Abstract
Multiple reaction monitoring (MRM) mass spectrometry identifies and quantifies specific peptides in a complex mixture with very high sensitivity and speed and thus has promise for the high throughput screening of clinical samples for candidate biomarkers. We have developed an interactive software platform, called MRMer, for managing highly complex MRM-MS experiments, including quantitative analyses using heavy/light isotopic peptide pairs. MRMer parses and extracts information from MS files encoded in the platform-independent mzXML data format. It extracts and infers precursor-product ion transition pairings, computes integrated ion intensities, and permits rapid visual curation for analyses exceeding 1000 precursor-product pairs. Results can be easily output for quantitative comparison of consecutive runs. Additionally MRMer incorporates features that permit the quantitative analysis experiments including heavy and light isotopic peptide pairs. MRMer is open source and provided under the Apache 2.0 license.
Multiple reaction monitoring-mass spectrometry is a state-of-the-art mass spectrometry mode for detecting the presence of particular molecules in a complex mixture. MRM provides a higher selectivity and sensitivity than is achievable by traditional LC-MS approaches and has been well established in the pharmaceutical industry for detecting small molecules (1–3) as well as in clinical laboratories for drug metabolites (4, 5). Given the large number of potential biomarkers, the need for MRM1-MS biomarker assays arises because clinical validation by immunoassay is simply not feasible because of the great cost and time of developing each assay individually (6).
MRM-MS assays are commonly carried out on electrospray ionization-coupled triple quadrupole mass spectrometers. The paired mass filters of the tandem quadrupoles, the first on the precursor ion and the second on the product ion, provide a highly selective, specific, and sensitive method for species identification with a dynamic range of about 104 (compared with 103 in shotgun proteomics) and sensitivities of detection in the attomolar range in uncomplicated mixtures (7).
MRM-MS is frequently used for quantitative analysis by calculating the area under the curve (AUC) of the transmitted signal for a single product ion. Recent applications have exploited this characteristic to measure concentrations of various analytes in complex mixtures such as human serum (8–13). Quantitative comparison between or within analyses is possible using isotopically heavy reference peptides, a strategy termed AQUA, with limits of detection in the ng/ml range. Stahl-Zeng et al. (7) and Zhang et al. (14) recently described a method for the detection of plasma proteins at concentrations in the ng/ml or sub-ng/ml range and their accurate quantification over 5 orders of magnitude using a glycopeptide capture technique. Using MRM-MS, Keshishian et al. (11) recently reported quantitative, multiplexed assays for six proteins in plasma that achieve limits of quantitation in the 1–10 ng/ml range using abundant protein depletion and strong cation exchange. This sample preparation strategy yielded a 1000-fold improvement compared with direct analysis of proteins in plasma by MS.
MRM-MS is performed using quadrupole instruments that take only a few milliseconds to switch between distinct MRM transitions and thus may detect and quantify hundreds to thousands of precursor-product pairs in a single experiment. Because of these capacities, the platform is uniquely suited to clinical biomarker discovery programs that are required to rapidly evaluate large numbers of putative biomarker targets in clinically relevant samples. The transition of the field of MRM-MS from studies focused on a small set of molecules (e.g. drug metabolites) to ones where many hundreds of potential biomarkers (including isotopically heavy/light pairs) may be measured in a single run requires the development of new analysis tools designed specifically for this task.
We sought to develop a new software platform to manage highly complex MRM-MS experiments, including quantitative analyses using heavy/light isotopic peptide pairs. We specified that the program be independent of any particular instrument data format, and thus it was built upon an extension of the existing mzXML standard (15). We also required the following: 1) automated extraction of product ions and correct association with the precursor mass, 2) functionality for simultaneously viewing a complete “family” of product ions derived from the same precursor to visually validate co-elution, 3) capacity to calculate the AUC to quantify a product ion, and 4) the ability to calculate the relative area under the curve for product ions derived from peptide pairs differentially labeled with stable isotopes. These goals have been achieved in the program we call “MRMer” (pronounced “murmur”).
