Abstract
Matrix metalloproteinase-9 (MMP-9) is implicated in tumor metastasis as well as a variety of inflammatory and pathological processes. Although many substrates for MMP-9, including components of the extracellular matrix, soluble mediators such as chemokines, and cell surface molecules have been identified, we undertook a more comprehensive proteomics-based approach to identify new substrates to further understand how MMP-9 might contribute to tumor metastasis. Previous proteomics approaches to identify protease substrates have depended upon differential labeling of each sample. Instead we used a label-free quantitative proteomics approach based on ultraperformance LC-ESI-high/low collision energy MS. Conditioned medium from a human metastatic prostate cancer cell line, PC-3ML, in which MMP-9 had been down-regulated by RNA interference was compared with that from the parental cells. From more than 200 proteins identified, 69 showed significant alteration in levels after depletion of the protease (>±2-fold), suggesting that they might be candidate substrates. Levels of six of these (amyloid-β precursor protein, collagen VI, leukemia inhibitory factor, neuropilin-1, prostate cancer cell-derived growth factor (PCDGF), and protease nexin-1 (PN-1)) were tested in the conditioned media by immunoblotting. There was a strong correlation between results by ultraperformance LC-ESI-high/low collision energy MS and by immunoblotting giving credence to the label-free approach. Further information about MMP-9 cleavage was obtained by comparison of the peptide coverage of collagen VI in the presence and absence of MMP-9 showing increased sensitivity of the C- and N-terminal globular regions over the helical regions. Susceptibility of PN-1 and leukemia inhibitory factor to MMP-9 degradation was confirmed by in vitro incubation of the recombinant proteins with recombinant MMP-9. The MMP-9 cleavage sites in PN-1 were sequenced. This study provides a new label-free method for degradomics cell-based screening leading to the identification of a series of proteins whose levels are affected by MMP-9, some of which are clearly direct substrates for MMP-9 and become candidates for involvement in metastasis.
Matrix metalloproteinase-9 (MMP-9)1 or gelatinase B was originally identified as a secreted enzyme that could degrade collagens, especially collagen IV (1, 2). These early observations have since been refined as it became apparent that denatured collagen I, IV as well as gelatin are efficiently cleaved but that intact collagen I or IV are poor substrates for the enzyme (3, 4). Indeed MMP-9 fails to promote cellular invasion of natural basement membranes (5). Although MMP-9 may not be a determining factor for invasion of collagen IV-rich basement membranes, collagen IV degradation by MMP-9 has been shown to be involved in angiogenesis. For instance, the reduced levels of antiangiogenic peptides derived from collagen IV in mice genetically deficient in MMP-9 reflect the decreased turnover of collagen IV in the absence of MMP-9 (6). Many non-collagenous substrates have also been identified as MMP-9 substrates including cell surface molecules and secreted proteins. These include molecules involved in cell adhesion, cell surface receptors, cytokines, protease inhibitors, angiogenic factors, and growth factors. The cleavage of many of these substrates affects the cellular phenotype and is associated with several pathological conditions in mice (7, 8).
In cancers MMP-9 produced by the host contributes to squamous cancer progression by activation of the angiogenic switch and recruitment of macrophages or hematopoietic stem cells to the pre-existing metastatic niche (9, 10). MMP-9 made by the tumor cell also contributes to cancer progression (11). In some cases metastatic potential is associated with increased MMP-9 levels, and down-regulation of MMP-9 depresses metastasis (11). However, a protective role for MMPs in cancer progression has also been demonstrated (12). For example, MMP-9 can also result in decreased angiogenesis because of the release of extracellular endostatin in a breast cancer model, therefore contributing to tumor regression (13).
Despite the abundant evidence supporting a role for MMP-9 in promotion of tumor metastasis, the nature of the substrates targeted by MMP-9, which contributes to this process, remain largely unknown. As a prelude to define the targets of MMP-9 activity in cancer metastasis, we developed a new method to identify MMP-9 substrates in cancer cells. MMP-9 is a member of the matrix metalloproteinase family of zinc-dependent endopeptidases that characteristically have a conserved catalytic domain with Zn2+ complexed to three histidines at the active site (11, 14, 15). A cysteine residue in a propeptide domain of MMPs interacts with the Zn2+ in the catalytic site inhibiting activity and maintaining latency (14). Although cleavage of the propeptide domain is a common mechanism for MMP activation and acquisition of enzymatic activity, evidence has shown that catalytic activity can be acquired without cleavage of the propeptide, in particular in pro-MMP-9 (16, 17). MMP-9 and its close homolog MMP-2 (gelatinase A) possess a gelatin-binding domain, which comprises three fibronectin-like repeats that provide binding to and specificity for denatured collagen (4). Like all MMPs, MMP-9 also contains a hemopexin-like domain, which is connected to the catalytic domain by a highly glycosylated hinge region. The function of the hemopexin-like domain of MMP-9 in catalysis has not been elucidated. However, the complex of pro-MMP-9 with TIMP-1 involves binding of the inhibitor to the hemopexin-like domain (18). The glycosylation of the hinge region affects activity indirectly through influences on cell surface binding and internalization (19, 20). Different cell lines catalyze different glycosylation patterns (21). These factors further complicate the ability to predict valid substrates in a cellular context. MMP-9 like many MMPs is secreted but can also bind the cell surface through collagen (α2) IV chains (2), CD44 (22), and through the lipoprotein receptor-related protein receptor (23).
