Abstract
The 20 S proteasomes play a critical role in intracellular homeostasis and stress response. Their function is tuned by covalent modifications, such as phosphorylation. In this study, we performed a comprehensive characterization of the phosphoproteome for the 20 S proteasome complexes in both the murine heart and liver. A platform combining parallel approaches in differential sample fractionation (SDS-PAGE, IEF, and two-dimensional electrophoresis), enzymatic digestion (trypsin and chymotrypsin), phosphopeptide enrichment (TiO2), and peptide fragmentation (CID and electron transfer dissociation (ETD)) has proven to be essential for identifying low abundance phosphopeptides. As a result, a total of 52 phosphorylation identifications were made in mammalian tissues; 44 of them were novel. These identifications include single (serine, threonine, and tyrosine) and dual phosphorylation peptides. 34 phosphopeptides were identified by CID; 10 phosphopeptides, including a key modification on the catalytically essential β5 subunit, were identified only by ETD; eight phosphopeptides were shared identifications by both CID and ETD. Besides the commonly shared phosphorylation sites, unique sites were detected in the murine heart and liver, documenting variances in phosphorylation between tissues within the proteasome populations. Furthermore the biological significance of these 20 S phosphoproteomes was evaluated. The role of cAMP-dependent protein kinase A (PKA) to modulate these phosphoproteomes was examined. Using a proteomics approach, many of the cardiac and hepatic 20 S subunits were found to be substrate targets of PKA. Incubation of the intact 20 S proteasome complexes with active PKA enhanced phosphorylation in both existing PKA phosphorylation sites as well as novel sites in these 20 S subunits. Furthermore treatment with active PKA significantly elevated all three peptidase activities (β1 caspase-like, β2 trypsin-like, and β5 chymotrypsin-like), demonstrating a functional role of PKA in governing these 20 S phosphoproteomes.
Proteasome complexes serve as the major molecular machinery for intracellular protein degradations; they are essential for maintaining cell homeostasis and adaptation to stress (1–4). Dysfunction of this system has severe pathological consequences, including cardiovascular diseases and malignancy (5, 6). With the appreciation of its vital importance, libraries of pharmaceutical agents have been screened for the potential to modulate this megaprotease (7, 8). However, to date, the regulatory mechanisms governing the function of this organelle remain poorly understood.
The mammalian proteasomes are composed of a heterogeneous population of multiprotein complexes (9). Recent investigations suggest that control of its proteolytic function may involve the recruiting of differential regulation partners (10, 11) and incorporation of inducible subunits (1, 3, 12). 20 S proteasomes1 have a long half-life of 8–9 days in the liver (13). The assembly of new complexes after subunit expression may take a minimum of 30 min (14, 15). A temporal delay would be expected to tune proteasome activities by means of regulating their levels of expression. Post-translational modifications (PTMs), therefore, may provide a venue to regulate the activity of the 20 S proteasome population, especially for an acute response. Accordingly phosphorylation (12, 16, 17) and acetylation (18) of proteasomes have attracted considerable attention. The advancement of proteomics has enabled us to systematically characterize PTMs (19). Amid the consensus that mammalian 20 S proteasomes are heavily modified post-translationally and the potential significance of PTMs in its functional regulation (12, 16, 17), the task of obtaining a comprehensive phosphorylation profile of proteasomes becomes increasingly important.
In this study, we conducted an investigation on the global phosphorylation profile of 20 S proteasome populations in the murine heart and liver. Significant efforts were made to broaden the dynamic range of phosphopeptide detection. The optimizations were conducted at the levels of sample fractionation, in-gel digestion, and phosphopeptide enrichment to balance specificity and sensitivity. A combined approach of peptide fragmentation (CID and electron transfer dissociation (ETD)) enabled a comprehensive characterization of all subunits in these large multiprotein complexes. A total of 52 phosphorylation identifications were made from either the heart or the liver 20 S proteasomes; 44 of them were novel to mammalian tissues. Dephosphorylation with calf intestinal alkaline phosphatase (CIAP) abolished the detection of these phosphorylations, substantiating their specificity. The biological significance of the 20 S phosphoproteomes was evaluated. Using proteomics approaches, we showed that many of the cardiac and liver 20 S proteasome subunits were targets of cAMP-dependent protein kinase A (PKA); furthermore both the cardiac and the hepatic 20 S peptidase activities were augmented after PKA phosphorylation. Taken together, these findings support PKA as a key regulatory element of the phosphoproteomes of the mammalian 20 S proteasomes.
MATERIALS AND METHODS
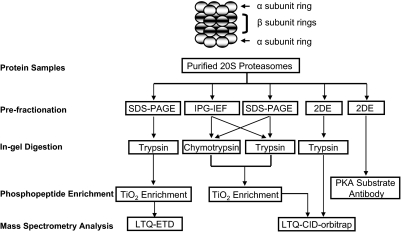
An overview of our experimental strategy is presented in Fig. 1 and in supplemental Fig. S1.
Fig. 1.
An overview of the experimental strategy for the global characterization of the phosphoproteome of the mammalian 20 S proteasome complexes. A comprehensive proteomics strategy that integrates parallel approaches of sample fractionation (IPG-IEF, SDS-PAGE, or 2DE), endoproteolytic digestion (trypsin or chymotrypsin), phosphopeptide enrichment (TiO2), and peptide fragmentation (CID or ETD) was implemented. Multiple novel identifications were made; data obtained in this study provide the dynamic profiles of mammalian 20 S proteasome phosphoproteomes.
Purification of Murine 20 S Proteasomes—
20 S proteasome complexes were purified from 8–10-week-old male ICR mouse tissues via multidimensional chromatography as described previously (11). 20 S proteasomes from 30 hearts were pooled to generate one biological sample; a total of three cardiac samples from 90 hearts were examined (three biological replicates). 20 S proteasomes from five livers were pooled to generate one biological sample; three hepatic samples from 15 livers were examined. The functional viability of the purified 20 S complexes was verified by the peptidase activity assay described below. Phosphorylation experiments were carried out using the intact and functionally viable 20 S proteasomes; CIAP (Promega) treatment (10 units/μg of proteasome, 50 mm Tris-HCl, 1 mm MgCl2, 0.1 mm ZnCl2 at pH 8.5 at 35 °C for 30 min) served as the negative control in all experiments.
Sample Fractionations: SDS-PAGE, Preparative IEF, and Two-dimensional Electrophoresis (2DE)—
Purified 20 S proteasome complexes were fractionated by three parallel approaches: 20 S samples were displayed by SDS-PAGE, preparative IEF, or 2DE. SDS-PAGE was performed according to the Laemmli protocol (20). The SDS-PAGE gel was stained with colloidal Coomassie Blue. Gel bands containing the 20 S proteasome subunits were excised for in-gel digestion. The plug containing the α7 subunit was extracted for separate analyses. Pro-Q Diamond staining and SYPRO Ruby poststaining (Invitrogen) were performed to visualize proteasome subunits. Gels were imaged with a Typhoon 8600 differential imager (GE Healthcare). For preparative IEF, 20 S proteasome complexes were first desalted via TCA/acetone precipitation and then resolved on 18-cm pH 3–10 non-linear IPG strips (GE Healthcare) following a cup loading protocol (150 V, 2 h; 300 V, 2 h; 600 V, 2 h; 8000 V, gradient, 50 min; 8000 V, for a total of 28,000 V-h) (21). The IPG strips were split. The first part of the IPG strips was subjected directly to digestion and LC-MS/MS; these strips were each cut into nine pieces, fixed, and intensively washed before in-gel digestion. The α7-containing piece was extracted for separate LC-MS/MS analyses. The second part of the IPG strips underwent further 2DE (12.5% PAGE). The proteasome subunits were visualized on the 2DE gel. The 2DE gel plugs containing the various subunits were extracted for separate LC-MS/MS analyses.
Endoproteolytic Digestion—
For the CID analyses, in-gel digestions were carried out with either trypsin (Promega; 1:50 as the ratio of enzyme versus proteasomes in 50 mm ammonium bicarbonate (AmBC), pH 8.0) overnight at 37 °C or chymotrypsin (Roche Applied Science; 1:50 as the ratio of enzyme versus proteasomes in 50 mm AmBC, pH 8.5) overnight at 25 °C. Peptides were extracted with 50 mm AmBC in 50% ACN and then with ACN only. For the ETD analyses, in-gel digestions were carried out with trypsin only as follows. The experiments were performed at a reduced amount of trypsin with a shorter incubation time to achieve partial digestion. A ratio of 1:100 enzyme versus proteasomes was used in solutions containing 50 mm AmBC, pH 8.0. The reactions were carried out at 37 °C for 3 h. Peptides were extracted with 50 mm AmBC in 50% ACN and then with ACN only.
Phosphopeptide Enrichment—
Phosphorylation generally occurs substoichiometrically in vivo; therefore enrichment procedures to concentrate phosphopeptides to a detectable level are required (22). Peptides extracted from gel pieces were dried and redissolved in 100 μl of 50% ACN solution containing 0.1% TFA (v/v). Phosphopeptides were enriched using MonoTip TiO2 (GL Sciences Inc.) optimized from a published protocol that favored shorter fully tryptic peptides, which were more ideal for CID analyses (23). Briefly TiO2 MonoTips were sequentially equilibrated with three solutions: 50 mm AmBC containing 25% ACN, 50% ACN solution containing 5% TFA (v/v), and lastly 50% ACN solution containing 0.1% TFA (v/v). Subsequently dissolved peptides were loaded onto the tip with 80 repetitions assuring adequate column binding. The loaded tip was then rinsed for 10 cycles with 100 μl of 50% ACN solution containing 0.1% TFA (v/v) repeated twice. Finally the enriched phosphopeptides were eluted sequentially with 100 μl of 50% ACN solution containing 5% TFA (v/v), 50 mm ammonium bicarbonate containing 20% ACN, and 1% ammonia. All elution fractions of each biological sample were dried and then reconstituted for subsequent mass spectrometric analyses.
