Abstract
Cytochrome P450 (CYP) inhibition often occurs in a strongly substrate- and inhibitor-dependent manner, with a given inhibitor affecting the metabolism of different substrates to differing degrees, and with a given substrate responding differently to different inhibitors. Traditionally, patterns of functional similarity and dissimilarity among substrates and inhibitors have been studied using clustering analysis of pairwise correlation coefficients. Principal component analysis (PCA) is a widely-used statistical technique that identifies the globally most significant independent trends in a set of data. Here, we show that PCA can be usefully applied to study the differential effects on a panel of CYP probe substrates by a panel of inhibitors, using published data on CYP3A4 (Kenworthy et al., 1999) and CYP2C9 (Kumar et al., 2006). PCA can detect functional similarities among substrates and inhibitors that are not readily apparent using pairwise clustering analysis. PCA also allows identification of the functionally typical and atypical substrates that might be used in combination to fully explore the CYP functional landscape.
The choice of optimal probe substrates for in vitro inhibition studies of cytochrome P450s (CYPs) – especially relevant to pharmacokinetic predictions and in vivo-in vitro correlation – is complicated by the diverse nature of substrate-inhibitor interactions for some of these enzymes (Kenworthy et al., 1999; Kumar et al., 2006; Foti and Wahlstrom, 2008). A single probe substrate can respond differently to various inhibitors; a single inhibitor can have different effects on a panel of probe substrates. The differential behavior of substrates and inhibitors with drug-metabolizing CYPs is presumably due to their promiscuity and catalytic allosterism (Guengerich, 2001; Atkins, 2006; Nath and Atkins, 2008).
Two recent studies provide valuable insight into the varied nature of substrate-inhibitor interactions, and into the patterns of similarity among substrates and inhibitors: Houston and co-workers (Kenworthy et al., 1999) studied the effects of 34 different inhibitors on the metabolism of 10 probe substrates by CYP3A4. Subsequently, Tracy and co-workers (Kumar et al., 2006) studied how 21 different inhibitors affected the metabolism of 5 probe substrates by the CYP2C9 variants *1 and *3. Both groups used hierarchical clustering to analyze relative similarity among their respective panels of substrates and inhibitors; here, we show how principal component analysis (PCA), applied to both groups' datasets, can serve as a powerful global alternative to clustering analysis. This application of PCA is similar to its previous use in functionally characterizing CYP102 variants generated by directed evolution (Bloom et al., 2007).
PCA (Wall et al., 2003) is a mathematical technique to extract the most significant trends in a set of observations. PCA relies on the assumption that the most significant trends (called ‘principal components’ or PCs) are those that show the greatest covariance between different observables over multiple observations. For a data set with n observables, PCs can be thought of as vectors in n-dimensional space, and each observation can be transformed from the original observable-space to PC-space, where its coordinates (scores) reflect how much each component contributes to the observation. PCs are ranked in order of variance (i.e., significance): because there are typically more observables than significant PCs, PCA serves as a multidimensional scaling technique. The locations of different observations in component-space provide a quick and global comparison of relative similarity and dissimilarity.
Our intent is to demonstrate how PCA can be used to empirically analyze functional patterns among CYP substrates and inhibitors. We should emphasize that a compound's score in a particular PC does not necessarily correlate with any one of its functional, physical or chemical characteristics. More advanced statistical techniques, such as common factor analysis or partial least squares regression, can be used to explain how specific characteristics of compounds contribute to their scores, but such analysis is beyond the scope of this paper.
Methods
CYP3A4 data from Kenworthy et al. consist of relative (%) inhibition of 12 probe reactions by single concentrations of 34 different inhibitors, and are presented in Table 1. CYP2C9 data from Kumar et al. consist of KI values measured for 21 inhibitors using 5 probe substrates, for allelic variants CYP2C9*1 and CYP2C9*3, presented in Table 2 respectively. To study the functional similarity of substrates, data were entered into a matrix with each row representing a probe substrate and each column representing an inhibitor. PCA was performed using a script, available upon request, in the Python programming language with the SciPy module (Jones et al., 2001). Briefly: columns were mean-centered, and compact singular value decomposition was performed on the resulting matrix using the linalg.svd () function of SciPy. The matrix of PC scores for all substrates is given by the product of the left singular vector matrix and the singular value matrix. To study the functional similarity of inhibitors, the matrix was transposed so that rows represented inhibitors and each column represented a probe substrate, and then PCA was performed as described above.
