Abstract
Introduction
The sodium glucose co-transporter (SGLT1) is responsible for all active intestinal glucose uptake. Hepatocyte nuclear factors HNF1α and β activate the SGLT1 promoter, while GATA-5 and CDX2 regulate transcription of other intestinal genes. We investigated SGLT1 regulation by these transcription factors using promoter studies and RNA-interference.
Methods
Chinese Hamster Ovary (CHO) cells were transiently co-transfected with an SGLT1-luciferase promoter construct and combinations of expression vectors for HNF1α, HNF1β, CDX2 and GATA-5. Caco-2 cells were stably transfected with knockdown vectors for either HNF1α or HNF1β. mRNA levels of HNF1α, HNF1β and SGLT1 were determined using qPCR.
Results
HNF1α, GATA-5, and HNF1β significantly activated the SGLT1 promoter (p<0.05). Co-transfection of GATA-5 with HNF1α had an additive effect, while HNF1β and CDX2 antagonized HNF1α and GATA-5. SGLT1 expression was significantly reduced in HNF1α or HNF1β knockdowns (p<0.001). HNF1α knockdown significantly reduced HNF1β expression and vice versa (p<0.005).
Conclusions
HNF1α and HNF1β are important transcription factors for endogenous SGLT1 expression by cultured enterocytes. GATA-5 and CDX2 also regulate SGLT1 promoter activity and show cooperativity with the HNF1s. We therefore propose a multifactorial model for SGLT1 regulation, with interactions between HNF1, GATA-5 and CDX2 modulating intestinal glucose absorption.
Keywords: HNF1, GATA-5, CDX2, Intestinal absorption, glucose uptake
Introduction
The intestinal sodium glucose co-transporter (SGLT1) is located on the brush border of enterocytes and plays a key role in glucose metabolism. SGLT1 is responsible for all active glucose uptake from the intestine, thereby regulating portal and liver glucose concentration 1, 2. The transport of sodium and glucose absorbs water from the intestinal lumen, the principle behind the use of oral rehydration therapy 3, 4. In addition, recent studies have shown an elevation of SGLT1 expression in the intestine of diabetic rodents and humans and over-expression of SGLT1 is associated with an obese phenotype 5–7.
Despite its importance in intestinal function, factors regulating SGLT1 expression in the intestine remain poorly characterized. Cis-regulatory elements for hepatocyte nuclear factors 1 α and β (HNF1α and HNF1 β) are present in the SGLT1 promoter, and are functional in transient promoter assays 8, 9. The ratio of α and β dimerization is particularly important in modulating the transcription of SGLT1 - homodimers of HNF1α are associated with increased SGLT1 transcription, while heterodimers of HNF1α /β decrease transcription 10. HNF1α interacts with caudal-type homeobox protein 2 (CDX2) and GATA-binding protein (GATA) family members to regulate the expression of other intestinal genes, such as sucrase isomaltase and lactase-phloridzin hydrolase 11, 12. Cis-binding elements for CDX2 and GATA are present in the SGLT1 promoter, suggesting that these may also co-regulate SGLT1 expression in the intestine.
We hypothesized that HNF1 is a transcription factor for SGLT1, and aimed to confirm this by knockdown of HNF1α and β in an in vitro enterocyte model. We further hypothesized that CDX2 and GATA-binding proteins co-regulate SGLT1 transcription, and tested this using promoter assays involving co-transfection of combinations of HNF1α, HNF1β, CDX2 and GATA-5. We show that HNF1 is an important transcription factor for endogenous SGLT1 expression in CaCo-2 cells. Moreover, co-regulation of SGLT1 promoter activity with CDX2 and GATA-5 suggests a complex, multifactorial control of SGLT1 transcription in the intestine.