Proteomics Analysis of 20 S Proteasome Phosphorylation—
Two types of peptide fragmentation were carried out in parallel. LC-MS/MS with CID fragmentation was performed on an LTQ Orbitrap (ThermoFisher Scientific) integrated with an Eksigent nano-LC instrument (12). A prepacked reverse-phase column (PicoFrit C18 with a dimension of 75 μm × 10 cm, New Objective) containing resin (Biobasic C18, 5-μm particle size, 300-Å pore size, New Objective) was used for both the ETD and CID analyses. ESI conditions for the LTQ-ETD instrument and LTQ Orbitrap were set as follows: capillary temperature of 190 °C, a spray voltage of 2.0 kV, and a nano-LC source. The flow rate of reverse-phase chromatography was set to 5 μl/min for loading and 220 nl/min for separation (buffer A: 0.1% formic acid, 2% ACN; buffer B: 0.1% formic acid, 80% ACN). Peptides were resolved by the following gradient: 2–50% buffer B over 100 min, then increased to 100% buffer B over 10 min, maintained at 100% for 10 min, and then reversed to 0% buffer B. The LTQ Orbitrap was operated in a data-dependent mode without using the FT module to assure the sensitivity and duty cycle of the analyses. One full MS scan was followed by five MS2 scans. MS3 was automatically triggered when a neutral loss signal of 98, 49, or 32.7 was detected among the top 10 most intense signals of an MS2 spectrum. Spectra were searched against the International Protein Index mouse database (Version 3.24 with 52,326 entries) using the Sequest algorithm (Bioworks Version 3.3.1 was used for the .dta file generation. For the CID analyses, the minimal ion threshold was set to 20, intensity threshold was 1000, precursor tolerance was 1.4 amu, and mass range was 400–6000). Other search parameters were set as follows: partial enzymatic digestion (trypsin or chymotrypsin); two miscleavages allowed; peptide tolerance of 2.0; fragment tolerance of 1.0; fixed modification of cysteine carboxyamidomethylation (+57 Da); and differential modifications of methionine oxidation (+16 Da), serine, threonine, and tyrosine phosphorylation (+80 Da), serine and threonine water loss (−18 Da), and protein N-terminal acetylation (+42 Da). Identified peptides were filtered according to the following criteria: Xcorr >2.5 (+2) or >3.5 (+3) (24). All reported peptides had a probability better than 0.001 (p < 0.001) in Sequest. Manual spectra inspection was conducted on all potential phosphopeptides to eliminate false positives. Each final data set was the result of four technical replicates for each of the three biological replicates combined. Each biological sample contained three elution fractions from the phosphopeptide enrichment step. Each identification was confirmed in at least two technical replicates.
LC-MS/MS with ETD fragmentation was performed on an LTQ-ETD instrument (ThermoFisher Scientific) integrated with a Surveyor nano-LC instrument. Sample loading and peptide separation chromatography were set identically to those used in the CID setup. The mass spectrometer was operated in a data-dependent mode. One full MS scan was set for every five MS2 scans. Supplemental activation was enabled to facilitate the fragmentation of doubly charged peptide precursors (25). The reaction time of peptide precursors with fluoranthene was set at 100 ms. The spectra were searched against the International Protein Index mouse database (Version 3.24 with 52,326 entries) using the Sequest algorithm (Bioworks Version 3.3.1 was used for the .dta file generation. For the ETD analyses (trypsin partial digestion), minimal ion threshold was set to 20, intensity threshold was 1000, precursor tolerance was 1.4 amu, and mass range was 600–7500). The other search parameters were set as follows: partial enzymatic digestion (trypsin); three miscleavages allowed; peptide tolerance of 2.0; fragment tolerance of 1.0; fixed modifications of cysteine carboxyamidomethylation (+57 Da); and dynamic modifications of methionine oxidation (+16 Da), serine, threonine, and tyrosine phosphorylation (+80 Da), serine and threonine water loss (−18 Da), and protein N-terminal acetylation (+42 Da). Identified peptides were filtered according to the following criteria: Xcorr >2.0 (+2), >2.5 (+3), or >3.0 (>+3) (26). Manual spectra inspection was conducted on all potential phosphorylation sites to eliminate false positives. Each data set was the result of four technical replicates for each of the three biological replicates. Each biological sample contained three elution fractions from the phosphopeptide enrichment step. Each identification was confirmed in at least two technical replicates.
20 S Proteasome Peptidase Activity Assay—
Substrate concentrations (0–500 μm) were established based on pilot studies that attempted to achieve Vmax for the endogenous 20 S proteasome extracted from heart and liver tissues. The three peptidase activities (β1, β2, and β5) of the 20 S proteasomes were assayed with specific fluorophore-tagged peptides (Z-LLE-AMC for β1, Boc-LSTR-AMC for β2, and Suc-LLVY-AMC for β5) (Bachem) based on a published protocol (11). The assay buffers contained detergents that act as activators for the peptidase activities. Peptidase activities were determined by the release of a free AMC group after cleavage detected by a Fluoroskan Ascent fluorometer (Thermo Fisher Scientific) (excitation at 390 nm and emission at 460 nm).
PKA and 20 S Proteasome Phosphorylation and Activities—
Both the cardiac and the hepatic 20 S proteasome complexes were treated with PKA (6 units/μg of proteasome) (Sigma) in buffer (50 mm Tris-HCl, pH 7.5, 20 mm MgCl2, 1 mm DTT, 2 mm ATP) at 35 °C for 15 min (11). The kinase-dead PKA served as control. The experimental assay mixtures, in parallel with controls, were then precipitated and resolved by 2D electrophoresis (11 cm, pH 3–10 non-linear). Proteasome subunits were transferred to a nitrocellulose membrane, then blocked with gelatin, and probed with anti-α7 antibody (Biomol) and an antibody specific for all generic PKA substrates (Cell Signaling Technology Inc.). Fluorophore-conjugated secondary antibodies (Rockland Immunochemicals) were used to differentiate primary antibodies. The Odyssey dual laser, dual detection scanner (LI-COR Biosciences) enabled the simultaneous detection of both fluorophores (700 and 800 nm).
In Silico Structural Analysis—
The prediction of the 3D structures of the 20 S proteasome phosphorylation sites were based on the resolved bovine liver 20 S proteasome structure (Protein Data Bank code 1iru) deposited in the Protein Data Bank (27). Illustration of these 20 S subunits in their predicted structure was aided by RasTop script (GeneInfinity).
RESULTS
Delineation of phosphorylation events of 20 S proteasome subunits has been challenging because of various technical difficulties including sample preparation and the sensitivity of detection methods (24). In this investigation, we organized a proteomics strategy that integrates parallel approaches of sample fractionation, in-gel digestion, phosphopeptide enrichment, and peptide fragmentation (Fig. 1). This strategy enabled multiple novel identifications and provided for the first time a global profiling of the dynamic phosphoproteome of mammalian 20 S proteasomes.
Sample Fractionation-Preparation of the 20 S Proteasome Complexes—
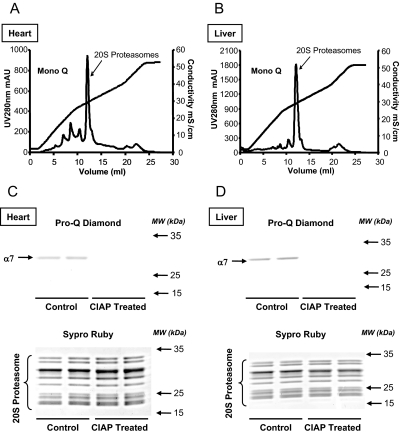
The first step in profiling the phosphoproteome begins with a highly purified preparation of 20 S proteasome complexes (∼95% purity; Fig. 2, A and B) obtained using a protocol established previously (11). Purified 20 S proteasome complexes retained their endogenous phosphorylation profiles as shown by Pro-Q Diamond stain (Fig. 2, C and D). A preliminary characterization confirmed that the α7 subunit was the strongly stained band in both heart and liver tissues; furthermore the phosphorylation signal could be diminished by CIAP treatment (Fig. 2, C and D). In parallel, the purified 20 S proteasomes were fractionated by IPG-IEF, SDS-PAGE, or 2DE.
Fig. 2.
Purification and characterization of the endogenous murine 20 S proteasome complexes. Murine 20 S proteasomes were purified through multidimensional chromatography. The extracted 20 S proteasome complexes (structurally and functionally intact) were more than 95% pure. Analytical Mono Q ion-exchange chromatograms indicate successful collection of the cardiac (A) and the liver (B) 20 S proteasomes. A shallow salt gradient was applied around the 20 S proteasome fractions to ensure a high purity in the preparation. Purified cardiac (C) and liver (D) 20 S proteasomes were displayed with SDS-PAGE, probed with Pro-Q Diamond, and sequentially stained with SYPRO Ruby stain for a loading control. A dominant signal was detected at ∼28 kDa in the 20 S proteasomes that was identified as the α7 subunit. Dephosphorylation of the 20 S proteasomes by CIAP abolished Pro-Q Diamond staining of the α7 subunit. mAU, milliabsorbance units.
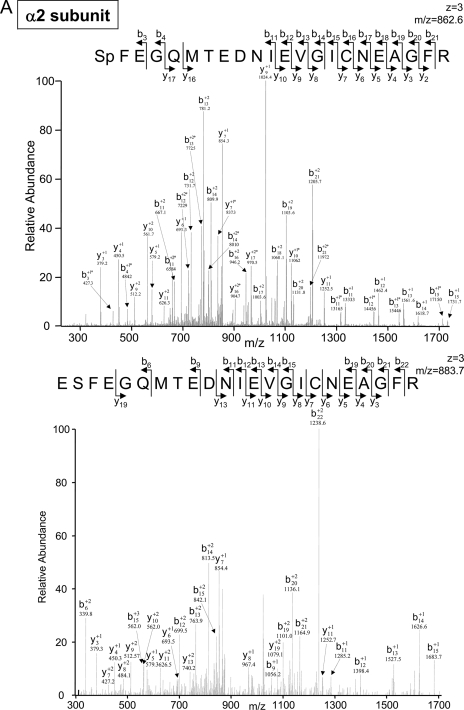
Fig. 4.
