to explore the functional consequences of LLT1 expression, and to describe the role of NKR-P1A on memory CD4+ and CD8 T+ cells, in addition to NK cells. We found that LLT1 is widely expressed on many B cell lines and on primary B cells activated by anti-CD40 or anti-IgM cross-linking or by treatment with the TLR9 ligand CpG DNA. We further demonstrated that stimulation of PBMC through TLR3, TLR4, TLR7, TLR8, or TLR9 induced the expression of LLT1. Because TLR9 ligands strongly induced the expression of LLT1 on B cells, we tested whether CpG-B DNA could also induce LLT1 expression on pDCs, which constitutively express TLR9. We found that TLR9-and TLR7-stimulated pDCs expressed LLT1 protein, as did TLR3, TLR4, or TLR8-stimulated Mo-DC. Overall, we observed LLT1 expression following stimulation via TLR3, TLR4, TLR7, TLR8, or TLR9. Interestingly, these TLRs are unique because they all induce activation of TRAF3 (28).
We examined LLT1 expression on fresh monocytes, resting and IL-2 activated T cells, and resting and IL-2 activated NK cells and found no evidence for LLT1 protein by flow cytometry and Western blot analysis (data not shown). This was surprising because a previous report claimed that NK cells express LLT1 and that cross-linking LLT1 on NK cells induced production of IFN-γ (25). However, using the L9.7 mAb (25) (provided by Dr. P. Mathew, University of North Texas Health Science Center, Fort Worth, TX), we failed to detect LLT1 on our LLT1-transfected Ba/F3 cells. It is also noteworthy that previous studies using this mAb reported a much higher m.w. of LLT1 protein as analyzed by SDS-PAGE than we observed in our studies (25). We cannot explain the discrepancy between our results and these previously reported findings, but are likely attributable to the nature of the mAbs used.
Prior studies from our laboratory and others have demonstrated that interactions between human NKR-P1A on NK cells and LLT1 on target cells inhibit NK cell-mediated cytotoxicity and cytokine production (9, 10). Our new findings that LLT1 is preferentially expressed on activated DC and B cells suggest that this molecule might regulate the interactions between NK cells and DC during viral infection or interactions between NK cells and activated B cells. pDCs can directly activate NK cells through mechanisms involving pDC-derived IFN-γ and glucocorticoid-induced TNFR-related protein ligand (29, 30); however, activated pDC are known to be resistant to NK lysis. As LLT1 is expressed on activated pDCs, it is possible that LLT1 up-regulation on activated pDC might contribute to the resistance of these cells to NK cell-mediated lysis. Further studies will be required to explore the relative contribution of MHC class I and LLT1 on activated DC in protection against NK cell-mediated attack. Similarly, activated B cells expressing LLT1 might be protected, in part, from NK cell-mediated cytotoxicity via interaction with NKR-P1A. We have shown the potential for transformed B cells (e.g., 721.221 and Raji) to be protected through LLT1-NKR-P1A interactions from NK cell attack. Another consequence of LLT1 expression might be protection of tumors, particularly those low in expression of MHC class I, from NK cell surveillance. In this regard, LLT1 was described recently as being expressed on malignant glioma cells (31). As LLT1 expression could represent a mechanism of immune evasion, it will be interesting to see whether LLT1 is aberrantly expressed in cancer.