Identification of substrates for proteases in general poses a series of technical problems. Prediction of substrates based on cleavage of phage or peptide libraries in vitro has often overestimated sites because many sequences that are capable of being cleaved are not accessible in the intact protein or exposed to the enzyme in its cellular locations (24–26). A more successful approach to identify new protease substrates includes the use of mass spectrometry and in particular quantitative proteomics approaches to compare protein levels after alterations in the protease activity because the cleavage of substrates is assessed under live conditions (26, 27). In these experiments, cell culture supernatants from cells with a differential expression level of the protease under study are examined for enrichment of proteins that can be considered as candidate substrates (28). Initial studies of this kind included the use of stable isotope labeling methodologies to quantify peptide and protein levels by mass spectrometry more accurately (27). For instance, isotope-coded affinity tags (ICAT) that react with cysteine residues were used to identify novel substrates for the matrix metalloproteinase MT1-MMP (29). A more recent study applied isobaric tags for relative and absolute quantitation (iTRAQ) and mass spectrometry to uncover novel substrates for MMP-2 (30). This approach revealed a significantly higher number of potential substrates because this mass tag is not restricted to cysteine but is reactive to free amines (peptide N termini) (30). However, both approaches require the attachment of isotope labels to peptide material prior to MS analysis (29, 30), which may introduce an uncertainty factor for quantitation because the tagging reactions may not be complete. As an alternative, peak height and ion counts observed in liquid chromatography runs can also be used to determine levels of biomolecules (31). This method has been problematic for proteomics analysis because of the low reproducibility of nano-LC runs and the requirement for MS to MS/MS switching to obtain sequence information, resulting in low quality chromatograms (32). The development of chromatographic columns with smaller particle size as well as LC pumps with higher pressure limits and nanoflow delivery capacity permitted the achievement of much improved chromatography performance, referred to as ultraperformance LC (UPLC) (31–33). The recent development of nano-UPLC coupled to ESI-MS/MS now allows the analysis of low abundance biological samples (31, 33). Initial results demonstrate that the enhanced chromatography performance leads to significant gains in the collection of information on biomolecules such as peptides and proteins (sequence coverage and post-translational modifications) (33, 34). In addition, the level of reproducibility in nano-UPLC allows quantitative assessment of changes between samples with higher precision without the need for using stable isotope-based techniques, in particular when combined with high speed MS-MS/MS switching capabilities (31, 32) or parallel parent and fragment ion analysis MSE (35, 36). In this study, we used nano-UPLC coupled to high/low collision energy MS (MSE)-based label-free quantitative shotgun proteomics to reveal novel MMP-9 substrates present in prostate cancer cell culture supernatants and confirm the susceptibility of several proteins to MMP-9 using independent biochemical methods.
EXPERIMENTAL PROCEDURES
Cell Culture and Conditioned Medium Collection—
PC-3ML cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum at 37 °C with 5% CO2. To collect conditioned medium, plated PC-3ML cells were washed in serum-free DMEM three times and then incubated with serum-free DMEM with or without 10 μm BB-94 (British Biotech), 100 nm phorbol 12-myristate 13-acetate (PMA; Sigma), or MMP-9 inhibitor I (Calbiochem) for 24 h. After harvest, the conditioned medium was centrifuged at 1000 × g to remove the cell debris and then concentrated 10–15 times through a Centricon concentrator device. For mass spectrometry, the proteins from concentrated conditioned medium were extracted through chloroform-methanol precipitation (37), and protein concentration was measured and normalized for in-solution tryptic digestion (38). For immunoblot and zymography, the loading of concentrated medium was normalized according to cell numbers and protein content.
shRNA—
Sequences coded for shRNAs directly against MMP-9 were cloned into either pcDNA3 or IMG-800 vectors and tested for the ability to reduce the expression and activity of MMP-9 in the PC-3ML cell line. In general, 1 μg of shRNA MMP-9 was transfected into PC-3ML cells using the FuGENE transfection reagent (Roche Applied Science) according to the manufacturer's protocol. The cell colonies were then selected by 400 μg/ml G418 for a number of passages until it was stably expressed. Conditioned medium was collected to test for the reduction of MMP-9 activity. The sequence of final applied shRNA MMP-9 for mass spectrometry was CAAGTGGCACCACCACAACAT (39).
Analysis by Nano-UPLC-MSE Tandem Mass Spectrometry—
Collected and precipitated material from cell supernatants was subjected to in-solution trypsin digestion as described previously (38). Digested samples were desalted and concentrated using Sep-Pak C18 column cartridges (Waters) according to the manufacturer's instructions. Samples were analyzed by nano-UPLC-MSE using a 75-μm-inner diameter × 25-cm C18 nanoAcquity™ UPLC™ column (1.7-μm particle size; Waters) and a 90-min gradient: 2–45% solvent B (solvent A: 99.9% H2O, 0.1% formic acid; solvent B: 99.9% acetonitrile, 0.1% formic acid) on a Waters nanoAcquity UPLC system (final flow rate, 250 nl/min; 7000 p.s.i.) coupled to a Waters QTOFpremier™ tandem mass spectrometer. Data were acquired in high definition MSE mode (low collision energy, 4 eV; high collision energy ramping from 15 to 40 eV, switching every 1.5 s) and processed with ProteinLynx Global Server (PLGS; version 2.2.5; Waters) to reconstruct MS/MS spectra by combining all masses with identical retention times. Each sample was analyzed in triplicate runs. The mass accuracy of the raw data was corrected using Glu-fibrinopeptide (200 fmol/μl, 700 nl/min flow rate, 785.8426 Da [M + 2H]2+) that was infused into the mass spectrometer as a lock mass during sample analysis. Low and high collision energy MS data were calibrated at intervals of 30 s. The raw data sets were processed including deisotoping and deconvolution, and peak lists were generated on the basis of assigning precursor ions and fragments based on similar retention times (35, 36). A Swiss-Prot database (release 51.0, October 2006, 241,242 entries) was used for database searches of each triplicate run with the following parameters: peptide tolerance, 15 ppm; fragment tolerance, 0.015 Da; trypsin missed cleavages, 1; and variable modifications, carbamidomethylation and Met oxidation. Alternatively MS/MS spectra (peak lists) were also searched against the Swiss-Prot database (release 54.0; July 2007; number of entries, 276,256) using Mascot version 2.2 (Matrix Science, London, UK) and the following parameters: peptide tolerance, 0.2 Da; 13C = 2; fragment tolerance, 0.1 Da; missed cleavages, 3; and instrument type, ESI-Q-TOF. The interpretation and presentation of MS/MS data were performed according to published guidelines (40). In addition, individual MS/MS spectra for peptides with a Mascot Mowse (molecular weight search) score lower than 40 (expect value <0.015) were inspected manually and included in the statistics only if a series of at least four continuous y or b ions were observed. Protein identification was also based on the assignment of at least two peptides. False positive rates for the data sets generated in this study were evaluated using Mascot and a randomized database based on Swiss-Prot (decoy function) and are illustrated in supplemental Table S3. The local “in-house” Mascot server used for this study is supported and maintained by the Computational Biology Research Group at the University of Oxford.