Methods
Knockdown vectors and sequences
Knockdown vectors were purchased containing short-hairpin RNA sequences against either HNF1α or HNF1β within a lentiviral plasmid vector (pLKO.1-puro, Sigma, St Louis, MO). All shRNA sequences were BLAST searched (NCBI, http://www.ncbi.nlm.nih.gov/BLAST/) against all human sequences deposited in the GenBank and RefSeq databases to exclude homology of the HNF1α shRNA to the HNF1β gene and vice versa. A scrambled oligonucleotide sequence was used as a negative control (Sigma, St Louis, MO).
Reporter and expression vectors
All reporter and expression vector backbones were purchased from Promega (Madison, WI) and Invitrogen (Carlsbad, CA) respectively. The firefly luciferase-containing reporter vector for the SGLT1 promoter was created by ligation of the −997/−1 fragment of the rat SGLT1 promoter into the pGL3 basic vector backbone. Expression vectors for HNF1α and HNF1β were created by ligation of their respective sequences into the pcDNA3.1(+) backbone. CDX2 and GATA-5 expression vectors were prepared in pcDNA3.1 and pRC-CMV respectively 11, 12. The pRL-TK reporter vector, with Renilla luciferase fused to the thymidine kinase promoter, was co-transfected into all wells to control for transfection efficiency.
Cell culture and transfections for reporter assays
Chinese Hamster Ovary cells (CHO) purchased from ATCC (Manassas, VA) were maintained in F12K medium supplemented with 10% fetal bovine serum and 1% penicillin- streptomycin (Invitrogen, Carlsbad, CA). CHO cells have no described endogenous expression of SGLT1, HNF1α, HNF1β, GATA-5 or CDX2. Cells between passages 3 and 4 were transfected at 80% confluence with combinations of reporter (SGLT1, pGL3 Basic empty vector) and expression vectors (HNF1α, HNF1 β, CDX2, GATA-5, empty pcDNA3.1(+) vector) using Effectene (Qiagen, Valencia, CA) according to manufacturer’s instructions (n=3 per transfection combination). Luciferase luminescence was quantified at 48 hours post-transfection using the Dual Luciferase Reporter Assay System (Promega, Madison, WI).
Cell culture and transfections for stable knockdowns
Caco-2 cells, which differentiate into small bowel epithelium and express SGLT1 on confluence, were purchased from ATCC (Manassas, VA) and maintained in Dulbecco’s Modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin- streptomycin (Invitrogen, Carlsbad, CA). Cells between passages 10 and 12 were transfected via electroporation (BioRad Gene Pulser, Biorad, Hercules, CA) in Cytomix buffer containing 240mM KCl, 0.3mM CaCl2, 20mM K2HPO43H2O and KH2PO4, 50mM HEPES, 4mM EGTA and 10mM MgCl2 , supplemented with 50mM ATP and fresh glutathione (3.08mg/ml). Transfections were carried out at 960uF and 300V. At 48 hours post-transfection antibiotic selection was commenced using 12µg/ml puromycin (Invivogen, San Diego, CA).
RNA extraction, reverse transcription and real-time PCR
RNA was extracted from Caco-2 cells at five days post-confluence using the miRVaNA kit (Ambion, Austin, TX). Reverse transcription was carried out with oligoDT priming using the Superscript III kit (Invitrogen, Carlsbad, CA). Real-time quantitative PCR was carried out using SYBR Green master mix (Applied Biosystems, Foster City, CA) and primers for HNF1α, HNF1 β, SGLT1 and actin (Invitrogen, Carlsbad, CA, Table 1). Primer sequences were BLAST searched (NCBI, http://www.ncbi.nlm.nih.gov/BLAST/) against all human sequences deposited in the GenBank and RefSeq databases. No homology of the HNF1α primer to the HNF1β gene was detected and vice versa.
Table 1.
