Abstract
Tandem mass spectrometry (MS/MS) is frequently used in the identification of peptides and proteins. Typical proteomic experiments rely on algorithms such as SEQUEST and MASCOT to compare thousands of tandem mass spectra against the theoretical fragment ion spectra of peptides in a database. The probabilities that these spectrum-to-sequence assignments are correct can be determined by statistical software such as PeptideProphet or through estimations based on reverse or decoy databases. However, many of the software applications that assign probabilities for MS/MS spectra to sequence matches were developed using training datasets from 3D ion-trap mass spectrometers. Given the variety of types of mass spectrometers that have become commercially available over the last five years, we sought to generate a dataset of reference data covering multiple instrumentation platforms to facilitate both the refinement of existing computational approaches and the development of novel software tools. We analyzed the proteolytic peptides in a mixture of tryptic digests of 18 proteins, named the “ISB standard protein mix”, using 8 different mass spectrometers. These include linear and 3D ion traps, two quadrupole time-of-flight platforms (qq-TOF) and two MALDI-TOF-TOF platforms. The resulting dataset, which has been named the Standard Protein Mix Database, consists of over 1.1 million spectra in 150+ replicate runs on the mass spectrometers. The data were inspected for quality of separation and searched using SEQUEST. All data, including the native raw instrument and mzXML formats and the PeptideProphet validated peptide assignments, are available at http://regis-web.systemsbiology.net/PublicDatasets/.
Keywords: Proteomics, reference dataset, database search software, standard protein mix, Standard Protein Mix Database
Introduction
The field of proteomics has come to play an important role in biological research. Tandem mass spectrometry (MS/MS) of peptides is the most frequently used approach to identify protein components in these samples (for reviews see1–3). State-of-the-art high-throughput mass spectrometry platforms now permit rapid and extensive interrogation of biological specimens. Typically, proteins are digested into peptides using the enzyme trypsin, separated by reverse phase HPLC, and introduced to the mass spectrometer either directly, through coupling to an electrospray ionization (ESI) source, or indirectly through spotting the eluate onto a stage for subsequent matrix-assisted laser desorption/ionization (MALDI). Within the mass spectrometer individual peptide ions are selected in a sequential fashion for collision-induced dissociation (CID), generating tandem mass spectra that contain fragments specific to the selected precursor peptide ion.
Several methods have been employed to identify peptides in a sample from their corresponding tandem mass spectra. SEQUEST4 was the first computer algorithm successfully developed specifically to identify peptides using only their uninterpreted tandem mass spectra; it is still widely used today. This algorithm correlates an experimentally measured tandem mass spectrum to the theoretically derived mass spectra of all peptides in a protein database that have the same precursor ion mass. Alternatives such as Mascot5 and others (reviewed in 6, 7) work in a similar manner using a variety of spectrum-to-peptide scoring algorithms.
Selection of the successful peptide to tandem mass spectrum assignment from amongst the thousands within a single experiment can be performed either by manual inspection or by applying a set of filtering criteria such as the number of observed tryptic termini or algorithm score, thresholds 8–10. Such approaches are problematic as experimenatl error rates are undefined and researchers often apply their own different subjective filtering criteria.
Recently, statistical modeling has been used to determine the probability that a particular peptide assignment is correct. PeptideProphet uses the scores returned by the search algorithm as well as features specific to the experiment, such as number of observed tolerable termini (based on the enzyme used), to assign a probability to the top peptide match of each searched tandem mass spectrum11. These numerical scores are combined into a single discriminant score using a linear function for which the coefficients were previously determined through analysis of multiple tandem mass spectrometry experiments performed on known mixtures of commercially available proteins. The bimodal distribution of this discriminant score for incorrect and correct assignments is then used to determine accurate probability, sensitivity and error rates. In their analysis demonstrating the performance of PeptideProphet, Keller et al searched tandem mass spectra from experiments on a mixture of commercially available proteins against a database comprised of the commercial proteins appended to a larger decoy protein sequence database of Drosophila or H. influenzae12. This allowed the authors to distinguish incorrect peptide assignments and determine false positive error rates under the hypothesis that incorrect assignments will be randomly assigned to peptides from the much larger decoy database. While these data were developed based on SEQUEST search results using a Thermo LCQ ion trap13, it has subsequently been optimized and extended to support other search tools such as MASCOT14, up-front fractionation strategies15, and instruments including TOF- and FT-based platforms. Likewise, ProteinProphet takes a statistical approach to determining the probability of a protein assignment, basing its assessment on the PeptideProphet output and modifying it based on supporting evidence from other peptides from the same protein16. By making the assessment of search results an objective process, these two programs have streamlined the tandem mass spectrometry data analysis pipeline and facilitated high throughput proteomic analyses. The error rate of spectral assignment in a data set can also be estimated by performing a search using a “reverse” or “decoy” database strategy (i.e. a database in which the sequences were scrambled or randomized to produce exclusively false positive identifications17–19). Such a database is concatenated to the normal database to estimate the confidence of the identified proteins. Unlike the PeptideProphet software, reversed/decoy database search results do not estimate the false negative error rate of a dataset.
