Abstract
Background and Purpose
Apolipoprotein E genotype (APOE) influences cholesterol levels and ischemic heart disease. Although there is no convincing overall association with ischemic stroke, APOE may influence large artery (atherothrombotic) stroke, for which carotid intima-media thickness (CIMT) is an informative intermediate phenotype. We therefore performed a systematic review and meta-analysis of the association between APOE and CIMT.
Methods
We sought all published studies assessing the association between APOE and CIMT. From each study, we extracted available data on study methods, subjects’ characteristics, and mean (and standard deviation) CIMT for each genotype or genotype group. We calculated study-specific and random effects pooled differences in mean CIMT between genotype groups, and assessed heterogeneity between studies and predefined subgroups using I2 and χ2 statistics.
Results
Meta-analysis of 22 published studies (30 879 subjects) showed a significant association between APOE and CIMT (pooled mean difference ε4-versus ε2-allele containing genotypes 46 μm, 95% CI 29 to 62, P<0.00001). We found evidence of small study (mainly publication) bias, with a diminished (but still highly statistically significant) association in studies of >1000 subjects (pooled mean difference 17 μm, 95% CI 12 to 23, P<0.00001). The association was larger among high vascular risk and eastern Asian populations, but this may simply reflect the smaller size of these studies.
Conclusion
Our results show a clear association of APOE with CIMT, even though publication bias means that this is overestimated by the published literature. These findings suggest the possibility of a specific association with large artery ischemic stroke.
Keywords: apolipoprotein E, association, carotid intimal medial thickness, genetics, meta-analysis
Twin and family history studies have shown that the incidence of ischemic stroke is likely to be influenced by genetic factors.1 However, case-control candidate gene association studies have so far had limited success in consistently identifying potentially causative genes.2-4 Inadequate study size, control selection bias, and lack of distinction between different ischemic stroke subtypes have all been suggested as reasons.4,5
The apolipoprotein E gene (apoE=protein, APOE=gene) is a widely studied gene in vascular and neurodegenerative diseases, including stroke.4,6 Its protein product, the glyco-protein apoE, has 3 common isoforms, E2, E3 and E4, encoded by the alleles ε2, ε3 and ε4, giving rise to 6 genotypes, with ε3/ε 3 occurring in about one half to two thirds of people in most populations. The 3 common protein isoforms interact differently with specific lipoprotein receptors, ultimately altering circulating levels of cholesterol through different effects on lipoprotein metabolism, mediated through the hepatic binding, uptake, and catabolism of chylomicrons, chylomicron remnants, very low density lipoprotein and high density lipoprotein subspecies. The ε4 allele is associated with increased total cholesterol levels and the ε2 allele with decreased levels, and so APOE genotype would be expected to influence the development of atherosclerosis and atherosclerotic vascular diseases.6 Possession of an ε4 allele increases risk of ischemic heart disease by about one third.7 Our recent meta-analysis found no convincing overall association between APOE and ischemic stroke (OR 1.11, 95% CI 1.01 to 1.22), but results from a few studies with information on ischemic stroke subtypes suggested that ε4 allele-containing genotypes may specifically increase the risk of large artery (atherothrombotic) ischemic stroke (OR 1.33), with no effect on other subtypes (ORs between 0.86 and 1.06).4
Another way of assessing the association between APOE (or other genes) and large artery atherothrombosis is to assess its effect on the quantitative intermediate phenotype of carotid intima-media thickness (CIMT).8 CIMT is a good surrogate measure for subclinical atherosclerosis, and increasing CIMT is directly associated with an increased risk both of myocardial infarction and of stroke. Each 1 SD (0.2 mm) increase in common carotid artery (CCA) IMT is associated with about a one third increase in the risk of myocardial infarction and stroke.9 The association with stroke may arise mainly from a specific association with large artery ischemic stroke, as CIMT has been found to be significantly higher in patients with large artery compared with small artery (lacunar) ischemic stroke.10 Lowering LDL cholesterol has also been shown to produce a reduction in CIMT.11 CIMT has reasonably high heritability, although estimates vary between 30% and 86%.12-14
Many studies have now investigated the association between APOE genotype and CIMT. We aimed to assess any association reliably, with a systematic review and meta-analysis.
Methods
Identification of Studies
We sought all available published studies of the association between APOE and CIMT in humans. We used a comprehensive search strategy in Medline (1966 to the end of 2006) and Embase (1980 to the end of 2006), which included MeSH terms and textwords for APOE and for CIMT (see Appendix). We also checked the reference lists of all relevant articles for further studies.
We included all studies that had measured the thickness of the intima-media of the carotid artery, but excluded studies which only reported the presence or extent of atheroma or plaque. We included studies among healthy subjects from the general population (low risk) or groups of subjects with existing vascular disease or vascular risk factors, such as hypertension, diabetes, hyperlipidemia, or coronary heart disease (high risk). We sought articles in all languages. We excluded from our analyses otherwise relevant studies from which the data required were not available in the relevant publication(s).
Data Extraction
For each included study we extracted information on year of publication, country in which the study was conducted, ethnicity of subjects, nature of the study population (eg, patients with diabetes), total number of subjects, mean age and gender distribution of the subjects, genotyping methodology, whether genotyping was done blind to CIMT and vice versa, and the definition of CIMT. In addition, we extracted information on (or where possible tested directly for) Hardy-Weinberg equilibrium. For each genotype or group of genotypes, we extracted the mean CIMT and its standard deviation (SD). Where possible, we treated studies that included both a high and a low risk group or more than one ethnic group as separate substudies.
