Abstract
Here we demonstrate rapid production of solubilized and functional membrane protein by simultaneous cell-free expression of an apolipoprotein and a membrane protein in the presence of lipids, leading to the self-assembly of membrane protein-containing nanolipoprotein particles (NLPs). NLPs have shown great promise as a biotechnology platform for solubilizing and characterizing membrane proteins. However, current approaches are limited because they require extensive efforts to express, purify, and solubilize the membrane protein prior to insertion into NLPs. By the simple addition of a few constituents to cell-free extracts, we can produce membrane proteins in NLPs with considerably less effort. For this approach an integral membrane protein and an apolipoprotein scaffold are encoded by two DNA plasmids introduced into cell-free extracts along with lipids. For this study reported here we used plasmids encoding the bacteriorhodopsin (bR) membrane apoprotein and scaffold protein Δ1–49 apolipoprotein A-I fragment (Δ49A1). Cell free co-expression of the proteins encoded by these plasmids, in the presence of the cofactor all-trans-retinal and dimyristoylphosphatidylcholine, resulted in production of functional bR as demonstrated by a 5-nm shift in the absorption spectra upon light adaptation and characteristic time-resolved FT infrared difference spectra for the bR → M transition. Importantly the functional bR was solubilized in discoidal bR·NLPs as determined by atomic force microscopy. A survey study of other membrane proteins co-expressed with Δ49A1 scaffold protein also showed significantly increased solubility of all of the membrane proteins, indicating that this approach may provide a general method for expressing membrane proteins enabling further studies.
A number of recent studies have demonstrated successful incorporation of membrane proteins into discoidal nanolipoprotein particles (NLPs)1 (1–4). NLPs have recently been used to understand and limit the oligomerization of membrane proteins, e.g. G-protein-coupled receptors, enabling further studies critical to understanding macromolecular recognition and cellular signaling (4–6). However, the number and type of membrane proteins studied in the context of NLPs have been limited thus far (2, 4–9). In NLPs, the apolipoproteins act as a supporting protein “scaffold” organizing the lipid into bilayers (10, 11). As we have reported previously, NLPs have an average height of 5.0 ± 0.5 nm with diameters ranging from 10 to 60 nm (±3%), depending upon the apolipoprotein and lipid used to make the NLP (10). These methods of in situ generation of NLPs have not been demonstrated previously in cell-free reactions, which may allow for a greater range of cofactors and additives for obtaining improved solubility and function. Such additions may also help expand the applicability of NLPs to a more diverse population of proteins.
Cell-free protein expression has been used to enable protein biochemistry studies for many years (12). Typically cell-based in vivo overexpression of membrane proteins results in cell toxicity, protein aggregation, misfolding, and low yield. However, only a small number of reports describe cell-free methodologies as applied to the expression of membrane proteins (13–19). Cell-free expression of membrane proteins can also be challenging, although the problems with both types of membrane protein expression may be principally due to the need for lipid bilayer insertion for proper folding and function. Successful incorporation of cell-free produced bacteriorhodopsin into liposomes has been reported previously using two different methods. The first involves lengthy refolding after cell-free expression, whereas the second involves cell-free protein expression in the presence of lipids (19, 20). Both methods produced functional bacteriorhodopsin (bR) liposomes but with limited solubility and stability (19, 20). The study of membrane proteins in liposomes is further limited in that membrane proteins inserted into liposomes have only one side accessible to the solvent, inherently limiting the ability to probe the membrane protein interactions with other molecules. The co-expression of NLPs and membrane proteins using cell-free methods described herein may overcome such liposome-based shortcomings and those of in vivo membrane protein expression methods. This method may provide a general cell-free approach for membrane protein expression, making a wider range of membrane proteins available for study compared with current techniques.
EXPERIMENTAL PROCEDURES
Plasmids—
The truncated form of apolipoprotein A-I (Δ1–49), or Δ49A1, was cloned using the following primers: forward, 5`-atgctaaagctccttgacaactgg-3`, and reverse, 5`-ttactgggtgttgagcttcttagtg-3`, from the full-length apolipoprotein A-I. The resulting PCR product was cloned into the vector pIVEX2.4d using NdeI and SmaI restriction sites. The resulting vector construct encodes a C-terminal penta-His tag for nickel affinity purification. The bacterioOpsin sequence (bOp), which encodes the bR protein, was amplified from the plasmid p72bop (19) using the following primers: 5`-ggggcatatgcaagctcaaat-3` and 5`-ggggatccaaaaaaaacgggcc-3`. The resulting PCR product was cloned directionally into the His-tagged pIVEX 2.4b vector using the Nde1 and BamHI restriction enzyme sites. All constructs were verified by DNA sequencing.
Cell-free Reactions—
Preparative 1-ml reactions were carried out using the Invitrogen Expressway Maxi kit or the Roche Applied Science RTS 500 ProteoMaster kit. In brief, lyophilized reaction components (lysate, reaction mixture, amino acid mixture, and methionine) were dissolved in Reconstitution Buffer and combined as specified by the manufacturer. For co-expression a total of 5 μg of each plasmid DNA (bOp and Δ49A1) was added to the lysate mixture along with added DMPC vesicles and retinal cofactor (see below). The reactions were incubated at 30 or 37 °C according to the manufacturer's procedure for the different lysates for 4–18 h, typically overnight for large scale reactions (1 ml). bR production was clearly observed (purple color) as early as 4 h. For membrane protein survey studies, 0.2 μg of Δ49A1-encoding plasmid DNA and 1 μg of each membrane protein-encoding plasmid DNA were added to the cell-free mixture where [35S]Met (135 mCi/mmol final concentration) (PerkinElmer Life Sciences) was added in place of methionine. The soluble fraction was obtained by centrifuging the completed reactions at 14,000 × g for 5 min. Proteins were then separated by SDS-PAGE (data not shown). Autoradiograms and the percentage of solubility was determined using ImageJ software (National Institutes of Health) to quantize labeled protein in the presence and absence of apolipoprotein Δ49A1.
Retinal Cofactor—
An all-trans-retinal (Sigma) solution was prepared with 100% ethanol at a stock concentration of 0.586 or 10 mm. The stock solution was diluted to achieve a final working concentration of 30–50 μm in cell-free reactions. The difference in the amount of ethanol added or the change in the retinal concentration did not significantly affect the cell-free reaction yield.