Much of the above described software was developed and tested using data acquired on a single type of mass spectrometer: a 3D Ion-trap. Other instrument platforms with different characteristics have become increasingly common in recent years. The development of new or improved search and/or validation software for use with these platforms often requires both test and validation datasets where the true peptide complements are known. At this time, there is no publicly available, comprehensive dataset which incorporates analysis of the same sample using multiple mass spectrometry platforms. We describe here such a dataset generated by performing repeat analyses of a standard sample of 18 trypsin-digested proteins on a variety of instruments available in our laboratory and in those of our collaborators.
Experimental Procedures
Preparation of the ISB standard 18 protein mixture
All reagents were purchased from Sigma Aldrich (St. Louis, MO) except as noted. The 18 proteins used to prepare the ISB standard protein mixture are listed in Table 1. One nanomole of each protein (based on manufacturer’s claimed purity) was dissolved in 20 mM ammonium bicarbonate pH 8.0 and 0.05% SDS to a final concentration of 1μM. The final protein concentration of the mixture was 970 μg/mL. The sample was reduced with 2.5mM TCEP at 50°C for 30 minutes and alkylated in the dark for 1 hour with 10mM iodoacetimide. The proteins were then digested using sequencing grade trypsin (Promega, Madison, WI) at a 1:40 (w/w) ratio. Digestion was performed either by incubation at 37°C overnight or high-intensity focused ultrasound sonication as per Lopez-Ferrer20. Briefly, sonication was performed at 4°C, 50% duty cycle, and an output control setting of six for 60 s using Branson Sonifier 250 with microtip. Digestion was confirmed by SDS-PAGE. Samples were dried in a Speed Vac and cleaned up using a Waters (Milford, MA) Oasis MCX cartridge per the manufacturer’s instruction. The final eluate was evaporated and re-suspended in 1ml of 0.1% formic acid, 1%ACN, in HPLC grade water (VWR, West Chester, PA). Over the course of the experiment, the standard mixture was consumed and subsequently prepared anew four times. These mixtures were named mix 1, mix 2, mix 3, and mix 4. Mix 1, mix 3 and mix 4 were digested using the overnight incubation method and mix 2 was digested using the Lopez-Ferrer sonication approach.
Table 1.
Contents of the standard mix
| Protein | Organism | Swiss- Prot AC | MW (kD) | Sigma # |
|---|---|---|---|---|
| Actin, aortic smooth muscle | Bovine | P62739 | 42.0 | A3653 |
| Alkaline phosphotase | E. coli | P00634 | 49.4 | 79377 |
| Alpha-amylase | B. licheniformis | P06278 | 58.5 | A4551 |
| Alpha-lactalbumin | Bovine | P00711 | 16.2 | L6010 |
| Beta-casein | Bovine | P02666 | 25.1 | C6905 |
| Beta-galactosidase | E. coli | P00722 | 116.5 | G5635 |
| Beta-lactoglobulin | Bovine | P02754 | 19.9 | L0130 |
| Carbonic anhydrase 2 | Bovine | P00921 | 29.1 | C2522 |
| Catalase | Bovine | P00432 | 59.9 | C40 |
| Cytochrome c | Bovine | P62984 | 11.7 | C2037 |
| Glyceraldehyde-3-phosphate dehydrogenase | Rabbit | P46406 | 35.8 | G2267 |
| Glycogen phosphorylase, muscle form | Rabbit | P00489 | 97.3 | P6635 |
| Mannose-6-phosphate isomerase | E. coli | P00946 | 42.9 | P2621 |
| Myoglobin | Horse | P68082 | 17.1 | M0630 |
| Myosin light chain 1, skeletal muscle isoform | Rabbit | P02602 | 20.9 | M9891 |
| Ovalbumin | Chicken | P01012 | 42.9 | A2512 |
| Serotransferrin | Bovine | Q29443 | 77.8 | T0178 |
| Serum albumin | Bovine | P02769 | 69.3 | A3059 |
LTQ, LCQ Deca, Q-TOF and QSTAR
For analysis on the Thermo Electron (Waltham, MA) LTQ, ABI (Foster City, CA) API QSTAR Pulsar i, and Thermo Electron LCQ DECA instruments, each sample was run on an automated mass spectrometry system using an in-house developed electrospray source as described21. Two microliters of standard mix, corresponding to approximately 200 fmol of total protein, was loaded onto a 75 μm internal diameter fused silica fritted capillary pre-column packed to a bed length of 2 cm with Magic C18Aq spherical silica resin (mean particle size, 5 μm; pore size, 200 Å; (Michrom Bioresources, Auburn, CA)). A separating column was made in house by pulling a tip on a 75 μm internal diameter fused silica capillary and packing the bed to 10cm with Magic C18Aq spherical silica resin (mean particle size, 5 μm, pore size, 100 Å (Michrom Bioresources)). The loaded pre-column was washed isocratically for 5 minutes with 0.1% formic acid (buffer A). Peptides were eluted using a linear gradient of 10–35% 0.1% formic acid, 100%ACN (buffer B) over 60 minutes (with the exception of mix 1 on the LCQ DECA, where 30 minute gradients were used as part of a pilot study) at a tip flow rate of 200 nl/min using an HP 1100 solvent delivery system (Agilent, Palo Alto, CA). Between runs, column were washed with 80% ACN for 10 min and reequilibrated with 5% ACN. The mixture was also analyzed on a Waters/Micromass (Milford Massachusetts) Q-TOF Ultima using a setup similar to the LTQ with the exception of the auto-sampler and pump, (Agilent 1100 series autosampler and Agilent 1100 series nano-pump flowing at 200nl per minute during elution, resptectivly).