Two authors (L.P., N.A.M.G.) independently reviewed study eligibility and extracted the information and data from each study, resolving any disagreements by discussion with a third author (C.S.). Some articles reported several different measures of CIMT (eg, from different carotid artery sites). Where a choice of measurement was available we chose the one that was closest to our ideal: the mean CIMT of the left and right far wall of the CCA. We chose this method because the CCA may give more reproducible results than the internal carotid artery (ICA) because of its accessibility, and far wall measurements are more accurate than near wall, which have to be performed at the trailing edge of the ultrasound pulse.15
We defined genotype groups as E2 (ε3/ε2 or ε2/ε2), E3 (ε3/ε3), and E4 (ε3/ε4 or ε4/ε4), and excluded any ε2/ε4 subjects from each study, as they could not be classified as E2 or E4. These were extremely small in number (2% of all subjects), making it unlikely that their inclusion or exclusion would affect the results.
Statistical Analysis
We first calculated the overall mean CIMT for each genotype group across all studies, and then carried out meta-analyses using Cochrane RevMan software (version 4.2) comparing the 2 genotype groups with the highest and lowest overall mean CIMT (E4 versus E2). We calculated the mean CIMT difference between the E4 and E2 genotype groups for each study, and pooled results using a random effects model. We used the I2 statistic to assess heterogeneity between studies, where I2 estimates the percentage of variation between studies that cannot be attributed to chance.16
We plotted the study-specific mean CIMT difference between E4 and E2 genotypes against the standard error of this difference (a funnel plot) to check for the possibility of small study bias. To assess the impact of study characteristics on the association we performed prespecified subgroup analyses. We grouped the studies according to risk status of individuals (those of high and low risk of vascular disease), study size (above or below the mean total number of subjects), ethnicity (Eastern Asian, White, or Black African), and method of measuring CIMT (ideal or not, where ideal measurements were those only from the far wall of the CCA). We assessed the significance of differences between subgroups by partitioning heterogeneity and using χ2 tests.
Results
Study Selection
Figure 1 shows the process of study selection and exclusion. Our search identified 490 articles, of which 32 were potentially relevant.17-48 Four studies were duplicates of other included studies and so were excluded.39-42 Six studies had insufficient data presented to be included in the analysis43-48 (including 2 that combined E2 and E3 genotype groups46,48). We therefore included 22 studies (increasing to 25 studies after 3 were split into 2 substudies) in a total of 30 879 subjects in our analyses.
Figure 1.
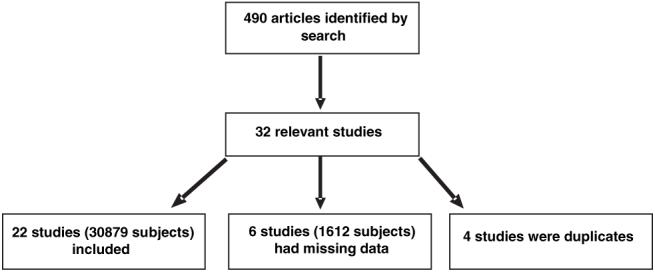
Flow diagram of study selection and exclusion process.
Study Characteristics
Details of the included studies are shown in the Table. Sample sizes ranged from 52 to 9304 with a mean of 1235. Subjects were mostly middle-aged to elderly. Most studies had approximately equal proportions of males and females, although 5 included only men.21,25,28,35,36 The studies were conducted in several European countries, the USA, Australia, Japan, and China. Most subjects were included in low risk studies (healthy subjects or general populations), whereas a smaller proportion were included in high-risk studies (see Table). Most studies used a polymerase chain reaction/restriction fragment length polymorphism method for genotyping, whereas 3 used isoelectric focusing,17,25,28 2 used polymerase chain reaction/allele specific oligonucleotide hybridization,23,31 and the 2 most recent studies used Taqman.37,38 Genotype group frequencies were fairly consistent across all studies. No studies were found to depart from Hardy-Weinberg equilibrium, although this was not reported and could not be tested directly in 5.20,23,24,33,34
Table. Characteristics of Included Studies.
