β-Lactamases are the most widespread cause of bacterial resistance to the β-lactam antibiotics, such as the penicillins and cephalosporins, and the mechanisms of these enzymes are intensely studied. For the class A β-lactamases, such as TEM, SHV, and the newly emergent extended-spectrum β-lactamase (ESBL) CTX-M enzymes,1 X-ray structures have been determined at every major step along the reaction coordinate (Figure 1a). This includes multiple apo, acylation transition-state analogue, acyl-enzyme, and deacylation transition state analogue complex structures.2–6 These structures have been accompanied by elegant mechanistic and mutagenesis studies and, increasingly, theoretical simulations of the reaction coordinate.7,8
Figure 1.
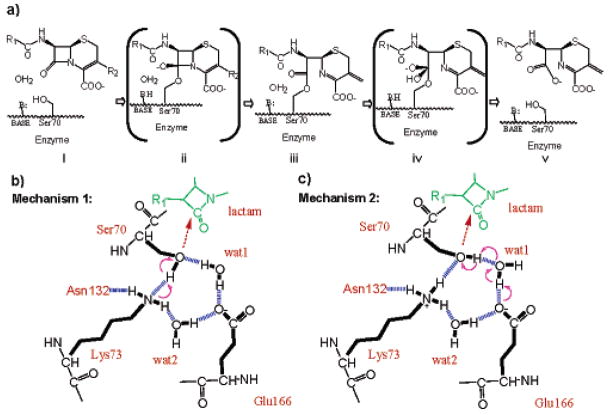
Possible acylation mechanisms of class A β-lactamases. (a) The overall reaction pathway for β-lactam hydrolysis. Beginning with a ground-state Michaelis complex (i), catalysis proceeds through a high-energy acylation transition state (ii) to an acyl-enzyme intermediate (iii). These steps constitute the acylation half of the reaction. Deacylation then proceeds through a high-energy deacylation transition state (iv) and a postcovalent product complex (v), after which the product leaves. (b) Acylation mechanism 1: a neutral Lys73 as the general base. Hydrogen bonds are indicated by blue dashed lines and electron transfer by purple arrows. (c) Acylation mechanism 2: Glu166 as the general base, through the catalytic water (wat1). Lys73 is positively charged in this model.
Despite this scrutiny, key aspects of the mechanism of class A β-lactamase remain controversial. Among them is the identity of the catalytic base for acylation.2,7–10 On the basis of mutagenesis studies and structural evidence, it was suggested that the conserved Lys73 acts in its neutral form as the general base to deprotonate the nucleophile Ser70, which then attacks the β-lactam substrate, forming the acyl intermediate (Figure 1b).2,9 More recently, a related mechanism was proposed where the Lys73 would be protonated and charged in the apo enzyme, but would transfer a proton to Glu166 on formation of the preacylation complex and then act as the catalytic base for acylation.11 Conversely, the pH dependence of mutant activities, and the observation of proton positions on key catalytic residues by ultrahigh-resolution X-ray crystallography of SHV β-lactamase, suggested that Glu166, acting through the catalytic water, is the general base for the acylation step (Figure 1c).3,5,12,13 The resolution of this controversy hinges on the protonation states and hydrogen-bonding patterns of the central protagonists: Ser70, the catalytic nucleophile; Lys73 and Ser130, one candidate for the general base and its partner; the catalytic water and Glu166, the alternative candidate as the general base, thought to act in a proton relay.
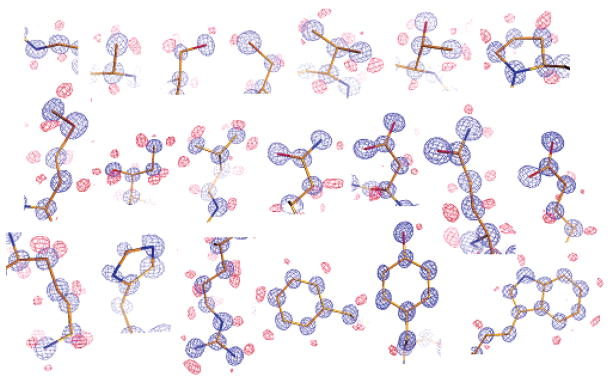
In an effort to differentiate between these two possibilities, the class A β-lactamase CTX-M-9 was crystallized at pH 8.8 and the structure refined to 0.88 Å. The high resolution measurements allowed for anisotropic temperature factor refinements using Shelx (Table 1). With R and Rfree values at 10.4% and 12.0%, respectively, this structure identifies alternate residue conformations and proton positions, providing an unusually clear snapshot of the apo enzyme (Figure 2). Correlated conformational changes of residue side chains can be observed in the protein core, and individual heavy atoms such as carbon and oxygen can be distinguished by the electron density volume. Hydrogen atoms can be identified not only for the protein backbone, but also for the side chains of nearly all twenty residue types, including the protons on polar functional groups (Figure 2). The hydrogen atoms are also visible on at least 24 well-ordered water molecules. In modeling nonriding hydrogen atoms, only those with positive peaks above 1.5βin the hydrogen-omitted Fo–Fc map were used.5
Table 1.