On the LCQ each MS scan triggered three MS/MS scans with collision energy of 35 percent. A 25 entry exclusion list was populated with peaks (+/− 1.5 Da) that were seen more than once within a 60 s window. Peaks were removed from this MS/MS exclusion list after 3 min. On the LTQ each MS scan triggered three MS/MS scans with collision energy of 35 percent. A 50 entry exclusion list was populated with peaks (+/− 1.5 Da) that were seen more than once within a 30 s window. Peaks were removed from this MS/MS exclusion list after 3 min. The QTOF was programmed to select the three most intense MS peaks for a MS/MS scan of 2.1 s after which mass was excluded for 60 s. The QSTAR was programmed to select the three most intense MS peaks for a single MS/MS scan of 2 s after which the selected precursor mass was excluded for 120 s. The collision energy profiles for both TOF instruments was optimized prior to this analysis by running a yeast extract and adjusting energies to yield the greatest number of peptide identifications. The profiles, which vary according to precursor mass, are provided on the standard mix web site. All runs were performed in a back-to-back fashion.
Agilent XCT Ultra
The standard mix was analyzed using an Agilent 1100 LC-Chip system coupled to a XCT Ultra ion trap. Buffer A was 0.1% formic acid in water and buffer B was 90% ACN with 0.1% formic acid in water. The chip consisted of a 40 nl trap column and a 15 cm analytical column, using Zorobax 300SB-C18 5μm particles as the stationary phase. The sample was loaded onto the pre-column using an Agilent 1100 Capillary pump with a flow of 4 μl/min of 5% buffer B. After sample loading, the pre-column was washed using a further 6 μL of 5% buffer B. For analysis, the pre-column was placed inline with the analytical column. A gradient of 5% to 55% buffer B was delivered over 50 minutes at 200nl/min by an Agilent 1100 nanopump. After the gradient, the system was washed for 10 min with 90% buffer B and subsequently equilibrated for 5 min with 5% buffer B. The spectra were acquired in the m/z-range from 400 to 1800 using the standard-enhanced scan mode. Each MS scan triggered five MS/MS scans. Precursor ion masses were added to an exclusion list for one minute after twice being selected for CID.
Applied Biosystems ABI 4800
For analysis using the ABI 4800 TOF-TOF, 2 μL of the standard mix was separated by reverse-phase chromatography and spotted directly onto a MALDI sample plate using an Ultimate HPLC system coupled with a Famos microautosampler (Dionex, Sunnyvale, CA). Buffer A was 2% ACN/0.1% TFA in water and buffer B was 80% ACN/0.1% TFA in water. A gradient of 0–50% buffer B over 82 min at a flow rate of 300 nl/min was used for peptide separation on a PepMap 100 C18 column (75 μm i.d. × 15 cm length, 3 μm, 100 Å, LC Dionex). The eluate from the capillary column was mixed with -cyano-4-hydroxycinnamic acid (Sigma-Aldrich, St. Louis, MO) matrix solution (2.5 mg/mL in 70% ACN-water/0.1% TFA in water) pumped at 800 nL/min flow-rate in a mixing tee during spotting onto the MALDI plate. The fractions were automatically collected at 10 s intervals on the MALDI plate using a Probot microfraction collector (Dionex, Sunnyvale, CA). The samples were analyzed by MALDI-TOF/TOF on the ABI 4800 mass spectrometer. Both MS and MS/MS data were acquired with a neodymium doped yttrium aluminum garnet laser (Nd:YAG) with a 200-Hz sampling rate. For MS spectra, 1000 laser shots per spot were used. In MS/MS mode, the CID spectra were generated using a collision energy of 1 keV with air as the collision gas. Typically 1500 laser shots were used for MS/MS acquisition. A maximum of 12 CID attempts were permitted per spot. The data acquisition time for one LC run took about 8–10 hr depending on the number of CID attempts. The total analysis time was about 100 hr for 10 LC runs. Both MS and MS/MS data were acquired using the instrument’s default calibration.
Applied Biosystems 4700
Peptides were separated on an Express LC-100 HPLC (Eksigent, Dublin, CA) using a 5 cm x 300 μm Zorbax C18 column (Agilent Technologies, Palo Alto, Ca). Buffer A was 0.1 % TFA acid in water and buffer B was 0.1 % TFA acid in ACN. A gradient was run from 5% to 35% buffer B over 30 min at a flow rate of 1.5 μL/min. In all cases, 10 μL of standard mix was injected using a Famos autosampler (LC Packing/Dionex, Sunnyvale, CA). Alpha-cyano-4-hydroxycinnamic acid (CHCA) matrix solution (Agilent Technologies, Palo Alto, CA) was mixed with the column eluate at a flow rate of 1.5 μL/min. Elution fractions were collected every 4 s on a 575 spot stainless steel MALDI target plate using a Probot microfraction collector (LC Packing/Dionex). Collected fractions were analyzed with an ABI MALDI TOF-TOF 4700 proteomics analyzer. The frequency-tripled Nd:YAG laser, operating at a wavelength of 355 nm, was fired at 200 Hz. MS data were acquired in reflectron mode by accumulating 1000 laser shots. MS/MS data were acquired by accumulating 2000 laser shots with a collision energy of 1 keV with air as the collision gas. A maximum of seven MS/MS events per spot were collected. The data acquisition time for one LC run took ~4 hrs depending on the number of CID attempts. The total analysis time was ~40 hrs for 10 LC runs.