| Primary Author | Year | Country | Subjects | Sample Size | Genotyping Method | Genotype Frequency |
HWE | ||
|---|---|---|---|---|---|---|---|---|---|
| E2 | E3 | E4 | |||||||
| Terry17 | 1996 | US | Referrals for coronary angiography (HR) | 254 | IEF | 0.13 | 0.61 | 0.26 | √ |
| Cattin18 | 1997 | Italy | Population sample (LR) | 254 | PCR/RFLP | 0.12 | 0.70 | 0.18 | √ |
| Kogawa19 NIDDM | 1997 | Japan | NIDDM patients (HR) | 349 | PCR/RFLP | 0.07 | 0.73 | 0.18 | √ |
| Kogawa19 controls | 1997 | Japan | Non-diabetic subjects at local check-up (LR) | 231 | PCR/RFLP | 0.09 | 0.75 | 0.14 | √ |
| Sass20 | 1998 | France | Population sample (LR) | 144 | PCR/RFLP | 0.17 | 0.62 | 0.21 | ? |
| Zhang21 | 1998 | China | CHD patients (HR) | 52 | PCR/RFLP | 0.08 | 0.73 | 0.19 | √ |
| Guz22 | 2000 | Turkey | Hemodialysis patients (HR) | 261 | PCR/RFLP | 0.13 | 0.77 | 0.11 | √ |
| Hanon23 | 2000 | France | Referrals to clinic - normal carotid walls (LR) | 312 | PCR/ASO | 0.12 | 0.67 | 0.21 | ? |
| Horejsi24 | 2000 | Czech Republic | Lipoprotein disorder patients (HR) | 112 | PCR/RFLP | 0.09 | 0.69 | 0.22 | ? |
| Ilveskoski25 | 2000 | Finland | Population sample (LR) | 189 | IEF | 0.11 | 0.58 | 0.32 | √ |
| Slooter26 | 2001 | Netherlands | Population sample (LR) | 5264 | PCR/RFLP | 0.14 | 0.58 | 0.26 | √ |
| Tabara27 | 2001 | Japan | Population sample (LR) | 202 | PCR/RFLP | 0.13 | 0.68 | 0.19 | √ |
| Haraki28 | 2002 | Japan | Healthy subjects at annual check-up (LR) | 95 | IEF | 0.11 | 0.68 | 0.21 | √ |
| Beilby29 | 2003 | Australia | Population sample (LR) | 1079 | PCR/RFLP | 0.14 | 0.59 | 0.26 | √ |
| Li30 | 2003 | China | Hypertensive patients (HR) | 92 | PCR/RFLP | 0.11 | 0.70 | 0.19 | √ |
| Xiang31 NIDDM | 2003 | China | NIDDM patients (HR) | 253 | PCR/ASO | 0.13 | 0.63 | 0.23 | √ |
| Xiang31 controls | 2003 | China | Healthy controls (LR) | 106 | PCR/ASO | 0.10 | 0.70 | 0.19 | √ |
| Elosua32 | 2004 | US | Population sample (LR) | 2723 | PCR/RFLP | 0.13 | 0.66 | 0.21 | √ |
| Fernandez33 | 2004 | Spain | CHD patients (HR) | 225 | PCR/RFLP | 0.08 | 0.70 | 0.22 | ? |
| Kahraman34 | 2004 | Turkey | Renal transplant recipients (HR) | 118 | PCR/RFLP | 0.10 | 0.78 | 0.12 | ? |
| Bednarska35 | 2005 | Poland | Heavy drinkers (HR) | 127 | PCR/RFLP | 0.13 | 0.70 | 0.17 | √ |
| Bleil36 | 2006 | US | Untreated hypertensives (HR) | 182 | PCR/RFLP | 0.13 | 0.66 | 0.21 | √ |
| Debette37 | 2006 | France | Elderly population sample (LR) | 5764 | Taqman | 0.12 | 0.67 | 0.19 | √ |
| Volcik38 blacks | 2006 | US | Black population sample (LR) | 3187 | Taqman | 0.16 | 0.59 | 0.25 | √ |
| Volcik38 whites | 2006 | US | White population sample (LR) | 9304 | Taqman | 0.20 | 0.45 | 0.35 | √ |
NIDDM indicates noninsulin dependent diabetes mellitus; CHD, coronary heart disease; LR, low risk; HR, high risk; PCR/RFLP, polymerase chain reaction/restriction fragment length polymorphism; IEF, isoelectric focusing; PCR/ASO, polymerase chain reaction/allele specific oligonucleotide hybridization; HWE, Hardy-Weinberg equilibrium.
All studies measured CIMT with B-mode ultrasound. The supplemental Table I, available online at http://stroke.ahajournals.org, shows which segment of the carotid artery was measured and how multiple measurements were combined to obtain the final CIMT value. Only 2 studies stated that ultrasonography staff were blind to genotype data,28,34 and only one stated that genotyping was carried out blind to the ultrasonography findings.29
Association Between APOE and CIMT
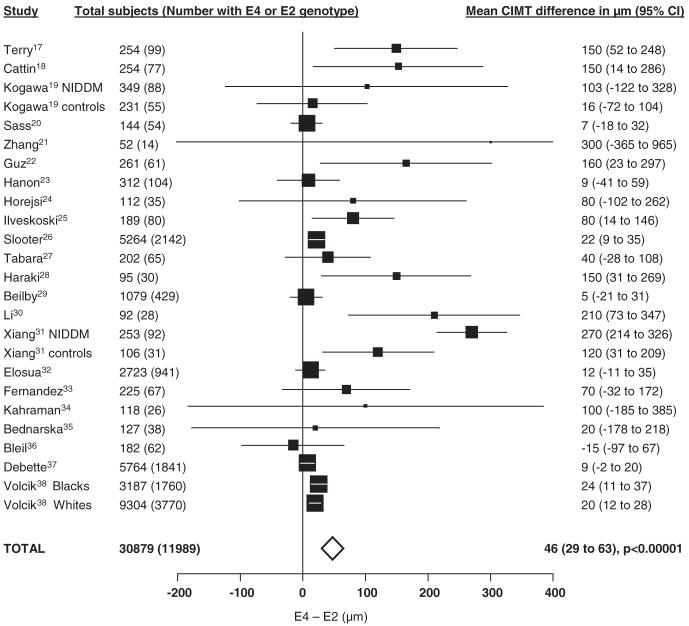
E4 genotypes had the highest mean CIMT across all studies (760 μm), the E3 genotype group had an intermediate mean CIMT (751 μm), and E2 genotypes had the lowest mean CIMT (743 μm; in keeping with their known effects on cholesterol levels). We therefore compared mean CIMT for E4 versus E2 genotypes in our meta-analyses. Figure 2 shows the study-specific and pooled results for the E4 versus E2 comparison in a total of 11989 subjects. Overall there was a highly significant pooled mean CIMT difference of 46 μm (95% CI 29 to 62, P<0.00001). There was substantial heterogeneity between the studies (I2=80%).