Although the interaction of LLT1 with NKR-P1A inhibits NK cell functions such as cytokine production and cytotoxicity, the functional consequences of LLT1 interacting with NKR-P1A on conventional effector or memory T cells had not been addressed previously. We used LLT1+ Raji cells and neutralizing mAbs against NKR-P1A to explore this question. Previous reports describing the function of NKR-P1A on human NKT cell clones and immature thymocytes using anti-NKR-P1A mAbs suggested a co-stimulatory role for this receptor on T cells (7, 8). In contrast, in our studies TCR-induced IFN-γ or TNF-α production by conventional peripheral blood NKR-P1A+ CD4+ T cell was not affected by the presence of LLT1 on the APC (Raji B cells) or by cross-linking with an anti-NKR-P1A mAb that inhibited NK cell-mediated cytotoxicity and cytokine production. Similarly, engaging NKR-P1A did not affect peripheral blood CD8+ T cell degranulation. Lastly, we demonstrated that although ligation of NKR-P1A did not affect CD8+ T cell IFN-γ production, it did decrease anti-CD3-induced CD8+ T cell TNF-α production in some donors, suggesting a preferential inhibitory role on different effector functions in a donor specific fashion. In contrast, NKR-P1A did not decrease SEB-induced CD8+ T cell TNF-α production, possibly because SEB and anti-CD3 stimulation differ or different cells were responsive to SEB vs anti-CD3. In comparing CD45RO+ effector/memory T cell subsets, we found that, in agreement with previous reports, NKR-P1A+ CD4+ T cells produce more IFN-γ and TNF-α than their NKR-P1A− counterparts (6, 32). We also found that NKR-P1A+ CD8+ T cells are capable of degranulating and producing more TNF-α than NKR-P1A− CD8+ effector/ memory T cells. Although NKR-P1A+ CD8+ T cells do produce IFN-γ, they produce less than NKR-P1A− CD8+ cells, in agreement with other studies (6).
Although previous experiments exploring the role of NKR-P1A on NKT cells have suggested a stimulatory role for this receptor, we have observed either an inhibitory role for this receptor on NK cell-mediated cytotoxicity or cytokine production or no effect of ligating this receptor on TCR-activated conventional T cells in most assays. There are several possible explanations for the discrepancy in these various studies. First, most of the commonly used anti-NKR-P1A mAbs block the interaction between LLT1 and NKR-P1A (9, 10). Therefore, what appears to be “stimulation” might actually be caused by the anti-NKR-P1A mAb blocking the interaction between the inhibitory NKR-P1A receptor and its ligand on an Ag-presenting cell or target cell. In the studies demonstrating that anti-NKR-P1A mAb augmented the activation of immature thymocytes (8) and invariant NKT cell clones (7), it is possible that the anti-NKR-P1A mAb was actually blocking the interaction between an inhibitory NKR-P1A receptor on these T cells and LLT1 ligand being expressed by the NKT cell clones or thymocytes. In this regard, we have detected LLT1 protein by Western blot analysis in thymocyte lysates (data not shown). Alternatively, it is possible that the NKP-P1A on certain cell types, such as NKT cells or immature thymocytes, might possess an activating function in certain situations. However, unlike rodents, humans have only a single NKR-P1 gene and there is no evidence of alternatively spliced isoforms that can mediate distinct functional outcomes. Aside from inhibiting anti-CD3 mAb induced CD8+ T cell TNF-α production, we found no evidence that NKR-P1A enhanced or suppressed the TCR-induced activation of T cells, as determined by evaluating cytokine production and cyto-toxicity. However, this does not rule out a role for NKR-P1A in other T cell functions. Other inhibitory C-type lectins, such as NKG2A, have been reported to inhibit TCR-induced apoptosis (33), thus it is possible that NKR-P1A serves a similar role. We also examined anti-CD3-induced T cell proliferation and detected no role for NKR-P1A, although, interestingly, expression of NKR-P1A on the cell surface decreased after stimulation (data not shown).