Quantitative Analysis—
The analysis of quantitative changes in protein abundance, which is based on measuring peptide ion peak intensities observed in low collision energy mode in a triplicate set, was carried out using Waters Expression Analysis Software (WEPS™), which is part of PLGS 2.2.5 (Expression version 2). For normalization, each sample was spiked with 125 fmol of tryptic digest of α-enolase as an internal standard. Included were all protein hits that were identified with a confidence of >95%. Identical peptides from each triplicate set per sample were clustered based on mass precision (<15 ppm, typically ∼5 ppm) and a retention time tolerance of <0.25 min using the clustering software included in PLGS 2.2.5. Protein scores were increased when the same peptide assignments were made in more than one replicate run. To avoid potential errors due to redundancies in assignments, searches were performed using the non-redundant Swiss-Prot database (as described above). Furthermore if two or more distinct proteins shared an identical peptide but were found to be regulated differently, then the quantitation algorithm did not include the peptide in question. To allow for this, peptide probabilities are always softened slightly by the PLGS software prior to quantitation. Because of this, the contributions from peptides with even 100% probability of presence were suppressed to avoid potential errors in quantitation. Normalization of the data sets was performed based on the spiked standard, but very similar results were obtained using the PLGS “autonormalization” function (not shown).
Protease Nexin-1 (PN-1) Peptide Mapping—
The determination of MMP-9 cleavage sites in PN-1 as described for Fig. 7 was performed by comparing in-solution tryptic digests of PN-1 control with PN-1 incubated with recombinant MMP-9. The presence of non-tryptic cleavage sites that were not observed in the control sample was ranked based on the individual Mascot peptide scores and divided into high probability (Mascot peptide score >40) and low probability (Mascot peptide score <40). The peak list files of one representative experiment are available as supplemental material (CTRL_MMP9KD1/2/3 and MMP9KD1/2/3).
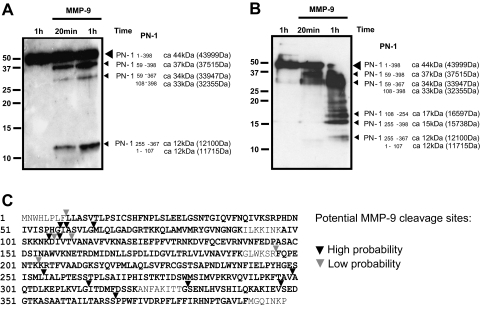
Fig. 7.
Mapping MMP-9 cleavage sites on PN-1. A, recombinant PN-1 (500 ng) was incubated with 100 ng of active MMP-9 at 37 °C for 20 min or 1 h. 5% of the cleaved products were resolved by 15% SDS-PAGE and immunoblotted with anti-PN-1 antibody. B, second independent experiment as described in A except using a different batch of recombinant MMP-9 (Calbiochem). C, sequence coverage obtained by in-solution trypsin digestion of the samples above followed by LC-MS/MS analysis as described under “Experimental Procedures.” Peptides detected by MS are highlighted, and potential MMP-9 cleavage sites are indicated by triangles.
Immunoblotting—
Whole cell lysates were extracted through lysing cell pellets in TNN buffer (50 mm Tris at pH 8.0, 120 mm NaCl, and 0.5% Nonidet P-40) containing protease inhibitor mixture (Calbiochem), 4 mm DTT, and 0.2 mm PMSF at 4 °C for 30 min. After removing the cell debris, protein aliquots were made and normalized by equal intracellular protein concentration. Conditioned medium was concentrated and normalized according to cell numbers and intracellular proteins. Immunoblots were carried out as described previously (41). Concisely cell lysates and concentrated conditioned medium were separated by SDS-PAGE, transferred to PVDF membrane, and probed with the indicated antibodies: anti-amyloid-β precursor protein (APP) (22C11) antibody (Chemicon), anti-leukemia inhibitory factor (LIF) (N-18), anti-neuropilin 1 (H-286) and anti-PC cell-derived growth factor (PCDGF) (N-19) (Santa Cruz Biotechnology), anti-PN-1 (R&D Systems), and anti-collagen VI (Abcam).