Figure 2.
Study-specific and pooled mean differences of the CIMT between E4 and E2 genotypes (ordered by publication date). The sizes of the squares are proportional to the statistical weight given to each study. The horizontal lines represent 95% CI. The width of the diamond represents the 95% CI of the pooled estimate. Heterogeneity between studies: I2=80%.
The funnel plot for the analysis was markedly asymmetrical, suggestive of small study (presumably mainly publication) bias (supplemental Figure I, available online at http://stroke.ahajournals.org).
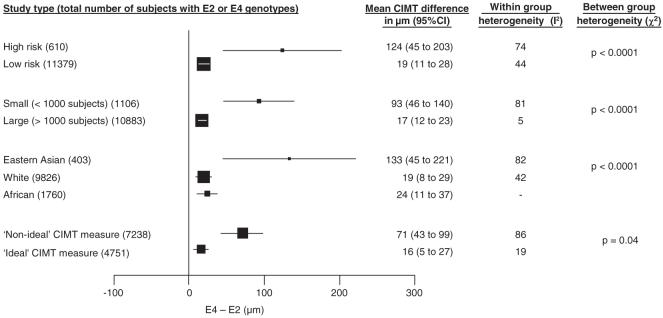
Results of our subgroup analyses are shown in Figure 3. We found a substantially larger pooled mean CIMT difference among high-risk compared with low-risk populations and in Eastern Asian compared with White or Black African populations. However, we also found that the pooled mean CIMT difference in smaller studies was more extreme than that in larger studies (smaller studies [mean number of subjects analyzed 58]:93 μm, 95% CI 46 to 140; larger studies [mean number of subjects analyzed 1814]:17 μm, 95% CI 12 to 23, P<0.00001; χ2 test for difference between the 2 subgroups: P<0.0001), suggesting the existence of small study bias, consistent with the appearance of the funnel plot (supplemental Figure I). There was little heterogeneity between the results of the larger studies (I2=5%). Study size could explain the apparent difference in size of association between high and low risk and between ethnic groups, because most studies in high risk populations and among Eastern Asian subjects were small (mean number of subjects analyzed in high-risk group 55, in low-risk group 813, in Eastern Asians 50, in other ethnic groups 682).
Figure 3.
Pooled mean differences of the CIMT between E4 and E2 genotypes: results for various subgroups. The size of the squares is proportional to the number of subjects. Horizontal lines represent 95% CI.
As CIMT was measured at a variety of sites within the carotid artery (supplemental Table I), we split the data into those studies which used our preferred method (measuring only the far wall of the CCA) and those that used any other method. Studies using our ideal CIMT measurement method yielded less heterogeneous results and a smaller pooled mean CIMT difference (16 μm, 95% CI 5 to 27), consistent with the estimated association in the larger studies.
Robustness to Missing Data
We considered the impact of excluding relevant studies without available data for the E4 versus E2 genotype group comparison. Two relevant studies combined E2 and E3 data and so could not be included in our analyses. One found no relationship between APOE and CIMT among patients with noninsulin dependent diabetes, but found that ε4 allele-containing genotypes increased CIMT by 0.14 mm compared with other genotypes in a nondiabetic population.48 The other found no significant relationship between APOE and CIMT.46 Four studies did not present mean and SD CIMT data by genotype or genotype groups and so could not be included in any of our analyses. Three found no significant association between APOE and CIMT,43-45 whereas the fourth found that E4 genotypes were significantly more frequent than E2 genotypes in the subjects with higher CIMT values.47 The total number of subjects in all genotype groups in all of these potentially relevant additional studies was 1612, bringing to 32 491 the total possible number of subjects that could have been included if data had been available. “Missing data” therefore comprised only 5% of the total, making it very unlikely that including these studies would have made a material difference to our results. Furthermore, the largest of these “missing data” studies included 511 subjects.46 Therefore, none would have been categorized as a large and so more reliable study (>1000 subjects).
Discussion
The results of our meta-analysis show a clear association between APOE and CIMT. In keeping with the known effects of APOE on cholesterol levels, E2 genotypes had the lowest CIMT, E3 genotypes an intermediate CIMT, and E4 genotypes the highest CIMT.
The overall pooled mean difference between E4 and E2 genotypes was 46 μm, but we found evidence of small study bias, both from a funnel plot and a subgroup analysis based on study size. Larger studies were far less heterogeneous, and found a substantially smaller—but still highly statistically significant—mean CIMT difference between E4 and E2 genotypes of 17 μm. Thus, although the published literature taken as a whole probably overestimates the size of the association, the consistency of direction of the association observed across the studies, and the highly statistically significant findings for the larger, more reliable studies, strongly suggests the presence of a true association.