EXPERIMENTAL PROCEDURES
Dilution Series Using a Yeast Enolase Digest—
We performed an MRM analysis using a serial dilution of a commercial preparation of trypsin-digested yeast enolase (Waters) to demonstrate the ability to reconstruct a complex MRM experiment and extract quantitative information. A 1 pmol/μl stock solution of enolase digest mixture was made using 1% ACN in HPLC grade water containing 0.1% formic acid. Dilutions were made using the same buffer. Data were acquired on a Waters Quattro Premier triple quadrupole instrument coupled with a Waters nanoAcquity ultraperformance LC pump fitted with a Waters Symmetry 5-μm-particle diameter C18 180-μm × 20-mm trap column and a 1.7-μm particle BEH130 C18 100-μm × 100-mm analytical column. After loading and washing for 5 min with 0.1% formic acid (buffer A), peptides were eluted using a linear gradient from 1 to 35% buffer B (0.1% formic acid, 100% ACN) over 12 min at a flow rate of 300 nl/min. Precursor-product ion pairs used in the three analyses are given in supplemental Table 1. For all experiments, the MS instrument was operated in the positive mode. MS source conditions for all experiments were evaluated for best response under positive mode nano-ESI conditions by infusing a standard solution, via a syringe pump, into the mobile phase on a regular basis. MS source parameters were as follows: capillary voltage, 2.9 kV; cone voltage, 36V; source temperature, 90 °C; cone gas flow rate, 40 liters/h at 4 p.s.i. Nitrogen (99.998% purity; Airgas, Seattle, WA) and argon (99.999% purity; Airgas) were used as cone and collision gases, respectively. The dwell time for each transition was 60 ms with a 10-ms interscan delay, 10-ms interchannel delay, and a scan width of 0 Da. Data acquisition for all experiments was carried out by MassLynx version 4.1 software. In the first analysis using varying numbers of product ions per precursor ion, 10 fmol of enolase digest was loaded. In the second series of experiments a single precursor-product was chosen for each of the six peptides. Serial 1:1 dilutions of the yeast enolase digest, from 20 to 0.625 fmol, were analyzed consecutively in order of increasing sample concentration. In a third experiment 10 precursor-product pairs were chosen for three of the six enolase peptides, and mass spectrometry runs were performed with loading of 125 and 62 fmol of enolase digest.
Crude Yeast Digest for Large Scale Analysis—
Saccharomyces cerevisiae strain JLY1 (Matα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 arg4:KANMx4) was grown to an A600 of 1.0 in minimal medium supplemented with arginine and lysine of normal isotopic distribution. Cells were frozen in liquid nitrogen and disrupted with a Retsch PM100 mixer mill. The powder was suspended in 25 mm ammonium bicarbonate and cleared by centrifugation. Protein was digested by the addition of trypsin (1:100, w/w), and digestion was checked by PAGE. The mixture was dried by evaporative centrifugation, and the sample was suspended in 1 ml of 1% ACN, 0.1% formic acid in water. The sample was loaded to a C18 cleanup column (Waters), washed with the loading buffer, and eluted in 45% ACN, 0.1% formic acid. After drying, 500 μg of the samples was suspended in water and separated using the OFF-GEL Fractionator (Agilent) according to the manufacturer's instructions using an IPG strip with pH range 3–10. All fractions were run on the Waters Q-Tof Premier instrument operating in MSE mode coupled with a Waters nanoAcquity HPLC pump using a 60-min gradient from 1 to 35% buffer B. HPLC buffers, columns, and flow rates were identical to those described above. Data were searched using the Waters Identity pipeline (16). We chose 1023 intense precursor-product pairs from a single fraction for MRM analysis on the Quattro Premier instrument (supplemental Table 1). A subset of 12 intense precursor-product ion pairs distributed across the elution profile were used to generate a plot of retention time on the Q-Tof Premier (60-min gradient) compared with the Quattro Premier (30-min gradient). A curve was fit relating the elution times on the two instruments, and a predicted elution time was calculated for each of the 310 peptides on the Quattro Premier. The precursor-product pairs were grouped according to elution time into 32 groups of overlapping time windows and programmed into 32 segments over the 30-min analysis. Instrument configuration was unchanged from that described above with the following exceptions: the dwell time established for all transitions in this experiment was 5 ms, and individual collision energy voltages were established using the following equation: Collision energy = precursor m/z × 0.034 + 3.314.
SILAC Sample Preparation—
S. cerevisiae strain JLY1 (Matα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 arg4:KANMx4) was grown to A600 in minimal medium supplemented with arginine and lysine of either normal isotopic distribution or isotopically heavy arginine (13C6,15N4-labeled) and lysine (13C6,15N2-labeled). Cells were suspended in 20 mm Tris, pH 8, 10 mm MgCl2, 1 mm EDTA, and 1 mm PMSF. Lysates were prepared by bead beating and clarified by centrifugation. Protein concentration was determined by BCA assay (Pierce). 2 mg (total) of lysate from yeast grown in heavy or light amino acids was mixed in a 1:4 heavy:light ratio (w/w), reduced with 2 mm TCEP for 30 min at room temperature, and alkylated with 10 mm iodoacetamide for 30 min at room temperature. Trypsin (Promega) was added at a ratio of 1:100 (w/w) for overnight digestion at 37 °C. Digestion was verified by PAGE. The mixture was dried by evaporative centrifugation, and the sample was suspended in 1 ml of 1% ACN, 0.1% formic acid in water. The sample was cleaned up using a C18 column as above. We chose 154 intense precursor-product pairs (46 individual precursors) from the earlier global analysis of yeast on the Q-Tof Premier (supplemental Table 3). Elution times were again predicted as described above. The m/z ratios of the heavy precursor and product ions were calculated based on the additional mass of the heavy amino acids. We programmed 308 (heavy and light) precursor-product ions into 32 groups of overlapping time windows for the 30-min analysis. Instrument configuration was unchanged from that described above with the following exception: the dwell time used for this experiment was 20 ms.