A shows CID-based identification of a novel serine 198 phosphorylation site of the cardiac α2 subunit after trypsin digestion. An LTQ-CID Orbitrap spectrum from a phosphorylated peptide of the α2 subunit (serine 198) is shown in the upper panel. The lower panel shows the unmodified form of the same peptide. The acidic residue-rich sequence of the phosphopeptide and fragments are displayed. * denotes a water loss peak. B and C show ETD-based identification of novel β5 phosphorylation sites in heart and liver. B shows an LTQ-ETD spectrum of a phosphorylated peptide from the cardiac β5 subunit (serine 204) following PKA phosphorylation. C shows the detection of the hepatic β5 subunit (serine 192) following PKA phosphorylation. Please note that the ETD fragmentation clearly distinguished the hepatic serine 192 phosphorylation site from that of the cardiac serine 204 phosphorylation site. The c and z ion fragment series acquired from ETD are presented.
Multiple fractionation approaches applied in parallel were beneficial to broaden the dynamic range of phosphopeptide detection. However, this was not feasible when limited samples were at hand, and one separation was selected. SDS-PAGE was most effective in obtaining high recovery of separated samples for the subsequent enrichment of phosphopeptides and the LC-MS/MS analyses. When a sample mixture contained a large dynamic range of phosphopeptides with minute difference in their molecular weight (such as the 20 S proteasomes), the IPG-IEF was effective because the protein with a strong phosphorylation signal (the α7 subunit) was separated from the other subunits with relatively weak phosphorylation signal; this step permitted the removal of the α7 subunit, which otherwise obfuscated the detection of phosphopeptides from the other subunits. The 2DE separation was effective to visualize various phosphorylated subunits (providing a reference map), but it fell short in its ability to recover sufficient proteins for the subsequent enrichment step.
Endoproteolytic Digestion and Phosphopeptide Enrichment—
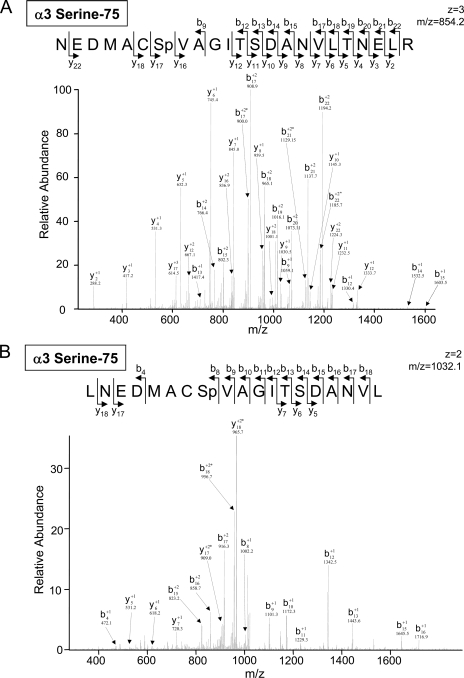
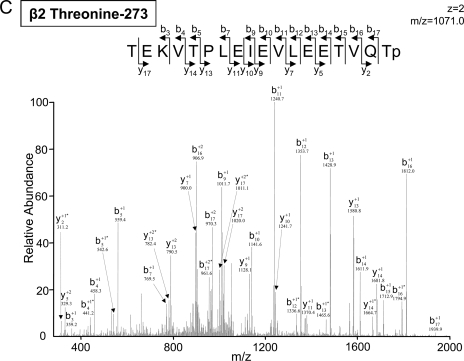
Both tryptic and chymotryptic digestions were carried out following fractionation by either IPG-IEF or SDS-PAGE (Fig. 1). The utilization of chymotrypsin in parallel with trypsin improved identification confidence, e.g. α3 serine 75 was identified by both tryptic and chymotryptic digestions (Fig. 3). It also expanded phosphoproteome coverage, e.g. β2 threonine 273 was identified only by chymotryptic digestion (Table I and Fig. 3).
Fig. 3.
Identification of the phosphopeptides using tryptic and chymotryptic digestions. Phosphopeptides obtained after trypsin in-gel digestion of cardiac proteasomes were scanned with the LTQ Orbitrap (A); the same peptides obtained after chymotrypsin in-gel digestion were scanned with the LTQ Orbitrap (B). This identification supports a previous report on the α3 subunit in HEK293 cells (36). Digestion by chymotrypsin identified the cardiac β2 threonine 273 phosphopeptide (C), which was not seen in trypsin-digested phosphopeptides. * denotes a water loss peak.
Table I.
Phosphopeptides of murine cardiac 20 S proteasomes identified by LC-MS/MS
A summary of endogenous phosphorylation sites identified from purified heart 20 S proteasomes is shown. The modified subunits (Subunit), Swiss-Prot accession number (Accession), experimentally identified phosphopeptide sequences (Phosphopeptide), phosphorylated amino acid residues (Residue), m/z of detected precursor (m/z), charge state of the precursor (z), cross-correlation score of the identification by Sequest (Xcorr), sample fractionation strategies (1DE, IEF, or 2DE), peptide fragmentation methods (CID, ETD, or both), novelty of identified sites (Novelty), and the postulated orientation of phosphorylation sites (Notes) are listed. Additional example spectra and sequences are shown in supplemental Figs. S2–S31.
| Subunit | Accession | Phosphopeptidea | Residueb | m/z | z | Xcorr | Preparationc | MS/MSd | Noveltye | Notes |
|---|---|---|---|---|---|---|---|---|---|---|
| α2 | P49722 | SpFEGQMTEDNIEVGICNEAGFR (T) | 198 | 862.5 | 3 | 5.9 | 1DE/IEF/2DE | CID | Novel | f |
| α3 | Q9R1P0 | NEDMACSpVAGITSDANVLTNELR (T) | 75 | 854.2 | 3 | 5.8 | 1DE/IEF/2DE | CID | 36 | g |
| LNEDMACSpVAGITSDANVL (CT) | 1032.08 | 2 | 3.3 | g | ||||||
| ATCIGNNSAAAVSpML (T) | 173 | 780.6/520.9 | 2/3 | 3.4/3.9 | 1DE/IEF/2DE | CID/ETD | Novel | f | ||
| α4 | Q9Z2U0 | ECQSHRLTpVEDPVTVEYITR (T) | 97 | 820.2 | 3 | 4.2 | 1DE/IEF | CID | Novel | f |
| α5 | Q9Z2U1 | GAMSpRPFGVALLFGGVDEK (T) | 134 | 684.03 | 3 | 4.3 | IEF | CID | Novel | f |
| LNATNIELATVQPGQNFHMFTp (T) | 230 | 809.81 | 3 | 5.5 | 1DE/IEF/2DE | CID | Novel | f | ||
| α7 | O70435 | ESLKEEDESpDDDNM (T) | 250 | 868.7 | 2 | 4.5 | 1DE/IEF/2DE | CID | 36 | f |
| AKESLKEEDESpDDDNM (CT) | 976.8 | 2 | 4.8 | f | ||||||
| β1 | Q60692 | SVPMGGMMVRQSpFAIGGSGS SYIYGYVDATYR (T) | 157 | 1174.8 | 3 | 3.7 | 1DE | CID | Novel | f |
| β2 | P70195 | TEKVTPLEIEVLEETVQTp (CT) | 273 | 1071.0 | 2 | 4.2 | 1DE | CID | Novel | f |
| β5 | O55234 | VIEINPYLLGTpMAGGAADCSFWER (T) | 48 | 923.2 | 3 | 3.6 | IEF | CID | Novel | g |
| β6 | O09061 | GSpASAMLQPLLDNQVGFK (T) | 167 | 978.9 | 2 | 3.9 | 1DE/IEF/2DE | CID | Novel | f |
The sequence of phosphopeptides. T means digested by trypsin; CT means digested by chymotrypsin.
The identity of phosphorylated amino acid residues.
The sample fractionation approaches applied in phosphorylation site identification.
The fragmentation mechanism applied to sequence phosphopeptides.
This column describes whether a phosphorylation site has been reported previously.
This phosphorylation site was located in an exposed motif according to a crystallized bovine 20 S proteasome structure.
This phosphorylation site was located in a flexible region of 20 S proteasome structure, the modification of which indicates a deviation from the crystallized conformation.
TiO2-based enrichment preceded the MS/MS characterization of phosphorylation (28). In this study, we optimized the enrichment protocol to balance both selectivity and sensitivity. We utilized the previously reported phosphopeptide of the α7 subunit (C-terminal ESLKEEDESpDDDNM where Sp is phosphoserine) as a positive control for enrichment. The loading (phosphopeptide capture), washing (removal of non-modified peptides), and elution steps (recovery of phosphopeptides) were all optimized. To overcome the issues concerning phosphopeptide loss, unmodified peptides were removed by washing in 50% ACN, 0.1% TFA without 2,5-dihydroxybenzoic acid. The implementation of sequential elution steps was found to be necessary for efficient phosphopeptide recovery, including that of the positive control (ESLKEEDESpDDDNM). The first elution (50% ACN, 5% TFA) reduced column affinity for phosphopeptides by inhibiting their deprotonation. The second elution (50 mm AmBC, 25% ACN) enhanced phosphopeptide retrieval by using ion competition, and a third elution (1% ammonia, pH >10.5) completed recovery.
The phosphorylation signals from various 20 S subunits covered a wide dynamic range because a particular modification can exist within a several thousand-fold range, e.g. the Ser-250-containing peptide of the α7 subunit (ESLKEEDESpDDDNM). In this case, signals from this particular α7 phosphopeptide obfuscated other phosphorylation signals. To overcome this issue, we extracted the 20 S α7 subunit in samples during the fractionation steps (IPG-IEF, 1DE, or 2DE). The identification of phosphorylation was subsequently conducted on the 20 S samples without the α7 subunit interfering with the others. Analysis of α7 subunit phosphorylation was carried out in parallel experiments. This strategy led to the successful identification of low stoichiometry phosphorylation sites (Tables I and II). Dephosphorylation with CIAP abolished the detection of these phosphorylations, substantiating their specificity. An example spectrum of the Ser-250 α7 subunit after CIAP treatment is shown in supplemental Fig. S31.
Table II.