Our subgroup analyses also suggested that the association between APOE genotype and CIMT might be larger in high-versus low-risk subjects and in Eastern Asian populations. A recent meta-analysis of the association between the angiotensin-converting enzyme insertion/deletion polymorphism and CIMT also found a larger association in high risk populations, and suggested that this may be attributable to an interaction with smoking.49,50 But, although the vascular risk and ethnicity subgroup differences in our meta-analysis could conceivably be real, they may have arisen from confounding by study size, because the studies in high-risk and Eastern Asian subjects were generally smaller than those in low-risk and other ethnic groups, respectively.
The results of studies using ideal CIMT measurement methods were far less heterogeneous than those using a nonideal measurement, suggesting that a consistent approach to CIMT measurement (using the mean of the right and left far wall measurements 1 cm below the bifurcation) may lead to more reliable results that are comparable between studies. Although the size of the association between APOE and CIMT varied with the location of measurement, the results of individual studies showed that the direction of association was consistent.
Several different genotyping methods were used in the studies, but any variations in the resulting genotyping accuracy should not introduce any systematic error, and indeed we did not find any influence of genotyping method on the size of the association between APOE and ischemic stroke in a previous meta-analysis.4
Although our results concur with what we would expect from the known effect of APOE on cholesterol levels, there may be other pathways through which APOE influences atherosclerosis and so CIMT. Some studies included in our meta-analysis adjusted for covariates, including cholesterol levels, in their analyses of the association between APOE and CIMT. Whereas in some this resulted in loss or diminution of the association between APOE and CIMT,25,32 in others the association between APOE and CIMT was preserved,17,18,26,28,35,37,38 suggesting that APOE genotype may also influence CIMT independently of its effects on cholesterol levels.
Summary
Our results have shown a modest association between APOE genotype and CIMT, even though small study bias means that the published literature tends to overestimate the size of this association. This suggests that APOE might modestly influence risk of large artery ischemic stroke, and would support large case-control studies specifically to examine this hypothesis. Further work is also required to determine whether or not the association we have observed varies among different types of subjects according to ethnicity and vascular risk status, and to further elucidate the mechanisms by which APOE may influence CIMT.
Supplementary Material
Acknowledgments
We thank Brenda Thomas for advice on compiling the search strategy, and Professors Joanna Wardlaw, Charles Warlow, and David Porteous for their helpful comments on previous drafts.
Sources of Funding
L.P. was funded by UK Medical Research Council, N.A.M.G. was funded by ConacYt-Mexico, and C.S. was funded by the Wellcome Trust, UK.
Appendix
Search strategy in Medline (similar strategy was designed for Embase)
carotid artery diseases/ or carotid artery thrombosis/ or carotid stenosis/ or moyamoya disease/
carotid artery disease/ge or carotid artery thrombosis/ge or carotid stenosis/ge or moyamoya disease/ge
carotid arteries/ or exp carotid artery, common/
(carotid adj5 [atherosclero$ or arteriosclero$ or steno$ or imt or cimt or intima media$ or ultrasound or plaque or sclero$ or atheroma$ or fatty streak or disease$ or disorder$]).tw.
1 or 3 or 4
apolipoproteins/ or apolipoproteins e/
([apolipoprotein$ adj e] or [apoprotein$ adj e] or apo-e or apo e or apoe).tw.
6 or 7
5 and 8
2 or 9
limit 10 to humans
Footnotes
Disclosures
None.
References
- 1.Flossmann E, Schulz UGR, Rothwell PM. Systematic review of methods and results of studies of the genetic epidemiology of ischemic stroke. Stroke. 2004;35:212–227. doi: 10.1161/01.STR.0000107187.84390.AA. [DOI] [PubMed] [Google Scholar]
- 2.Casas JP, Hingorani AD, Bautista LE, Sharma P. Meta-analysis of genetic studies in ischemic stroke - Thirty-two genes involving approximately 18000 cases and 58000 controls. Arch Neurol. 2004;61:1652–1661. doi: 10.1001/archneur.61.11.1652. [DOI] [PubMed] [Google Scholar]
- 3.Hassan A, Markus HS. Genetics and ischaemic stroke. Brain. 2000;123:1784–1812. doi: 10.1093/brain/123.9.1784. [DOI] [PubMed] [Google Scholar]