RESULTS
The MRMer code is based on an extension to msInspect (17–19) and is written in Java using Java Swing components, high level JFreeChart objects, and SwiXml XML-based graphical user interface description and display. The software is freely disseminated using the unrestrictive Apache 2.0 free software license (contact authors). The application also may be acquired from the distribution site using the Java Web Start package, which permits automatic downloading of the most up-to-date version.
MRMer functions in an instrument-agnostic fashion using an updated standard of the mzXML format. In the mzXML file format MRM scans are annotated using the “scanType” attribute, labeling them as “MRM” for both the mzStar (Applied Biosystems) and MassWolf (Waters) converters and “SRM” (single reaction monitoring) for the ReAdW (Thermo) converter. The Thermo converter additionally adds a “filterLine” attribute to the scan element of the mzXML file that encodes Thermo-specific scan information into a text string; the m/z of the product ions are parsed from this text string. The mzXML file format converters have been updated to encode MRM experiments and are freely available at SourceForge, Inc. Because some instrument types/work flows include precursor ion scans, MS1 data may be provided as an optional element in the file, and MRMer can make use of these scans when available. All other fields in the file retain their original definition in the mzXML format. Following conversion of three proprietary formats (from Applied Biosystems, Waters, and Thermo) into standardized mzXML format, the precursor-product pairings were identical to those displayed by the manufacturer's software. Because MRMer derives precursor and product ion masses from the mzXML file, there are occasionally slight differences between the “programmed” masses and the inferred masses.
Analysis using MRMer begins when the user launches the program and designates a particular mzXML file as input. The user can also specify a precursor and product mass tolerance that is used to infer precursor-product pairs. MRMer creates a precursor-product centric view from the scancentric mzXML file by associating successive scans with precursor and product masses falling within the specified tolerances. The selection of mass tolerance for signal extraction depends on instrument accuracy and calibration as well as sample complexity. MRMer automatically groups precursor-product pairs according to precursor mass. Each member of a group of product ions associated with a precursor ion is assigned the same start and stop time (see below), and the total ion current AUC of each product ion is calculated.
MRMer determines elution start and end times and calculates AUCs using an interchangeable signal processing module that has not been optimized to any specific instrument. The default “basic” strategy identifies the start of elution when product ion intensity exceeds 2% of the base peak for three consecutive scans; elution ends when three consecutive scans fall below this threshold. An optional “low intensity” strategy requires eight consecutive scans with intensity of at least 2% of the base peak and above 500 counts per scan. This threshold is more likely to automatically reject spectra with low intensity peaks and can be used to speed curation of large files with many low intensity MRM transitions. When multiple elution peaks are observed for a single precursor-product pair, MRMer groups product ions using the start and stop times of the peak with greatest AUC (calculated using a trapezoidal approximation). After grouping product ions according to precursor ions, MRMer defines the group start time as the earliest start time observed for all members; the group stop time is the latest start time observed. The visual data presented are determined by these values. MRMer allows one to replace or add additional signal processing components without the need to change the remaining parsing and display code.
Following these automated steps, MRMer presents the inferred results to the user with precursor-product groups organized for visual inspection. The user can alter the start and stop times for a group of product ions associated with a specific precursor ion by dragging the mouse across the elution peak graph. This initiates the recalculation of all AUCs for the group. The user can also label a product ion for “rejection.” The results of the curated analysis can be exported in a tab-delimited file for analysis in a spreadsheet format. For analysis of samples where isotopically heavy and light peptide pairs are used, MRMer utilizes a tab-delimited input that associates each precursor mass with a peptide sequence. Also provided in this input file is information regarding which amino acids are isotopically heavy and by how many Da.