Data Collection and Refinement Statistics
| Data Collection | |
| space group | P21 |
| cell dimensions | |
| a, b, c (Å) | 45.162, 106.796, 47.785 |
| α, β, γ(°) | 90.00, 102.09, 90.00 |
| resolution (Å) | 22.00–0.88 (0.91–0.88)a |
| no. reflections | 310 765 (28 059) |
| Rmerge (%)b | 3.2 (32.6) |
| I/σI | 18.1 (2.06) |
| completeness (%) | 88.9 (80.6) |
| Redundancy | 5.5 (2.1) |
| Refinement | |
| resolution (Å) | 10.00–0.88 |
| Rwork/Rfree | 10.0%/11.4% |
| no. heavy atoms | |
| protein | 4357 |
| ligand/ion | 37 |
| water | 1002 |
| no. residues with double conformations | 70c |
| B-factors (Å2) | |
| protein | 10.2 |
| ligand/ion | 16.8 |
| water | 24.0 |
| rms deviationsd | |
| bond lengths | 0.015 Å |
| angle distances | 0.032 Å |
| ramanchandran plot e | |
| most favored region (%) | 91.6 |
| additionally allowed (%) | 8.0 |
| generously allowed (%) | 0.4 |
Figure 2.
Structural details revealed by ultrahigh-resolution diffraction, showing characteristic electron density for all 20 residue types, each observed in this structure. 2Fo–Fc map (blue) and hydrogen-omitted Fo–Fc map (red) are both contoured at 3σ. Carbon, nitrogen, and oxygen atoms are colored in gold, blue, and red, respectively.
These peaks were present in the map calculated with the full range of diffraction data (0.88–10 Å), but we found that using only the high-resolution portion of the data (0.88–2 Å) at this final stage of refinement reduced the noise of the difference map. Thus our final model for the hydrogen positions was built with these highest resolution measurements. This resembles the high-angle data weighing scheme commonly used in small molecule X-ray crystallography14,15 and the high-order refinement protocol used in the refinement of a 0.66 Å aldose reductase structure.16
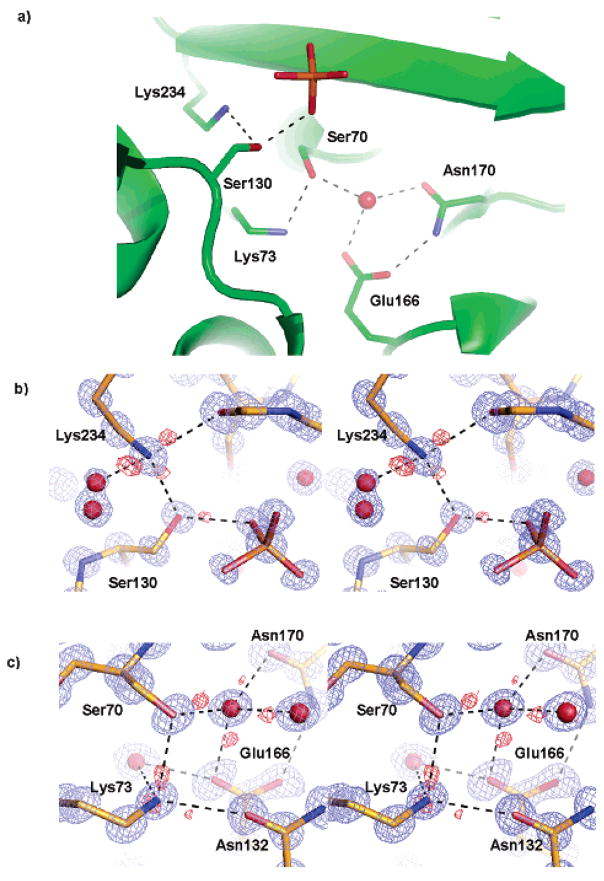
These ultrahigh-resolution features reveal the protonation states and hydrogen-bonding partners of all of the active site residues; all such states and patterns are shared by both monomers in the asymmetric unit (Figure 3). Nearly all hydrogen atoms are identified for Ser70, Lys73, Ser130, Glu166, Lys234, and the catalytic water in the Fo–Fc map at contour levels above 3β, allowing us to determine the role of each residue in a particular hydrogen bond (Figure 3b,c). In the apo protein, Ser70 accepts a proton from Lys73 and donates its own proton to the catalytic water, which in turn donates a proton to Glu166. Glu166 itself appears unprotonated and anionic in this structure, but Lys73 is fully protonated and apparently cationic: features in the hydrogen-omitted Fo–Fc difference map are consistent with three protons on this lysine, acting as hydrogen-bond donors to Ser70, Asn132, and a water molecule. The proton donated to the water (Nζ···O distance 3.01 Å and Hζ··· O distance 2.0 Å) may be shared with Glu166 (Nζ ···Oε1 distance 2.93 Å and Hζ···Oε1 distance 2.5 Å). These protonation states and hydrogen-bonding patterns are consistent with the role of Glu166, together with the catatlytic water, as the general base for deprotonating Ser70 during the acylation step of class A β -lactamase.