Table 1.
Data adapted with permission from Table 1 in Kenworthy et al. (1999), showing the percent inhibition achieved by 34 effectors for 11 different CYP3A4 substrates. Italicized values in parentheses represent percent activation.
| DX | DZ | MZ | TZ | TFA | TFZ | ER | CY | TS | NF | BROD | EROD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Astemizole | 65 | 74 | 78 | 48 | 70 | 70 | 90 | 80 | 83 | 94 | (221) | 3 |
| Budesonide | 30 | 48 | 71 | 56 | 73 | 77 | 91 | 77 | 64 | 49 | (3509) | (3) |
| Caffeine | 8 | (2) | 0 | (22) | (2) | 1 | 3 | 2 | (7) | (3) | 34 | 9 |
| Cisapride | 41 | 72 | 48 | 66 | 43 | 52 | 24 | 52 | 54 | 78 | (70) | 11 |
| Clotrimazole | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 64 |
| Clozapine | 20 | 23 | 26 | 9 | 15 | 14 | 31 | 19 | 43 | 50 | 49 | 11 |
| Cyclosporin | 90 | 87 | 75 | 85 | 68 | 77 | 90 | 93 | 90 | 82 | 90 | 2 |
| Dextromethorphan | 9 | 5 | 34 | 15 | 15 | (17) | 9 | 5 | 9 | 28 | 44 | 3 |
| Diazepam | (62) | 17 | 25 | 1 | 11 | 9 | 12 | 20 | 22 | 6 | (159) | (3) |
| Digitoxin | 31 | 27 | 21 | (7) | 26 | 33 | 57 | 28 | 49 | 47 | 35 | 8 |
| Disopyramide | 46 | 54 | 44 | 32 | 6 | 9 | 35 | 38 | 55 | 50 | 50 | (3) |
| Erythromycin | 71 | 63 | 44 | 33 | 23 | 24 | 60 | 64 | 28 | 37 | 71 | 5 |
| Ethionamide | 6 | 11 | (2) | (9) | 27 | 21 | 0 | 0 | 15 | 2 | 39 | 5 |
| Fluconazole | 42 | 59 | 65 | 46 | 35 | 42 | 42 | 53 | 37 | 44 | 90 | 11 |
| Gentamycin | 14 | 5 | (11) | (21) | (10) | (18) | 11 | 2 | (1) | 1 | 16 | 3 |
| Haloperidol | (20) | 3 | 2 | 52 | 17 | 27 | 33 | 36 | 29 | 96 | (33) | 1 |
| Ibuprofen | 16 | (2) | (13) | (9) | (1) | 6 | 0 | 0 | (17) | (1) | 25 | 10 |
| Itraconazole | 75 | 80 | 86 | 73 | 85 | 88 | 85 | 92 | 97 | 91 | 89 | (4) |
| Ketoconazole | 96 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 97 | 7 |
| Metoclopramide | 7 | (5) | (8) | 2 | (7) | 6 | 1 | 9 | 6 | 10 | 36 | 27 |
| Metronidazole | (3) | 10 | 0 | (6) | (7) | (4) | (3) | 3 | (15) | 3 | 27 | 2 |
| Nifedipine | 68 | 47 | 54 | 39 | 41 | 50 | 62 | 70 | 77 | 55 | 71 | 89 |
| Nimodipine | 69 | 89 | 60 | 74 | 78 | 76 | 100 | 96 | 96 | 85 | 85 | 27 |
| Nitrendipine | 74 | 63 | 60 | 60 | 43 | 51 | 79 | 84 | 82 | 66 | 28 | 3 |
| Phenacetin | 11 | (13) | 5 | (30) | 4 | 2 | 2 | 24 | 0 | 26 | 27 | 18 |
| Phenytoin | 1 | 4 | 6 | (77) | (4) | 6 | (16) | 3 | 16 | 3 | 36 | (1) |
| Piroxicam | 24 | 3 | 22 | (13) | (3) | 3 | 9 | 5 | 14 | 4 | 37 | 6 |
| Procainamide | 16 | 11 | 2 | (15) | 3 | 0 | 17 | (15) | (7) | 1 | 34 | 7 |
| Propofol | (3) | 18 | 3 | 11 | 37 | 17 | 7 | (7) | 19 | 30 | 51 | 8 |
| Quinidine | 30 | 29 | 47 | 34 | 16 | 20 | 37 | 32 | 49 | 88 | 61 | 13 |
| Roxithromycin | 52 | 37 | 40 | 7 | 11 | 14 | 30 | 37 | 20 | 20 | 71 | 0 |
| Salbutamol | 7 | 3 | 1 | (9) | (6) | (2) | 8 | 3 | 6 | (1) | 31 | 15 |
| Terfenadine | 64 | 67 | 77 | 77 | 86 | 86 | 94 | 89 | 88 | 84 | (472) | 9 |
| Testosterone | (83) | (7) | 28 | (39) | 35 | 29 | 74 | 60 | 38 | 3 | (364) | 2 |
Table 2.