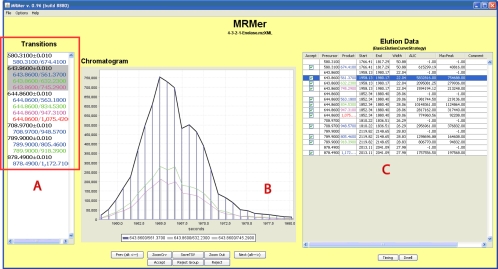
MRMer was designed specifically to foster intuitive visualization of the results of highly multiplexed experiments. It presents the user with the results from a single MRM experiment in three frames (Fig. 1). Frame A, called the “Transitions Pane,” groups all discovered MRM transitions in the file. Frame B, called the “Chromatogram Pane,” shows superimposed single elution profiles for products assigned to the same precursor (each distinct product having a different color). Frame C, named the “Elution Data Pane,” displays the computed information, including computed start and end times of the detected ions, total ion current AUC, and optional comments or annotations. As will be discussed below, additional information is included for experiments using pairs of peptides incorporating isotopically heavy amino acids. For rapid visualization, buttons (and associated keyboard shortcuts) allow the sequential viewing of each product ion as well as the acceptance (or rejection) of the information recorded in the data pane for that particular entry or the entire group of product ions associated with a single precursor. The accept/reject value in the tab-delimited output allows rapid filtering of results using a spreadsheet. Two buttons allow automated entry of comments indicating the need to adjust the dwell time or timing of an MRM transition in a scheduled run. The “Dwell” button inserts a “D” into the comment cell; the “Timing” button inserts a “T.”
Fig. 1.
A screen shot of MRMer showing precursor-product pairs from a commercial digest of yeast enolase. Frame A shows the grouping of all precursor-product pairs according to precursor mass. Frame B shows the co-elution of all three product ions from the precursor at 643.860. Frame C displays a summary of quantitative information for the entire data set including the start and end times used for AUC calculation as well as the calculated AUC. Buttons allow easy navigation and streamline the process of curation.
We evaluated the ability of MRMer to correctly extract and display information from a complex MRM experiment on a yeast enolase digest. The complexity of the experiment arises from the fact that the number of product ions vary from precursor to precursor and that two of the precursors are quite close in mass. As can be seen in the Transitions Pane in Fig. 1A, the software has correctly identified that the user has programmed different numbers of MRM transitions for each of the six precursor ions (ranging from 1 to 4 products) and has differentiated between two precursor masses that differ in mass by 1 m/z (643.86 from 644.86). The three MRM transitions associated with precursor 643.86 are selected for detailed display in the Chromatogram Pane (Fig. 1B). In this view all three of the single ion chromatograms for all three products may be seen simultaneously. Each single ion chromatogram is drawn in a color that matches the font color in the Transitions Pane (Fig. 1A). As can be seen here all three product ions co-elute and present comparable chromatograms. Their AUCs are shown in the data pane (Fig. 1C) at right. This file is available for download from the MRMer Website.
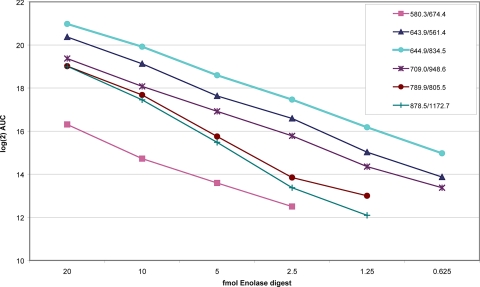
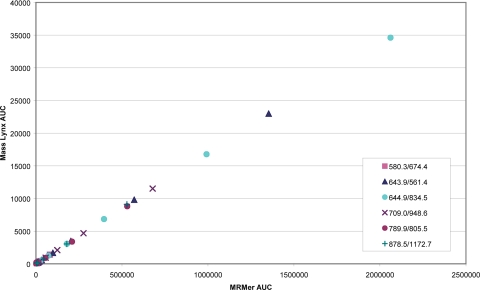
We conducted a more extensive experiment using the yeast enolase digest to benchmark the AUC calculation by MRMer against 1) that expected by serial dilution and 2) that obtained using the commercial software (MassLynx version 4.1) from Waters. The results of the first analysis are summarized in Fig. 2. The log of the integrated AUC (vertical axis) for each of the six precursor-product ion pairs (supplemental Table 1) is shown plotted against the protein concentration (horizontal axis). In every instance the calculated AUC tracks the dilution series. The more intense precursor-product pairs (644.9, 643.9, and 709.0) were detected throughout the entire dilution series down to 625 amol. These same data are used to compare the AUC calculation by MRMer and MassLynx (Fig. 3). Although the units of the AUC calculated by MRMer and MassLynx software are not comparable, the integrated intensities of MRMer and MassLynx software are highly correlated, demonstrating functional equivalence between the platforms. A spreadsheet comparing MRMer and MassLynx using these data is provided in supplemental Table 2.
Fig. 2.
Calculation of AUCs for a dilution series of peptides from a tryptic digestion of enolase. The precursor-product transitions indicated in the legend were followed across the dilution series, and each was run analyzed by MRMer.
Fig. 3.
Comparison of integrated ion intensities produced by MRMer (horizontal axis) and MassLynx (vertical axis), the Waters commercial analysis system.