Primer sequences used for real-time PCR
| Primer | Sequence |
|---|---|
| HNF1α forward | 5’- ACGCCCTCTACAGCCACAAGC |
| HNF1α reverse | 5’- GGTGATGAGCATAGTCTGCG |
| HNF1β forward | 5’- CAGCGGGCGGAGGTGGACCGG |
| HNF1β reverse | 5’- CCCTTGATCATTTTAGCAGCC |
| SGLT1 forward | 5’ - CCTCTTCGCCATTTCTTTCATC |
| SGLT1 reverse | 5’ - ATGCACATCCGGAATGGGT |
| β-actin forward | 5’ - AGCAGGAGTATGACGAGTCCG |
| β-actin reverse | 5’ - AGTCATAGTCCGCCTAGAAGCA |
All samples underwent reverse transcription and real-time PCR simultaneously to minimize variations in reaction efficiencies. PCR reaction mixtures contained 2ul experimental cDNA and 12.5pmol each of forward and reverse primers in a total volume of 12.5ul. Amplification was carried out for 2 minutes at 50°C, 10 minutes at 95°C, then 40 cycles of 15 seconds 95°C and 1 minute at 60°C. Fluorescence was measured at the end of each 60°C step. Expression of HNF1α, HNF1β and SGLT1 was expressed as a ratio to the stably expressed housekeeping gene actin. Melting curve analysis confirmed that all samples were free of non-specific products, multiple amplicons or contaminants.
Statistical analysis
Data are presented as means ± standard error of the mean. p-values were estimated using ANOVA with post-hoc Tukey’s HSD test for comparisons between multiple groups and Student’s t-test for comparisons between 2 groups, with p<0.05 taken as significant. The effect of co-transfection of two expression vectors was compared to the effect of individual expression vectors by a previously published method termed “interaction response” 11, 12. This was determined by the mean logarithm of the ratio of the effect observed on co-transfection to the sum of effects observed on single transfection (e.g. log {(GATA-5 + HNF-1 α)/ [(GATA-5) + (HNF-1 α)]}). Interaction values from −0.1 to +0.1 were defined as additive, with values >+0.1 considered synergistic and values <−0.1 considered antagonistic. Computations were performed using a commercially available statistical package (Statistica V4.3, StatSoft, Tulsa, OK).
Results
Knockdown of HNF1α and HNF1β significantly decreases SGLT1 expression in vitro
Real-time PCR results confirmed that controls bearing scrambled shRNA vectors expressed 2.1-fold higher HNF1α mRNA expression compared to HNF1α knockdown cells (p<0.05, Fig 1). Similarly, controls expressed 2.7-fold higher HNF1β mRNA expression compared to HNF1β knockdown cells (p<0.05, Fig 1). SGLT1 mRNA expression was 2.0-fold and 2.5-fold higher in negative controls compared to HNF1α knockdowns and HNF1β knockdowns respectively (p<0.001, Fig 1).
Fig 1.
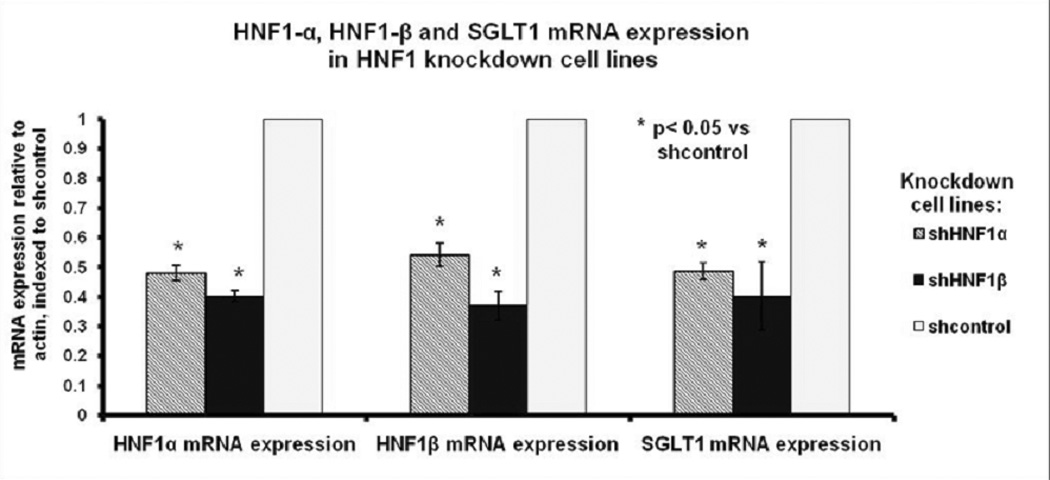
HNF1α, HNF1β and SGLT1 mRNA expression in HNF1α, HNF1β and negative control knockdown cell lines (labeled shHNF1α, shHNF1β and shcontrol respectively). mRNA was extracted from Caco-2 cells at 5- days post confluence and quantified by real-time PCR. Values are expressed as means +/− SEM, normalized to actin and indexed to the negative control (n=5 per group).