Thermofinnigan LTQ-FT
Three instruments were used from contributing labs. For the LTQ-FT analysis of mix 1, the gradient was performed exactly as described above for the LTQ except for the use of JT Baker LC/MS grade water, and Thermo Betasil C-18 columns (100 x 1 mm). The flow rate was 65 μl/min. Each MS1 (FT, Resolution 100000) scan triggered 5 MS/MS scans. A 50 entry exclusion list was populated with masses (+/− 1.5 Da) that were seen more than once within a 30 s window. Peaks were removed from the MS/MS exclusion list after 3 s. The LTQ-FT analyses of mix 3 were performed using an Agilent 1100 system in micro mode at a flow rate of 1.2 μl/min. A 60 min gradient was used starting from 2% buffer B (98% acetonitrile) to 30% buffer B. Each MS scan (FT, Resolution 100000) triggered 3 MS/MS scans on the LTQ. The 150 μm diameter columns, length 10.5 cm were packed in house with Michrom C18-Magic particles (5μm, 200 Å pore). The LTQ-FT analyses of mix 4 were performed using a system identical to that described for the LTQ above except for an analytical column length of 25 cm. LC/ESI-MS/MS was performed with a nano2D LC (Eksigent) at 300 nL/min with gradient elutions from 2% B to 40% B over 60 min using 0.1% formic acid in water (buffer A) and 0.1% formic acid in acetonitrile (buffer B). Each MS (FT, Resolution 100000) triggered 5 MS/MS scans. Normalized collision energy of 30% and an isolation width of 3.0 were used for MS/MS events. An isolation width of 3.0 was used for MS/MS events and dynamic exclusion was enabled with a repeat count of 1, a repeat duration of 30 seconds, an exclusion duration of 60 seconds, and an exclusion mass width of 2
Data Analysis
To ensure a comprehensive and divisible dataset, ten replicate analyses were performed with each mass spectrometer employed in this study. Each standard mix preparation (mix 1, mix 2, mix 3) was run on a subset of the mass spectrometers as described below. Native instrument data files were converted into mzXML format22 and searched, using SEQUEST, against a Haemophilus influenzae database containing the 18 proteins of interest, common contaminating proteins such as keratin and trypsin, and trace level contaminants (see table 2) which were detected in small amount in the samples. Trace level contaminants were identified by searching the LTQ data from mix 2 against version 50.4 of the Uniprot/SwissProt database. Results from both searches were analyzed using PeptideProphet software and the Trans-Proteomic Pipeline23 to confirm the quality of the data generated. After checking for quality, the raw data and mzXML files were moved to a public repository (excepting two Applied Biosystems 4800 TOF-TOF files and all the mix 1 raw data files for the Applied Biosystems API QSTAR Pulsar i, which were lost due to hard drive failure). Analysis of LTQ data for determining the characteristics of each mix was performed using in-house generated PERL scripts in combination with the Trans-Proteomic Pipeline data. Xcaliber 2.0 was use for examining single ion chromatograms from the LTQ analysis.
Table 2.
Contaminant proteins
| Protein | Organism | Swiss- Prot AC |
|---|---|---|
| Transthyretin | Bovine | O46375 |
| Aldehyde dehydrogenase, mitochondrial | Bovine | P81178 |
| Troponin I, fast skeletal muscle | Rabbit | P02643 |
| Myosin regulatory light chain 2, skeletal muscle isoform type 2 | Rabbit | P02608 |
| Glucoamylase | Aspergillus niger | P69327 |
| Hemoglobin subunit alpha-1\2 | Rabbit | P01948 |
| Hemoglobin subunit beta-1\2 | Rabbit | P02057 |
| UPF0076 protein yjgF | E. coli | P0AF93 |
| Ubiquitin | Rabbit | P62975 |
| Fructose-bisphosphate aldolase A | Rabbit | P00883 |
| Alpha-actinin-3 | Bovine | Q08043 |
| Troponin C, skeletal muscle | Rabbit | P02586 |
| Glycerol kinase | E. coli | P0A6F3 |
| Tropomyosin 1 alpha chain | Rabbit | P58772 |
| Trypsin\factor XIIA inhibitor | Maize | P01088 |
Results
A mix of 18 proteins was digested using trypsin and the resulting peptides were analyzed using eight different mass spectrometry platforms. Each analysis consists of ten consecutively run replicates using the same chromatography column. The resulting mass spectrometry data have been assembled into a dataset which we have named the ISB Standard Protein Mix Database. The instruments used in the analysis were two linear ion traps (Thermo LTQ and LTQ-FT), two 3D ion traps (Thermo LCQ Deca and Aglient XCT Ultra), two quadrupole time-of-flight platforms (Waters/Micromass Q-TOF Ultima and ABI Pulsar i), and two MALDI-TOF-TOF platforms (ABI 4700 and 4800). In each analysis, approximately 200 fmol of the standard mixture (based on back calculation and assuming no loss during digestion and clean up) was analyzed following separation by HPLC for ESI or MALDI. In the case of the ABI 4700, 1 pmol was used to ensure a robust dataset.