Lipid Preparation—
Small unilamellar vesicles of DMPC (liposomes) were prepared by probe-sonicating a 68 mg/ml aqueous solution of DMPC until optical clarity was achieved, typically 15 min on ice. Two minutes of centrifugation at 13,700 relative centrifugal force was used to remove any metal contamination from the sonication probe tip. DMPC small unilamellar vesicles were added to the cell-free reaction at a concentration of 2 mg/ml.
Affinity Purification of NLP Complexes—
Immobilized metal affinity chromatography was used to isolate the proteins of interests (truncated Δ49A1 and bR) from the cell-free reaction mixture. The soluble fraction (∼1 ml) was mixed with nickel-nitrilotriacetic acid Superflow resin (1 ml of equilibrated 50% slurry (Qiagen)) according to the manufacturer's protocol using native purification conditions with the following modifications: 10 mm imidazole in PBS buffer was used for washing the column with 12 column volumes of buffer. A total of 6 ml of elution buffer (400 mm imidazole in PBS) were used to elute the bound protein from the column in 1-ml aliquots. All of the elutions were combined, concentrated, and buffer-exchanged into TBS using 100,000 molecular weight sieve filters (Vivascience) to achieve a final volume of ∼200 μl. This material was used for characterization.
Conventional Assembly of “Empty” NLPs and bR·NLPs—
Conventional assembly of NLPs is described elsewhere (1, 10). Briefly the truncated form of ApoA1 (Δ1–55), called MSP1T2 or Δ55A1, purchased from Nanodisc Inc. was combined with DMPC liposomes in a mass ratio of 1:4 in TBS buffer. The mixture was then incubated at room temperature for 2 h. The NLPs were then purified by size exclusion chromatography. For assembly of bR·NLPs, Δ55A1 was mixed with DMPC in a mass ratio of 1:4 in TBS buffer. Sodium cholate was then added to a final concentration of 20 mm. Purple membrane bacteriorhodopsin (21) was then added in a 0.67 mass ratio to the Δ55A1 apolipoprotein and incubated at room temperature for 2 h followed by dialysis in TBS for detergent removal. The NLPs were then purified by size exclusion chromatography.
Size Exclusion Chromatography—
The NLPs made with and without incorporated membrane protein were purified from “free protein” and “free lipid” by HPLC (Shimadzu) using a Superdex 200 10/300 GL column (GE Healthcare) with TBS running buffer at a flow rate of 0.5 ml/min. The column was calibrated with four protein standards (HMW Gel Filtration Calibration kit, GE Healthcare) of known molecular weight and Stokes diameter that span the separation range of the column and the NLP samples. The void volume was established with blue dextran. The NLP fraction was concentrated about 10-fold to ∼1.0 mg/ml using molecular weight sieve filters (Vivascience) with 50-kDa molecular mass cutoffs. Protein concentration was determined using the ADV01 protein concentration kit (Cytoskeleton).
SDS-PAGE—
1-μl aliquots of the total cell-free reaction, soluble fraction, and resuspended pellet were diluted with 1× lithium dodecyl sulfate sample buffer with reducing agents (Invitrogen), heat-denatured, and loaded onto a 4–12% gradient premade Bis-Tris gel (Invitrogen) along with the molecular weight standard SeeBlue plus2 (Invitrogen). The running buffer was 1× MES-SDS (Invitrogen). Samples were electrophoresed for 38 min at 200 V. Gels were stained with Coomassie Brilliant Blue.
Native PAGE—
Equal amounts of NLP samples (0.5–1.0 μg) were diluted with 2× native gel sample buffer (Invitrogen) and loaded onto 4–20% gradient premade Tris-glycine gels (Invitrogen). Samples were electrophoresed for 2 h at a constant 125 V. After electrophoresis, gels were incubated with SYPRO Ruby protein gel stain (Bio-Rad) for 2 h and then destained using 10% MeOH, 7% acetic acid. Following a brief wash with double distilled H2O, gels were imaged using the green laser (532 nm) of a Typhoon 9410 (GE Healthcare) with a 610-nm bandpass 30 filter. Molecular weights were determined by comparing migration versus log molecular weight of standard proteins found in the NativeMark standard (Invitrogen).
Atomic Force Microscopy (AFM)—
NLPs were imaged using an Asylum MFP-3D-CF atomic force microscope. Images were captured in tapping mode with minimal contact force and scan rates of 1 Hz. Asylum software was used for cross-sectional analysis to measure NLP height and diameter. Height and diameter were measured from 182 empty NLPs produced by cell-free expression. Height and diameter were also measured for 430 total NLPs produced by cell-free co-expression; 185 were found to be empty NLPs, and 255 were found to contain bR. Two-tailed Student's t tests were run to compare both the height and diameter of the empty NLP population in the sample co-expressed with bR compared with the sample with no bR expressed. A p value of <0.01 was considered significant.
FTIR Spectroscopy—
Time-resolved FTIR difference spectra were recorded in the rapid scan mode similar to earlier studies (22). Briefly a bR·NLP film was dried on a CaF2 window, rehydrated via the vapor phase, and then sealed in a temperature-controlled IR cell (Model TFC, Harrick Scientific Corp., Ossining, NY) using a second CaF2 window. Spectra were recorded with a Bruker IFS 66 v/s FTIR spectrometer (Bruker Optics, Ettlingen, Germany) at 4 cm−1 spectral resolution and 240-kHz scanner velocity, corresponding to the data acquisition window of 18 ms. Four single beam spectra were recorded before the laser flash and were used as the “dark” background. A neodymium-doped yttrium aluminum garnet (Nd/YAG) laser pulse initiated the photocycle, and 80 “light” spectra were recorded with each acquisition taking 18 ms. Spectra taken in this sequence were co-added 40,000 times to achieve a high signal-to-noise ratio.