KI values (in μM) calculated for 21 inhibitors and 5 probe substrates of CYP2C9. Taken with permission from Table 1 of Kumar et al. (2006).
| CYP2C9.1 | CYP2C9.3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Flurbiprofen | Warfarin | Phenytoin | Tolbutamide | Diclofenac | Flurbiprofen | Warfarin | Phenytoin | Tolbutamide | Diclofenac | |
| Amiodarone | 2.1 | 2.99 | 4.03 | 0.69 | 1.89 | 3.64 | 1.66 | 1.87 | 0.81 | 2.1 |
| Benzbromarone | 0.004 | 0.001 | 0.04 | 0.02 | 0.01 | 1.54 | 0.01 | 0.71 | 0.03 | 0.04 |
| Clozapine | 4.13 | 3.46 | 12.88 | 11 | 11.43 | 10.49 | 2.8 | 5.07 | 5.92 | 8.04 |
| Fluvoxamine | 0.63 | 0.58 | 2.46 | 2.74 | 4.12 | 4.19 | 0.67 | 2.37 | 1.57 | 2.53 |
| Gemfibrozil | 12.43 | 0.79 | 2.38 | 2.83 | 3.64 | 14.33 | 1.65 | 4.95 | 7.21 | 14.41 |
| (S)-Ibuprofen | 4.27 | 3.06 | 4.02 | 3.95 | 4.46 | 9.78 | 3.15 | 5.5 | 14.03 | 14.68 |
| Indomethacin | 53.41 | 0.66 | 15.76 | 14.24 | 14.47 | 38.74 | 5.04 | 12.66 | 16.87 | 25.62 |
| Ketoconazole | 0.38 | 0.08 | 1.81 | 1.79 | 1.52 | 1.51 | 0.08 | 0.69 | 0.7 | 0.79 |
| Mibefradil | 11.1 | 1.04 | 6.75 | 6.63 | 13.79 | 13.08 | 1.8 | 6.9 | 6.9 | 14.3 |
| Miconazole | 0.03 | 0.01 | 0.11 | 0.05 | 0.04 | 0.1 | 0.02 | 0.12 | 0.06 | 0.07 |
| α-Naphthoflavone | 0.78 | 0.29 | 0.75 | 0.34 | 0.41 | 1.13 | 0.18 | 0.39 | 0.24 | 0.44 |
| Nicardipine | 0.07 | 0.01 | 0.33 | 0.03 | 0.03 | 0.29 | 0.01 | 0.02 | 0.02 | 0.02 |
| Nifedipine | 1.14 | 0.34 | 1.35 | 0.84 | 0.57 | 1.53 | 0.28 | 0.61 | 0.28 | 1.18 |
| Omeprazole | 5.33 | 0.64 | 2.16 | 0.92 | 0.41 | 16.24 | 0.9 | 1.6 | 1.46 | 0.3 |
| Progesterone | 1.72 | 1.41 | 4.3 | 4.01 | 5.2 | 11.03 | 1.97 | 3.27 | 2.58 | 7.91 |
| Quercetin | 1.18 | 0.25 | 0.27 | 0.14 | 0.13 | 0.25 | 0.11 | 0.29 | 0.08 | 0.13 |