Interdependence of HNF1α and HNF1β expression
Knockdown of HNF1α reduced expression of HNF1β and vice versa. Controls expressed 1.9-fold greater HNF1β mRNA expression than HNF1α knockdown cells, and 2.5-fold greater HNF1α mRNA expression than HNF1β knockdown cells (p<0.005, Fig 1).
GATA-5 stimulates SGLT1 promoter activity in vitro
GATA-5 and HNF1α had stimulatory effects on the SGLT1 promoter, producing luciferase levels 2.3-times and 6.4-times that of the empty vector respectively (p<0.05, Fig 2). HNF1β produced a much smaller increase in SGLT1 promoter activity, with a 1.9-fold increase in luciferase expression compared to the empty vector (p<0.05, Fig 2).
Fig 2.
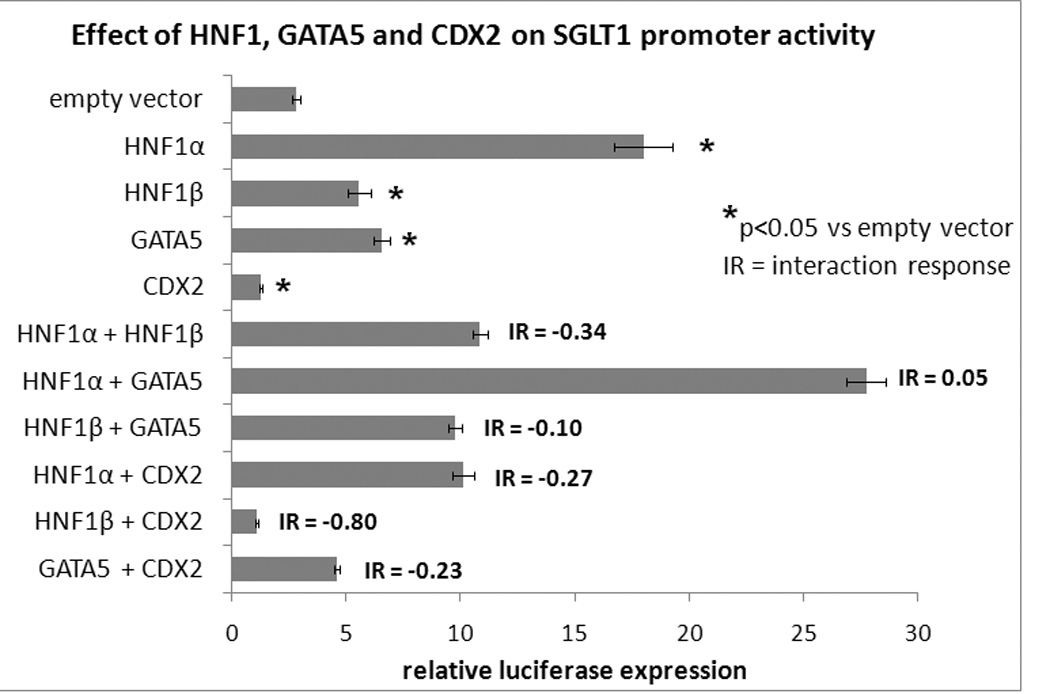
SGLT 1 promoter activity on co-transfection of the SGLT1-luciferase promoter with individual or multiple expression vectors. Firefly luciferase expression was used as a surrogate measure of SGLT1 promoter activity and normalized to Renilla luciferase to correct for variability in transfection efficiency. Results are expressed as means +/− SEM, n=3 per group, and are representative of 2 individual experiments. Transfection of the SGLT1-luciferase promoter with the pcDNA3.1 empty vector served as the control.