- 4.Sudlow C, Gonzalez NAM, Kim J, Clark C. Does apolipoprotein E genotype influence the risk of ischemic stroke, intracerebral hemorrhage, or subarachnoid hemorrhage? Systematic review and meta-analyses of 31 studies among 5961 cases and 17965 controls. Stroke. 2006;37:364–370. doi: 10.1161/01.STR.0000199065.12908.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dichgans M, Markus HS. Genetic association studies in stroke: methodological issues and proposed standard criteria. Stroke. 2005;36:2027–2031. doi: 10.1161/01.STR.0000177498.21594.9e. [DOI] [PubMed] [Google Scholar]
- 6.Eichner JE, Dunn ST, Perveen G, Thompson DM, Stewart KE, Stroehla BC. Apolipoprotein E polymorphism and cardiovascular disease: A HuGE review. Am J Epidemiol. 2002;155:487–495. doi: 10.1093/aje/155.6.487. [DOI] [PubMed] [Google Scholar]
- 7.Song Y, Stampfer MJ, Liu S. Meta-analysis: Apolipoprotein E genotypes and risk for coronary heart disease. Ann Intern Med. 2004;141:137–147. doi: 10.7326/0003-4819-141-2-200407200-00013. [DOI] [PubMed] [Google Scholar]
- 8.Humphries SE, Morgan L. Genetic risk factors for stroke and carotid atherosclerosis: insights into pathophysiology from candidate gene approaches. Lancet Neurol. 2004;3:227–235. doi: 10.1016/S1474-4422(04)00708-2. [DOI] [PubMed] [Google Scholar]
- 9.O’Leary DH, Polak JF, Kronmal RA, Manolio TA, Burke GL, Wolfson SK. Carotid-artery intima and media thickness as a risk factor for myocardial infarction and stroke in older adults. N Engl J Med. 1999;340:14–22. doi: 10.1056/NEJM199901073400103. [DOI] [PubMed] [Google Scholar]
- 10.Pruissen MO, Gerritsen SAM, Prinsen TJ, Dijk JM, Kappelle LJ, Algra A. Carotid intima-media thickness is different in large- and small-vessel ischaemic stroke: the SMART study. Stroke. 2007;38:1371–1373. doi: 10.1161/01.STR.0000260220.37016.88. [DOI] [PubMed] [Google Scholar]
- 11.Taylor AJ, Kent SM, Flaherty PJ, Coyle LC, Markwood TT, Vernalis MN. ARBITER: arterial biology for the investigation of the treatment effects of reducing cholesterol - a randomized trial comparing the effects of atorvastatin and pravastatin on carotid intima medial thickness. Circulation. 2002;106:2055–2060. doi: 10.1161/01.cir.0000034508.55617.65. [DOI] [PubMed] [Google Scholar]
- 12.Duggirala R, Gonzalez VC, O’Leary DH, Stern MP, Blangero J. Genetic basis of variation in carotid artery wall thickness. Stroke. 1996;27:833–837. doi: 10.1161/01.str.27.5.833. [DOI] [PubMed] [Google Scholar]
- 13.Fox CS, Polak JF, Chazaro I, Cupples A, Wolf PA, D’Agostino RA, O’Donnell CJ. Genetic and environmental contributions to atherosclerosis phenotypes in men and women: heritability of carotid intima-media thickness in the Framingham Heart Study. Stroke. 2003;34:397–401. doi: 10.1161/01.str.0000048214.56981.6f. [DOI] [PubMed] [Google Scholar]
- 14.Juo SH, Lin HF, Rundek T, Sabala EA, Boden-Albala B, Park N, Lan M-Y, Sacco RL. Genetic and environmental contributions to carotid intima-media thickness and obesity phenotypes in the Northern Manhattan Family Study. Stroke. 2004;35:2243–2247. doi: 10.1161/01.STR.0000142132.20442.d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Bortel LM. What does intima-media thickness tell us? J Hypertens. 2005;23:37–39. doi: 10.1097/00004872-200501000-00009. [DOI] [PubMed] [Google Scholar]
- 16.Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terry JG, Howard G, Mercuri M, Bond MG, Crouse JR. Apolipoprotein E polymorphism is associated with segment-specific extracranial carotid artery intima-media thickening. Stroke. 1996;27:1755–1759. doi: 10.1161/01.str.27.10.1755. [DOI] [PubMed] [Google Scholar]
- 18.Cattin L, Fisicaro M, Tonizzo M, Valenti M, Danek GM, Fonda M, Da Col PG, Casagrande S, Pincetti E, Bovenzi M, Baralle F. Polymorphism of the apolipoprotein E gene and early carotid atherosclerosis defined by ultrasonography in asymptomatic adults. Arterioscler Thromb Vasc Biol. 1997;17:91–94. doi: 10.1161/01.atv.17.1.91. [DOI] [PubMed] [Google Scholar]
- 19.Kogawa K, Nishizawa Y, Hosoi M, Kawagishi T, Maekawa K, Shoji T, Okuno Y, Morii H. Effect of polymorphism of apolipoprotein E and angiotensin-converting enzyme genes on arterial wall thickness. Diabetes. 1997;46:682–687. doi: 10.2337/diab.46.4.682. [DOI] [PubMed] [Google Scholar]