The proteins in the standard mixture, including SwissProt accession number, Sigma catalog number, and molecular weight are listed in Table 1. A consistent presence of contaminant proteins in the 18 mix preparations was detected after searching the tandem mass spectra from the LTQ runs of mix 2 against a more expansive protein sequence database (uniprot_sprot_v50.4.fasta). High confidence peptide assignments corresponding to contaminant proteins were individually inspected. The observed contaminant proteins are given in Table 2. With the exception of glucoamylase precursor from Aspirgillis niger, all contaminants identified appear to be derived from the same species used to prepare the individual standard proteins. Glucoamylase precursor protein was identified in the analyses performed on machines in multiple laboratories, thus it did not arise from work done in our laboratory or from carry-over from other runs. Because all the genomes of each of the organisms used to prepare the individual proteins (with the exception of rabbit) are fully or nearly fully sequenced, it is likely that the glucoamylase precursor was a contaminant present in one of the protein standards (or perhaps added as part of sample preparation). We are in the process of individually examining each of the constituent proteins to determine their source. Analysis of the peptides identified from the contaminant proteins shows that, as expected, their peak intensities are substantially below those of the purified proteins used to formulate the standard mixture.
The standard mixture was prepared anew on four separate occasions during the course of data acquisition when it was noted to be either degrading in quality or when the prior sample was consumed. The mix 1, 3 and 4 batches were made using an overnight trypsin digestion while the mix 2 batch was made using trypsin digestion assisted by sonication as per Lopez-Ferrer20. Data from each analysis were searched using SEQUEST, against a database consisting of our standard proteins plus contaminants appended to the Haemophilus influenzae database. The resulting peptide identifications were then assigned a probability of being correct using PeptideProphet. The count of unique peptide identifications with a PeptideProphet probability not less than 0.9 was used as a benchmark to assess the quality of the data with respect to reproducibility and instrument performance. The number of unique identifications varied across instrument platforms and the different mixtures. A summary of these results is shown in table 3. These results are not intended to represent a comparison of the performance of the platform. While each instrument was operating under normal conditions, none was optimized specifically for the comparison. In addition, it is possible that our database analysis could have introduced bias favoring one instrument over another.
Table 3.
Mean peptide number identified by each instrument series
| Instrument | Mean (FDR 2.5%) | |
|---|---|---|
| Mix 1 | LCQ DECA | 323.0 |
| LTQ | 738.2 | |
| LTQ-FT | 530.3 | |
| QSTAR | 640.5 | |
| Mix 2 | LCQ DECA | 783.2 |
| LTQ | 1033.1 | |
| QSTAR | 485.6 | |
| QTOF1 | 519.9 | |
| QTOF2 | 556.3 | |
| 4800 | 687.8 | |
| XCT | 604.4 | |
| Mix 3 | LCQ DECA | 397.5 |
| LTQ | 645.9 | |
| QTOF | 249.1 | |
| 4700 | 210.1 | |
| XCT | 349.9 | |
| LTQ-FT | 603.3 | |
| Mix 4 | LTQ | 468.2 |
| QSTAR | 277.3 | |
| QTOF | 182.9 | |
| LTQ-FT | 573.8 | |
| LCQ DECA | 299.9 |
A comparison of the results for the four standard mix preparations was made using the data acquired on the same LTQ instrument which highlights some differences between them. Table 4 shows the mean sequence coverage of the ten runs performed on each mix. These data indicates that the first two mix preparations yielded excellent sequence coverage, 72 and 76 percent on average, while mixes 3 and 4 were lower with averages of 55 and 49 percents, respectively. The percent sequence coverage is indicated for each protein in the mixture along with the mean spectrum count. For each protein individually, the number of unique peptides identified in each of the mixtures as a whole, and the percentage of peptides with two, one, and zero termini corresponding to proteolysis with trypsin is shown in Table 5. Within these data, mix 2 stands out as having substantially more unique peptides than the others: 698, 1075, 625, 455 for mixes one through four respectively. When the intensity of the single ion chromatograms for a sample of peptides seen in all four runs was examined, a trend was noted wherein the intensity in mix 2 was substantially higher than mixes 1 and 3; the corresponding peak intensity in mix 4 was found to be lower than that seen in mixes one and three (data not shown). Hence the protein coverage and unique peptides identified generally correlate with sample load which varied as a consequence of sample preparation. Because the additional unique peptides found in mix 2 do not translate to additional protein coverage, we checked for evidence of in-source decay by examining a subset of peptides from mix 2 with sequences originating from preexisting tryptic peptides, i.e. those peptides subsumed by others identified. Within this subclass of identified peptide, most did not co-elute with a larger fully tryptic peptide (data not shown), indicating that their origin was from non-tryptic proteolytic activity rather than in-source decay. Finally we determined the efficiency of cysteine alkylation by using a dynamic modification on this residue for database searching of the LTQ dataset. In mixes 1–4 the percent of cysteine residues that were modified were 96.4, 99.7, 98.6, and 93.8 percent respectively.
Table 4.