Multiple transcription factors co-regulate SGLT1 promoter activity in vitro
We also sought to determine the effects of co-transfection of multiple expression vectors transcription factors on SGLT1 promoter activity. GATA-5 had an additive effect on the SGLT1 promoter when co-transfected with HNF1α (interaction response of 0.05, Fig 2 and 3) but an antagonistic effect with HNF1β and CDX2 (interaction response of −0.10 and −0.23 respectively, Fig 2 and 3). CDX2 had antagonistic effects on the SGLT1 promoter on co-transfection with either HNF1α or HNF1β (interaction response of −0.27 and −0.80 respectively, Fig 2 and 3). We confirmed previous reports showing that HNF1β antagonizes the stimulatory effects of HNF1α on the SGLT1 promoter (interaction response −0.34, Fig 2 and 3) 9.
Fig 3.
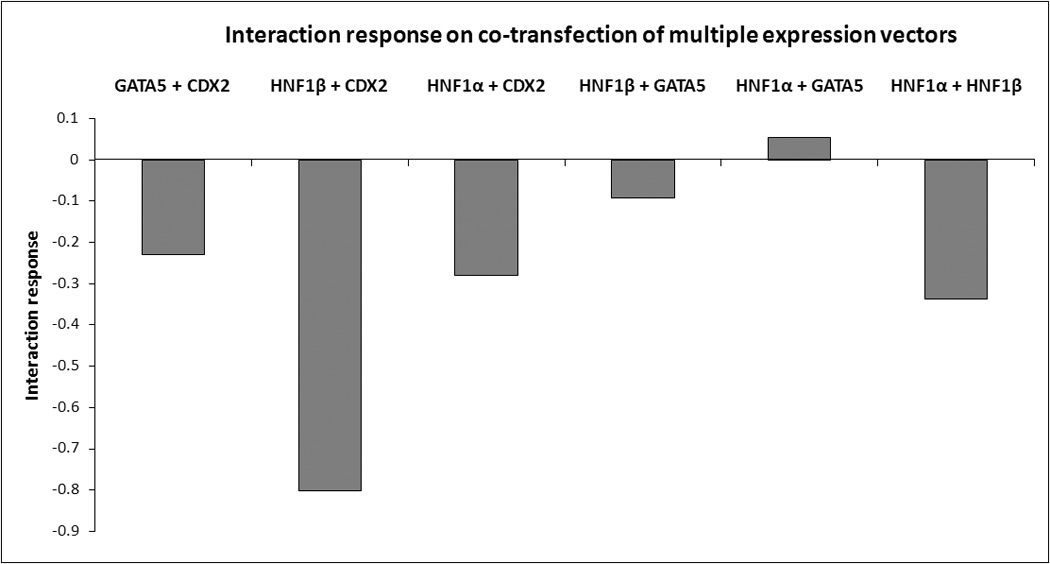
An overview of the interaction response on co-transfection of multiple expression vectors. An additive interaction was noted between HNF1α and GATA-5, while antagonistic interactions were observed in all other co-transfections. Additive interactions are defined as values between 0.1 and −0.1, and antagonistic interactions as values <−0.1.
Discussion
We show for the first time that HNF1α and HNF1β are important transcription factors for the expression of endogenous SGLT1 in CaCo2 enterocytes. Moreover, we found activation of the SGLT1 promoter by GATA-5 and CDX2, which revealed a functional cooperativity between HNF1, GATA-5 and CDX2.