We conducted a second analysis to determine the reproducibility of AUC calculation using MRMer. In this analysis, we collected data for up to 10 product ions per peptide for three peptides (supplemental Table 1) from the yeast enolase digest and compared two of the concentrations, 62 and 125 fmol. Specifically 10 precursor-product pairs were evaluated for precursors 580.31, 708.86, and 789.90 (Table I). Following visual curation of the results, nine, 10, and eight ion ratios were calculated, respectively. Within each precursor ion group, the multiple products display a high degree of agreement with coefficients of variation of 5, 1.7, and 2.3%, respectively. Note that although a single tube containing all three peptides was diluted 1:1 with buffer, the three peptides do not have the identical expected ratio of 2. The observed value of 1.87 for the precursor at m/z 708.86 indicates that the dilution was slightly less than 1:1. The values of 2.27 and 2.12 for the precursors at m/z 580.31 and 789.90, respectively, indicate that some of the sample was lost during the analysis. This is likely because of heterogeneous adsorption to surfaces (e.g. tubes, pipette tips, autosampler vials, and HPLC tubing) dependent on their specific physicochemical properties. Peptide adsorption is a well known problem and has been seen in other studies using standard peptides to quantify an unknown analyte (20). The consistency among precursor-product pairs for each peptide clearly demonstrates that the calculated ratio represents the quantity of peptide that reached the mass spectrometer rather than an error in sample preparation or software function.
Table I.
MRMer output in a tabular format for three precursor ions and collected product ions identified for these precursors
Rejections are shown in bold in the Accept column.
| Accept | Precursor | Product | AUC
|
Ratio | Mean | S.D. | |
|---|---|---|---|---|---|---|---|
| 62 fmol | 125 fmol | ||||||
| TRUE | 580.31 | 374.06 | 1,108,364 | 2,565,692 | 2.31 | ||
| TRUE | 580.31 | 486.24 | 139,913 | 337,016 | 2.41 | ||
| TRUE | 580.31 | 511.26 | 1,690,455 | 3,850,488 | 2.28 | ||
| TRUE | 580.31 | 523.96 | 102,159 | 214,488 | 2.10 | ||
| TRUE | 580.31 | 571.42 | 38,002 | 94,416 | 2.48 | ||
| TRUE | 580.31 | 674.41 | 1,346,554 | 2,965,865 | 2.20 | ||
| TRUE | 580.31 | 773.3 | 447,835 | 986,017 | 2.20 | ||
| FALSE | 580.31 | 786.11 | |||||
| TRUE | 580.31 | 902.37 | 97,174 | 221,241 | 2.28 | ||
| TRUE | 580.31 | 1,046.41 | 315,017 | 681,624 | 2.16 | 2.27 | 0.12 |
| TRUE | 708.86 | 451.24 | 4,127,918 | 7,688,695 | 1.86 | ||
| TRUE | 708.86 | 478.26 | 2,875,978 | 5,569,263 | 1.94 | ||
| TRUE | 708.86 | 591.35 | 2,250,577 | 4,323,125 | 1.92 | ||
| TRUE | 708.86 | 623.33 | 53,603,640 | 97,665,712 | 1.82 | ||
| TRUE | 708.86 | 699.86 | 3,546,440 | 6,587,226 | 1.86 | ||
| TRUE | 708.86 | 720.38 | 3,974,371 | 7,519,658 | 1.89 | ||
| TRUE | 708.86 | 819.45 | 4,807,088 | 9,002,428 | 1.87 | ||
| TRUE | 708.86 | 948.49 | 12,386,119 | 22,807,092 | 1.84 | ||
| TRUE | 708.86 | 1,047.57 | 6,315,388 | 11,771,412 | 1.86 | ||
| TRUE | 708.86 | 1,148.62 | 2,496,834 | 4,646,178 | 1.86 | 1.87 | 0.03 |
| TRUE | 789.9 | 490.21 | 5,460,347 | 11,316,034 | 2.07 | ||
| TRUE | 789.9 | 605.12 | 3,117,007 | 6,841,995 | 2.20 | ||
| TRUE | 789.9 | 675.13 | 801,183 | 1,672,939 | 2.09 | ||
| TRUE | 789.9 | 718.29 | 2,559,989 | 5,381,282 | 2.10 | ||
| TRUE | 789.9 | 805.46 | 16,069,040 | 34,231,080 | 2.13 | ||
| FALSE | 789.9 | 861.05 | |||||
| TRUE | 789.9 | 918.39 | 9,559,924 | 20,561,278 | 2.15 | ||
| FALSE | 789.9 | 956.07 | |||||
| TRUE | 789.9 | 1,031.53 | 5,079,041 | 10,495,289 | 2.07 | ||
| FALSE | 789.9 | 1,089.58 | 2.12 | 0.05 | |||
We next demonstrated the ability of MRMer to facilitate analysis with mzXML files encoding the largest possible data set achievable using currently available commercial triple quadrupole platforms. To do so, we utilized an instrument capable of analyzing more than 1000 precursor-product ion pairs (Waters Quattro Premier) and programmed runs up to the instrument limit of 1024 pairs. In runs with large numbers of precursor-product ion pairs, scans were scheduled to occur during one of 32 programmable overlapping time segments such that the mass spectrometer performs analyses on only a subset of the precursor-product pairs at any given time. This large scale analysis was performed using 1023 scheduled precursor-product pairs chosen from a yeast lysate (supplemental Table 1). Briefly a tryptic digest of yeast was separated according to pI using the OFF-GEL Fractionator (Agilent) and analyzed using a Waters Q-tof Premier in MSE mode and searched using the Waters Identity software (16). From a single OFF-GEL fraction containing peptides of pH ∼3.9–4.2, we selected 1023 product ions from 310 unique precursor ions. For this analysis, the dwell time was 5 ms/scan and the gradient lasted 30 min. Using MRMer, a single researcher was able to visually inspect and curate all pairs from this analysis (available on the MRMer Website) in ∼2 h.