Mean sequence coverage and counts of identified CID spectra
| Mean % sequence coverage (mean spectrum count)
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Protein | Mix 1 | Mix 2 | Mix 3 | Mix 4 | ||||
| Actin, aortic smooth muscle | 44.2 | (48.0) | 48.9 | (54.5) | 38.5 | (51.8) | 30.9 | (28.5) |
| Alkaline phosphotase | 86.2 | (218.7) | 82.8 | (161.8) | 91.7 | (150.0) | 83.5 | (95.8) |
| Alpha-amylase | 56.6 | (68.4) | 63.4 | (58.1) | 42.0 | (51.2) | 35.2 | (24.6) |
| Alpha-lactalbumin | 73.9 | (32.7) | 67.9 | (37.8) | 50.5 | (14.2) | 43.6 | (13.9) |
| Beta-casein | 37.1 | (28.0) | 66.8 | (51.0) | 30.4 | (15.5) | 22.9 | (5.9) |
| Beta-galactosidase | 79.1 | (292.4) | 87.4 | (273.7) | 51.8 | (126.9) | 46.0 | (78.4) |
| Beta-lactoglobulin | 77.4 | (54.4) | 80.8 | (47.7) | 55.9 | (36.1) | 53.8 | (21.5) |
| Carbonic anhydrase 2 | 78.2 | (69.6) | 83.2 | (85.3) | 74.5 | (76.0) | 53.9 | (27.7) |
| Catalase | 72.6 | (108.7) | 75.4 | (99.9) | 57.7 | (75.2) | 53.1 | (57.8) |
| Cytochrome c | 67.3 | (40.8) | 66.0 | (32.6) | 69.6 | (24.6) | 60.5 | (14.4) |
| Glyceraldehyde-3-phosphate dehydrogenase | 73.2 | (261.2) | 78.3 | (173.7) | 69.6 | (126.7) | 59.2 | (62.7) |
| Glycogen phosphorylase, muscle form | 71.5 | (230.2) | 76.5 | (180.0) | 54.0 | (115.3) | 52.1 | (82.1) |
| Mannose-6-phosphate isomerase | 80.4 | (88.2) | 91.5 | (122.6) | 42.9 | (34.0) | 38.1 | (20.2) |
| Myoglobin | 74.6 | (35.8) | 79.4 | (43.0) | 48.2 | (12.6) | 46.0 | (9.4) |
| Myosin light chain 1, skeletal muscle isoform | 87.8 | (61.5) | 85.1 | (55.8) | 44.3 | (19.1) | 43.3 | (16.3) |
| Ovalbumin | 70.6 | (92.8) | 80.4 | (112.5) | 31.2 | (37.8) | 23.1 | (17.7) |
| Serotransferrin | 74.0 | (167.8) | 70.4 | (139.5) | 67.6 | (170.9) | 61.6 | (102.3) |
| Serum albumin | 72.8 | (155.0) | 74.0 | (145.8) | 64.2 | (75.0) | 65.3 | (83.9) |
|
| ||||||||
| Average coverage (total mean spectra count) | 71.5 | (2054.2) | 75.5 | (1875.3) | 54.7 | (1212.9) | 48.5 | (763.1) |
Table 5.
Mean unique peptide sequence and percentage of observed of enzyme termini
| Unique peptides (NTT2, NTT1, NTT0)a | ||||||||
|---|---|---|---|---|---|---|---|---|
| Protein | Mix 1 | Mix 2 | Mix 3 | Mix 4 | ||||
| Actin, aortic smooth muscle | 16.1 | (68,32,0) | 33.2 | (31,56,13) | 25.0 | (44,55,0) | 15.0 | (62,38,0) |
| Alkaline phosphotase | 59.8 | (51,49,0) | 77.2 | (33,65,3) | 64.0 | (56,44,0) | 49.2 | (63,37,0) |
| Alpha-amylase | 29.9 | (71,29,1) | 37.7 | (43,49,8) | 19.6 | (82,11,7) | 15.5 | (85,10,5) |
| Alpha-lactalbumin | 16.4 | (67,33,0) | 24.8 | (19,48,33) | 9.8 | (74,26,0) | 9.2 | (91,9,0) |
| Beta-casein | 10.2 | (19,79,2) | 39.0 | (10,52,38) | 9.7 | (64,36,0) | 4.9 | (63,35,2) |
| Beta-galactosidase | 97.9 | (56,44,1) | 161.6 | (30,58,12) | 67.2 | (60,40,0) | 49.6 | (74,26,0) |
| Beta-lactoglobulin | 18.3 | (64,36,0) | 28.6 | (41,56,4) | 18.6 | (53,47,0) | 15.0 | (69,31,0) |
| Carbonic anhydrase 2 | 29.2 | (45,55,0) | 45.7 | (26,67,6) | 45.0 | (38,62,0) | 20.2 | (53,47,0) |
| Catalase | 44.1 | (69,30,0) | 58.3 | (43,48,9) | 50.5 | (50,50,0) | 36.4 | (61,39,0) |
| Cytochrome c | 19.2 | (80,20,0) | 13.9 | (69,31,0) | 14.1 | (81,19,0) | 9.9 | (81,19,0) |
| Glyceraldehyde-3-phosphate dehydrogenase | 44.0 | (43,58,0) | 65.0 | (28,70,2) | 51.5 | (44,56,0) | 31.4 | (57,42,1) |
| Glycogen phosphorylase, muscle form | 84.0 | (78,22,0) | 103.7 | (54,43,3) | 72.2 | (75,25,0) | 58.6 | (79,21,0) |
| Mannose-6-phosphate isomerase | 38.3 | (61,37,1) | 79.8 | (22,64,13) | 18.4 | (72,27,1) | 12.8 | (77,23,0) |
| Myoglobin | 14.1 | (74,26,0) | 24.7 | (39,60,1) | 9.6 | (81,19,0) | 8.0 | (93,08,0) |
| Myosin light chain 1, skeletal muscle isoform | 22.9 | (73,27,0) | 33.9 | (40,57,3) | 11.5 | (71,29,0) | 9.3 | (86,14,0) |
| Ovalbumin | 26.3 | (58,42,0) | 71.0 | (23,70,7) | 16.2 | (51,49,0) | 8.8 | (68,32,0) |
| Serotransferrin | 63.2 | (80,20,0) | 87.9 | (42,54,4) | 71.6 | (75,25,0) | 54.8 | (83,17,0) |
| Serum albumin | 63.7 | (76,24,0) | 88.7 | (51,48,1) | 50.8 | (81,19,0) | 46.4 | (88,12,0) |
| Total | 697.6 | (64,35,0) | 1074.7 | (35,56,8) | 625.3 | (62,38,0) | 455.0 | (74,26,0) |
Arithmetic mean of the number of unique peptide sequences. Unique peptides are defined as distinct contiguous sequences regardless of peptide-ion charge state. NTT denotes the number of tolerable termini, in this case the number of peptide termini corresponding to proteolysis with trypsin, expressed as a percentage of the total number of unique contiguous peptide sequence observations; NTT2 – full-tryptic, NTT1 – semi-tryptic, NTT0 non-tryptic.