Previous studies have identified HNF1-mediated stimulation of the SGLT1 promoter in vitro 8–10. There have been no data to date on the effect of HNF1 on SGLT1 expression in the native (endogenous) state. The significant reduction in SGLT1 expression on knockdown of HNF1α or HNF1β shows that HNF1 regulates enterocyte SGLT1 transcription in vitro. Homodimers of HNF1α stimulate SGLT1 transcription, while HNF1α /β heterodimers reduce SGLT1 transcription 10. This is supported by our reporter assays which show a significant increase in SGLT1 promoter activity by HNF1α, an effect that decreases on addition of both HNF1α and HNF1 β.
Knockdown of either HNF1α or HNF1β reduced SGLT1 expression. However, knockdown of HNF1β reduced HNF1α expression and vice versa, making it difficult to distinguish the relative importance of HNF1β vs. HNF1α in vitro. Our results suggest that the transcription of each isoform is dependent on expression of the other. Choice of non-conserved regions in the HNF1s as shRNA or PCR primer targets (also confirmed by BLAST searches) makes it unlikely that cross-reactivity produced non-specific knockdown.
Cooperative regulation of brush border hydrolases sucrase isomaltase and lactase phloridzin hydrolase by HNF1, CDX2 and GATA-5 in vitro has been previously described. We demonstrate that GATA-5 and CDX2 also regulate SGLT1 promoter activity. GATA-4, 5 and 6 regulate the expression of several intestinal genes – intestinal fatty acid binding protein (IFABP), lactase phloridzin hydrolase (LPH) and sucrose isomaltase (SI) 11, 13–15. GATA-4 and 6 null mice exhibit early embryonic lethality 16, 17. Female GATA-5 null mutant mice display severe genitourinary abnormalities; however intestinal gene expression in this model has not been determined 18. CDX2 is a homolog of the Caudal gene in Drosophila, and plays a key role in intestinal epithelial development and maintenance 19.
The SGLT1 promoter contains several putative binding sites for each of these transcription factors; however their exact functional binding sites on the SGLT1 promoter remain to be determined. Physical interaction between HNF1α and CDX2 or GATA-5 mediates the cooperative regulation of the LPH gene promoter. These proteins also act in concert with co-factors such as CBP to drive sucrase transcription 11, 20, 21. Physical interaction and co-factors may similarly mediate the regulation of SGLT1.
These findings may be relevant in modulating the change in SGLT1 expression as enterocytes mature along the crypt-villus axis. HNF1α and HNF1β are expressed at high levels in the crypt and at low levels at the villus tips 22. CDX2 is expressed all along the crypt-villus axis 23, while GATA-5 is localized to the villus tip 13. SGLT1 mRNA expression increases with distance from the crypt, with the highest level of expression at the villus tips where nutrient exposure is highest 24. We hypothesize that HNF1α and β initiate SGLT1 transcription in the lower villus, while GATA-5 maintains SGLT1 expression in differentiated cells at the villus tip. CDX2 may negatively modulate SGLT1 expression along the length of the crypt-villus axis.
In summary our data show that HNF1α and HNF1β are essential transcription factors for SGLT1 expression in vitro. We also identify activation of the SGLT1 promoter by GATA-5 and CDX2, and determine functional cooperativity between HNF1, GATA-5 and CDX2 on SGLT1 promoter activity. Our findings suggest complex regulation of SGLT1 transcription by multiple transcription factors and raise the possibility that a group of intestine-specific transcription factors interact to regulate the expression of numerous transporters and enzymes expressed by differentiated enterocytes. Understanding the exact mechanisms underlying this may reveal new treatments for the modulation of SGLT1 expression in diseases such as malabsorption, diabetes and obesity.
Acknowledgment
The authors are grateful to Dr S.D. Krasinski (Children’s Hospital, Boston) for providing the CDX2 and GATA-5 expression vectors.