Zymography—
MMP-9 and MMP-2 enzyme activity was assayed by gelatin zymography as described previously (41). In brief, concentrated conditioned medium was separated by electrophoresis, under non-reducing condition, through an 8% polyacrylamide gel co-polymerized with 1 mg/ml gelatin (Sigma). Gels were washed in 2.5% Triton X-100 solution three times and then incubated overnight at room temperature in developing buffer containing 50 mm Tris-HCl, pH 7.5, 5 mm CaCl2, 5 μm ZnCl2, and 1% Triton X-100. Gels were subsequently stained with 0.25% Coomassie Brilliant Blue R-250 and then destained with 50 mm Tris-HCl (pH 7.4) and 2% Triton X-100. Enzymatic activity attributed to MMP-9 and MMP-2 was visualized in the zymogram as clear bands against dark background. Human recombinant MMP-9 and MMP-2 proteins (10 ng) (Calbiochem) were loaded as controls.
In Vitro Cleavage Assay—
Human active MMP-9, MMP-2, or pro-MMP-9 (Calbiochem) was tested for the ability to cleave human recombinant LIF (Chemicon) or PN-1 protein (R&D Systems) in vitro. Briefly recombinant proteins were mixed in the assay buffer (50 mm Tris/HCl, pH 7.5, 150 mm NaCl, 10 mm CaCl2, 0.05% Brij35, 100 μg/ml BSA, 0.5 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 1 μg/ml pepstatin, and 1 μg/ml leupeptin) and incubated at 37 °C. At different time points, reactions were terminated by 10 mm EDTA. The digestive products were subjected to 15% polyacrylamide electrophoresis, transferred to PVDF membranes, and immunoblotted with anti-LIF or anti-PN-1 antibodies. Alternatively the resulting cleavage products were analyzed by peptide mapping using LC-MS/MS as described above. Data analysis was performed using Mascot using the parameters as described above with the exception of using no restriction for enzyme cleavage.
RESULTS
MMP-9 Levels in Tumor Cells—
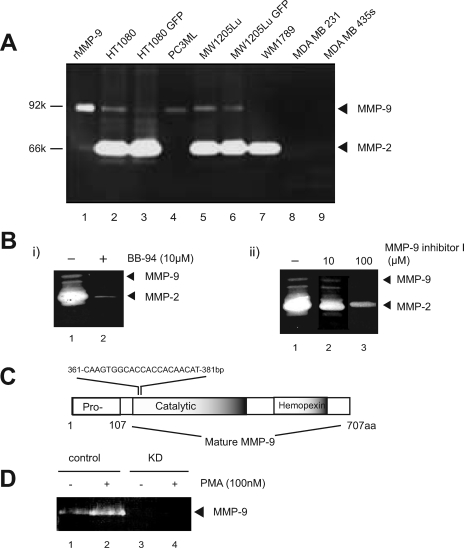
To select a tumor cell system to use for identification of potential substrates for MMP-9, several invasive tumor cell lines were screened for the expression and activity of MMP-9 through gelatin zymography (Fig. 1A). From this screening, we selected PC-3ML, a highly invasive and metastatic subline of PC-3 (human prostate cancer cell line), which express MMP-9 but little MMP-2 (Fig. 1A).
Fig. 1.
MMP-9 knockdown and pharmacological inhibition in tumor cells: initial characterization. A, conditioned medium collected from the indicated human tumor cell lines was screened through gelatin zymography for the expression and activity of gelatinases. Lane 1 represents 10 ng of recombinant MMP-9. B, equal aliquots of conditioned medium collected from HT1080 cells were resolved by gelatin SDS-PAGE. To test the chemical prevention of MMP-9 activity, BB-94 (10 μm; i, lane 2) or MMP-9 inhibitor I (10 or 100 μm; ii, lanes 2 and 3) were added while developing the gelatin gel. C, schematic illustration of the target sites of shRNA against MMP-9. D, conditioned medium collected from PC-3ML (control) or MMP-9 KD cell lines, treated with or without 100 nm PMA, were analyzed by zymography. aa, amino acids. GFP, green fluorescent protein.
Two sets of strategies were undertaken to regulate MMP-9 activity in PC-3ML cells, pharmacological inhibition and RNA interference expression. An expression vector coding for shRNA against MMP-9 was transfected and stably expressed (Fig. 1C); cells displayed significant loss of MMP-9 (Fig. 1D). Even after treatment with PMA, a potent transcriptional activator for several MMPs including MMP-9 (42), the level of MMP-9 released by the knockdown (KD) line remained undetectable, whereas it was significantly increased in PC-3ML cells (Fig. 1D). Two commercial compounds, BB-94 and an MMP-9 inhibitor, were tested for inhibition of MMP-9 activity in vitro. Both reduced the activity of MMP-2 and MMP-9. In particular, BB-94 almost completely ablated the activity of both gelatinases at a concentration of 10 μm (Fig. 1B).
Identification of MMP-9 Substrate Candidates by Nano-UPLC-MSE Analysis—
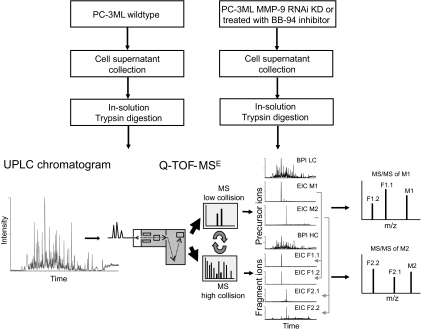
Serum-free conditioned medium was collected from PC-3ML cells after extensive washing (five times) to remove as much serum as possible. Medium was collected from control cells, cells treated with BB-94, and cells stably expressing shRNA against MMP-9 followed by sample concentration using a 10-kDa molecular mass cutoff spin filter. Concentrated material was precipitated and subjected to in-solution trypsin digestion. Digested material was analyzed using a label-free quantitative proteomics approach (Fig. 2). This method entails separation by nano-UPLC and analysis by an on-line Q-TOF tandem mass spectrometer. The presence of several hundred proteins in cell culture supernatants gives rise to thousands of peptides after proteolytic digestion that usually requires multiple chromatographic steps for efficient separation, such as MudPIT (33). In our approach, the use of a 25-cm reversed phase column packed with small particles (1.7 μm) combined with ultrahigh pressure (∼7000 p.s.i.) allowed for adequate separation in a single chromatography run (Figs. 2 and 3A).