The cytoplasmic domain of NKR-P1A contains the amino acid sequence AIYAEL, which is similar to an ITIM, (Ile/Val/Leu/Ser)-X-Tyr-X-X-(Leu/Val), where X denotes any amino acid, except that NKR-P1A contains an alanine in the –2 position relative to the tyrosine residue (34). Previous data have suggested that this –2 position is important for inhibitory function and mutating the –2 position to an alanine in an ITIM resulted in decreased inhibitory potential (35). This noncanonical ITIM likely explains the weak inhibitory potential of NKR-P1A. In mAb-redirected cytotoxicity experiments using the Fc receptor-bearing P815 target cell, cross-linking NKR-P1A inhibited anti-2B4-induced degranulation of human NK cells; however, cross-linking NKR-P1A did not significantly inhibit anti-CD16-induced degranulation of NK cells (data not shown), suggesting that the inhibitory potential of NKR-P1A can be masked by a strong activating stimulus. Thus, although NKR-P1A largely failed to modulate high-affinity anti-CD3 or SEB-induced responses, it remains possible that NKR-P1A does function in T cells physiologically to modulate low affinity TCR interactions, possibly against self-peptides. Collectively, our findings demonstrate that LLT1 can be induced on activated human B cells and DC and that the expression of LLT1 on B cell lines can inhibit NK cell-mediated cytotoxicity and cytokine production. Although under certain circumstances NKR-P1A might affect NKR-P1A-bearing T cell responses, it apparently does not suppress or augment activation induced by high affinity TCR engagement.
Acknowledgments
We thank Susan Watson, Art Weiss, and Bill Seaman for useful discussions and insights and Jonathan Benjamin, Mei Peng, Andy Gross, Tri Phan, and Erica Straus for phlebotomy.
Footnotes
This work was supported by National Institutes of Health Grant AI068129. L.L.L. is an American Cancer Society Research Professor, and D.B.R. was supported by a Genentech Graduate Student Fellowship and a University of California Chancellor’s Research Fellowship. J.P.H. is employed and supported by R&D Systems, Inc.
Abbreviations used in this paper: LLT1, lectin-like transcript-1; AICL, activation-induced C-type lectin; MOI, multiplicity of infection; pDC, plasmacytoid dendritic cell; Mo-DC, monocyte-derived dendritic cell; PNGase F, protein N-glycanase F; DC, dendritic cell.
Disclosures J.P.H. is employed and supported by R&D Systems.
References
- 1.Lanier LL, Chang C, Phillips JH. Human NKR-P1A: a disulfide linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. J Immunol. 1994;153:2417–2428. [PubMed] [Google Scholar]
- 2.Bennett IM, Zatsepina O, Zamai L, Azzoni L, Mikheeva T, Perussia B. Definition of a natural killer NKR-P1A+/CD56−/CD16− functionally immature human NK cell subset that differentiates in vitro in the presence of interleukin 12. J Exp Med. 1996;184:1845–1856. doi: 10.1084/jem.184.5.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azzoni L, Zatsepina O, Abebe B, Bennett IM, Kanakaraj P, Perussia B. Differential transcriptional regulation of CD161 and a novel gene, 197/15a, by IL-2, IL-15, and IL-12 in NK and T cells. J Immunol. 1998;161:3493–3500. [PubMed] [Google Scholar]
- 4.Poggi A, Costa P, Tomasello E, Moretta L. IL-12-induced up-regulation of NKRP1A expression in human NK cells and consequent NKRP1A-mediated down-regulation of NK cell activation. Eur J Immunol. 1998;28:1611–1616. doi: 10.1002/(SICI)1521-4141(199805)28:05<1611::AID-IMMU1611>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi T, Nieda M, Koezuka Y, Nicol A, Porcelli SA, Ishikawa Y, Tadokoro K, Hirai H, Juji T. Analysis of human V α 24+ CD4+ NKT cells activated by α-glycosylceramide-pulsed monocyte-derived dendritic cells. J Immunol. 2000;164:4458 – 4464. doi: 10.4049/jimmunol.164.9.4458. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi T, Dejbakhsh-Jones S, Strober S. Expression of CD161 (NKR-P1A) defines subsets of human CD4 and CD8 T cells with different functional activities. J Immunol. 2006;176:211–216. doi: 10.4049/jimmunol.176.1.211. [DOI] [PubMed] [Google Scholar]