Grant support: This study was funded by the NIH grant 5 R01 DK047326 (SWA), March of Dimes Grant#1-FY99-221 (DBR), the Harvard Clinical Nutrition Research Center grant (AT) P30-DK040561, the Nutricia Research Foundation (AB) and the Berkeley Fellowship (ATS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lane JS, Whang EE, Rigberg DA, Hines OJ, Kwan D, Zinner MJ, McFadden DW, Diamond J, Ashley SW. Paracellular glucose transport plays a minor role in the unanesthetized dog. Am J Physiol. 1999;276:G789–G794. doi: 10.1152/ajpgi.1999.276.3.G789. [DOI] [PubMed] [Google Scholar]
- 2.Hediger MA, Rhoads DB. Molecular physiology of sodium-glucose cotransporters. Physiol Rev. 1994;74:993–1026. doi: 10.1152/physrev.1994.74.4.993. [DOI] [PubMed] [Google Scholar]
- 3.Loo DD, Zeuthen T, Chandy G, Wright EM. Cotransport of water by the Na+/glucose cotransporter. Proc Natl Acad Sci U S A. 1996;93:13367–13370. doi: 10.1073/pnas.93.23.13367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright EM, Hirayama BA, Loo DF. Active sugar transport in health and disease. J Intern Med. 2007;261:32–43. doi: 10.1111/j.1365-2796.2006.01746.x. [DOI] [PubMed] [Google Scholar]
- 5.Dyer J, Garner A, Wood IS, Sharma AK, Chandranath I, Shirazi-Beechey SP. Changes in the levels of intestinal Na+/glucose co-transporter (SGLT1) in experimental diabetes. Biochem Soc Trans. 1997;25:479S. doi: 10.1042/bst025479s. [DOI] [PubMed] [Google Scholar]
- 6.Dyer J, Wood IS, Palejwala A, Ellis A, Shirazi-Beechey SP. Expression of monosaccharide transporters in intestine of diabetic humans. Am J Physiol Gastrointest Liver Physiol. 2002;282:G241–G248. doi: 10.1152/ajpgi.00310.2001. [DOI] [PubMed] [Google Scholar]
- 7.Osswald C, Baumgarten K, Stumpel F, Gorboulev V, Akimjanova M, Knobeloch KP, Horak I, Kluge R, Joost HG, Koepsell H. Mice without the regulator gene Rsc1A1 exhibit increased Na+-D-glucose cotransport in small intestine and develop obesity. Mol Cell Biol. 2005;25:78–87. doi: 10.1128/MCB.25.1.78-87.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vayro S, Wood IS, Dyer J, Shirazi-Beechey SP. Transcriptional regulation of the ovine intestinal Na+/glucose cotransporter SGLT1 gene. Role of HNF-1 in glucose activation of promoter function. Eur J Biochem. 2001;268:5460–5470. doi: 10.1046/j.0014-2956.2001.02488.x. [DOI] [PubMed] [Google Scholar]
- 9.Martin MG, Wang J, Solorzano-Vargas RS, Lam JT, Turk E, Wright EM. Regulation of the human Na(+)-glucose cotransporter gene, SGLT1, by HNF-1 and Sp1. Am J Physiol Gastrointest Liver Physiol. 2000;278:G591–G603. doi: 10.1152/ajpgi.2000.278.4.G591. [DOI] [PubMed] [Google Scholar]
- 10.Rhoads DB, Rosenbaum DH, Unsal H, Isselbacher KJ, Levitsky LL. Circadian periodicity of intestinal Na+/glucose cotransporter 1 mRNA levels is transcriptionally regulated. J Biol Chem. 1998;273:9510–9516. doi: 10.1074/jbc.273.16.9510. [DOI] [PubMed] [Google Scholar]
- 11.Krasinski SD, Van Wering HM, Tannemaat MR, Grand RJ. Differential activation of intestinal gene promoters: functional interactions between GATA-5 and HNF-1 alpha. Am J Physiol Gastrointest Liver Physiol. 2001;281:G69–G84. doi: 10.1152/ajpgi.2001.281.1.G69. [DOI] [PubMed] [Google Scholar]