Fig. 2.
A label-free quantitative proteomics approach for MMP-9 substrate degradomics. Cell culture supernatants of PC3-ML control and those treated either with BB-94 inhibitor or induction of MMP-9 RNA interference (RNAi) knockdown were harvested and subjected to in-solution trypsin digestion. Samples were separated by reversed phase nano-UPLC using a 25-cm 1.7-μm-particle size C18 column coupled to a Q-TOF tandem mass spectrometer (shown as base peak intensity liquid chromatogram (BPI LC)). MS data acquisition was performed using MSE technology, which is based on continuous switching between low and high collision energy to collect precursor ion masses (low collision) and fragment ions (high collision) at intervals as short as 0.5 s. Low collision MS provides information about precursor ions (indicated as M1 and M2) and their retention times revealed by extracted ion chromatograms (EIC). High collision energy MS provides the masses for fragment ions (indicated as F1 and F2). Association of fragment ions to their corresponding precursor ion is determined by their similar retention times (obtained from extracted ion chromatograms) and allows the creation of MS/MS spectra that can be used to obtain sequences and protein identification.
Fig. 3.
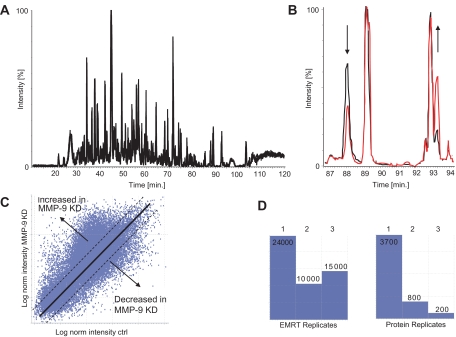
Principle of label-free relative quantitation by UPLC-MSE. A, chromatographic separation of cell culture supernatants digested with trypsin on a nano-UPLC C18 reversed phase column using a 90-min gradient (base peak intensity chromatogram). B, relative quantitation is based on peak intensities observed in base peak intensity chromatograms as shown for peaks in the range of 87–92 min (black and red). C, a comparison between proteolytic digests obtained from PC3-ML control (ctrl) and MMP-9 knockdown cell culture supernatants reveals the presence of ∼30,000 mass peaks defined as EMRTs with different intensities obtained in a single chromatographic run. Along the diagonal axis are the log values of peptide ion intensities that were present at equal levels between the two samples, and differentially expressed peaks are present above or below the axis, representing tryptic peptides from proteins that were either increased or reduced upon MMP-9 knockdown, respectively. The adequacy of relative quantitation is refined by analyzing the samples in triplicate runs, allowing the detection of changes in expression above the relative ratios of 1.1 and below 0.9 with high significance (p < 0.01; dotted lines). norm, normalized; KD, knockdown. D, the availability of triplicate data sets provides information about the data quality, such as replicate rates between repeated runs (left panel) and the confidence and reproducibility of protein identification (right panel). Under these conditions, 200 proteins were identified in all three runs (triplicates) and are therefore of high confidence. 800 proteins were assigned in two of the three repeats and are therefore of intermediate confidence, and 3700 proteins were identified in one replicate, representing low confidence hits (see also supplemental material for additional information).
A current limitation of data collection is determined by the acquisition speed in data-dependent acquisition (DDA) mode, which requires switching from MS to MS/MS to obtain both ion counts of precursor ions and fragmentation information in a tandem mass spectrometer. This usually leads to insufficient collection of data points to accurately define peak shapes in MS mode because of fragment data collection in MS/MS mode at the same time (43). In addition, for a complex mixture, a fraction of precursor ions will be selected for sequencing only because of the sequential acquisition of MS- and MS/MS-based spectra (43). To overcome this, we used the recently developed MSE technology, which is based on continuous switching between low and high collision energy to collect precursor ion masses (low collision) and fragment ions (high collision) at intervals as short as 1 s (35, 36). In this way, approximately 5–10 times more information can be obtained from a single LC run as compared with data-dependent switching modes (43). The assignment of precursor ions to their associated fragment ions is based on their equivalent retention time profiles, allowing the recreation of MS/MS spectra (Fig. 2). In addition, in the same chromatography run, sufficient data points can be collected in low collision energy mode to define a precursor ion peak shape that is adequate for quantitation by measuring its peak height. The principle of relative quantitation without previous incorporation of stable isotope tags of amino acids is based on measuring peak intensities of individual peptide masses obtained in separate chromatography runs for each sample (Fig. 3, A and B). Nano-UPLC is now sufficiently reproducible to accomplish this task (Fig. 3B). Each mass peak (precursor or fragment ion) is therefore defined by its exact mass and retention time (EMRT).