Figure 3.
Hydrogen bonding (dashed lines) in the active site. 2Fo–Fc map (blue) and Fo–Fc map (red) are both contoured at 3σ. (a) Active site configuration showing the key catalytic residues, the catalytic water, and a phosphate molecule. (b) Protons of Lys234Nζand Ser130Oγ, identified by Fo–Fc map, shown in stereoview. (c) Hydrogen bonds among Ser70, Lys73, Glu166, and the catalytic water shown in stereoview.
A mechanism that reconciles all of the structural measurements and many of the previous mechanistic and computational studies may now be proposed. In the apo enzyme at physiological pH values, Glu166 is an anion that accepts a proton from the catalytic water, which in turn accepts a proton from the nucleophilic Ser70. Possibly co-incident with substrate binding, Ser70 donates its proton to the catalytic water, which then transfers its proton to Glu166. The now anionic Ser70 is free to attack the electrophilic carbonyl of the β-lactam substrate. This is consistent with an acylation transition-state analogue complex of class A β-lactamase TEM-1 at 0.85 Å resolution.5 In this complex, the transfer from Ser70 → water → Glu166 has occurred and a proton is observed on Glu166Oε1, donated to the catalytic water. This mechanism is also consistent with a 0.91 Å apo structure of SHV-2 where the proton on Ser70 was identified and shared with the water, although other polar-group hydrogen atoms of the catalytic residues were not determined in this model.3
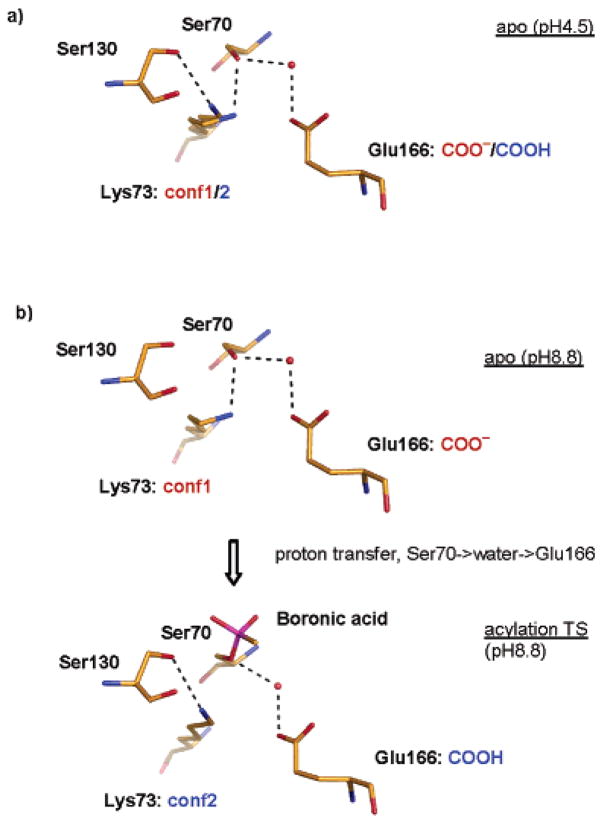
The second critical step in the acylation mechanism is the activation of the leaving-group lactam nitrogen. Frere, Mobashery, Strynadka, and others have suggested a role for Lys73 and Ser130 in this activation.2,7,19–21 In the structure determined here, Ser130 donates its proton, clearly visible in the Fo–Fc map at 3σ, to a phosphate ion that fortuitously binds in the same site occupied by oxygen atoms of boronic acid and phosphonate transition-state analogues. In the transition-state analogue structures, these oxygen atoms represent the leaving-group nitrogen, and the Ser130 proton is well placed to stabilize this leaving group.4,5 In the current pH 8.8 apo structure, however, Lys73 is 4.3 Å away from Ser130Oγ, too far to directly hydrogen bond to and activate the serine residue. Intriguingly, in a 0.98 Å structure of the same CTX-M-9 apo enzyme, determined previously at pH 4.5, Lys73 adopts two conformations. Conformation 1 is identical to that in the current structure and does not interact with Ser130, whereas conformation 2 rotates to establish a hydrogen bond with Ser130 (Figure 4a).22 Consistent with a state after accepting the Ser70 proton, conformation 2 may reflect the partial protonation of Glu166 at the acidic pH.12 This conformation 2 of Lys73 