RESULTS
Cell-free Membrane Protein·NLP Self-assembly—
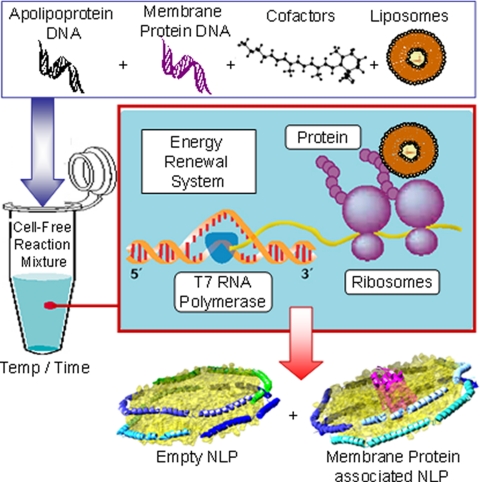
The overall aim of this study was to develop cell-free methods for functional membrane protein production that would both stabilize and solubilize the membrane protein in a native-like environment where both sides of the membrane protein are accessible. Our strategy is based on the ability of membrane proteins to insert into lipid bilayers during cell-free synthesis (20, 23), the ability of apolipoprotein to sequester lipid bilayer patches (10), and the demonstrated ability of NLPs to solubilize the membrane proteins (2, 5, 7, 24). Plasmid DNA encoding both the membrane protein bR from Halobacterium salinarium and a version of apolipoprotein A-I where the first 49 amino acids were deleted (Δ49A1) were added to cell-free reactions along with phospholipids and cofactors to produce membrane protein-associated NLPs in a single reaction as shown in Fig. 1. bR, a seven-transmembrane (TM) protein, has been considered as a structural model protein for rhodopsin and other seven-TM proteins such as the G-coupled receptor family of proteins.
Fig. 1.
Schematic of single step cell-free co-expression and stabilization of integral membrane proteins using an apolipoprotein scaffold. Constituents (DNA, lipid vesicles, cofactors, and cell-free lysates) are added together in a single reaction vial. The cell-free lysates take advantage of the T7-coupled transcription and translation system to produce a mixed population of self-assembled NLPs with and without associated integral membrane protein.
Enhanced bR Solubility When Complexed with NLPs—
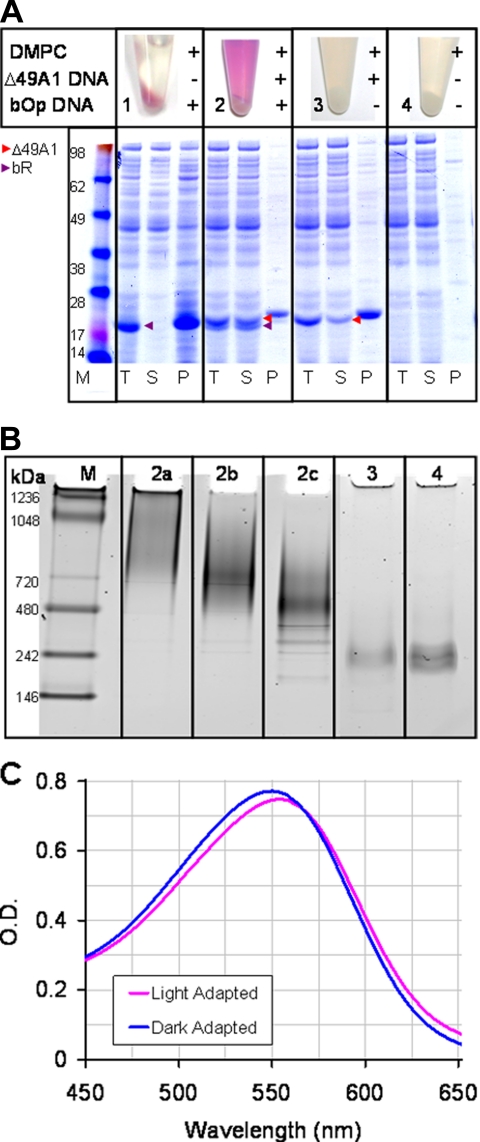
Single step co-expression and assembly of the soluble bR·NLP complex was observed within 4 h. Purification of the soluble fraction after 18 h yielded 0.1 mg/ml functional bR. The levels of bR obtained were comparable to or better than previous published findings (19) (Fig. 2A and supplemental Fig. S1). Simultaneous cell-free protein expression of both bR and Δ49A1 in the presence of DMPC in a single reaction produced a functional bR·NLP complex (Fig. 2, B and C). This extremely rapid approach was also applicable to a variety of other transmembrane proteins (Fig. 3) indicating the general applicability of this method.
Fig. 2.
Single step production, purification, and characterization of MP·NLP complexes. A, Coomassie-stained SDS-PAGE gel image of total (T), soluble (S), and pellet (P) fractions from cell-free produced bR in the presence and absence of co-expressed apolipoprotein (Δ49A1). 1 μl of total, soluble, and resuspended pellet fractions were used for the gel. + indicates the addition of either DMPC, Δ49A1 DNA, or bOp DNA to the cell-free reaction; − denotes absence of additive. Red arrows indicate Δ49A1, and purple arrows indicate bR. Sample 1 (bOp and DMPC), bR is insoluble in the absence of co-expression of Δ49A1; Sample 2 (bOp and Δ49A1 co-expressed in the presence of DMPC), bR remains in the soluble fraction with co-expressed Δ49A1; Sample 3 (Δ49A1 and DMPC), production of empty NLPs; Sample 4, control cell-free reaction (no DNA) in the presence of DMPC only. All were expressed in the presence of 30–50 μm all-trans-retinal and 2 mg/ml DMPC. Purple color development observed in Samples 1 and 2 indicates incorporation of retinal into the bOp transcript representing proper folding of bR. B, native gel of size exclusion-purified NLPs prepared with Δ49A1 or other similar apolipoprotein as noted with and with out bR. Lane M, molecular weight marker; Lane 2a–2c, fractions from SEC-purified cell-free co-expressed bR·NLPs; Lane 2a, lipid-rich first fraction; Lane 2b, bR·NLP second fraction; Lane 2c, bR·NLP third fraction; Lane 3, cell-free produced empty NLPs; Lane 4, conventional assembly of empty NLPs with purchased Δ1–55 apolipoprotein A-I (Δ55A1). C, light/dark adaptation of affinity-purified bR·NLPs concentrated with a 100-kDa molecular mass cutoff filter and buffer-exchanged into TBS, pH 7.4. Blue, dark-adapted bR·NLPs with a λmax = 549 nm; magenta, light-adapted bR·NLP with a λmax = 554 nm. Dark-adapted spectra were collected after overnight adaptation. Light-adapted spectra were collected upon 15-min exposure to a white light-emitting diode source. Spectra were collected in a 50-μl masked quartz cuvette with a 1-cm path length. Absorbance maxima differ by a 5-nm shift between light- and dark-adapted bR.