A comparison between proteolytic digests obtained from media of control and MMP-9 knockdown PC-3ML cells revealed the presence of ∼30,000 EMRTs with different intensities (Fig. 3C) obtained in a single chromatographic separation step. Along the diagonal axis are the peptide ions that were present at similar levels in each of the two samples, and differentially expressed peaks are present above or below the axis, representing tryptic peptides from proteins that were either increased or reduced upon MMP-9 knockdown, respectively. The adequacy of relative quantitation was refined by analyzing the samples in triplicate runs, allowing the detection of changes in expression above the relative ratios of 1.1 and below 0.9 with high significance (p < 0.01; Fig. 3C, Table I, and supplemental material). The availability of triplicate data sets can also provide information about the data quality, such as replicate rates between repeated runs (Fig. 3D, left panel) and the confidence and reproducibility of protein identification (Fig. 3D, right panel). Under these conditions, 200 proteins were identified in all three runs (triplicates) and are therefore of high confidence. 800 proteins were assigned in two of the three repeats and are therefore of intermediate confidence.
Table I.
Proteins with different abundance levels in supernatants of control versus MMP-9 siRNA- or BB-94-treated PC-3ML cells identified by tandem mass spectrometry
ND, not detected; ECM, extracellular matrix; Ctrl, control; Seq. cov., sequence coverage. — indicates that a p value could not be calculated because the indicated protein was not detected in one of the paired samples.
| Protein name | Ref. | Accession no. (Swiss-Prot)a | Scoreb (PLGS) | Seq. cov. | No. of MS/MS spectra | MMP-9 siRNA:Ctrl | p valuec | BB-94:Ctrl | p valuec | Scored (Mascot) |
|---|---|---|---|---|---|---|---|---|---|---|
| % | ||||||||||
| Known substrates | ||||||||||
| Alzheimer disease amyloid A4 protein (APP)e | 47–49 | P05067 | 222f | 17 | 10 | 1.1 | 0.36 | 0.3 | <0.01 | 79f |
| Galectin 3-binding protein precursor | 45 | Q08380 | 102g | 22 | 11 | ND | — | 0.8 | <0.01 | 53f |
| Fibronectin | 64 | Q28275 | 128f | 7 | 4 | 0.7 | <0.01 | 0.6 | <0.01 | 152g |
| Collagen α1 XI chain precursor | 58 | Q61245h | 79f | 13.4 | 24 | 0.24 | <0.01 | ND | — | ND |
| Collagen α4 IV chain precursor | 1, 2 | P53420 | 93f | 6.3 | 8 | 0.3 | 0.02 | ND | — | ND |
| Collagen α1 II chain precursor | 4 | P02458 | 79f | 8.2 | 8 | Ctrl | — | ND | — | 0f |
| Integrin β1 precursor | 44 | P05556 | 42f | 1 | 2 | Ctrl | — | ND | — | ND |
| TIMPs | ||||||||||
| TIMP-1 | P01033 | 135f | 30 | 5 | 0.7 | <0.01 | 0.6 | <0.01 | 58f | |
| TIMP-2 | P16035 | 62g | 35 | 7 | ND | — | 0.7 | <0.01 | ND | |
| Novel substrate candidates | ||||||||||
| Cell surface | ||||||||||
| CD166 | Q13740 | 96f | 7.4 | 6 | 3.2 | <0.01 | ND | — | 34f | |
| Neuropilin-1 precursor vascular endothelial celle | O14786 | 123f | 12 | 9 | 1.9 | <0.01 | ND | — | ND | |
| Galectin 1 β-galactoside-binding lectin L 14 | P09382 | 65f | 13 | 2 | 1.75 | 0.16 | 0.8 | <0.01 | ND | |
| Saposin-A | P07602 | 254f | 21 | 11 | 1.5 | <0.01 | 0.5 | <0.01 | 260f | |
| Semaphorin 7A | O75326 | 111g | 6 | 3 | 0.9 | 0.1 | Ctrl | — | 35f | |
| Integrin α3 galactoprotein B3 | P26006 | 64f | 7.7 | 8 | MMP-9 KD | — | ND | — | ND | |
| Angiopoietin 1 receptor precursor Tie-2 | Q02763 | 49f | 2 | 2 | MMP-9 KD | — | ND | — | ND | |
| Clathrin heavy chain CLH17 | Q00610 | 72f | 6.5 | 7 | MMP-9 KD | — | ND | — | ND | |
| Secreted | ||||||||||
| Laminin β1 chain precursor, laminin B1 chain | P07942 | 76f | 5.4 | 7 | MMP-9 KD | — | ND | — | ND | |
| Nexin-1 (PN-1)e | P07093 | 81g | 19 | 7 | >10 | <0.01 | 2.0 | <0.01 | 40f | |
| LIFe | P09056h | 55f | 24 | 5 | 2.6 | <0.01 | ND | — | ND | |
| Granulins precursor acrogranin (PCDGF)e | P28799 | 185f | 8.6 | 5 | 1.4 | <0.01 | ND | — | 54f | |
| HSP-90 | P08238 | 357g | 27 | 19 | 1.2 | <0.01 | 1.3 | <0.01 | 67f | |
| Urokinase type plasminogen activator precursor | P00749 | 254g | 35 | 12 | 0.6 | <0.01 | 0.8 | <0.01 | 135f | |
| Tissue type plasminogen activator precursor | P00750 | 142f | 29.8 | 13 | Ctrl | <0.01 | 0.7 | <0.01 | ND | |
| ECM | ||||||||||
| Collagenase 3 MMP-13 | P33435h | 33f | 5.1 | 3 | Ctrl | — | ND | — | ND | |
| Collagen α1 VI chain precursore | P12109 | 854g | 37 | 34 | 1.5 | <0.01 | 0.9 | <0.01 | 582f | |
| Collagen α2 VI chain precursor | P12110 | 252f | 13 | 13 | 1.5 | <0.01 | 0.8 | <0.01 | 34f | |
| Procollagen-lysine 2-oxoglutarate 5-dioxygenase 1 | Q02809 | 107f | 3.3 | 2 | 1.5 | <0.01 | 1.2 | <0.01 | ND |
Accession number from Swiss-Prot.