Although much of the data presented thus far demonstrate the ability to quantify peaks between runs, we have also incorporated into MRMer the capacity for quantitative analysis within a single run that utilizes isotopic dilution strategies such as SILAC (21) or AQUA (22). When coupled to a high throughput MRM platform with very high sensitivity, these quantitative applications have the potential to alter the current paradigm of mass spectrometry-based proteomics. In the case of a SILAC-based experiment where samples are grown in medium containing either heavy or light arginine/lysine, MRM-based analyses at the scale described above could be used for rapid, in-depth profiling of the entire proteome for which unique MS/MS peptide identifications exist. In the case of an AQUA type application in either a clinical or research setting, the ability to quantify against an internal heavy peptide standard of known quantity can substantially streamline the time and effort needed to determine the absolute analyte concentration in clinical or biological samples. To handle this experiment type, MRMer was designed to allow identification of peptides that differ through the inclusion of an isotopically heavy amino acid by providing a file that 1) identifies which amino acids are isotopically heavy and by what mass difference and 2) provides an amino acid sequence for each precursor mass. With this information MRMer pairs heavy and light precursor-product pairs, calculates the ratio of their AUCs, and displays this information in the Elution Data Pane. The user is able to select the appropriate elution windows by dragging across the Chromatogram Pane; this action recalculates the AUCs of all product ions associated with both the heavy and light precursor, and the relative ratio is also recalculated.
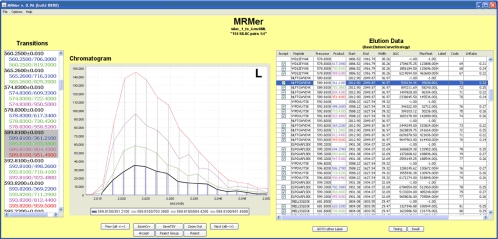
To demonstrate the utility of the quantification tool of MRMer, we created a test sample by growing S. cerevisiae strain JLY1 (strain Matα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 arg4:KANMx4), which is deficient in the enzymes required for the production of arginine and lysine, in medium supplemented with arginine and lysine of a normal isotopic distribution (“light”) or an isotopically heavy arginine (13C6,15N4-labeled) and lysine (13C6,15N2-labeled). A crude lysate was made for both light and heavy labeled samples, and protein was quantified. The samples were then mixed in a light:heavy ratio of 1:4 (w/w) and digested with trypsin. Peptides were introduced to the mass spectrometer by reversed phase chromatography without further sample fractionation. A list of 154 precursor-product ion pairs was made for the abundant peptides identified in the yeast lysate described above using the Waters Q-tof Premier. These 308 (light plus heavy) precursor-product pairs were then programmed into the Waters Quattro Premier. A screen shot of the MRMer analysis is shown in Fig. 4. Data were then curated using the MRMer visualization features to confirm that the appropriate start and end times were used for AUC calculation for each precursor peak and correct the errors when identified. Fig. 4 shows the additional columns now populated in the data pane including a peptide sequence for each precursor, a label (either heavy or light), a code unique to each heavy/light precursor-product pair, and a calculated heavy to light ratio. As can be seen in this figure (and in supplemental Table 3), nearly all heavy:light ratios are at the expected 1:4 ratio. The mzXML file for this run is also available for download from the MRMer Website.
Fig. 4.
Screen shot of MRMer during analysis of a sample containing pairs of isotopically heavy and light peptides at a ratio of 1:4 (w/w). The elution pane is populated with a peptide sequence for each precursor mass. Each precursor group is labeled as heavy or light, and each product ion is given a code that matches the heavy and light products. The LHratio column relates the ratios of AUCs of each heavy/light product ion pair.