| Quinine | 3.45 | 19.8 | 85.53 | 76.35 | 100 | 21.2 | 18.89 | 39.68 | 86.52 | 96.7 |
| Sulfamethizole | 17.9 | 2.22 | 13.08 | 7.17 | 14.69 | 35.04 | 3.87 | 24.74 | 25.93 | 31.86 |
| Sulfaphenazole | 0.06 | 0.12 | 0.23 | 0.22 | 0.15 | 0.72 | 0.15 | 0.39 | 0.33 | 0.36 |
| Tamoxifen | 3.44 | 0.66 | 3.77 | 3.32 | 4.67 | 9.77 | 0.58 | 6.69 | 2.72 | 8.36 |
| Thiobendazole | >100 | 17.22 | 36.21 | 33.09 | 41.08 | 100 | 11.02 | 26.92 | 24.54 | 69.85 |
Results and Discussion
CYP3A4 Substrates
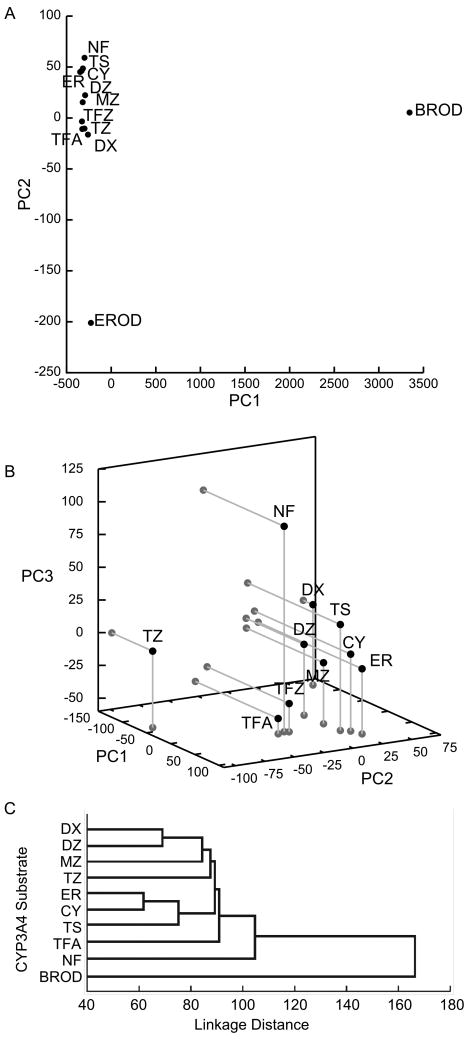
PCA of CYP3A4 probe substrate responses to inhibitors (Fig. 1a) shows all 11 probe substrates plotted in terms of their scores in the first two PCs. (There are 12 points because Kenworthy et al. monitored two different products from terfenadine – C-hydroxylation (TFA) and N-demethylation (TFZ). Additionally, the authors used seven of their probe substrates (TS: testosterone, CY: cyclosporine, ER: erythromycin, DZ: diazepam, DX: dextromethorphan, NF: nifedipine, and terfenadine) as inhibitors as well, approximating the extent of ‘inhibition’ of a probe substrate by itself as the percent maximal activity at 30 μM substrate concentration. Any resulting errors should be minor in a global analysis such as PCA.) It is immediately obvious that the two fluorescent substrates ethoxyresorufin and benzyloxyresorufin (EROD and BROD) are markedly different in their response from the other nine probe substrates. This is borne out by the raw data in Table 1, with BROD in particular showing marked activation by several compounds that inhibit all or most of the other probe substrates, and EROD showing a weaker response in general to most compounds than the nine other probes.