- 12.van Wering HM, Huibregtse IL, van der Zwan SM, de Bie MS, Dowling LN, Boudreau F, Rings EH, Grand RJ, Krasinski SD. Physical interaction between GATA-5 and hepatocyte nuclear factor-1alpha results in synergistic activation of the human lactase-phlorizin hydrolase promoter. J Biol Chem. 2002;277:27659–27667. doi: 10.1074/jbc.M203645200. [DOI] [PubMed] [Google Scholar]
- 13.Gao X, Sedgwick T, Shi YB, Evans T. Distinct functions are implicated for the GATA-4, -5, and -6 transcription factors in the regulation of intestine epithelial cell differentiation. Mol Cell Biol. 1998;18:2901–2911. doi: 10.1128/mcb.18.5.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzgerald K, Bazar L, Avigan MI. GATA-6 stimulates a cell line-specific activation element in the human lactase promoter. Am J Physiol. 1998;274:G314–G324. doi: 10.1152/ajpgi.1998.274.2.G314. [DOI] [PubMed] [Google Scholar]
- 15.Fang R, Olds LC, Santiago NA, Sibley E. GATA family transcription factors activate lactase gene promoter in intestinal Caco-2 cells. Am J Physiol Gastrointest Liver Physiol. 2001;280:G58–G67. doi: 10.1152/ajpgi.2001.280.1.G58. [DOI] [PubMed] [Google Scholar]
- 16.Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 17.Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–732. [PubMed] [Google Scholar]
- 18.Molkentin JD, Tymitz KM, Richardson JA, Olson EN. Abnormalities of the genitourinary tract in female mice lacking GATA5. Mol Cell Biol. 2000;20:5256–5260. doi: 10.1128/mcb.20.14.5256-5260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silberg DG, Swain GP, Suh ER, Traber PG. Cdx1 and cdx2 expression during intestinal development. Gastroenterology. 2000;119:961–971. doi: 10.1053/gast.2000.18142. [DOI] [PubMed] [Google Scholar]
- 20.Boudreau F, Rings EH, van Wering HM, Kim RK, Swain GP, Krasinski SD, Moffett J, Grand RJ, Suh ER, Traber PG. Hepatocyte nuclear factor-1 alpha, GATA-4, and caudal related homeodomain protein Cdx2 interact functionally to modulate intestinal gene transcription. Implication for the developmental regulation of the sucrase-isomaltase gene. J Biol Chem. 2002;277:31909–31917. doi: 10.1074/jbc.M204622200. [DOI] [PubMed] [Google Scholar]
- 21.Boudreau F, Zhu Y, Traber PG. Sucrase-isomaltase gene transcription requires the hepatocyte nuclear factor-1 (HNF-1) regulatory element and is regulated by the ratio of HNF-1 alpha to HNF-1 beta. J Biol Chem. 2001;276:32122–32128. doi: 10.1074/jbc.M102002200. [DOI] [PubMed] [Google Scholar]
- 22.Serfas MS, Tyner AL. HNF-1 alpha and HNF-1 beta expression in mouse intestinal crypts. Am J Physiol. 1993;265:G506–G513. doi: 10.1152/ajpgi.1993.265.3.G506. [DOI] [PubMed] [Google Scholar]
- 23.James R, Erler T, Kazenwadel J. Structure of the murine homeobox gene cdx-2. Expression in embryonic and adult intestinal epithelium. J Biol Chem. 1994;269:15229–15237. [PubMed] [Google Scholar]
- 24.Hwang ES, Hirayama BA, Wright EM. Distribution of the SGLT1 Na+/glucose cotransporter and mRNA along the crypt-villus axis of rabbit small intestine. Biochem Biophys Res Commun. 1991;181:1208–1217. doi: 10.1016/0006-291x(91)92067-t. [DOI] [PubMed] [Google Scholar]