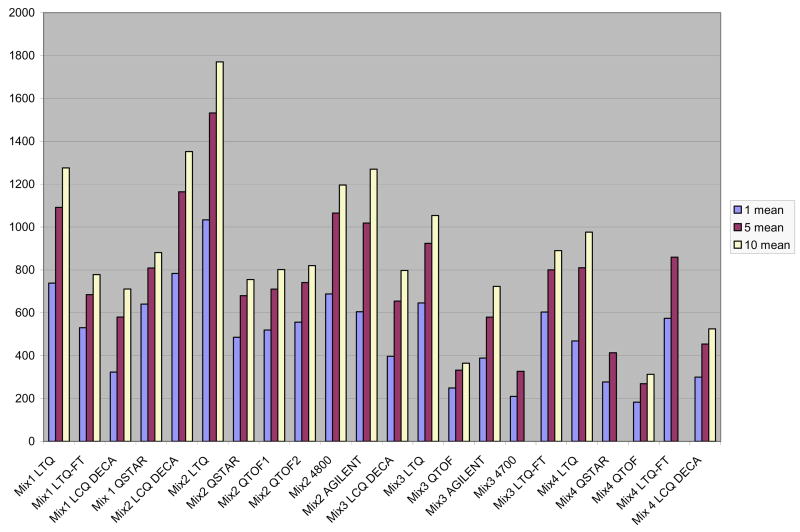
Ten replicate analyses were acquired on each mass spectrometer (unless otherwise noted) to give sufficient data to construct both control and validation datasets. To test the overlap within each subset of samples, we averaged the number of peptides identified (with a false positive rate of 2.5% or less based on PeptideProphet estimations) in a single run as well as all possible combinations of five runs, and compared this to the total identification number for all ten runs. On average, 86.5% of the unique high probability spectra were accounted for in the first 5 runs. This number ranged from 80% to 92% for different mass spectrometers. The data are summarized in Figure 1.
Figure 1.
Mean number of unique peptides found in combinations of up to ten runs. The number of unique peptides found in each data sub-set and the degree of overlap between successive runs on the same instrument is shown. Comparison was made by computing the mean number of unique peptides identified in all single runs, all possible combinations of 5 runs, and all 10 runs. The abbreviations for the mass spectrometers are as above.
Discussion
The development of software tools for the analysis of mass spectrometric proteomic data relies on the availability of good quality annotated datasets. To date, such datasets have typically been generated on only a single type of mass spectrometer24, 25. The current analysis substantially enlarges the pool of data available for software development by providing replicate datasets of a clearly defined mixture of commercially available proteins analyzed using a collection of state-of-the-art mass spectrometry platforms. While many aspects of proteomic data analysis may be generalizable between instruments, there are many instrument-specific factors such as sampling rate, mass resolution and accuracy, ionization techniques, and peptide ion fragmentation that can differ greatly. By systematically acquiring and analyzing data from the same standard sample on a variety of instruments, we have created a dataset that can be used to develop software that is optimized for individual platforms rather than based on a general set of observations. While the peptide “universe” of the standard mixture is somewhat limited, the fact that the components of the mixture are precisely defined provides a substantial advantage for software development because it provides the ability to calculate rather than estimate error rates.
The datasets produced with each of the four preparations of the standard mixture are very similar with the exception of an excess of partly- and non-tryptic peptides in mix 2. This mixture was the most concentrated based on single ion traces and thus most likely to allow identification of low abundance peaks. It was also the only one prepared using sonication (as per Lopez-Ferrer20) to aid digestion. The relative contribution of each of these factors is not known, however the identification of abundant partial- and non-tryptic peptides in our relatively simple mixtures is consistent with a recent report by Picotti et al26, which demonstrates the presence of numerous low intensity partial- and semi- tryptic peptide peaks in a mixture of similar complexity prepared using standard digestion methods.