- 7.Exley M, Porcelli S, Furman M, Garcia J, Balk S. CD161 (NKR-P1A) costimulation of CD1d-dependent activation of human T cells expressing invariant Vα24JαQ T cell receptor α chains. J Exp Med. 1998;188:867– 876. doi: 10.1084/jem.188.5.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poggi A, Costa P, Morelli L, Cantoni C, Pella N, Spada F, Biassoni R, Nanni L, Revello V, Tomasello E, et al. Expression of human NKRP1A by CD34+ immature thymocytes: NKRP1A-mediated regulation of proliferation and cytolytic activity. Eur J Immunol. 1996;26:1266 –1272. doi: 10.1002/eji.1830260613. [DOI] [PubMed] [Google Scholar]
- 9.Aldemir H, Prod’homme V, Dumaurier MJ, Retiere C, Poupon G, Cazareth J, Bihl F, Braud VM. Cutting edge: lectin-like transcript 1 is a ligand for the CD161 receptor. J Immunol. 2005;175:7791–7795. doi: 10.4049/jimmunol.175.12.7791. [DOI] [PubMed] [Google Scholar]
- 10.Rosen DB, Bettadapura J, Alsharifi M, Mathew PA, Warren HS, Lanier LL. Cutting edge: lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J Immunol. 2005;175:7796 –7799. doi: 10.4049/jimmunol.175.12.7796. [DOI] [PubMed] [Google Scholar]
- 11.Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. 2003;3:304 –316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 12.Welte S, Kuttruff S, Waldhauer I, Steinle A. Mutual activation of natural killer cells and monocytes mediated by NKp80-AICL interaction. Nat Immunol. 2006;7:1334 –1342. doi: 10.1038/ni1402. [DOI] [PubMed] [Google Scholar]
- 13.Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- 14.Kinsella TM, Nolan GP. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 15.Rosen DB, Araki M, Hamerman JA, Chen T, Yamamura T, Lanier LL. A structural basis for the association of DAP12 with mouse, but not human, NKG2D. J Immunol. 2004;173:2470 –2478. doi: 10.4049/jimmunol.173.4.2470. [DOI] [PubMed] [Google Scholar]
- 16.Voehringer D, Rosen DB, Lanier LL, Locksley RM. CD200 receptor family members represent novel DAP12-associated activating receptors on basophils and mast cells. J Biol Chem. 2004;279:54117–54123. doi: 10.1074/jbc.M406997200. [DOI] [PubMed] [Google Scholar]
- 17.Cao W, Rosen DB, Ito T, Bover L, Bao M, Watanabe G, Yao Z, Zhang L, Lanier LL, Liu YJ. Plasmacytoid dendritic cell-specific receptor ILT7-FcvarεRIγ inhibits Toll-like receptor-induced interferon production. J Exp Med. 2006;203:1399 –1405. doi: 10.1084/jem.20052454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Miles MF, Aldape KD. A model of molecular interactions on short oligonucleotide microarrays. Nat Biotechnol. 2003;21:818 – 821. doi: 10.1038/nbt836. [DOI] [PubMed] [Google Scholar]
- 19.Azuma M, Cayabyab M, Buck D, Phillips JH, Lanier LL. CD28 interaction with B7 co-stimulates primary allogeneic proliferative responses and cytotoxicity mediated by small, resting T lymphocytes. J Exp Med. 1992;175:353–360. doi: 10.1084/jem.175.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdorfer B, Giese T, Endres S, Hartmann G. Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol. 2002;168:4531– 4537. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- 21.Bourke E, Bosisio D, Golay J, Polentarutti N, Mantovani A. The toll-like receptor repertoire of human B lymphocytes: inducible and selective expression of TLR9 and TLR10 in normal and transformed cells. Blood. 2003;102:956 –963. doi: 10.1182/blood-2002-11-3355. [DOI] [PubMed] [Google Scholar]