Phosphopeptides of murine liver 20 S proteasomes identified by LC-MS/MS
A summary of endogenous phosphorylation sites identified from purified liver 20 S proteasomes is shown. The modified subunits (Subunit), Swiss-Prot accession number (Accession), experimentally identified phosphopeptide sequences (Phosphopeptide), phosphorylated amino acid residues (Residue), m/z of detected precursor (m/z), charge state of the precursor (z), cross-correlation score of the identification by Sequest (Xcorr), sample fractionation strategies (1DE, IEF, or 2DE), peptide fragmentation methods (CID, ETD, or both), novelty of identified sites (Novelty), and postulated orientation of phosphorylation sites (Notes) are listed. Additional example spectra and sequences are shown in supplemental Figs. S2–S31.
| Subunit | Accession | Phosphopeptidea | Residueb | m/z | z | Xcorr | Preparationc | MS/MSd | Noveltye | Notes |
|---|---|---|---|---|---|---|---|---|---|---|
| α2 | P49722 | SpFEGQMTEDNIEVGICNEAGFR (T) | 198 | 862.9 | 3 | 5.6 | 1DE/2DE | CID | Novel | f |
| α3 | Q9R1P0 | MEAIGHAGTpCLGILANDGVLLAAER (T) | 33 | 859.6 | 3 | 4.6 | 1DE/2DE | CID | Novel | g |
| NEDMACSpVAGITSDANVLTNELR (T) | 75 | 1281.2 | 2 | 4.0 | 1DE/2DE | CID | 36 | g | ||
| ATCIGNNSAAAVSpML (T) | 173 | 520.7/520.7 | 3/3 | 3.5/4.0 | 1DE | CID/ETD | Novel | f | ||
| α4 | Q9Z2U0 | ECQSHRLTpVEDPVTVEYITR (T) | 97 | 818.9 | 3 | 4.0 | 1DE | CID | Novel | f |
| α5 | Q9Z2U1 | LNATNIELATVQPGQNFHMFTp (T) | 230 | 810.0 | 3 | 6.1 | 2DE | CID | Novel | f |
| α7 | O70435 | SSIGTGYDLSASpTFSPDGR (T) | 13 | 667.1 | 3 | 3 | 1DE | ETD | Novel | f |
| MTpCRDVVKEVAK (T) | 186 | 380.4 | 4 | 4.6 | 1DE | ETD | Novel | f | ||
| ESLKEEDESpDDDNM (T) | 250 | 876.8 | 2 | 4.0 | 1DE/2DE | CID | 36 | f | ||
| β4 | Q9R1P3 | MSpEKILLLCVGEAGDTVQFAEYIQK (T) | 39 | 958.8 | 3 | 3.5 | 1DE/2DE | CID | Novel | g |
| β6 | O09061 | GSpASAMLQPLLDNQVGFK (T) | 167 | 653.1/653.1 | 3/3 | 4.8/3.7 | 1DE/2DE | CID/ETD | Novel | f |
| β7 | P99026 | IMRVNDSpTMLGASGDYADFQYLK (T) | 93 | 678.3 | 4 | 3.0 | 1DE | ETD | Novel | g |
The sequence of phosphopeptides.
The identity of phosphorylated amino acid residues.
The sample fractionation approaches applied in phosphorylation site identification.
The fragmentation mechanism applied to sequence phosphopeptides.
This column describes whether a phosphorylation site has been reported previously.
This phosphorylation site was located in an exposed motif according to a crystallized bovine 20 S proteasomes structure.
This phosphorylation site was located in a flexible region of 20 S proteasome structure, the modification of which indicates a deviation from the crystallized conformation.
Characterization of 20 S Phosphoproteome Complemented by Both ETD and CID—
The implementation of ETD fragmentation significantly broadened the phosphorylation profile made available by CID. Ten novel phosphorylation sites were identified by ETD only. The labile phosphate attachment was preserved during electron transfer dissociation, aiding in the identification of phosphorylated residues (25).
In this study, the advantage provided by CID was shown with its ability to detect peptides with a greater negative net charge (e.g. hepatic and cardiac α2 SpFEGQMTEDNIEVGICNEAGFR), whereas ETD can tolerate and even favor peptides with positively charged internal residues. Fig. 4 illustrates several examples where CID and ETD mutually aided the characterization of the 20 S phosphoproteomes. The acidic residue-rich sequence of the cardiac α2 phosphopeptide was fragmented by CID. A novel serine 198 phosphorylation site of the cardiac α2 subunit was identified only by CID (Fig. 4A and Table I); a similar phosphorylation site was also identified in the hepatic α2 subunits (Table II).
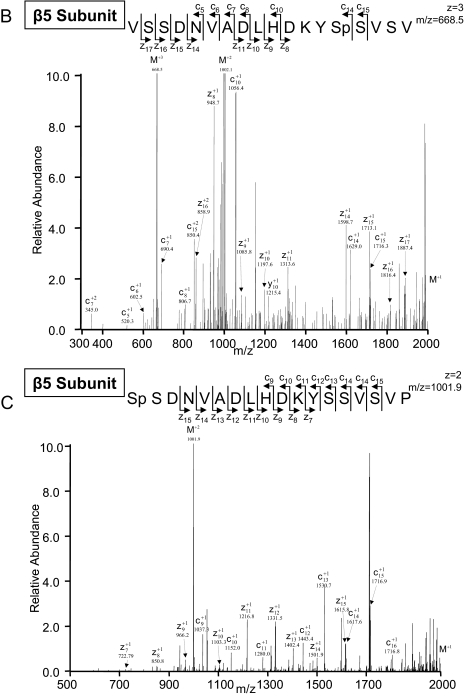
ETD-based identification of novel β5 phosphorylation sites was accomplished in both heart and liver. Fig. 4B shows an LTQ-ETD spectrum of a phosphorylated peptide of the cardiac β5 subunit (serine 204) following PKA phosphorylation and the detection of the hepatic β5 subunit (serine 192) following PKA phosphorylation. Both the serine 192 and the serine 204 sites are located in the C terminus of this proteolytically active β5 subunit and are conserved in human, bovine, rat, and mouse. Furthermore serine 192 represents a consensus site for PKA phosphorylation. ETD fragmentation clearly distinguished the hepatic serine 192 phosphorylation site from that of the cardiac serine 204 phosphorylation site. The c and z fragment ion series were acquired from ETD (Fig. 4, B and C).
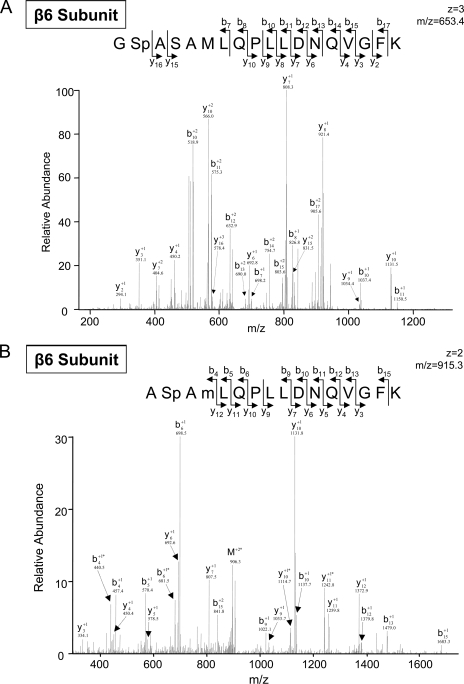
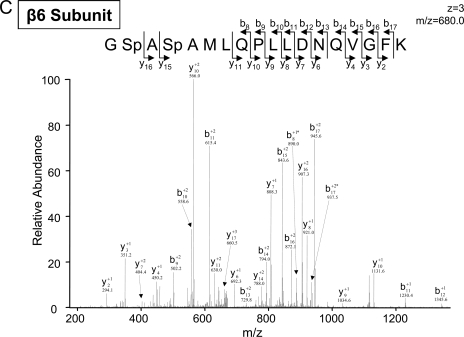
The technological strength of the combined CID and ETD approach was also evident in our identification of novel dual phosphorylation sites on the β6 subunits (Fig. 5). We identified peptides with two independent novel phosphorylation sites (either serine 167 or serine 169) in the cardiac β6 subunits (Tables I and III). In addition, we identified phosphopeptides with dual phosphorylation sites (both serine 167 and serine 169) in the cardiac β6 subunits (Table III). These two sites were independently detected by both the LTQ-CID Orbitrap and the LTQ-ETD instrument. Interestingly we only identified serine 167 in the hepatic β6 subunits (Tables II and IV), whereas the dual phosphopeptides were not detected in the hepatic β6 subunits. The reported 3D structure of the bovine liver 20 S proteasomes places these two phosphorylation sites at the surface of the β6 subunit in a loop motif adjacent to an α-helix (Protein Data Bank code 1iru (27)).
Fig. 5.
Identification of a peptide with two independent novel phosphorylation sites in the cardiac and hepatic β6 subunits. A, an LTQ-CID Orbitrap spectrum of a phosphorylated peptide from the cardiac β6 subunit (serine 167) is shown. The same phosphopeptide was also identified via LTQ-ETD after trypsin digestion (data shown in supplement Fig. S26). B, an LTQ-CID Orbitrap spectrum of a phosphorylated peptide from the cardiac β6 subunit (serine 169) is shown after trypsin digestion. C, an LTQ-CID Orbitrap spectrum of the dually phosphorylated peptide from the cardiac β6 (both serine 167 and serine 169) is shown. The same phosphopeptide was also identified via LTQ-ETD after trypsin digestion (data shown in supplement Fig. S28). The reported 3D structure of the bovine liver 20 S proteasomes predicts the placement of these two phosphorylation sites at the surface of the β6 subunit in a loop motif adjacent to an α-helix structure (Protein Data Bank code 1iru (27)). * denotes a water loss peak.
Table III.