Our composite dataset is large enough to support the development of new software tools as it includes four independent replicates of standard mix production as well as technical replicates of each mass spectrometry analysis. The size of each individual dataset, ten consecutive runs, allows partition into non-overlapping training and validation sets which can be useful in developing new applications where distinct sets of data are necessary. Our analysis demonstrates that for most of the acquired datasets, groups of five runs identify ~85% of the high confidence peptides seen in then entire group of ten.
This dataset is available for download at http://regis-web.systemsbiology.net/PublicDatasets/. It contains in excess of 1.1 million MS/MS spectra in both mzXML formatted and native instrument data files. Also available at this site is reference material including details of sample preparation, the databases searched and parameters used in the database searches. It is envisioned that with a growing awareness of such a database, contributions from other research groups will expand the available datasets with other instrument types and peptide fragmentation methods. The standard protein mixture used in this work is used in our facility as one determinant of mass spectrometry performance. Hence, it is produced regularly and freely available to collaborators wishing to contribute to the ISB Standard Protein Mix Database.
Acknowledgments
This work was supported by National Cancer Institute grant K08 CA097282 to D.B. Martin and contract N01-HV-28179 from the National Heart, Lung, and Blood Institute to R. Aebersold.
References
- 1.Ferguson PL, Smith RD. Proteome analysis by mass spectrometry. Annu Rev Biophys Biomol Struct. 2003;32:399–424. doi: 10.1146/annurev.biophys.32.110601.141854. [DOI] [PubMed] [Google Scholar]
- 2.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312(5771):212–7. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 3.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 4.Eng JK, McCormack AL, Yates JRr., III An approach to correlate tamdem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;(5):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 5.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 6.MacCoss MJ. Computational analysis of shotgun proteomics data. Curr Opin Chem Biol. 2005;9(1):88–94. doi: 10.1016/j.cbpa.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Eng JK, Martin DB, Aebersold R. Tandem mass spectrometry database searching. In: Dunn M, Jorde L, Little P, Subramaniam S, editors. Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics. John Wiley & Sons, Ltd; 2004. [Google Scholar]
- 8.Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat Biotechnol. 2001;19(10):946–51. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17(7):676–82. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 10.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 11.Keller A, Eng J, Zhou H, Aebersold R. Quantitative profiling of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):946–51. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 12.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 13.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1:2005 0017. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nesvizhskii AI. Protein identification by tandem mass spectrometry and sequence database searching. Methods Mol Biol. 2006;367:87–120. doi: 10.1385/1-59745-275-0:87. [DOI] [PubMed] [Google Scholar]
- 15.Malmstrom J, Lee H, Nesvizhskii AI, Shteynberg D, Mohanty S, Brunner E, Ye M, Weber G, Eckerskorn C, Aebersold R. Optimized peptide separation and identification for mass spectrometry based proteomics via free-flow electrophoresis. J Proteome Res. 2006;5(9):2241–9. doi: 10.1021/pr0600632. [DOI] [PubMed] [Google Scholar]
- 16.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 17.Moore RE, Young MK, Lee TD. Qscore: an algorithm for evaluating SEQUEST database search results. J Am Soc Mass Spectrom. 2002;13(4):378–86. doi: 10.1016/S1044-0305(02)00352-5. [DOI] [PubMed] [Google Scholar]
- 18.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2(9):667–75. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 19.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Ferrer D, Capelo JL, Vazquez J. Ultra fast trypsin digestion of proteins by high intensity focused ultrasound. J Proteome Res. 2005;4(5):1569–74. doi: 10.1021/pr050112v. [DOI] [PubMed] [Google Scholar]
- 21.Yi EC, Lee H, Aebersold R, Goodlett DR. A microcapillary trap cartridge-microcapillary high-performance liquid chromatography electrospray ionization emitter device capable of peptide tandem mass spectrometry at the attomole level on an ion trap mass spectrometer with automated routine operation. Rapid Commun Mass Spectrom. 2003;17(18):2093–8. doi: 10.1002/rcm.1150. [DOI] [PubMed] [Google Scholar]
- 22.Pedrioli PG, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, Pratt B, Nilsson E, Angeletti RH, Apweiler R, Cheung K, Costello CE, Hermjakob H, Huang S, Julian RK, Kapp E, McComb ME, Oliver SG, Omenn G, Paton NW, Simpson R, Smith R, Taylor CF, Zhu W, Aebersold R. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22(11):1459–66. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 23.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1:1–8. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purvine S, Picone AF, Kolker E. Standard mixtures for proteome studies. Omics. 2004;8(1):79–92. doi: 10.1089/153623104773547507. [DOI] [PubMed] [Google Scholar]
- 25.Keller A, Purvine S, Nesvizhskii AI, Stolyar S, Goodlett DR, Kolker E. Experimental Protein Mixture for Validating Tandem Mass Spectral Analysis. OMICS: A Journal of Integrative Biology. 2002;6(2):207–212. doi: 10.1089/153623102760092805. [DOI] [PubMed] [Google Scholar]
- 26.Picotti P, Aebersold R, Domon B. The Implications of Proteolytic Background for Shotgun Proteomics. Mol Cell Proteomics. 2007 doi: 10.1074/mcp.M700029-MCP200. [DOI] [PubMed] [Google Scholar]