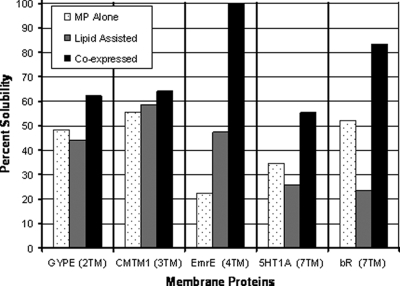
Fig. 3.
Co-expression of membrane proteins with apolipoprotein Δ49A1 in the presence of lipid increases solubility of multiple membrane proteins. A comparison was made between the membrane protein expressed alone (MP Alone; gray), expression of the membrane protein in the presence of DMPC vesicles (Lipid Assisted; striped), and membrane protein co-expressed with apolipoprotein (Δ49ApoA1) in the presence of DMPC vesicles (Co-expressed; black). Membrane proteins with the number of transmembrane domain in parentheses are: glycophorin B (GYPE) (MNS blood group) (two TMs), chemokine-like factor (CKLF-like) MARVEL transmembrane domain (CMTM1) (three TMs), Escherichia coli small multidrug resistance (SMR) efflux transporter (EmrE) (four TMs), 5-hydroxytryptamine (serotonin) receptor (5HT1A) (seven TMs), and bR (seven TMs). In all cases the solubility of the membrane protein increased with co-expression of Δ49ApoA1. Data were generated from autoradiograms by the incorporation of [35S]methionine into the cell-free reaction (data not shown) quantified from SDS-PAGE using ImageJ software (National Institutes of Health).
Cell-free synthesis of bR in the presence of liposomes and retinal produced insoluble but functional bR (purple color) (Fig. 2A and supplemental Fig. S2). Although the bR purple color was observed in the presence of DMPC without Δ49A1, very little of the material was soluble compared with when the Δ49A1 was co-expressed in the reaction mixture (Fig. 2A and supplemental Fig. S2) as indicated in the soluble (S) and pelleted (P) lanes with and without Δ49A1 co-expression. Two methods for refolding of cell-free expressed bR into lipid vesicles have been reported previously (19, 20). However, these two approaches required multiple steps over a lengthy period of time and were further encumbered by limited membrane protein accessibility because of the nature of liposomes. In contrast, the co-expressed bR·NLPs were soluble using our procedure.
bR Produced by Co-expression Is Functional—
Size exclusion chromatography identified a size shift in the bR·NLP complex compared with empty NLPs or liposomes. The bR·NLP complexes eluted primarily before empty NLPs and after liposomes (supplemental Figs. S3 and S4). A size range of ∼470–680 kDa was observed for bR·NLP complexes; this was 160–370 kDa larger than the empty self-assembled NLPs (Fig. 2B). bR·NLP complex heterogeneity was also observed by both native gel electrophoresis and SEC. This heterogeneity may have been due to multiple factors such as number of lipids per NLP, bR oligomerization or multiple molecules of bR within the NLPs, and/or generation of NLPs with varying diameters. Particle diameters measured by AFM (25) support the latter.
Functional activity of the soluble affinity-purified, self-assembled, co-expressed bR·NLP complex was determined by light-dark adaptation (Fig. 2C). The light-dark adaptation yielded a 5-nm shift with a dark absorption maxima of 549 nm and light absorption maxima of 554 nm. These results indicated that the majority of active bR is likely in a monomeric form in the presence of NLPs formed around a DMPC bilayer (26, 27). This is in agreement with other studies that used prepurified apolipoprotein scaffolds to solubilize native forms of bR (2). The yield of active bR isolated from the soluble fraction was ∼0.1 mg from a 1-ml cell-free reaction. The major advantage of our approach is that we obtained folded light-active bR·NLP assemblies in less than 4 h that were self-assembled in a single step, thereby eliminating the need for isolation of membrane protein, protein purification, dialysis, and refolding protocols prior to the formation of NLP·membrane protein complexes.
NLPs Increase Solubility of a Variety of Membrane Proteins—
To determine whether the co-expression method is generally applicable to membrane proteins, a series of membrane proteins with varying numbers of transmembrane domains were expressed using cell-free co-expression. Cell-free expression of the membrane protein alone and lipid-assisted expression of the membrane protein were used for comparison. The reactions included [35S]methionine to quantify the protein. Autoradiograms (not shown) were generated from total and soluble fractions separated by SDS-PAGE. Densitometry using ImageJ software (National Institutes of Health) was used to analyze the autoradiograms. The fraction of solubilized membrane protein compared with the total membrane protein was plotted in Fig. 3. In all cases co-expression with the truncated apolipoprotein Δ49A1 was greater than the expression of the membrane protein alone or lipid-assisted membrane protein expression (Fig. 3).