- 20.Sass C, Zannad F, Herbeth B, Salah D, Chapet O, Siest G, Visvikis S. Apolipoprotein E4, lipoprotein lipase C447 and angiotensin-I converting enzyme deletion alleles were not associated with increased wall thickness of carotid and femoral arteries in healthy subjects from the Stanislas cohort. Atherosclerosis. 1998;140:89–95. doi: 10.1016/s0021-9150(98)00120-8. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, You K, Zhang L, Jiang Y. Effect of apolipoprotein E polymorphism on serum lipids, coronary heart disease, and carotid artery atherosclerosis. Chinese J Cardiol. 1998;26:443–447. [Google Scholar]
- 22.Guz G, Nurhan OF, Sezer S, Isiklar I, Arat Z, Turan M, Haberal M. Effect of apolipoprotein E polymorphism on serum lipid, lipoproteins, and atherosclerosis in hemodialysis patients. Am J Kidney Dis. 2000;36:826–836. doi: 10.1053/ajkd.2000.17682. [DOI] [PubMed] [Google Scholar]
- 23.Hanon O, Girerd X, Luong V, Jeunemaitre X, Laurent S, Safar ME. Association between the apolipoprotein E polymorphism and arterial wall thickness in asymptomatic adults. J Hypertens. 2000;18:431–436. doi: 10.1097/00004872-200018040-00012. [DOI] [PubMed] [Google Scholar]
- 24.Horejsi B, Spacil J, Ceska R, Vrablik M, Haas T, Horinek A. The independent correlation of the impact of lipoprotein(a) levels and apolipoprotein E polymorphism on carotid artery intima thickness. Int Angiol. 2000;19:331–336. [PubMed] [Google Scholar]
- 25.Ilveskoski E, Loimaala A, Mercuri MF, Lehtimaki T, Pasanen M, Nenonen A, Oja P, Bond MG, Koivula T, Karhunen PJ, Vuori I. Apolipoprotein E polymorphism and carotid artery intima-media thickness in a random sample of middle-aged men. Atherosclerosis. 2000;153:147–153. doi: 10.1016/s0021-9150(00)00383-x. [DOI] [PubMed] [Google Scholar]
- 26.Slooter AJC, Bots ML, Havekes LM, Iglesias dS, Cruts M, Grobbee DE, Hofman A, van Broeckhoven C, Witteman JCM, van Duijn CM. Apolipoprotein E and carotid artery atherosclerosis: The Rotterdam study. Stroke. 2001;32:1947–1952. doi: 10.1161/hs0901.095377. [DOI] [PubMed] [Google Scholar]
- 27.Tabara Y, Kohara K, Nakura J, Miki T. Risk factor-gene interaction in carotid atherosclerosis: effect of gene polymorphisms of renin-angiotensin system. J Hum Genet. 2001;46:278–284. doi: 10.1007/s100380170079. [DOI] [PubMed] [Google Scholar]
- 28.Haraki T, Takegoshi T, Kitoh C, Wakasugi T, Saga T, Hirai JI, Aoyama T, Inazu A, Mabuchi H. Carotid artery intima-media thickness and brachial artery flow-mediated vasodilation in asymptomatic Japanese male subjects amongst apolipoprotein E phenotypes. J Intern Med. 2002;252:114–120. doi: 10.1046/j.1365-2796.2002.01011.x. [DOI] [PubMed] [Google Scholar]
- 29.Beilby JP, Hunt CCJ, Palmer LJ, Chapman CML, Burley JP, McQuillan BM, Thompson PL, Hung J. Apolipoprotein E gene polymorphisms are associated with carotid plaque formation but not with intima-media wall thickening: Results from the Perth Carotid Ultrasound Disease Assessment Study (CUDAS) Stroke. 2003;34:869–874. doi: 10.1161/01.STR.0000062901.54157.12. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Du Y, Du Y, Huang X. Association of apolipoprotein E gene polymorphism with essential hypertension and its complications. Clin Exp Med. 2003;2:175–179. doi: 10.1007/s102380300003. [DOI] [PubMed] [Google Scholar]
- 31.Xiang GD, Hu TH, Wang YL. Apolipoprotein E genotypes and carotid artery atherosclerosis in type 2 diabetes mellitus. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2003;20:66–68. [PubMed] [Google Scholar]
- 32.Elosua R, Ordovas JM, Cupples LA, Fox CS, Polak JF, Wolf PA, D’Agostino RA, O’Donnell CJ. Association of APOE genotype with carotid atherosclerosis in men and women: the Framingham Heart Study. J Lipid Res. 2004;45:1868–1875. doi: 10.1194/jlr.M400114-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Fernandez-Miranda C, Aranda JL, Martin MA, Arenas J, Nunez V, Gomez DLC. Apolipoprotein E polymorphism and carotid atherosclerosis in patients with coronary disease. Int J Cardiol. 2004;94:209–212. doi: 10.1016/j.ijcard.2003.04.030. [DOI] [PubMed] [Google Scholar]
- 34.Kahraman S, Kiykim AA, Altun B, Genctoy G, Arici M, Gulsun M, Erdem Y, Yasavul U, Turgan C, Caglar S. Apolipoprotein E gene polymorphism in renal transplant recipients: effects on lipid metabolism, atherosclerosis and allograft function. Clin Transplant. 2004;18:288–294. doi: 10.1111/j.1399-0012.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 35.Bednarska-Makaruk M, Rodo M, Markuszewski C, Rozenfeld A, Swiderska M, Habrat B, Wehr H. Polymorphisms of apolipoprotein E and angiotensin-converting enzyme genes and carotid atherosclerosis in heavy drinkers. Alcohol & Alcoholism. 2005;40:274–282. doi: 10.1093/alcalc/agh157. [DOI] [PubMed] [Google Scholar]