Scores based on the software PLGS 2.2.5.
p values calculated from individual peak intensities derived from all tryptic peptides detected per protein (see “Experimental Procedures”).
As a comparison, Mascot scores are included where applicable (Mascot version 2.2, MudPIT protein score).
Confirmed by immunoblotting (Fig. 4).
PLGS score, sequence coverage, and number of MS/MS spectra derived from the MMP-9 siRNA experiment.
PLGS score, sequence coverage, and number of MS/MS spectra derived from the BB-94 inhibitor experiment.
Accession number for the mouse protein sequence.
A subset of the identified proteins including known MMP-9 substrates and novel substrate candidates is described in Table I. For instance, known substrates such as collagens I, II, and IV and integrin β1 (1, 2, 44) were found to be reduced in media from MMP-9 knockdown PC-3ML cells. Galectin 3, which was shown previously to be processed by MMP-9 (45), was found to be significantly reduced when MMP activity was inhibited pharmacologically. In the same screen, a number of novel substrate candidates were detected. Collagen VI (α1 and α2), laminin β1, MMP-13, protease nexin-1, neuropilin, PCDGF, clathrin, integrin α3, and CD166 appeared to accumulate when MMP-9 levels were reduced. In addition, collagen α2 I and fibronectin were found to be decreased in the absence of active MMP-9, suggesting that they may be candidates for MMP-9-dependent cell surface shedding (Table I). TIMP-1, a naturally occurring inhibitor of MMP-9 that tightly binds to it, may have been reduced in amount simply because of decreased MMP-9.
Validation of Changes in Protein Levels after Down-regulation of MMP-9—
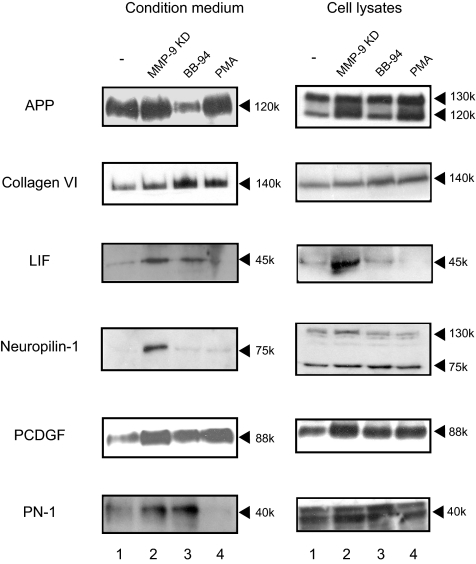
We used several criteria to select candidate proteins for biochemical validation as MMP-9 substrates. First, we chose proteins that have been identified as secreted, membrane-bound, or on the cell surface, recognizing that this might exclude proteins for which current information is incomplete. Second, only proteins that scored above 40 for significance were used. Third, an MMP-9 siRNA:control (KD/C) ratio >1 was set for selecting candidate substrates as the implication was that if they were truly substrates they would accumulate when MMP-9 activity was inhibited. This would exclude any surface protein that is released or shed into the medium by MMP-9, and this ratio would decrease in level after MMP-9 down-regulation. We did not choose candidates from the BB-94-generated list. Fourth, we required that good commercial antibody be available to facilitate validation. Fifth, we chose proteins whose levels had large differences in amount as well as those with only modest changes. These criteria led us to test APP, collagen VI, LIF, neuropilin-1, PCDGF, and PN-1 for protein levels in the conditioned medium generated from control and treated cells by Western blotting.
In this set of experiments, we sought to compare the results from the proteomics determination of differences with changes seen on immunoblotting. APP pathologically accumulates and forms aggregates in patients with Alzheimer disease (46). Interestingly MMP-9 and MMP-2 have been shown to cleave APP in vitro and in vivo (47–49). APP was found at a slightly increased level in the conditioned medium from the MMP-9 KD cell line compared with the control (Fig. 2) consistent with the MS KD/C ratio of 1.1 (Table I). There was a more marked decrease in the amount in conditioned medium from the BB-94-treated cell line also consistent with the ratio of 0.3. Collagen IV, whose denatured form has been well established as a classic substrate for MMP-9, appeared in the list. We chose, however, to test the potentially novel substrate collagen VI, a less well characterized collagen. α1 and α2 chains of collagen VI accumulated in the medium from KD cells consistent with the ratio of 1.5. The proteomics results in the BB-94-treated cells with a ratio of ∼0.8 were the only example of proteomics data not confirmed by Western blotting with more and not less collagen VI detected (Table I and Fig. 4). LIF scored at 55.69 and had KD/C ratio of 2.6, consistent with immunoblotting results. LIF is a cytokine that promotes the long term maintenance of embryonic stem cells by suppressing spontaneous differentiation. Neuropilin-1, a transmembrane protein with a large extracellular domain (50), accumulated in the conditioned medium of the MMP-9 KD cell line (Fig. 4), consistent with an MS score of 123 and KD/C ratio of 1.9. The fragment (75 kDa) released into conditioned medium appears to be a soluble isoform of neuropilin-1 (51). Neuropilin-1 was predicted by the proteomics screen to be unaltered in conditioned medium after BB-94 treatment, and this was confirmed by Western blotting. PCDGF, a secreted protein also known as granulins precursor or GP88, increased in both conditioned medium and whole cell lysates from MMP-9 KD cells and in conditioned medium from BB-94-treated cells. PN-1 also accumulated in the conditioned medium from both MMP-9 KD cells (score, 40.83; ratio, >10) and BB-94-treated cells (score, 81.46; ratio, 2.0) by proteomics analysis (Table I). Thus, the changes in levels noted in the proteomics screens were largely confirmed by immunoblotting.