The bR Protein Is Inserted within NLP Membrane—
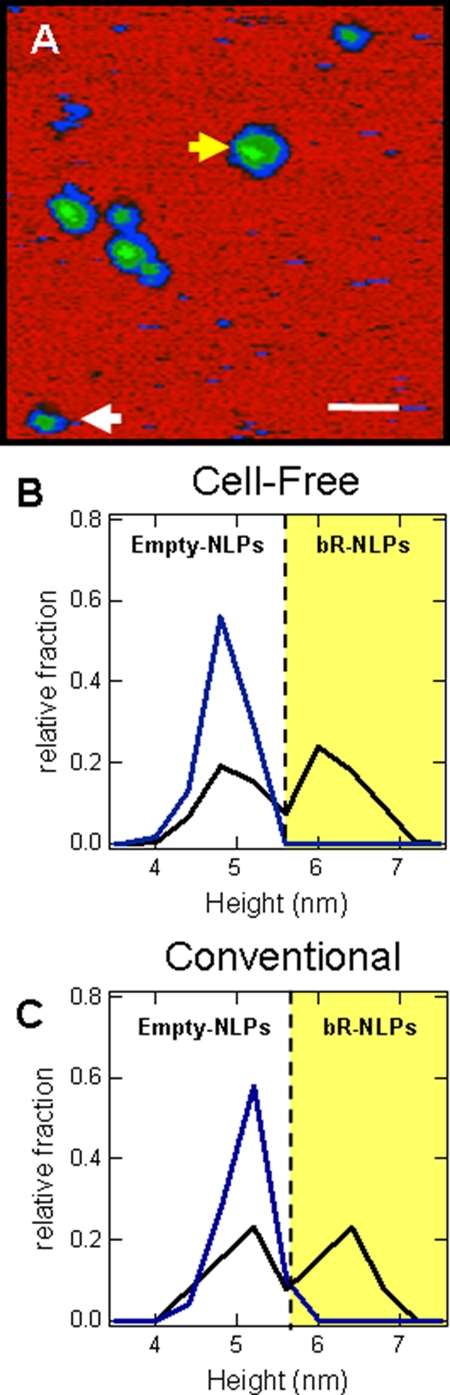
bR·NLPs from cell-free co-expression and NLPs assembled by conventional means (10, 25) were both examined by AFM to assess NLP size and shape and to demonstrate the association between bR with NLPs. For comparison with cell-free produced bR·NLP complexes, both bR·NLPs and empty NLPs were also prepared using methods described previously (1, 10). These NLPs and bR·NLP complexes were made with apolipoprotein A-I (Δ1–55) or Δ55A1 (MSP1T2, Nanodisc Inc.), purple membrane bR, and DMPC liposomes (1, 10). Both the co-expressed and conventionally assembled bR·NLPs showed similar 1.4-nm increases in particle height relative to an empty NLP indicating likely association of bR protein within the NLPs (Fig. 4).
Fig. 4.
AFM confirms the association between NLPs and bR. A, AFM image of NLPs produced through cell-free co-expression of Δ49A1 and bR in the presence of DMPC. The brighter green regions are NLPs with a higher height indicating the insertion and plausible location of bR in the lipid bilayer. Scale bars, 50 nm. The white arrow indicates expression of empty NLPs; the yellow arrow indicates the bR·NLP complex. B, height histogram of NLPs produced through conventional assembly of Δ55A1 with DMPC alone (blue) or in the presence of purple membrane bR and DMPC (black). The yellow shaded areas indicate populations with an increased height. C, height histogram of NLPs produced through cell-free expression of Δ49A1 with DMPC alone (blue) or co-expression of bR and Δ49A1 in the presence of DMPC (black). The yellow shaded areas indicate populations with an increased height. NLP heights were analyzed through cross-sectional analysis.
Empty NLPs produced either by conventional or cell-free methods displayed heights of ∼5.0 ± 0.3 nm (S.D.) as determined by AFM (Fig. 4, B and C, respectively). The NLPs produced by either conventional assembly of Δ55A1 and bR (Fig. 4B) or co-expression of Δ49A1 and bR (Fig. 4C) appeared as two distinct discoidal populations when examined by AFM cross-sectional height analysis. The first population had a height of ∼5.1 ± 0.3 nm (S.D.), analogous to empty NLPs (Fig. 4C). The second population, which was not observed in control experiments lacking bR, had a height of ∼6.4 ± 0.3 nm (S.D.) (Fig. 4C). The increased height observed in the presence of bR is located in the center region of the NLP (bright green dot, pseudocolor) and is consistent with the bR being contained within the NLP lipid bilayer (Fig. 4A). Additionally the increased height particles produced in the presence of bR also had an associated increased mean diameter where bR·NLPs had an average diameter of 27.8 ± 5.8 nm (S.D.) and the empty Δ49A1·NLPs had an average diameter of 22.0 ± 5.1 nm (S.D.) and 20.3 ± 5.2 (S.D.) in the presence and absence of bR, respectively (Table I). Using solely the increase in height as a basis for distinguishing bR·NLPs from empty Δ49A1·NLPs, we were able to determine an overall yield of NLPs with bR incorporation of 58% (Table I). Two-tailed Student's t tests indicated that there was no statistically significant difference between the diameter and height of the empty Δ49A1·NLPs produced by cell-free methods in the presence (n = 185; 2a) and absence (n = 182; 1) of bR (Table I) with p values of 0.94 and 0.04 respectively. However, a statistically significant increase in diameter and height was observed between the bR·NLPs (n = 255; 2b) and empty Δ49A1·NLPs (n = 185, 2a) with p values of 1.8e−24 and 3.9e−155, respectively (Table I). AFM was also used to visualize the first SEC fraction where high molecular weight lipid complexes were observed, consistent with results reported in Chromy et al. (10) (supplemental Figs. S3 and S4). The majority of this material was distinctly different in size, ranging from 35 to 60 nm in diameter and from 6.5 to 20 nm in height indicating that the majority of the material was large lipid complexes such as liposomes or membrane patches (28–31) (supplemental Fig. S5).
Table I.
Summary of analysis of cell-free expressed NLPs with and without co-expressed bR
| Sample | Height ± S.D. | Diameter | NLP | |
|---|---|---|---|---|
| nm | nm | % | ||
| 1 | Empty Δ49A1·NLPs | 5.0 ± 0.3 | 20.3 ± 5.2 | 100 |
| 2a | Co-expressed empty Δ49A1·NLPs | 5.1 ± 0.3 | 22.0 ± 5.1 | 42 |
| 2b | Co-expressed Δ49A1/bR·NLPs | 6.4 ± 0.3a | 27.6 ± 4.8a | 58 |
Statistically significant; see text for specific values.