Phosphopeptides of 20 S proteasomes from murine heart identified by LC-MS/MS after treatment with PKA
A summary of the PKA-altered phosphorylation profile of heart 20 S proteasomes is shown. The modified subunits (Subunit), Swiss-Prot accession number (Accession), experimentally identified phosphopeptide sequences (Phosphopeptide), phosphorylated amino acid residues (Residue), m/z of detected precursor (m/z), charge state of the precursor (z), cross-correlation score of the identification by Sequest (Xcorr), sample fractionation strategies (1DE, IEF, or 2DE), peptide fragmentation methods (CID, ETD, or both), novelty of identified sites (Novelty), whether the phosphorylation site was detected only after PKA treatment (In vitro), and postulated orientation of phosphorylation sites (Notes) are listed. Additional example spectra and sequences are shown in supplemental Figs. S2–S31.
| Subunit | Accession | Phosphopeptidea | Residueb | m/z | z | Xcorr | MS/MSc | Noveltyd | In vitro | Notes |
|---|---|---|---|---|---|---|---|---|---|---|
| α2 | P49722 | SpFEGQMTEDNIEVGICNEAGFR (T) | 198 | 862.7 | 3 | 5.9 | CID | Novel | e | |
| SpFEGQMTpEDNIEVGICNEAGFR (T) | 198/204 | 889.1 | 3 | 5.7 | CID | Novel/novel | Yes | e | ||
| α3 | Q9R1P0 | NEDMACSpVAGITSDANVLTNELR (T) | 75 | 1293.6 | 2 | 5.5 | CID | 36 | f | |
| LNEDMACSVAGITSpDAN (T) | 81 | 933.1 | 2 | 4.1 | CID | Novel | Yes | f | ||
| HYGFQLYQSDPSpGNYpGGW (T) | 153/156 | 1119.3 | 2 | 3.8 | CID | Novel/novel | Yes | f | ||
| ATCIGNNSAAAVSpML (T) | 173 | 521.1 | 3 | 3.5 | CID | Novel | e | |||
| α5 | Q9Z2U1 | GAMSpRPFGVALLFGGVDEK (T) | 134 | 683.7 | 3 | 5.0 | CID | Novel | e | |
| LNATNIELATVQPGQNFHMFTp (T) | 230 | 809.6 | 3 | 4.9 | CID | Novel | e | |||
| LNATNIELATpVQPGQNFHMFTp (T) | 219/230 | 836.4 | 3 | 4.0 | CID | Novel/novel | Yes | f | ||
| α7 | O70435 | MTpCRDVVKEVAK (T) | 186 | 380.2 | 4 | 4.8 | ETD | Novel | e | |
| ESLKEEDESpDDDNM (T) | 250 | 876.6 | 2 | 4.0 | CID | 36 | e | |||
| β5 | O55234 | VSSDNVADLHDKYSpSVSV (T) | 204 | 668.5 | 3 | 2.8 | ETD | Novel | Yes | e |
| β6 | O09061 | FSPYAFNGGTpVLAIAGEDFSpIVASDTR (T) | 38/48 | 990.3 | 3 | 4.2 | CID | Novel/novel | Yes | f |
| GSpASAMLQPLLDNQVGFK (T) | 167 | 653.3/653.6 | 3/3 | 4.6/3.9 | CID/ETD | Novel | e | |||
| ASpAMLQPLLDNQVGFK (T) | 169 | 915.3 | 2 | 3.9 | CID | Novel | Yes | e | ||
| GSpASpAMLQPLLDNQVGFK (T) | 167/169 | 1019.3/680.0 | 2/3 | 3.7/3.9 | CID/ETD | Novel | Yes | e |
The sequence of phosphopeptides.
The identity of phosphorylated amino acid residues.
The fragmentation mechanism applied to sequence phosphopeptides.
This column describes whether a phosphorylation site has been reported previously.
This phosphorylation site was located in an exposed motif according to a crystallized bovine 20 S proteasomes structure.
This phosphorylation site was located in a flexible region of 20 S proteasome structure, the modification of which indicates a deviation from the crystallized conformation.
Table IV.
Phosphopeptides of 20 S proteasome from murine liver identified by LC-MS/MS after treatment with PKA
A summary of the PKA-altered phosphorylation profile of liver 20 S proteasomes is shown. The modified subunits (Subunit), Swiss-Prot accession number (Accession), experimentally identified phosphopeptide sequences (Phosphopeptide), phosphorylated amino acid residues (Residue), m/z of detected precursor (m/z), charge state of the precursor (z), cross-correlation score of the identification by Sequest (Xcorr), sample fractionation strategies (1DE, IEF, or 2DE), peptide fragmentation methods (CID, ETD, or both), novelty of identified sites (Novelty), whether the phosphorylation site was detected only after PKA treatment (In vitro), and postulated orientation of phosphorylation sites (Notes) are listed. Additional example spectra and sequences are shown in supplemental Figs. S2–S31.
| Subunit | Accession | Phosphopeptidea | Residueb | m/z | z | Xcorr | MS/MSc | Noveltyd | In vitro | Notes |
|---|---|---|---|---|---|---|---|---|---|---|
| α2 | P49722 | SpFEGQMTEDNIEVGICNEAGFR (T) | 198 | 862.6 | 3 | 6.2 | CID | Novel | e | |
| α3 | Q9R1P0 | NEDMACSpVAGITSDANVLTNELR (T) | 75 | 854.6 | 3 | 5.4 | CID | (36) | f | |
| ATCIGNNSAAAVSpML (T) | 173 | 521.4/521.1 | 3/3 | 3.6/3.3 | CID/ETD | Novel | e | |||
| α5 | Q9Z2U1 | LNATNIELATVQPGQNFHMFTp (T) | 230 | 1214.0/607.6 | 2/4 | 5.6/5.4 | CID/ETD | Novel | e | |
| α7 | O70435 | MTpCRDVVKEVAK (T) | 186 | 506.8 | 3 | 2.6 | ETD | Novel | e | |
| ESLKEEDESpDDDNM (T) | 250 | 876.7 | 2 | 4.3 | CID | (36) | e | |||
| β1 | Q60692 | GMTpKDECLQFTANALALAMER (T) | 181 | 818.0 | 3 | 4.6 | CID | Novel | Yes | e |
| β2 | P70195 | VTPLEIEVLEETVQTMDTSp (T) | 277 | 1108.0 | 2 | 6.8 | CID | Novel | Yes | e |
| β3 | Q9R1P1 | IKPYTpLMSMVANLLYEK (T) | 86 | 704.5 | 3 | 4.2 | ETD | Novel | Yes | e |
| KPYpTpLMSMVANLLYEKR (T) | 85/86 | 559.5 | 4 | 4.4 | ETD | Novel | Yes | e | ||
| β5 | O55234 | SpSDNVADLHDKYSSVSVP (T) | 192 | 1001.9 | 2 | 2.4 | ETD | Novel | Yes | e |
| β6 | O09061 | GSpASAMLQPLLDNQVGFK (T) | 167 | 653.3/653.2 | 3/3 | 4.8/4.4 | CID/ETD | Novel | e | |
| β7 | P99026 | MRVNDSpTMLGASGDYADFQYLK (T) | 93 | 855.5 | 3 | 3.7 | ETD | Novel | f |
The sequence of phosphopeptides.
The identity of the phosphorylated amino acid residue.
The fragmentation mechanism applied to sequence phosphopeptides.
This column describes whether a phosphorylation site has been reported previously.
This phosphorylation site was located in an exposed motif according to a crystallized bovine 20 S proteasomes structure.
This phosphorylation site was located in a flexible region of 20 S proteasome structure, the modification of which indicates a deviation from the crystallized conformation.
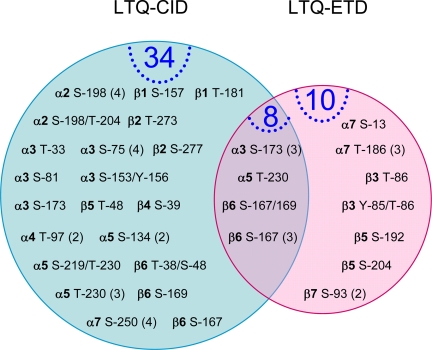
In summary, our global characterization of the 20 S phosphoproteome led to the identification of a total of 52 phosphorylation events. These phosphoproteins constitute a dynamic phosphoproteome in both the cardiac and hepatic 20 S proteasomes (Fig. 6). Among these identifications, 44 phosphorylation sites were novel (never identified before in any species or cell type). In the cardiac 20 S proteasome complexes, a total of nine subunits possessed phosphorylation sites: α2, α3, α4, α5, α7, β1, β2, β5, and β6 (Tables I and III). In the hepatic 20 S proteasome complexes, a total of 12 subunits possessed phosphorylation sites: α2, α3, α4, α5, α7, β1, β2, β3, β4, β5, β6, and β7 (Tables II and IV). Among them, 34 phosphopeptides were identified by CID; 10 phosphopeptides, including a key modification on the catalytically essential β5 subunit, were identified only by ETD; and eight phosphopeptides were shared identifications by both CID and ETD. A partial digestion by trypsin was used to aid the yield of longer peptides for the ETD analyses; however, it may have generated a lesser quantity of identifiable phosphopeptides. In addition, phosphopeptides were enriched using MonoTip TiO2 (GL Sciences Inc.) optimized from a published protocol that favored shorter peptides, which were more ideal for CID analyses. These factors contributed to a somewhat lesser number of phosphopeptide identified by ETD. Additional information on all phosphopeptides identified is summarized in supplemental Tables S1A–S1D.
Fig. 6.
Phosphoproteomes of the mammalian 20 S proteasomes. An overview of LTQ-ETD and LTQ-CID Orbitrap enabled the identification of phosphorylation sites/peptides. Identifications from distinct data sets (cardiac 20 S, hepatic 20 S, PKA-treated cardiac 20 S, and PKA-treated hepatic 20 S proteasomes) were combined. CID and ETD provided both unique and complimentary identifications. For example, the β5 Ser-204 was only identified by LTQ-ETD in the PKA-treated heart, whereas the α2 Ser-198 was identified by LTQ-CID Orbitrap in all four data sets (cardiac 20 S, hepatic 20 S, PKA-treated cardiac 20 S, and PKA-treated hepatic 20 S proteasomes).
The Biological Significance of the 20 S Phosphoproteomes—
20 S proteasomes play essential roles in protein degradation in nearly all mammalian cell types. Our previous studies demonstrated phosphorylation as a key regulatory mechanism to the 20 S proteolytic function (9). In this investigation, we hypothesized that the phosphoproteomes of 20 S proteasomes are governed by many kinases in the heart and liver. To provide a first assessment of the functional significance of these 20 S phosphoproteomes, we examined these phosphorylation events under the regulation of PKA, a primary signaling kinase in the β-adrenergic receptor pathway and a well established regulatory enzyme in both cardiac and hepatic function in vivo.