DISCUSSION
We have developed a new instrument-independent software platform to manage highly complex MRM-MS experiments, including quantitative analyses using heavy/light isotopic peptide pairs. Starting with a standard mzXML file, MRMer automatically extracts and groups precursor-product pairs for visual validation of co-elution and calculates absolute and relative AUCs for standard and AQUA/SILAC type experiments, respectively. MRMer is open source and easily updated. We anticipate that MRMer will be adapted to meet the needs of the research community, including optional signal extraction components for instrument-specific performance, optimizing the signal processing components, and the addition and extension of its visualization capabilities.
Quantitative MRM analyses incorporating many hundreds of MRM transitions may soon become the method of choice for conducting high throughput global analyses of model organisms in the laboratory and for assessing biomarkers in the clinical setting. MRMer facilitates both such work flows by providing the ability to quickly assess both small and large scale MRM-MS studies. The grouping and visualization tools of MRMer allow easy assessment of co-elution and relative strength of peaks that can permit determination of which to retain and which to discard. In addition, using MRMer the researcher can 1) evaluate whether the selected dwell time is adequate for the intensity of each of the peaks targeted, 2) evaluate the adequacy of a data acquisition window for scheduled studies, and 3) quickly compare studies acquired across a range of adjustable parameters such as collision energy and source voltages. The latter feature is critical because at this time the parameters that determine optimal signal for each precursor-product pair are not determined; for maximal sensitivity, the instrument settings must be determined for each peptide individually. MRMer facilitates the evaluation optimization analyses through the ability to export calculated AUC in a tabular format whereby successive runs (e.g. with increasing collision energy) can be compared in a single spread sheet.
MRMer will also provide substantial assistance in quantitative experiments performed using isotopic dilution strategies. We have successfully demonstrated the capacity for the software to facilitate a high throughput analysis using SILAC labeling in yeast. Numerous studies are ongoing to use MRM-MS for quantitative analysis through the addition of heavy internal standard peptides. Many such studies are in the clinical realm where the sensitivity of MRM is hoped to provide additional power to monitor biomarker peptides in complex mixtures such as plasma and urine. The number of precursor-product pairs that can be effectively monitored is determined by the relationship between the dwell time per transition (5–40 ms) and the typical chromatographic elution peak width. In our laboratory, tight scheduling of precursor-product pairs enabled through the use of a nanoflow ultraperformance LC system permits the monitoring of many hundreds of targets. When combined with the features of MRMer that permit rapid visual and quantitative evaluation, high throughput quantitative studies of many samples may soon be easily accomplished. This should allow expanded biomarker target monitoring for a large number of clinical samples as well as whole proteome-targeted analysis by MRM in model organisms, which will substantially advance the developing field of systems biology.
Supplementary Material
Footnotes
Published, MCP Papers in Press, July 18, 2008, DOI 10.1074/mcp.M700504-MCP200
The abbreviations used are: MRM, multiple reaction monitoring; AUC, area under the curve; AQUA, absolute quantification; SILAC, stable isotope labeling by amino acids in cell culture.
This work was supported, in whole or in part, by National Institutes of Health Grants R21-CA126216 and P50 GM076547 (to D. M.) and Grant (U01 CA111273-04S1) by NCI, National Institutes of Health (to M. M.). This work was also supported by the Allen Family Foundation, the Department of Defense Grant W81XWH-06-1-0100, and in part by the University of Washington's Proteomics Resource (to J. E.) as well as the Canary Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Kostiainen, R., Kotiaho, T., Kuuranne, T., and Auriola, S. ( 2003) Liquid chromatography/atmospheric pressure ionization-mass spectrometry in drug metabolism studies. J. Mass Spectrom. 38, 357–372 [DOI] [PubMed] [Google Scholar]
- 2.Sannino, A., Bolzoni, L., and Bandini, M. ( 2004) Application of liquid chromatography with electrospray tandem mass spectrometry to the determination of a new generation of pesticides in processed fruits and vegetables. J. Chromatogr. A 1036, 161–169 [DOI] [PubMed] [Google Scholar]