Functional Validation of the bR Photocycle—
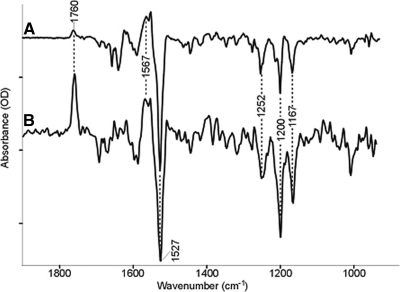
Time-resolved FTIR difference spectroscopy of wild type and bR·NLPs are shown in Fig. 5. Positive bands characteristic of the M2 photointermediate appear at 1760 and 1567 cm−1 (32). The 1760 cm−1 band arises from protonation of the Schiff base counterion, Asp-85 from the Schiff base at this stage of the photocycle (33). The positive band at 1567 cm−1 is assigned the ethylenic stretch of the chromophore in the M intermediate of the photocycle and indicates that the bR·NLP is able to form a normal M intermediate. Negative bands, which reflect vibrations of the ground state of the light-adapted form of the retinylidene chromophore, are seen at 1527 (C=C, ethylenic stretch mode), 1252, 1200, and 1167 cm−1 (C–C, polyene stretch modes) are identical to frequencies appearing in the FTIR difference spectra of native bR in purple membrane (34). The latter three bands are indicative of the normal all-trans-retinal chromophore configuration of light-adapted bR that are depleted during the photocycle giving rise to the negative bands (34). Note that the 1760 cm−1 peak in the bR·NLP spectrum (Fig. 5B) has a much larger intensity relative to native bR in purple membrane. This can be explained as a reduction of polarization effects that arise from the loss of orientation of the bR·NLP relative to the native purple membrane (35, 36).
Fig. 5.
FTIR difference spectra of bR wild type (A) and bR·NLP (B). The largest peaks are 9.4 and 0.34 milli-optical density units, respectively. The positive bands represent vibrations in the M state, and the negative bands represent the ground state. Despite the smaller signal, the spectrum of BR·NLP clearly indicates functional protein that is stable over ∼104 laser flashes.
DISCUSSION
We have developed a new cell-free co-expression method for rapid self-assembly of discoidal nanolipoprotein particles containing membrane protein (MP·NLPs). Our results combine integral membrane protein insertion into lipid bilayers during cell-free synthesis (20), the ability of apolipoproteins to sequester lipid bilayer patches (37), and the demonstrated ability of NLPs to solubilize membrane proteins (1) within in a single reaction, which has not been demonstrated previously. Validation of this technique using bR and a truncated form of apolipoprotein A-I (Δ49A1) produced soluble bR·NLP complexes (Fig. 2 and supplemental Figs. S1 and S2) that were discoidal in shape (Fig. 4) and light-active (Figs. 2 and 5). Distinct purple coloration, an indication of properly folded functional bR protein, was observed when all-trans-retinal and phospholipid were included in the reaction mixtures (Fig. 2 and supplemental Figs. S2 and S3). Solubility survey results indicate that this rapid approach may also be applicable to a wide variety of other transmembrane proteins (Fig. 3). Atomic force microscopy of samples containing bR separated by SEC (supplemental Fig. S3) revealed that 58% of the particles had an increased height (Fig. 4 and Table I) confirming bR incorporation and in agreement with previous AFM characterization of bR structure in native membrane sheets (38). The weak electrostatic interactions that could potentially hold bR to the surface without insertion into the NLP would likely be disrupted during AFM scanning, and it is therefore unlikely that bR is just surface-associated. Time-resolved FTIR difference spectroscopy further corroborated that the cell-free produced bR·NLPs contain functional bR. FTIR spectra of bR·NLPs showed the characteristics indicative of protein and chromophore conformational changes that occur during the bR → M transition of the bR photocycle. Differences seen in the FTIR spectra are likely because of orientation effects of the bR·NLPs that could have been disordered in the bulk NLP as compared with the liquid crystalline planar trimeric order of bR observed in purple membrane (35, 36). Importantly the chromophore was able to undergo the conformational changes necessary for the proper function of bR.
Cell-free production of proteins has become accepted as a means to overcome bottlenecks associated with protein expression and purification in support of high throughput structural proteomics of soluble proteins (17, 18, 39, 40). Our results provide a novel approach to extend cell-free technologies to make a wide range of membrane proteins accessible for future studies and may enable high throughput proteomics of membrane proteins. Importantly our approach also provides a single step process for the production of soluble membrane proteins that eliminates the need for cell growth, cell lysis, and subsequent refolding and purification (14, 41). Planned future experiments with NLPs made by cell-free co-expression will demonstrate novel capabilities for labeling/tagging membrane proteins not readily available to whole cell protein synthesis systems (39, 41, 42). These labeling and tagging methods will enable novel characterization strategies for membrane proteins. Further characterization of these NLP·membrane protein complexes should be possible as is demonstrated by recent results describing protein structural studies (13, 15), (43). Our cell-free self-assembled NLPs will also enable optimization strategies for high throughput screening for membrane protein expression using a variety of additives that may augment protein production and function; example additives include chaperonins (44), lipids (45), redox factors (46), and protease inhibitors (45).
As mentioned previously, a limited number of prepurified detergent-solubilized G-protein-coupled receptors and model proteins such as bR have been reconstituted into NLPs using various lipids, DMPC alone (2), 1-palmitoyl-2-oleoylphosphatidylcholine alone (5, 9), or a mixture of 1-palmitoyl-2-oleoylphosphatidylcholine and 1-palmitoyl-2-oleoylphosphatidylglycerol (6). Using our cell-free based approach, detergents could be avoided by using single step addition of lipids and other molecules important to protein function. The advantages realized from this single step assembly of integral membrane proteins will expand the repertoire of applications for cell-free protein expression and lead to high throughput production of properly folded soluble membrane proteins supported by a nanolipoprotein particle.
Footnotes
Published, MCP Papers in Press, July 4, 2008, DOI 10.1074/mcp.M800191-MCP200
The abbreviations used are: NLP, nanolipoprotein particle or nanodisc; MP, membrane protein; bR, bacteriorhodopsin; bOp, bacterioOpsin; DMPC, dimyristoylphosphatidylcholine; Δ49A1, apolipoprotein A-I with amino acids 1–49 deleted; Δ55A1, apolipoprotein A-I with amino acids 1–55 deleted; AFM, atomic force microscopy; IR, infrared; TM, transmembrane; Bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; SEC, size exclusion chromatography.