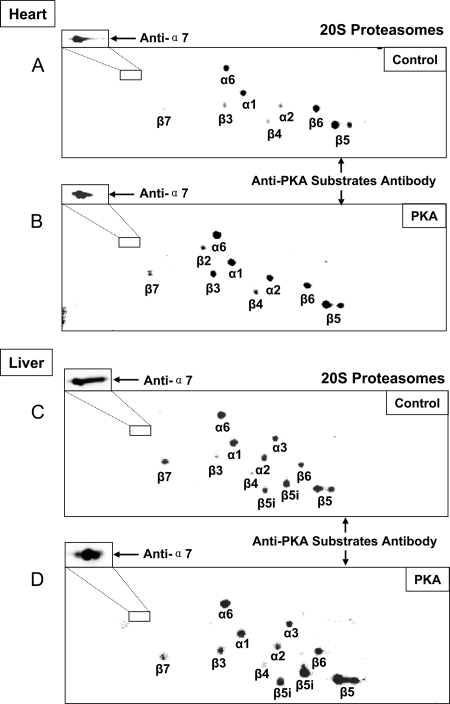
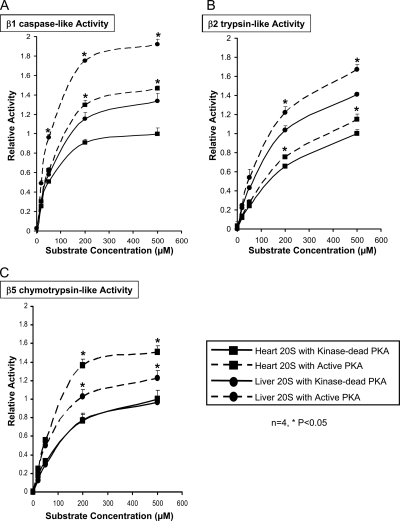
We first determined that the 20 S subunits are kinase targets of PKA. The cardiac (Fig. 7, A and B) and hepatic (Fig. 7, C and D) 20 S proteasome complexes were displayed by 2DE; phosphoantibodies against pan-PKA substrates were used for the 2DE immunoblotting, and LC-MS/MS was used to identify the subunits. Our data showed that many 20 S subunits are endogenous targets/substrates of PKA (Fig. 7, A and C). Addition of PKA to the purified 20 S proteasomes significantly enhanced the phosphorylation signals of multiple 20 S subunits (Fig. 7, B and D). Moreover PKA augmented the peptidase activity of these 20 S proteasomes; particularly the Vmax of β1 caspase-like, β2 trypsin-like, and β5 chymotrypsin-like activities were all significantly enhanced in both the heart and liver (Fig. 8). This functional significance of PKA-dependent regulation of the 20 S phosphoproteome was also supported by the analysis of the three proteolytically active proteasome subunits, β1, β2, and β5. The combined CID and ETD approach enabled the identification of phosphorylation sites on all three β subunits; furthermore these phosphorylation sites (e.g. serine 192 on β5) were regulated by PKA (Tables III and IV). Collectively we present data in this study to show that phosphorylation is an important mechanism in regulating the 20 S proteolytic function, that many 20 S subunits are substrate targets of PKA, and that PKA-dependent phosphorylation of 20 S proteasomes modulates proteolytic activities in both the heart and liver.
Fig. 7.
Dynamics of the 20 S phosphoproteome during PKA signaling. PKA has been identified previously as a 20 S proteasome-associating partner. 20 S proteasomes were treated with either active or heat-inactivated PKA and displayed via 2DE. The endogenous phosphorylation and PKA-induced phosphorylations of 20 S subunits were probed with an antibody recognizing the generic phosphorylation sites of PKA (PKA substrate antibody). A dual laser scanner differentiated anti-α7 conjugated to a different fluorophore for internal standardization. 20 S proteasome subunits were identified by LTQ-CID Orbitrap. A, the cardiac α1, α2, α6, β3, β4, β5, β6, and β7 subunits exhibited endogenous levels of PKA phosphorylation, indicating that these cardiac 20 S subunits are substrates of PKA. B, upon PKA treatment, fluorescent signals corresponding to the cardiac 20 S subunits (α1, α2, α6, β3, β4, and β7) were enhanced. C, the hepatic α1, α2, α3, α6, β3, β4, β5, β6, β7, and β5i subunits exhibited endogenous levels of PKA phosphorylation, indicating that these hepatic 20 S subunits are substrates of PKA. D, PKA treatment enhanced fluorescent signals corresponding to the hepatic 20 S subunits (α1, α6, β3, and β5).
Fig. 8.
PKA-dependent regulation of 20 S peptidase activities in heart and in liver. To assess the biological significance of the 20 S phosphoproteome, we examined the effect of PKA on three proteolytic activities (β1 caspase-like, β2 tryptic-like, and β5 chymotryptic-like) of the 20 S proteasomes in both heart and liver. A, the hepatic 20 S proteasomes exhibited a higher β1 caspase-like activity than that of the cardiac 20 S proteasomes. PKA significantly augmented both cardiac and hepatic β1 caspase-like activities to a similar magnitude. B, the hepatic 20 S proteasomes also exhibited a higher β2 trypsin-like activity than that of the cardiac 20 S proteasomes. PKA enhanced both cardiac and hepatic β2 trypsin-like activities to modest but statistically significant levels. C, hepatic and cardiac 20 S proteasomes maintained a similar level of β5 chymotrypsin-like activity. PKA drastically elevated the cardiac β5 chymotrypsin-like activity (Vmax), and the hepatic β5 chymotrypsin-like activity was also significantly increased but to a lesser degree than that of the heart. The relative activities were displayed as average values with their corresponding standard error bars. *, p < 0.05.
DISCUSSION
This investigation achieved a global characterization of the mammalian 20 S proteasome phosphoproteomes. The biological discoveries were enabled by an integrated technology platform encompassing parallel approaches in sample fractionation (SDS-PAGE, IPG-IEF, or 2DE), endoproteolytic digestion (trypsin or chymotrypsin), phosphopeptide enrichment (TiO2), and peptide fragmentation (CID or ETD). A total of 44 novel phosphorylation sites/peptides were identified in various proteasome subunits in both the liver and heart. The characterized 20 S phosphoproteomes manifested many novel single and dual phosphorylation sites/peptides from serine, threonine, and tyrosine residues. The cardiac and hepatic 20 S phosphoproteomes maintain unique and shared phosphoproteins. Furthermore the biological significance and dynamics of these 20 S phosphoproteomes were investigated. In silico analysis revealed that 20 S proteasomes contain many phosphorylation consensus sequences of various kinases, including that of PKA. Our results showed that many 20 S proteasome subunits were substrate targets of PKA as phosphorylation by PKA augmented the proteolytic function of the 20 S in heart and in liver. Taken together these findings demonstrate PKA as a key regulator of these 20 S phosphoproteomes.
Methodology Considerations—
Phosphorylation is increasingly recognized as a key regulatory event to many biological processes. Radioisotope labeling ([γ-32P]ATP) is a classical strategy and has been used to monitor phosphorylation events (29). The development of specific dyes, such as Pro-Q Diamond, offers non-radioactive alternatives (30). More recently, phosphoantibodies have shed new light on this line of investigation. The general pan-antibodies for Ser(P), Thr(P), and Tyr(P) have aided in the illumination of many phosphoproteins, whereas the characterization of site-specific phosphorylation has remained a challenge (31). Recent developments in mass spectrometry have facilitated phosphoproteome analyses (31, 32) and enabled global characterization of phosphorylation sites (25, 26, 33). However, the complexity and dynamics of biological samples, in particular the heterogeneity in the various magnitudes of phosphorylation signals displayed in the samples, remain as insurmountable technical difficulties. A number of approaches have been developed to address the issues relating to sample fractionation and enrichment procedures, such as peptide isoelectric focusing (34), IMAC (35), and TiO2 affinity pulldown (23).
The investigation into proteasome phosphorylation has been a long pursuit with groups such as Huang and coworkers (36) successfully pioneering the mass spectrometry-based approach. Using 2DE, several phosphorylated 20 S subunits have been visualized by either isotopic labeling ([γ-32P]ATP), immunoblotting, or phosphoprotein dye (11, 37, 38). Mutagenesis has also been used to validate the α2 Tyr-121, α3 Tyr-153, and α7 Ser-243/250 phosphorylation sites (39–42). During the last 20 years, a total of 15 unique sites have been identified in at least eight different reports from eight subunits of the mammalian 20 S proteasomes (33, 36, 39, 40, 42–45). In this study, a total of 52 phosphorylation events were identified; among which, 44 events were novel identifications. This study exceeded the total number of 20 S phosphorylation sites identified in the last 2 decades combined. The murine hepatic phosphorylation site (α7 Ser-243) and murine brain phosphorylation sites (α1 Tyr-159, α2 Tyr-76, and β6 Tyr-149) identified in two recent studies were not confirmed possibly because of the different age, tissue origin, and strain of mice used in this study (33, 45). The success achieved in this study is a direct demonstration of the technological strength afforded by parallel approaches in sample fractionation and peptide fragmentation (combined CID and ETD approaches).
The phosphorylation of serine 250 in the 20 S α7 subunit has been reported most frequently (19, 36). Importantly our pilot studies illustrated that this phosphopeptide exists in high abundance in vivo. In the presence of the dominating phosphate signal from the α7 (serine 250), very few other phosphopeptides displayed sufficient signals to be identified. Experiments were subsequently conducted to separate the α7 subunit during sample fractionation steps (SDS-PAGE, IEF, or 2DE). This strategy supported the recovery of low abundance phosphopeptides while sparing the phosphate signals of many other subunits. These results demonstrated sample fractionation as a critical step in achieving the global characterization of 20 S phosphoproteomes.