Figure 1.
a) Scores in the 1st and 2nd-most significant PCs for 12 CYP3A4 probe reactions. (The units of both axes do not have direct physical relevance, and should be taken to represent only the relative similarity of the various probe reactions.) Fluorescent substrates BROD and EROD are markedly different functionally from non-fluorescent substrates. b) PCA with fluorescent substrates BROD and EROD omitted, showing scores for the 1st, 2nd and 3rd-most significant PCs. DX, TS, CY, ER and MZ form a central cluster that may comprise the best-representative substrates of CYP3A4. c) Hierarchical clustering analysis for pairwise correlation coefficients of inhibition, adapted with permission from Fig. 3 of Kenworthy et al. (1999).
Kenworthy et al. correctly recognized that these two fluorescent substrates are highly dissimilar from the other nine probe substrates, and therefore may not be representative of CYP3A4 substrates in general. To more closely examine the relationships between the nine remaining probe substrates (i.e., the ten remaining probe reactions), we therefore eliminated all data for EROD and BROD from the dataset. The first three PCs of the remaining data (Fig. 1b) together describe 56.1% of the variance in the data – meaning that analysis of even less significant PCs can provide additional useful information. Nonetheless, scores in PCs 1-3 show a central cluster (TS, CY, ER, MZ: midazolam, and DZ). Slightly removed, and similar to each other, are the terfenadine reactions (TFA, TFZ). Further out are DX, NF and triazolam (TZ), all about equidistant from the central cluster. The PCA results suggest that the experimental use of any of the central cluster (as representative substrates) could be complemented by also using DX, NF and/or TZ to better explore CYP3A4 functional space.
Compared to the hierarchical clustering (Fig. 1c) based on pairwise correlation coefficients presented by Kenworthy et al., PCA provides a more global and comprehensive picture of the patterns of similarity and dissimilarity between various probe substrates. For example, pairwise clustering fails to capture the similarity in responses between TS and MZ, or the distance between DX and DZ. (Part of the reason that the authors underestimate the dissimilarity of DX and TZ from other substrates is that their clustering calculations ignored activation, although DX and TZ were activated by several of the ‘inhibitors’ in the panel.)
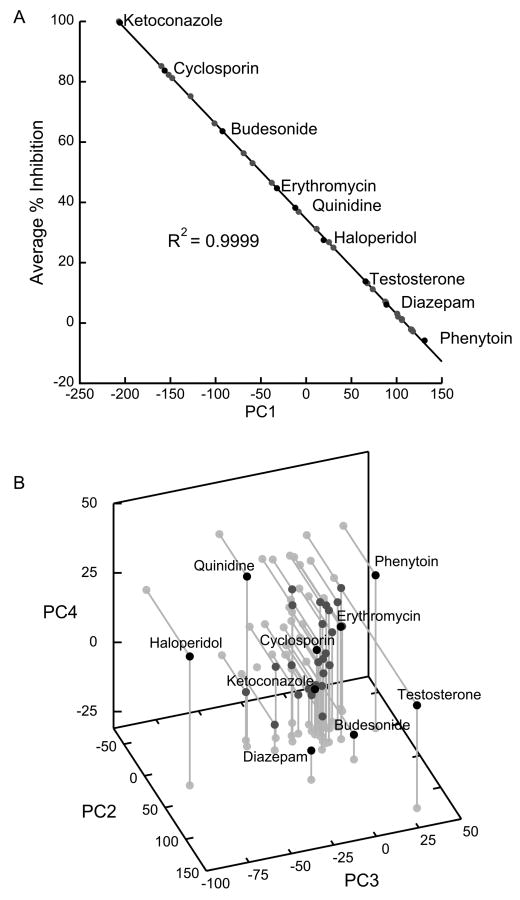
CYP3A4 Inhibitors
When the input data are transposed, PCA illustrates how various CYP3A4 inhibitors/effectors relate to each other functionally. As one might expect, the greatest amount of variance in the transposed data set is explained by the average extent of inhibition by each member of the inhibitor panel – the relevant [I]/KI with [I] = 30 μM, under assay conditions. This is clearly demonstrated by the very high correlation (R2 = 0.9999, Fig. 2a) between the average inhibition % and inhibitors' scores in the first PC. Therefore, the most useful information about functional differences is contained in PCs 2-4. (PCs 1-4 account for 75.6% of variance.) From plots (Fig. 2b) of these three PCs, we see most members of the inhibitor panel cluster together, with testosterone and haloperidol (and perhaps diazepam and quinidine) as outliers. Compounds in the center of the cluster, such as cyclosporine or ketoconazole, may be good prototypical CYP3A4 inhibitors, while outliers can be used as probes of atypical heterotropic effects.