During the process of proton translocation, bacteriorhodopsin cycles through several distinct photointermediates as follows, starting with the light adapted state, LA: LA → K → L → M → N → O → LA.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM069969 (to K. J. R.). This work was also performed under the auspices of the United States Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344 with support from the Laboratory Directed Research and Development Office (06-SI-003, LLNL-JRNL-401933 to P. D. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Bayburt, T. H., Carlson, J. W., and Sligar, S. G. ( 1998) Reconstitution and imaging of a membrane protein in a nanometer-size phospholipid bilayer. J. Struct. Biol. 123, 37–44 [DOI] [PubMed] [Google Scholar]
- 2.Bayburt, T. H., Grinkova, Y. V., and Sligar, S. G. ( 2006) Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch. Biochem. Biophys. 450, 215–222 [DOI] [PubMed] [Google Scholar]
- 3.Bayburt, T. H., and Sligar, S. G. ( 2003) Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 12, 2476–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leitz, A. J., Bayburt, T. H., Barnakov, A. N., Springer, B. A., and Sligar, S. G. ( 2006) Functional reconstitution of β2-adrenergic receptors utilizing self-assembling Nanodisc technology. BioTechniques 40, 601–602, 604, 606, passim. [DOI] [PubMed] [Google Scholar]
- 5.Bayburt, T. H., Leitz, A. J., Xie, G., Oprian, D. D., and Sligar, S. G. ( 2007) Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J. Biol. Chem. 282, 14875–14881 [DOI] [PubMed] [Google Scholar]
- 6.Whorton, M. R., Bokoch, M. P., Rasmussen, S. G., Huang, B., Zare, R. N., Kobilka, B., and Sunahara, R. K. ( 2007) A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl. Acad. Sci. U. S. A. 104, 7682–7687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alami, M., Dalal, K., Lelj-Garolla, B., Sligar, S. G., and Duong, F. ( 2007) Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 26, 1995–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boldog, T., Grimme, S., Li, M., Sligar, S. G., and Hazelbauer, G. L. ( 2006) Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc. Natl. Acad. Sci. U. S. A. 103, 11509–11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz, F., and Edmondson, D. E. ( 2007) Kinetic properties of recombinant MAO-A on incorporation into phospholipid nanodisks. J. Neural. Transm. 114, 699–702 [DOI] [PubMed] [Google Scholar]
- 10.Chromy, B. A., Arroyo, E., Blanchette, C. D., Bench, G., Benner, H., Cappuccio, J. A., Coleman, M. A., Henderson, P. T., Hinz, A. K., Kuhn, E. A., Pesavento, J. B., Segelke, B. W., Sulchek, T. A., Tarasow, T., Walsworth, V. L., and Hoeprich, P. D. ( 2007) Different apolipoproteins impact nanolipoprotein particle formation. J. Am. Chem. Soc. 129, 14348–14354 [DOI] [PubMed] [Google Scholar]
- 11.Jonas, A. ( 1986) Reconstitution of high-density lipoproteins. Methods Enzymol. 128, 553–582 [DOI] [PubMed] [Google Scholar]
- 12.Katzen, F., Chang, G., and Kudlicki, W. ( 2005) The past, present and future of cell-free protein synthesis. Trends Biotechnol. 23, 150–156 [DOI] [PubMed] [Google Scholar]
- 13.Klammt, C., Lohr, F., Schafer, B., Haase, W., Dotsch, V., Ruterjans, H., Glaubitz, C., and Bernhard, F. ( 2004) High level cell-free expression and specific labeling of integral membrane proteins. Eur. J. Biochem. 271, 568–580 [DOI] [PubMed] [Google Scholar]
- 14.Klammt, C., Schwarz, D., Eifler, N., Engel, A., Piehler, J., Haase, W., Hahn, S., Dotsch, V., and Bernhard, F. ( 2007) Cell-free production of G protein-coupled receptors for functional and structural studies. J. Struct. Biol. 158, 482–493 [DOI] [PubMed] [Google Scholar]
- 15.Koglin, A., Klammt, C., Trbovic, N., Schwarz, D., Schneider, B., Schafer, B., Lohr, F., Bernhard, F., and Dotsch, V. ( 2006) Combination of cell-free expression and NMR spectroscopy as a new approach for structural investigation of membrane proteins. Magn. Reson. Chem. 44, S17–S23 [DOI] [PubMed] [Google Scholar]
- 16.Mori, M., Morris, S. M., Jr., and Cohen, P. P. ( 1979) Cell-free translation and thyroxine induction of carbamyl phosphate synthetase I messenger RNA in tadpole liver. Proc. Natl. Acad. Sci. U. S. A. 76, 3179–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawasaki, T., Hasegawa, Y., Tsuchimochi, M., Kamura, N., Ogasawara, T., Kuroita, T., and Endo, Y. ( 2002) A bilayer cell-free protein synthesis system for high-throughput screening of gene products. FEBS Lett. 514, 102–105 [DOI] [PubMed] [Google Scholar]
- 18.Schwarz, D., Klammt, C., Koglin, A., Lohr, F., Schneider, B., Dotsch, V., and Bernhard, F. ( 2007) Preparative scale cell-free expression systems: new tools for the large scale preparation of integral membrane proteins for functional and structural studies. Methods 41, 355–369 [DOI] [PubMed] [Google Scholar]
- 19.Sonar, S., Patel, N., Fischer, W., and Rothschild, K. J. ( 1993) Cell-free synthesis, functional refolding, and spectroscopic characterization of bacteriorhodopsin, an integral membrane protein. Biochemistry 32, 13777–13781 [DOI] [PubMed] [Google Scholar]
- 20.Kalmbach, R., Chizhov, I., Schumacher, M. C., Friedrich, T., Bamberg, E., and Engelhard, M. ( 2007) Functional cell-free synthesis of a seven helix membrane protein: in situ insertion of bacteriorhodopsin into liposomes. J. Mol. Biol. 371, 639–648 [DOI] [PubMed] [Google Scholar]
- 21.Oesterhelt, D., and Stoeckenius, W. ( 1974) Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 31, 667–678 [DOI] [PubMed] [Google Scholar]
- 22.Kralj, J. M., Bergo, V. B., Amsden, J. J., Spudich, E. N., Spudich, J. L., and Rothschild, K. J. ( 2008) Protonation state of Glu142 differs in the green- and blue-absorbing variants of proteorhodopsin. Biochemistry 47, 3447–3453 [DOI] [PubMed] [Google Scholar]