CID fragments selected peptide ions by random protonation along the peptide bond (yielding b and y ion products). The resonant vibration of precursor peptides and collision with helium atoms in gas phase lead to efficient fragmentation and peptide identification, yet these conditions may cause a loss of labile post-translational modifications such as phosphorylation. Preservation of these modifications may be achieved by the ETD method of peptide fragmentation (25, 26). In this method, an electron from a donor compound such as fluoranthene reacts with the peptide, cleaving the backbone after the amide group (yielding c and z ion products). Our approach combined the strengths of both technologies as 34 of the identified phosphopeptides (e.g. SpFEGQMTEDNIEDNIEVGICNEAGFR of α2) could only be identified with CID and 10 of the phosphopeptides (e.g. KPYpTpLMSMVANLLYEKR (where Yp is phosphotyrosine and Tp is phosphothreonine) of β3) could only be identified with ETD. Eight phosphopeptides of the 20 S proteasomes (e.g. GSpASpAMLQPLLDNQVGFK of β6) were confirmed through both strategies, thus highlighting the complementary benefits of both techniques.
It is interesting to note that within this reported data set in this study, only the C-terminal peptide of the α7 subunit was identified by MS3 with the observation of neutral loss. This finding was consistent with a previous report where two phosphorylation sites, the α7 serine 250 and the α3 serine 75, were characterized by CID with the observation of neutral loss (36). Both of these phosphorylation sites were confirmed in our study; however, there was no significant neutral loss observed for the α3 serine 75. The underlying mechanism for this difference remains to be investigated. It is possible that the occurrence of neutral loss may depend upon the peptide precursors. In this notion, neutral loss peak was favored for precursors within the mass range of 500–1500 Da but was suppressed in peptide precursors of extended length, multiple charges, and multiple basic amino acids (46).
The Biological Significance of 20 S Proteasome Phosphoproteome—
Proteasomes are ubiquitously expressed in all mammalian cell types. Despite a rapid progress in proteasome research, molecular mechanisms underlying the regulation of proteasome function (proteolytic activities) are poorly understood. Previous investigations from our group and others have implicated phosphorylation as a key regulatory signal to modulate proteolytic function (3, 14, 47). Accordingly we postulated that many kinases/phosphatases may serve as active regulators of 20 S proteasome function. We hypothesized that the subunits of 20 S proteasomes are targeted substrates of these kinases. In silico analyses revealed that the 20 S subunits host many phosphorylation consensus sites for a plethora of signaling kinases including protein kinase C, protein kinase A (PKA), casein kinase II, and protein kinase G (Table V). In the current study, we elected to examine the effect of PKA on the phosphoproteomes and the proteolytic function of 20 S proteasomes using a pan-PKA substrate antibody combined with 2DE and LC-MS/MS. Our data demonstrated that multiple 20 S subunits are substrates of PKA. Several of the 20 S phosphorylation sites identified are consistent with the predicted phosphorylation consensus motif of PKA (Tables III, IV, and V). These include the phosphorylation sites of the cardiac α3 serine 81 (GGITSpDAN), cardiac β1 serine 157 (RQSp), hepatic β4 serine 39 (KMSpEK), hepatic β5 serine 192 (RVSp), and both the cardiac and hepatic β6 serine 167 (FKAGGSp).
Table V.
Detected phosphorylation sites and predicted phosphorylation consensus motifs
This table lists the 20 S subunits (Subunit), experimentally identified phosphopeptides (Phosphopeptide), experimentally identified phosphorylation residue (Residue), names of kinases whose predicted phosphorylation consensus motif match the experimentally identified phosphorylation sites (Kinase), and tissues (heart, liver, or both) from which the 20 S proteasome complexes were purified. Please note that the predicted kinase phosphorylation motif was analyzed with the aid of NetphosK 1.0 Server at ExPASy and Protein Kinase Phosphorylation Site Sequences and Consensus Specificity Motif: Tabulations (48). PKG, protein kinase G; CKII, casein kinase II; PKC, protein kinase C.
| Subunit | Phosphopeptide | Residue | Kinase | Tissue |
|---|---|---|---|---|
| α2 | SpFEGQMTEDNIEVGICNEAGFR | 198 | PKG/PKC | Heart/liver |
| α2 | SpFEGQMTpEDNIEVGICNEAGFR | 204 | CKII | Heart |
| α3 | LNEDMACSVAGITSpDAN | 81 | PKA | Heart |
| α4 | ECQSHRLTpVEDPVTVEYITR | 97 | PKG/PKC | Heart/liver |
| α5 | LNATNIELATVQPGQNFHMFTp | 230 | CKII | Heart/liver |
| α7 | MTpCRDVVKEVAK | 186 | PKG/PKC | Heart/liver |
| α7 | ESLKEEDESpDDDNM | 250 | CKII | Heart/liver |
| β1 | SVPMGGMMVRQSpFAIGGSGSSYIYGYVDATYR | 157 | PKA/PKG/PKC | Heart |
| β1 | GMTpKDECLQFTANALALAMER | 181 | PKG/CKII | Liver |
| β2 | TEKVTPLEIEVLEETVQTp | 273 | CKII | Heart |
| β3 | KPYTpLMSMVANLLYEKR | 86 | PKG/PKC | Liver |
| β4 | MSpEKILLLCVGEAGDTVQFAEYIQK | 39 | PKA/PKG/PKC | Liver |
| β5 | SpSDNVADLHDKYSSVSVP | 192 | PKA/PKG/PKC/CKII | Liver |
| β5 | VSSDNVADLHDKYSpSVSV | 204 | PKG/PKC | Heart |
| β6 | GSpASAMLQPLLDNQVGFK | 167 | PKA/PKG | Heart/liver |
| β7 | MRVNDSpTMLGASGDYADFQYLK | 93 | PKG | Liver |
Tissue heterogeneity of 20 S proteasome subunits was evident in the murine 20 S proteasomes by the 2DE patterns. Distinct populations could be observed and fractionated with high resolution non-denaturing IEF. The variance in pI between cardiac and liver 20 S proteasomes indicated that they possess distinct biochemical properties (9). With phosphoantibodies, it was shown that phosphorylation contributed to this diversity. A total of 28 phosphorylation sites were shared by both the cardiac and hepatic 20 S subunits (such as α7 Ser-250). With Pro-Q Diamond, one dominant fluorescent band was observed in both heart and liver proteasomes (Fig. 2), indicating that this subunit was highly phosphorylated compared with other subunits in both tissues. As shown in Tables I–IV, 24 phosphorylation sites were different in these two organs between normal and PKA-stimulated conditions. The dynamics of tissue-specific phosphorylation profiles were further revealed by comparing cardiac and liver phosphoproteomes under PKA stimulation. Without PKA, the cardiac 20 S proteasomes displayed four unique phosphorylation sites, and the hepatic 20 S proteasomes displayed five unique phosphorylation sites. However, with PKA stimulation, there were nine unique phosphorylation sites in the heart and six in the liver. The distinction in phosphorylation profiles was paralleled by their functional differences. Purified liver 20 S proteasomes showed significantly higher β1 and β2 activities compared with that exhibited by the cardiac 20 S proteasomes. Despite a similar β5 activity in normal tissue, PKA provoked a more dramatic elevation of the cardiac β5 activity than that in the hepatic tissue. Taken together, these findings underscore the biological significance of PKA regulation of the 20 S proteasome phosphoproteomes and afford great promises to advance our understanding of this important organelle in health and disease.
Footnotes
Published, MCP Papers in Press, June 25, 2008, DOI 10.1074/mcp.M800064-MCP200
The abbreviations used are: 20 S proteasomes, proteolytic core particles of proteasomes; PTM, post-translational modification; PKA, cAMP-dependent protein kinase A; CIAP, calf intestinal alkaline phosphatase; ETD, electron transfer dissociation; 1DE, one-dimensional electrophoresis; 2DE, two-dimensional electrophoresis; AmBC, ammonium bicarbonate; Z, benzyloxycarbonyl; Boc, t-butoxycarbonyl; Suc, succinyl; AMC, amino-4-methylcoumarin; 3D, three-dimensional.