also closely resembles that adopted in the complex between CTX-M-9 and an acylation transition-state analogue6 and that adopted by Lys73 in an E166A mutant of TOHO-1, another CTX-M group ESBL.23 Taken together, these structures suggest that Lys73 conformation may be coupled to the protonation state of Glu166: When Glu166 is negatively charged, Lys73 adopts conformation 1 and interacts with both Glu166 and Ser70; when Glu166 is protonated, Lys73 moves away from Glu166 and closer to Ser130. This is consistent with the prediction of a proton-shuttle pathway from Lys73 to Ser130 and to the substrate leaving group as proposed previously2,7,21,24 and suggests that the second proton transfer can be activated by the protonation of Glu166 by Ser70 through the catalytic water.
Figure 4.
Conformation change during acylation catalysis. (a) The apo active site configuration observed at pH 4.5. The two conformations observed for Lys73 may correspond to the dual protonation states of Glu166. The putative partial protonation of Glu166 is indicated by the anionic (red) and neutral (blue) text. (b) Lys73 switches conformation from 1 in the apo-active site to 2 in the acylation transition-state analogue structure. Only the tetrahedral boronic acid group (magenta) is shown for the acylation transition-state analogue complex; the rest of the transition-state analogue is pointed outside the plane of the paper.
In conclusion, the ultrahigh-resolution apo-structures of CTX-M-9, determined both at pH 8.8 and pH 4.5, support the role of Glu166, acting in concert with an active site water, as the catalytic base for acylation in class A β-lactamases. This step has been the focus of much study,2,7,11,24,25 and we admit that even these structures only represent snapshots of low-energy states. They cannot rule out adoption of other states that would support alternative mechanisms. Indeed, one that invokes Lys73 as the catalytic base proposes that this residue is only deprotonated on formation of the precovalent complex, and in fact is fully protonated in the apo state, exactly as we observe.7 We certainly cannot discount this mechanism, though we continue to favor a direct role for Glu166 as the catalytic base. Certainly, future mechanistic studies in what has become a model system–the reaction coordinate of class A β-lactamases–must be reconciled with the extraordinary level of structural detail revealed in these structures. Correspondingly, they afford us an opportunity to prosecute genuinely atomic resolution inhibitor discovery campaigns against these enzymes, which remain key targets for antibiotic drug discovery.
Data Deposition
The coordinates and structural factors have been deposited in the Protein Data Bank with access code 2P74.
Acknowledgments
This work was supported by NIH Grant GM63813 (to B.K.S). We thank Johannes Hermann and Veena Thomas for insightful discussions and Kerim Babaoglu and Sarah Boyce for reading the manuscript.