- 22.Bernasconi NL, Onai N, Lanzavecchia A. A role for Toll-like receptors in acquired immunity: up-regulation of TLR9 by BCR triggering in naive B cells and constitutive expression in memory B cells. Blood. 2003;101:4500 – 4504. doi: 10.1182/blood-2002-11-3569. [DOI] [PubMed] [Google Scholar]
- 23.Hu YS, Zhou H, Myers D, Quinn JM, Atkins GJ, Ly C, Gange C, Kartsogiannis V, Elliott J, Kostakis P, et al. Isolation of a human homolog of osteoclast inhibitory lectin that inhibits the formation and function of osteoclasts. J Bone Miner Res. 2004;19:89 –99. doi: 10.1359/JBMR.0301215. [DOI] [PubMed] [Google Scholar]
- 24.Eichler W, Ruschpler P, Wobus M, Drossler K. Differentially induced expression of C-type lectins in activated lymphocytes. J Cell Biochem. 2001;(Suppl 36):201–208. doi: 10.1002/jcb.1107. [DOI] [PubMed] [Google Scholar]
- 25.Mathew PA, Chuang SS, Vaidya SV, Kumaresan PR, Boles KS, Pham HT. The LLT1 receptor induces IFN-γ production by human natural killer cells. Mol Immunol. 2004;40:1157–1163. doi: 10.1016/j.molimm.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 26.Lanier LL, Corliss B, Phillips JH. Arousal and inhibition of human NK cells. Immunol Rev. 1997;155:145–154. doi: 10.1111/j.1600-065x.1997.tb00947.x. [DOI] [PubMed] [Google Scholar]
- 27.Lanier LL, O’Fallon S, Somoza C, Phillips JH, Linsley PS, Okumura K, Ito D, Azuma M. CD80 (B7) and CD86 (B70) provide similar co-stimulatory signals for T cell proliferation, cytokine production, and generation of CTL. J Immunol. 1995;154:97–105. [PubMed] [Google Scholar]
- 28.Kawai T, Akira S. Signaling to NF-κB by Toll-like receptors. Trends Mol Med. 2007;13:460 – 469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Hanabuchi S, Watanabe N, Wang YH, Ito T, Shaw J, Cao W, Qin FX, Liu YJ. Human plasmacytoid predendritic cells activate NK cells through glucocorticoid-induced tumor necrosis factor receptor-ligand (GITRL) Blood. 2006;107:3617–3623. doi: 10.1182/blood-2005-08-3419. [DOI] [PubMed] [Google Scholar]
- 30.Gerosa F, Gobbi A, Zorzi P, Burg S, Briere F, Carra G, Trinchieri G. The reciprocal interaction of NK cells with plasmacytoid or myeloid dendritic cells profoundly affects innate resistance functions. J Immunol. 2005;174:727–734. doi: 10.4049/jimmunol.174.2.727. [DOI] [PubMed] [Google Scholar]
- 31.Roth P, Mittelbronn M, Wick W, Meyermann R, Tatagiba M, Weller M. Malignant glioma cells counteract antitumor immune responses through expression of lectin-like transcript-1. Cancer Res. 2007;67:3540 –3544. doi: 10.1158/0008-5472.CAN-06-4783. [DOI] [PubMed] [Google Scholar]
- 32.Iliopoulou EG, Karamouzis MV, Missitzis I, Ardavanis A, Sotiriadou NN, Baxevanis CN, Rigatos G, Papamichail M, Perez SA. Increased frequency of CD4+ cells expressing CD161 in cancer patients. Clin Cancer Res. 2006;12:6901– 6909. doi: 10.1158/1078-0432.CCR-06-0977. [DOI] [PubMed] [Google Scholar]
- 33.Gunturi A, Berg RE, Forman J. Preferential survival of CD8 T and NK cells expressing high levels of CD94. J Immunol. 2003;170:1737–1745. doi: 10.4049/jimmunol.170.4.1737. [DOI] [PubMed] [Google Scholar]
- 34.Ravetch JV, Lanier LL. Immune inhibitory receptors. Science. 2000;290:84 – 89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 35.Burshtyn DN, Yang W, Yi T, Long EO. A novel phospho-tyrosine motif with a critical amino acid at position −2 for the SH2 domain-mediated activation of the tyrosine phosphatase SHP-1. J Biol Chem. 1997;272:13066 –13072. doi: 10.1074/jbc.272.20.13066. [DOI] [PubMed] [Google Scholar]