This work was supported, in whole or in part, by National Institutes of Health Grants HL R01-63901, HL R01-65431, and HL P01-80111 (to P. P. as a principal investigator) and HL BRG 088640 (to P. P. as a co-investigator). This work was also supported by American Heart Association Grants 0715004Y (to G. W. Y.) and 0625062Y (to O. D.) and by an endowment from Theodore C. Laubisch at UCLA (to P. P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Dahlmann, B. ( 2007) Role of proteasomes in disease. BMC Biochem. 8, Suppl. 1, S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drews, O., Zong, C., and Ping, P. ( 2007) Exploring proteasome complexes by proteomic approaches. Proteomics 7, 1047–1058 [DOI] [PubMed] [Google Scholar]
- 3.Goldberg, A. L. ( 2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899 [DOI] [PubMed] [Google Scholar]
- 4.Huang, L., and Burlingame, A. L. ( 2005) Comprehensive mass spectrometric analysis of the 20S proteasome complex. Methods Enzymol. 405, 187–236 [DOI] [PubMed] [Google Scholar]
- 5.Loda, M., Cukor, B., Tam, S. W., Lavin, P., Fiorentino, M., Draetta, G. F., Jessup, J. M., and Pagano, M. ( 1997) Increased proteasome-dependent degradation of the cyclin-dependent kinase inhibitor p27 in aggressive colorectal carcinomas. Nat. Med. 3, 231–234 [DOI] [PubMed] [Google Scholar]
- 6.Ozaki, K., Sato, H., Iida, A., Mizuno, H., Nakamura, T., Miyamoto, Y., Takahashi, A., Tsunoda, T., Ikegawa, S., Kamatani, N., Hori, M., Nakamura, Y., and Tanaka, T. ( 2006) A functional SNP in PSMA6 confers risk of myocardial infarction in the Japanese population. Nat. Genet. 38, 921–925 [DOI] [PubMed] [Google Scholar]
- 7.Kitada, T., Asakawa, S., Hattori, N., Matsumine, H., Yamamura, Y., Minoshima, S., Yokochi, M., Mizuno, Y., and Shimizu, N. ( 1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 [DOI] [PubMed] [Google Scholar]
- 8.Nazif, T., and Bogyo, M. ( 2001) Global analysis of proteasomal substrate specificity using positional-scanning libraries of covalent inhibitors. Proc. Natl. Acad. Sci. U. S. A. 98, 2967–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drews, O., Wildgruber, R., Zong, C., Sukop, U., Nissum, M., Weber, G., Gomes, A. V., and Ping, P. ( 2007) Mammalian proteasome subpopulations with distinct molecular compositions and proteolytic activities. Mol. Cell. Proteomics 6, 2021–2031 [DOI] [PubMed] [Google Scholar]
- 10.Guerrero, C., Tagwerker, C., Kaiser, P., and Huang, L. ( 2006) An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol. Cell. Proteomics 5, 366–378 [DOI] [PubMed] [Google Scholar]
- 11.Zong, C., Gomes, A. V., Drews, O., Li, X., Young, G. W., Berhane, B., Qiao, X., French, S. W., Bardag-Gorce, F., and Ping, P. ( 2006) Regulation of murine cardiac 20S proteasomes: role of associating partners. Circ. Res. 99, 372–380 [DOI] [PubMed] [Google Scholar]
- 12.Gomes, A. V., Zong, C., Edmondson, R. D., Li, X., Stefani, E., Zhang, J., Jones, R. C., Thyparambil, S., Wang, G. W., Qiao, X., Bardag-Gorce, F., and Ping, P. ( 2006) Mapping the murine cardiac 26S proteasome complexes. Circ. Res. 99, 362–371 [DOI] [PubMed] [Google Scholar]
- 13.Cuervo, A. M., Palmer, A., Rivett, A. J., and Knecht, E. ( 1995) Degradation of proteasomes by lysosomes in rat liver. Eur. J. Biochem. 227, 792–800 [DOI] [PubMed] [Google Scholar]
- 14.Glynne, R., Kerr, L. A., Mockridge, I., Beck, S., Kelly, A., and Trowsdale, J. ( 1993) The major histocompatibility complex-encoded proteasome component LMP7: alternative first exons and post-translational processing. Eur. J. Immunol. 23, 860–866 [DOI] [PubMed] [Google Scholar]
- 15.Thomson, S., and Rivett, A. J. ( 1996) Processing of N3, a mammalian proteasome β-type subunit. Biochem. J. 315, 733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuckelkorn, U., Frentzel, S., Kraft, R., Kostka, S., Groettrup, M., and Kloetzel, P. M. ( 1995) Incorporation of major histocompatibility complex-encoded subunits LMP2 and LMP7 changes the quality of the 20S proteasome polypeptide processing products independent of interferon-γ. Eur. J. Immunol. 25, 2605–2611 [DOI] [PubMed] [Google Scholar]
- 17.Mason, G. G., Murray, R. Z., Pappin, D., and Rivett, A. J. ( 1998) Phosphorylation of ATPase subunits of the 26S proteasome. FEBS Lett. 430, 269–274 [DOI] [PubMed] [Google Scholar]
- 18.Lilley, K. S., Davison, M. D., and Rivett, A. J. ( 1990) N-terminal sequence similarities between components of the multicatalytic proteinase complex. FEBS Lett. 262, 327–329 [DOI] [PubMed] [Google Scholar]
- 19.Villen, J., Beausoleil, S. A., Gerber, S. A., and Gygi, S. P. ( 2007) Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U. S. A. 104, 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. ( 1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 21.Gorg, A., Weiss, W., and Dunn, M. J. ( 2004) Current two-dimensional electrophoresis technology for proteomics. Proteomics 4, 3665–3685 [DOI] [PubMed] [Google Scholar]
- 22.Annan, R. S., Huddleston, M. J., Verma, R., Deshaies, R. J., and Carr, S. A. ( 2001) A multidimensional electrospray MS-based approach to phosphopeptide mapping. Anal. Chem. 73, 393–404 [DOI] [PubMed] [Google Scholar]
- 23.Pinkse, M. W., Uitto, P. M., Hilhorst, M. J., Ooms, B., and Heck, A. J. ( 2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 76, 3935–3943 [DOI] [PubMed] [Google Scholar]
- 24.Cantin, G. T., Venable, J. D., Cociorva, D., and Yates, J. R., III ( 2006) Quantitative phosphoproteomic analysis of the tumor necrosis factor pathway. J. Proteome Res. 5, 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Good, D. M., Wirtala, M., McAlister, G. C., and Coon, J. J. ( 2007) Performance characteristics of electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 6, 1942–1951 [DOI] [PubMed] [Google Scholar]
- 26.Syka, J. E., Coon, J. J., Schroeder, M. J., Shabanowitz, J., and Hunt, D. F. ( 2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unno, M., Mizushima, T., Morimoto, Y., Tomisugi, Y., Tanaka, K., Yasuoka, N., and Tsukihara, T. ( 2002) The structure of the mammalian 20S proteasome at 2.75 Å resolution. Structure (Camb.) 10, 609–618 [DOI] [PubMed] [Google Scholar]
- 28.Larsen, M. R., Thingholm, T. E., Jensen, O. N., Roepstorff, P., and Jorgensen, T. J. ( 2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 29.Ludemann, R., Lerea, K. M., and Etlinger, J. D. ( 1993) Copurification of casein kinase II with 20 S proteasomes and phosphorylation of a 30-kDa proteasome subunit. J. Biol. Chem. 268, 17413–17417 [PubMed] [Google Scholar]
- 30.Hopper, R. K., Carroll, S., Aponte, A. M., Johnson, D. T., French, S., Shen, R. F., Witzmann, F. A., Harris, R. A., and Balaban, R. S. ( 2006) Mitochondrial matrix phosphoproteome: effect of extra mitochondrial calcium. Biochemistry 45, 2524–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, X., Huston, E., Lynch, M. J., Houslay, M. D., and Baillie, G. S. ( 2006) Phosphodiesterase-4 influences the PKA phosphorylation status and membrane translocation of G-protein receptor kinase 2 (GRK2) in HEK-293β2 cells and cardiac myocytes. Biochem. J. 394, 427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia, B. A., Shabanowitz, J., and Hunt, D. F. ( 2005) Analysis of protein phosphorylation by mass spectrometry. Methods 35, 256–264 [DOI] [PubMed] [Google Scholar]
- 33.Beausoleil, S. A., Villen, J., Gerber, S. A., Rush, J., and Gygi, S. P. ( 2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 34.Maccarrone, G., Kolb, N., Teplytska, L., Birg, I., Zollinger, R., Holsboer, F., and Turck, C. W. ( 2006) Phosphopeptide enrichment by IEF. Electrophoresis 27, 4585–4595 [DOI] [PubMed] [Google Scholar]
- 35.Chi, A., Huttenhower, C., Geer, L. Y., Coon, J. J., Syka, J. E., Bai, D. L., Shabanowitz, J., Burke, D. J., Troyanskaya, O. G., and Hunt, D. F. ( 2007) Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 104, 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, X., Chen, C. F., Baker, P. R., Chen, P. L., Kaiser, P., and Huang, L. ( 2007) Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry 46, 3553–3565 [DOI] [PubMed] [Google Scholar]
- 37.Bose, S., Mason, G. G., and Rivett, A. J. ( 1999) Phosphorylation of proteasomes in mammalian cells. Mol. Biol. Rep. 26, 11–14 [DOI] [PubMed] [Google Scholar]
- 38.Iwafune, Y., Kawasaki, H., and Hirano, H. ( 2002) Electrophoretic analysis of phosphorylation of the yeast 20S proteasome. Electrophoresis 23, 329–338 [DOI] [PubMed] [Google Scholar]
- 39.Benedict, C. M., and Clawson, G. A. ( 1996) Nuclear multicatalytic proteinase subunit RRC3 is important for growth regulation in hepatocytes. Biochemistry 35, 11612–11621 [DOI] [PubMed] [Google Scholar]
- 40.Bose, S., Stratford, F. L., Broadfoot, K. I., Mason, G. G., and Rivett, A. J. ( 2004) Phosphorylation of 20S proteasome α subunit C8 (α7) stabilizes the 26S proteasome and plays a role in the regulation of proteasome complexes by γ-interferon. Biochem. J. 378, 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castano, J. G., Mahillo, E., Arizti, P., and Arribas, J. ( 1996) Phosphorylation of C8 and C9 subunits of the multicatalytic proteinase by casein kinase II and identification of the C8 phosphorylation sites by direct mutagenesis. Biochemistry 35, 3782–3789 [DOI] [PubMed] [Google Scholar]
- 42.Liu, X., Huang, W., Li, C., Li, P., Yuan, J., Li, X., Qiu, X. B., Ma, Q., and Cao, C. ( 2006) Interaction between c-Abl and Arg tyrosine kinases and proteasome subunit PSMA7 regulates proteasome degradation. Mol. Cell 22, 317–327 [DOI] [PubMed] [Google Scholar]
- 43.Beausoleil, S. A., Jedrychowski, M., Schwartz, D., Elias, J. E., Villen, J., Li, J., Cohn, M. A., Cantley, L. C., and Gygi, S. P. ( 2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U. S. A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rush, J., Moritz, A., Lee, K. A., Guo, A., Goss, V. L., Spek, E. J., Zhang, H., Zha, X. M., Polakiewicz, R. D., and Comb, M. J. ( 2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 45.Ballif, B. A., Carey, G. R., Sunyaev, S. R., and Gygi, S. P. ( 2008) Large-scale identification and evolution indexing of tyrosine phosphorylation sites from murine brain. J. Proteome Res. 7, 311–318 [DOI] [PubMed] [Google Scholar]
- 46.Schlosser, A., Pipkorn, R., Bossemeyer, D., and Lehmann, W. D. ( 2001) Analysis of protein phosphorylation by a combination of elastase digestion and neutral loss tandem mass spectrometry. Anal. Chem. 73, 170–176 [DOI] [PubMed] [Google Scholar]
- 47.Feng, Y., Longo, D. L., and Ferris, D. K. ( 2001) Polo-like kinase interacts with proteasomes and regulates their activity. Cell Growth Differ. 12, 29–37 [PubMed] [Google Scholar]
- 48.Pearson, R. B., and Kemp, B. E. ( 1991) Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 200, 62–81 [DOI] [PubMed] [Google Scholar]