- 3.Tai, S. S., Bunk, D. M., White, E. V., and Welch, M. J. ( 2004) Development and evaluation of a reference measurement procedure for the determination of total 3,3`,5-triiodothyronine in human serum using isotope-dilution liquid chromatography-tandem mass spectrometry. Anal. Chem. 76, 5092–5096 [DOI] [PubMed] [Google Scholar]
- 4.Tiller, P. R., Cunniff, J., Land, A. P., Schwartz, J., Jardine, I., Wakefield, M., Lopez, L., Newton, J. F., Burton, R. D., Folk, B. M., Buhrman, D. L., Price, P., and Wu, D. ( 1997) Drug quantitation on a benchtop liquid chromatography-tandem mass spectrometry system. J. Chromatogr. A 771, 119–125 [DOI] [PubMed] [Google Scholar]
- 5.Lee, M. S., and Kerns, E. H. ( 1999) LC/MS applications in drug development. Mass Spectrom. Rev. 18, 187–279 [DOI] [PubMed] [Google Scholar]
- 6.Rifai, N., Gillette, M. A., and Carr, S. A. ( 2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 24, 971–983 [DOI] [PubMed] [Google Scholar]
- 7.Stahl-Zeng, J., Lange, V., Ossola, R., Eckhardt, K., Krek, W., Aebersold, R., and Domon, B. ( 2007) High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol. Cell. Proteomics 6, 1809–1817 [DOI] [PubMed] [Google Scholar]
- 8.Anderson, L., and Hunter, C. L. ( 2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 9.Roschinger, W., Olgemoller, B., Fingerhut, R., Liebl, B., and Roscher, A. A. ( 2003) Advances in analytical mass spectrometry to improve screening for inherited metabolic diseases. Eur. J. Pediatr. 162, Suppl. 1, S67–S76 [DOI] [PubMed] [Google Scholar]
- 10.Streit, F., Armstrong, V. W., and Oellerich, M. ( 2002) Rapid liquid chromatography-tandem mass spectrometry routine method for simultaneous determination of sirolimus, everolimus, tacrolimus, and cyclosporin A in whole blood. Clin. Chem. 48, 955–958 [PubMed] [Google Scholar]
- 11.Keshishian, H., Addona, T., Burgess, M., Kuhn, E., and Carr, S. A. ( 2007) Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 6, 2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn, E., Wu, J., Karl, J., Liao, H., Zolg, W., and Guild, B. ( 2004) Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics 4, 1175–1186 [DOI] [PubMed] [Google Scholar]
- 13.Barnidge, D. R., Goodmanson, M. K., Klee, G. G., and Muddiman, D. C. ( 2004) Absolute quantification of the model biomarker prostate-specific antigen in serum by LC-Ms/MS using protein cleavage and isotope dilution mass spectrometry. J. Proteome Res. 3, 644–652 [DOI] [PubMed] [Google Scholar]
- 14.Zhang, H., Li, X. J., Martin, D. B., and Aebersold, R. ( 2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 [DOI] [PubMed] [Google Scholar]
- 15.Pedrioli, P. G., Eng, J. K., Hubley, R., Vogelzang, M., Deutsch, E. W., Raught, B., Pratt, B., Nilsson, E., Angeletti, R. H., Apweiler, R., Cheung, K., Costello, C. E., Hermjakob, H., Huang, S., Julian, R. K., Kapp, E., McComb, M. E., Oliver, S. G., Omenn, G., Paton, N. W., Simpson, R., Smith, R., Taylor, C. F., Zhu, W., and Aebersold, R. ( 2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 16.Silva, J. C., Denny, R., Dorschel, C. A., Gorenstein, M., Kass, I. J., Li, G. Z., McKenna, T., Nold, M. J., Richardson, K., Young, P., and Geromanos, S. ( 2005) Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 77, 2187–2200 [DOI] [PubMed] [Google Scholar]
- 17.Bellew, M., Coram, M., Fitzgibbon, M., Igra, M., Randolph, T., Wang, P., May, D., Eng, J., Fang, R., Lin, C., Chen, J., Goodlett, D., Whiteaker, J., Paulovich, A., and McIntosh, M. ( 2006) A suite of algorithms for the comprehensive analysis of complex protein mixtures using high-resolution LC-MS. Bioinformatics 22, 1902–1909 [DOI] [PubMed] [Google Scholar]
- 18.Du, P., Sudha, R., Prystowsky, M. B., and Angeletti, R. H. ( 2007) Data reduction of isotope-resolved LC-MS spectra. Bioinformatics 23, 1394–1400 [DOI] [PubMed] [Google Scholar]
- 19.May, D., Fitzgibbon, M., Liu, Y., Holzman, T., Eng, J., Kemp, C. J., Whiteaker, J., Paulovich, A., and McIntosh, M. ( 2007) A platform for accurate mass and time analyses of mass spectrometry data. J. Proteome Res. 6, 2685–2694 [DOI] [PubMed] [Google Scholar]
- 20.Brun, V., Dupuis, A., Adrait, A., Marcellin, M., Thomas, D., Court, M., Vandenesch, F., and Garin, J. ( 2007) Isotope-labeled protein standards: toward absolute quantitative proteomics. Mol. Cell. Proteomics 6, 2139–2149 [DOI] [PubMed] [Google Scholar]
- 21.Ong, S. E., Blagoev, B., Kratchmarova, I., Kristensen, D. B., Steen, H., Pandey, A., and Mann, M. ( 2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 22.Kirkpatrick, D. S., Gerber, S. A., and Gygi, S. P. ( 2005) The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods 35, 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.