Figure 2.
PCA of transposed CYP3A4 inhibition/activation data from ref. (Kenworthy et al., 1999), showing how inhibitors from the panel cluster in functional space. Only selected inhibitors (black dots) are labeled. a) Inhibitor score PC1 is almost entirely determined by the average inhibition, a function of the average [I]/KI for each inhibitor ([I] = 30 μM in the experimental conditions). b) Functionally relevant characteristics of the various inhibitors are encapsulated in the 2nd through 4th PCs. Under experimental conditions, testosterone is the most atypical inhibitor in the panel, followed by haloperidol.
CYP2C9 Substrates
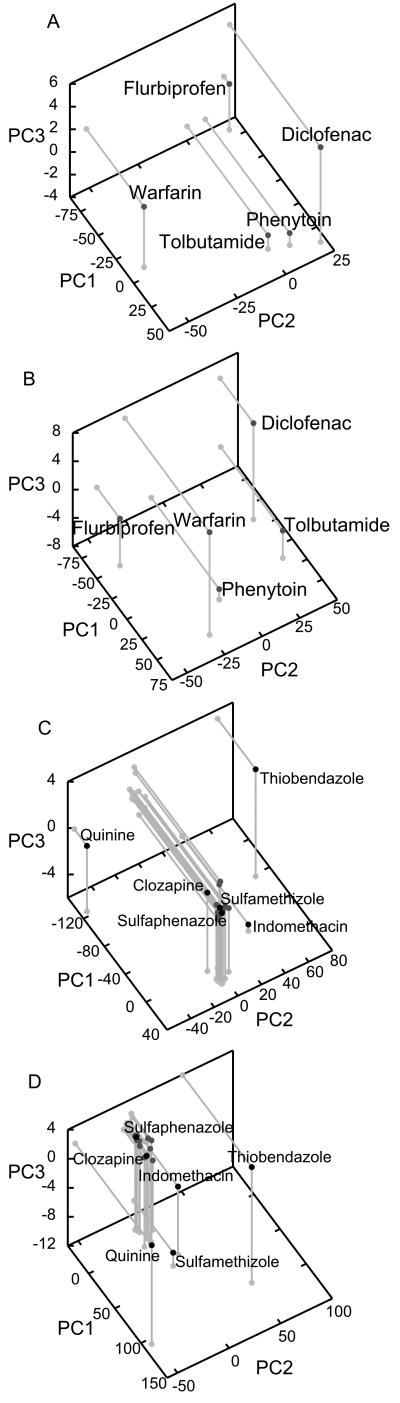
KI values of 21 substrates using five probe substrates of CYP2C9.1 and .3 presented by Kumar et al. are shown in Table 2. PCs 1-3 (>95% of variance) from these data, showing how probe substrates cluster in terms of their response to the inhibitor panel, are presented in Fig. 3 a and b. Interestingly, the single sequence alteration (Ile359Leu) between the *1 and *3 allelic variants results in markedly different probe substrate responses: for CYP2C9.1, phenytoin, tolbutamide and diclofenac form a distinct cluster with flurbiprofen and warfarin as outliers, as observed using pairwise clustering by Kumar et al. In contrast, the five substrates do not form a distinct cluster for CYP2C9.3 and are more evenly distributed in functional space.
Figure 3.
PCA of 21 KI values using 5 probe substrates for CYP2C9. a,b) PCA showing relationships between probe substrates for allelic variant CYP2C9.1 and .3 respectively. Tolbutamide, phenytoin and diclofenac form an apparent cluster for .1, but all five substrates are more evenly dispersed for .3. c,d) PCA of transposed data sets, showing the relative positions of inhibitors in functional space for CYP2C9 allelic variants .1 and .3 respectively. The recovered functional mapping is quite similar for both variants: quinine and thiobendazole are distinct outliers in both cases.