References
- 1.Bonnet R. Antimicrob Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strynadka NC, Adachi H, Jensen SE, Johns K, Sielecki A, Betzel C, Sutoh K, James MN. Nature. 1992;359:700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 3.Nukaga M, Mayama K, Hujer AM, Bonomo RA, Knox JR. J Mol Biol. 2003;328:289–301. doi: 10.1016/s0022-2836(03)00210-9. [DOI] [PubMed] [Google Scholar]
- 4.Maveyraud L, Pratt RF, Samama JP. Biochemistry. 1998;37:2622–2628. doi: 10.1021/bi972501b. [DOI] [PubMed] [Google Scholar]
- 5.Minasov G, Wang X, Shoichet BK. J Am Chem Soc. 2002;124:5333–5340. doi: 10.1021/ja0259640. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Shoichet B, Bonnet R. J Am Chem Soc. 2005;127:5423–5434. doi: 10.1021/ja042850a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meroueh SO, Fisher JF, Schlegel HB, Mobashery S. J Am Chem Soc. 2005;127:15397–15407. doi: 10.1021/ja051592u. [DOI] [PubMed] [Google Scholar]
- 8.Hermann JC, Ridder L, Mulholland AJ, Holtje HD. J Am Chem Soc. 2003;125:9590–9591. doi: 10.1021/ja034434g. [DOI] [PubMed] [Google Scholar]
- 9.Gibson RM, Christensen H, Waley SG. Biochem J. 1990;272:613–619. doi: 10.1042/bj2720613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adachi H, Ohta T, Matsuzawa H. J Biol Chem. 1991;266:3186–3191. [PubMed] [Google Scholar]
- 11.Golemi-Kotra D, Meroueh SO, Kim C, Vakulenko SB, Bulychev A, Stemmler AJ, Stemmler TL, Mobashery S. J Biol Chem. 2004;279:34665–34673. doi: 10.1074/jbc.M313143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escobar WA, Tan AK, Lewis ER, Fink AL. Biochemistry. 1994;33:7619–7626. doi: 10.1021/bi00190a015. [DOI] [PubMed] [Google Scholar]
- 13.Lietz EJ, Truher H, Kahn D, Hokenson MJ, Fink AL. Biochemistry. 2000;39:4971–4981. doi: 10.1021/bi992681k. [DOI] [PubMed] [Google Scholar]
- 14.Dunitz JD, Seiler P. Acta Crystallogr, Sect B. 1973;29:589–595. [Google Scholar]
- 15.Sheldrick GM, Schneider TR. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 16.Muzet N, Guillot B, Jelsch C, Howard E, Lecomte C. Proc Natl Acad Sci USA. 2003;100:8742–8747. doi: 10.1073/pnas.1432955100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 19.Lamotte-Brasseur J, Dive G, Dideberg O, Charlier P, Frere JM, Ghuysen JM. Biochem J. 1991;279:213–221. doi: 10.1042/bj2790213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atanasov BP, Mustafi D, Makinen MW. Proc Natl Acad Sci USA. 2000;97:3160–3165. doi: 10.1073/pnas.060027897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas VL, Golemi-Kotra D, Kim C, Vakulenko SB, Mobashery S, Shoichet BK. Biochemistry. 2005;44:9330–9338. doi: 10.1021/bi0502700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Delmas J, Sirot J, Shoichet B, Bonnet R. J Mol Biol. 2005;348:349–362. doi: 10.1016/j.jmb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Ibuka A, Taguchi A, Ishiguro M, Fushinobu S, Ishii Y, Kamitori S, Okuyama K, Yamaguchi K, Konno M, Matsuzawa H. J Mol Biol. 1999;285:2079–2087. doi: 10.1006/jmbi.1998.2432. [DOI] [PubMed] [Google Scholar]
- 24.Hermann JC, Hensen C, Ridder L, Mulholland AJ, Holtje HD. J Am Chem Soc. 2005;127:4454–4465. doi: 10.1021/ja044210d. [DOI] [PubMed] [Google Scholar]
- 25.Damblon C, Raquet X, Lian LY, Lamotte-Brasseur J, Fonze E, Charlier P, Roberts GC, Frere JM. Proc Natl Acad Sci USA. 1996;93:1747–1752. doi: 10.1073/pnas.93.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]