- 36.Bleil ME, Ferrell RE, Sutton-Tyrrell K, Muldoon MF, Manuck SB. Apolipoprotein E polymorphism and preclinical carotid artery disease in untreated hypertensive men. Eur J Cardiovasc Prev Rehab. 2006;13:98–100. doi: 10.1097/01.hjr.0000194418.16638.d9. [DOI] [PubMed] [Google Scholar]
- 37.Debette S, Lambert J-C, Gariépy J, Fievet N, Tzurio C, Dartigues J-F, Ritchie K, Dupuy A-M, Alpérovitch A, Ducimetière P, Amouyel P, Zureik M. New insight into the association of apolipoprotein E genetic variants with carotid plaques and intima-media thickness. Stroke. 2006;37:2917–2923. doi: 10.1161/01.STR.0000249011.94055.00. [DOI] [PubMed] [Google Scholar]
- 38.Volcik KA, Barkley RA, Hutchinson RG, Mosley TH, Heiss G, Sharrett AR, Ballantyne CM, Boerwinkle E. Apolipoprotein E polymorphisms predict low density lipoprotein cholesterol levels and carotid wall thickness but not incident coronary heart disease in 12,491 ARIC study participants. Am J Epidemiol. 2006;164:342–348. doi: 10.1093/aje/kwj202. [DOI] [PubMed] [Google Scholar]
- 39.De Andrade M, Thandi I, Brown S, Gotto A, Jr., Patsch W, Boerwinkle E. Relationship of the apolipoprotein E polymorphism with carotid artery atherosclerosis. Am J Hum Genet. 1995;56:1379–1390. [PMC free article] [PubMed] [Google Scholar]
- 40.Loimaala A, Rontu R, Vuori I, Mercuri M, Lehtimäki T, Nenonen A, Bond MG. Blood leukocyte count is a risk factor for intima-media thickening and subclinical carotid atherosclerosis in middle-aged men. Atherosclerosis. 2006;188:363–369. doi: 10.1016/j.atherosclerosis.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 41.Zannad F, Visvikis S, Guegen R, Sass C, Chapet O, Herbeth B, Siest G. Genetics strongly determines the wall thickness of the left and right carotid arteries. Hum Genet. 1998;103:183–188. doi: 10.1007/s004390050804. [DOI] [PubMed] [Google Scholar]
- 42.Visvikis S, Sass C, Pallaud C, Grow MA, Zannad F, Siest G, Erlich H, Cheng S. Familial studies on the genetics of cardiovascular diseases: the stanislas cohort. Clin Chem Lab Med. 2000;38:827–832. doi: 10.1515/CCLM.2000.119. [DOI] [PubMed] [Google Scholar]
- 43.Asakimori Y, Yorioka N, Tanaka J, Kohno N. Effect of polymorphism of the endothelial nitric oxide synthase and apolipoprotein E genes on carotid atherosclerosis in hemodialysis patients. Am J Kidney Dis. 2003;41:822–832. doi: 10.1016/s0272-6386(03)00030-1. [DOI] [PubMed] [Google Scholar]
- 44.Brenner D, Labreuche J, Touboul P-J, Schmidt-Petersen K, Poirer O, Perret C, Schönfelder J, Combadière C, Lathrop M, Cambien F, Brand-Herrmann S-M, Amarenco P. Cytokine polymorphisms associated with carotid intima-media thickness in stroke patients. Stroke. 2006;37:1691–1696. doi: 10.1161/01.STR.0000226565.76113.6c. [DOI] [PubMed] [Google Scholar]
- 45.Junyent M, Cofán M, Núñez I, Gilabert R, Zambón D, Ros E. Influence of HDL cholesterol on preclinical carotid atherosclerosis in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2006;26:1107–1113. doi: 10.1161/01.ATV.0000218507.95149.42. [DOI] [PubMed] [Google Scholar]
- 46.Karvonen J, Kauma H, Kervinen K, Ukkola O, Rantala M, Paivansalo M, Savolainen MJ, Kesaniemi YA. Apolipoprotein E polymorphism affects carotid artery atherosclerosis in smoking hypertensive men. J Hypertens. 2002;20:2371–2378. doi: 10.1097/00004872-200212000-00015. [DOI] [PubMed] [Google Scholar]
- 47.Olmer M, Renucci JE, Planells R, Bouchouareb D, Purgus R. Preliminary evidence for a role of apolipoprotein E alleles in identifying haemodialysis patients at high vascular risk. Nephrol Dial Transplant. 1997;12:691–693. doi: 10.1093/ndt/12.4.691. [DOI] [PubMed] [Google Scholar]
- 48.Vauhkonen I, Niskanen L, Ryynanen M, Voutilainen R, Partanen J, Toyry J, Mercuri M, Rauramaa R, Uusitupa M. Divergent association of apolipoprotein E polymorphism with vascular disease in patients with NIDDM and control subjects. Diabet Med. 1997;14:748–756. doi: 10.1002/(SICI)1096-9136(199709)14:9<748::AID-DIA469>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 49.Sayed-Tabatabaei FA, Houwing-Duistermaat JJ, Van Duijn CM, Witteman JC. Angiotensin-converting enzyme gene polymorphism and carotid artery wall thickness: a meta-analysis. Stroke. 2003;34:1634–1639. doi: 10.1161/01.STR.0000077926.49330.64. [DOI] [PubMed] [Google Scholar]
- 50.Sayed-Tabatabaei FA, Schut AF, Hofman A, Bertoli-Avella AM, Vergeer J, Witteman JC, van Duijn CM. A study of gene-environment interaction on the gene for angiotensin converting enzyme: a combined functional and population based approach. J Med Genet. 2004;41:99–103. doi: 10.1136/jmg.2003.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.