CYP2C9 Inhibitors
PCs 1-3 (>95% of variance) of the transposed CYP2C9 data (Fig. 3 c and d) illustrate the functional relationships between the various inhibitors. Analogous to the CYP3A4 result, the greatest fraction of variance is explained by each inhibitor's potency: this is reflected in the high correlation between KI and PC1 scores: R2 = 0.9906 and 0.9997 respectively for CYP2C9.1 and .3. Both isoforms show a large central cluster comprising most of the panel, implying relatively similar patterns of inhibition; our analysis supports the continued use of sulfaphenazole as a prototypical CYP2C9 inhibitor, since it is located in this central cluster. Quinine and thiobendazole (and perhaps indomethacin) are outliers. There is in general less variation in inhibitor function between allelic variants than was observed with probe substrates.
In conclusion, we have shown how PCA can be used to globally compare the functional characteristics of CYP substrates and inhibitors. The global nature of PCA allows the recognition of similarities or dissimilarities that may not be evident in traditional pairwise clustering analysis; similarly, mapping a new drug-like compound into functional space (using known substrate or inhibitor panels) could augment predictions of allosteric behavior and drug-drug interactions. As an important caveat, PCA is sensitive to systematic biases in the original data, and its meaningful application to in vitro metabolism studies relies on the type of high-quality, consistent and extensive datasets generated by Kenworthy et al. and Kumar et al.
Acknowledgments
We are grateful to Dr. R. Scott Obach (Pfizer Inc.), Prof. Tim Tracy (University of Minnesota) and Prof. Jeff P. Jones (Washington State University) for their insightful comments.
This work was supported by National Institutes of Health Grant GM-32165.
Abbreviations
- CYP
cytochrome P450
- PCA
principal component analysis
- PC
principal component
- DX
dextromethorphan
- DZ
diazepam
- MZ
midazolam
- TZ
triazolam
- TFA
terfenadine (C-hydroxylation)
- TFZ
terfenadine (N-demethylation)
- ER
erythromycin
- CY
cyclosporin
- TS
testosterone
- NF
nifedipine
- BROD
benzyloxyresorufin
- EROD
ethoxyresorufin
References
- Atkins WM. Current views on the fundamental mechanisms of cytochrome P450 allosterism. Expert Opin Drug Metab Toxicol. 2006;2:573–579. doi: 10.1517/17425255.2.4.573. [DOI] [PubMed] [Google Scholar]
- Bloom JD, Romero PA, Lu Z, Arnold FH. Neutral genetic drift can alter promiscuous protein functions, potentially aiding functional evolution. Biol Direct. 2007;2:17. doi: 10.1186/1745-6150-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti RS, Wahlstrom JL. CYP2C19 inhibition: the impact of substrate probe selection on in vitro inhibition profiles. Drug Metab Dispos. 2008;36:523–528. doi: 10.1124/dmd.107.019265. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- Jones E, Oliphant T, Peterson P, et al. SciPy: Open Source Scientific Tools for Python. 2001 URL: http://www.scipy.org.
- Kenworthy KE, Bloomer JC, Clarke SE, Houston JB. CYP3A4 drug interactions: correlation of 10 in vitro probe substrates. Br J Clin Pharmacol. 1999;48:716–727. doi: 10.1046/j.1365-2125.1999.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Wahlstrom JL, Rock DA, Warren CJ, Gorman LA, Tracy TS. CYP2C9 inhibition: impact of probe selection and pharmacogenetics on in vitro inhibition profiles. Drug Metab Dispos. 2006;34:1966–1975. doi: 10.1124/dmd.106.010926. [DOI] [PubMed] [Google Scholar]
- Nath A, Atkins WM. A quantitative index of substrate promiscuity. Biochemistry. 2008;47:157–166. doi: 10.1021/bi701448p. [DOI] [PubMed] [Google Scholar]
- Wall ME, Rechtsteiner A, Rocha LM. Singular value decomposition and principal component analysis. In: Berrar DP, Dubitzky W, Granzow M, editors. A Practical Approach to Microarray Data Analysis. Kluwer; Norwell, MA: 2003. pp. 91–109. [Google Scholar]