- 23.Nomura, S. M., Kondoh, S., Asayama, W., Asada, A., Nishikawa, S., and Akiyoshi, K. ( 2008) Direct preparation of giant proteo-liposomes by in vitro membrane protein synthesis. J. Biotechnol. 133, 190–195 [DOI] [PubMed] [Google Scholar]
- 24.Whorton, M. R., Jastrzebska, B., Park, P. S., Fotiadis, D., Engel, A., Palczewski, K., and Sunahara, R. K. ( 2008) Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer. J. Biol. Chem. 283, 4387–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanchette, C. D., Law, R., Benner, W. H., Pesavento, J. B., Cappuccio, J. A., Walsworth, V., Kuhn, E. A., Corzett, M., Chromy, B. A., Segelke, B. W., Coleman, M. A., Bench, G., Hoeprich, P. D., and Sulchek, T. A. ( 2008) Quantifying size distributions of nanolipoprotein particles with single-particle analysis and molecular dynamic simulations. J. Lipid Res. 49, 1420–1430 [DOI] [PubMed] [Google Scholar]
- 26.Dencher, N. A., Kohl, K. D., and Heyn, M. P. ( 1983) Photochemical cycle and light-dark adaptation of monomeric and aggregated bacteriorhodopsin in various lipid environments. Biochemistry 22, 1323–1334 [DOI] [PubMed] [Google Scholar]
- 27.Wang, J., Link, S., Heyes, C. D., and El-Sayed, M. A. ( 2002) Comparison of the dynamics of the primary events of bacteriorhodopsin in its trimeric and monomeric states. Biophys. J. 83, 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller, C. A., Glasmastar, K., Zhdanov, V. P., and Kasemo, B. ( 2000) Formation of supported membranes from vesicles. Phys. Rev. Lett. 84, 5443–5446 [DOI] [PubMed] [Google Scholar]
- 29.Radler, J. O., Feder, T. J., Strey, H. H., and Sackmann, E. ( 1995) Fluctuation analysis of tension-controlled undulation forces between giant vesicles and solid substrates. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics 51, 4526–4536 [DOI] [PubMed] [Google Scholar]
- 30.Reviakine, I., and Brisson, A. ( 2000) Formation of supported phospholipid bilayers from unilamellar vesicles investigated by atomic force microscopy. Langmuir 16, 1806–1815 [Google Scholar]
- 31.Richter, R. P., and Brisson, A. R. ( 2005) Following the formation of supported lipid bilayers on mica: a study combining AFM, QCM-D, and ellipsometry. Biophys. J. 88, 3422–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rothschild, K. J., Zagaeski, M., and Cantore, W. A. ( 1981) Conformational changes of bacteriorhodopsin detected by Fourier transform infrared difference spectroscopy. Biochem. Biophys. Res. Commun. 103, 483–489 [DOI] [PubMed] [Google Scholar]
- 33.Braiman, M. S., Mogi, T., Marti, T., Stern, L. J., Khorana, H. G., and Rothschild, K. J. ( 1988) Vibrational spectroscopy of bacteriorhodopsin mutants: light-driven proton transport involves protonation changes of aspartic acid residues 85, 96, and 212. Biochemistry 27, 8516–8520 [DOI] [PubMed] [Google Scholar]
- 34.Rothschild, K. J., Marrero, H., Braiman, M., and Mathies, R. ( 1984) Primary photochemistry of bacteriorhodopsin: comparison of Fourier transform infrared difference spectra with resonance Raman spectra. Photochem. Photobiol. 40, 675–679 [DOI] [PubMed] [Google Scholar]
- 35.Kelemen, L., and Ormos, P. ( 2001) Structural changes in bacteriorhodopsin during the photocycle measured by time-resolved polarized Fourier transform infrared spectroscopy. Biophys. J. 81, 3577–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothschild, K. J., and Clark, N. A. ( 1979) Anomalous amide I infrared absorption of purple membrane. Science 204, 311–312 [DOI] [PubMed] [Google Scholar]
- 37.Jonas, A., Kezdy, K. E., and Wald, J. H. ( 1989) Defined apolipoprotein A-I conformations in reconstituted high density lipoprotein discs. J. Biol. Chem. 264, 4818–4824 [PubMed] [Google Scholar]
- 38.Muller, D. J., Kessler, M., Oesterhelt, F., Moller, C., Oesterhelt, D., and Gaub, H. ( 2002) Stability of bacteriorhodopsin α-helices and loops analyzed by single-molecule force spectroscopy. Biophys. J. 83, 3578–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kigawa, T., Yabuki, T., and Yokoyama, S. ( 1999) Large-scale protein preparation using the cell-free synthesis. Tanpakushitsu Kakusan Koso 44, 598–605 [PubMed] [Google Scholar]
- 40.Segelke, B. W., Schafer, J., Coleman, M. A., Lekin, T. P., Toppani, D., Skowronek, K. J., Kantardjieff, K. A., and Rupp, B. ( 2004) Laboratory scale structural genomics. J. Struct. Funct. Genomics 5, 147–157 [DOI] [PubMed] [Google Scholar]
- 41.Coleman, M. A., Lao, V. H., Segelke, B. W., and Beernink, P. T. ( 2004) High-throughput, fluorescence-based screening for soluble protein expression. J. Proteome Res. 3, 1024–1032 [DOI] [PubMed] [Google Scholar]
- 42.Doi, N., Takashima, H., Kinjo, M., Sakata, K., Kawahashi, Y., Oishi, Y., Oyama, R., Miyamoto-Sato, E., Sawasaki, T., Endo, Y., and Yanagawa, H. ( 2002) Novel fluorescence labeling and high-throughput assay technologies for in vitro analysis of protein interactions. Genome Res. 12, 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keppetipola, N., and Shuman, S. ( 2006) Mechanism of the phosphatase component of Clostridium thermocellum polynucleotide kinase-phosphatase. RNA 12, 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frydman, J., and Hartl, F. U. ( 1996) Principles of chaperone-assisted protein folding: differences between in vitro and in vivo mechanisms. Science 272, 1497–1502 [DOI] [PubMed] [Google Scholar]
- 45.Klammt, C., Schwarz, D., Fendler, K., Haase, W., Dotsch, V., and Bernhard, F. ( 2005) Evaluation of detergents for the soluble expression of α-helical and β-barrel-type integral membrane proteins by a preparative scale individual cell-free expression system. FEBS J. 272, 6024–6038 [DOI] [PubMed] [Google Scholar]
- 46.Jewett, M. C., and Swartz, J. R. ( 2004) Rapid expression and purification of 100 nmol quantities of active protein using cell-free protein synthesis. Biotechnol. Prog. 20, 102–109 [DOI] [PubMed] [Google Scholar]