Fig. 4.
Six candidate substrates of MMP-9 identified by MS were validated through immunoblot. Conditioned medium and whole cell lysates collected from PC-3ML cells treated as indicated were subjected to electrophoresis, transferred to PVDF membranes, and probed with the appropriate antibodies. Cell lysate aliquots were made and normalized by intracellular protein concentration. The loading of concentrated conditioned medium was normalized according to cell numbers and intracellular proteins.
We examined the complexity of the system in response to other manipulations that perturb protease activity. BB-94 as indicated above is a nonspecific inhibitor of MMPs. Thus in a simple system the results of down-regulation of MMP-9 might be paralleled by its inhibition by a nonspecific inhibitor such as BB-94. In fact, however, these results did not track together as measured either by proteomics or by immunoblotting. Similarly overexpression of MMP-9 induced by PMA, a treatment that has many effects in addition to up-regulation of MMP-9, might in a simple system exaggerate the difference with the MMP-9 down-regulated cells. However, this was not the case indicating that the regulation allowing accumulation of proteins in the conditioned medium was much more complex. Thus to determine the validity of proteins as MMP-9 substrates, we sought to determine whether candidates could be cleaved by MMP-9 through biochemical means.
Processing of Collagen VI Is Dependent on MMP-9—
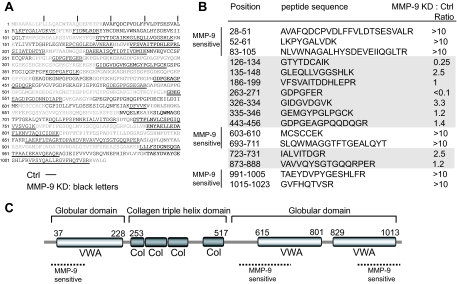
One of the most abundant proteins found in PC3-ML cell culture supernatants was collagen VI (Table I). This collagen type appears to affect early mammary tumor progression by modulating the tumor/stroma microenvironment (52). We noted a wide range of relative abundance rates for individual tryptic peptides derived from collagen VI that correlated with their location within different domains (Fig. 5). Peptides located at the N and C termini and outside the triple helix domain strongly accumulated when MMP-9 levels were reduced, suggesting MMP-9-dependent processing under normal conditions (Fig. 5, A and B).
Fig. 5.
Cleavage topology of collagen VI by MMP-9. A, collagen VI sequence coverage obtained by nano-UPLC-MSE analysis of trypsin-digested cell culture supernatants from control (underlined) and MMP-9 KD samples (black letters). B, relative quantitation of individual tryptic peptides derived from collagen VI observed in control (Ctrl) and MMP-9 KD cell culture supernatants. MMP-9-sensitive areas were defined in which tryptic peptides with relative abundance ratios (MMP-9 KD/control (Ctrl)) were more than 10. Mass intensity values were chosen only from peptides that were identified with a high confidence PLGS score (>95%) and that were observed in at least two of the triplicate runs. C, MMP-9-sensitive areas of collagen VI are predominantly located in the C- and N-terminal globular regions (VWA, von Willebrand factor domain), whereas the triple helix domain appears to be more stable (Col, collagen triple helix domain), indicating differential processing of collagen VI by MMP-9. The domain structure of human collagen VI (accession number P12109) was obtained from the Pfam Website.
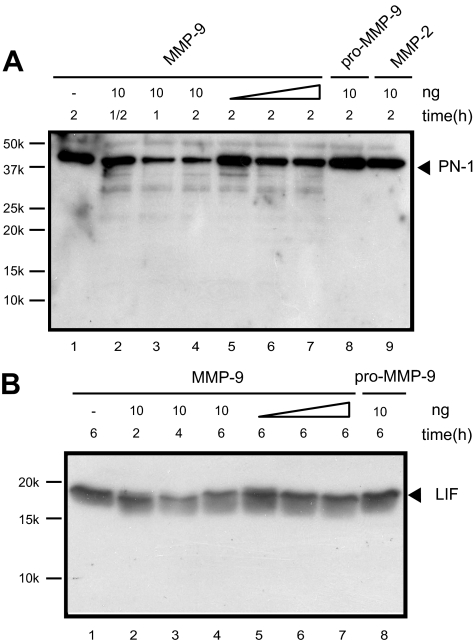
MMP-9 Cleaves LIF and PN-1 in Vitro—
To test whether the candidate substrates could be directly digested by MMP-9, cleavage assays were set up in vitro using active MMP-9 enzyme and recombinant proteins. MMP-9 cleaved LIF and PN-1 (Fig. 6) in a dose- and time-dependent manner. PN-1 remained intact after incubation with either pro-MMP-9 or active MMP-2 (Fig. 6A), demonstrating the specificity of the MMP-9 cleavage. The MMP-9-specific cleavage sites on PN-1 were identified through proteomics approaches. The cleavage products (5% of total reaction) were resolved by SDS-PAGE; gels showed three predominant PN-1-reactive fragments at around 37, 33, and 12 kDa (Fig. 7A) but also a number of intermediate breakdown fragments (Fig. 7B). To map candidate cleavage sites, the reaction mixture (50%) was processed by tryptic in-solution digestion. The sequence coverage obtained was up to 89%, and potential MMP-9-specific cleavage sites were identified as peptides without tryptic cleavage sites (C-terminal Arg or Lys or not preceded by these residues) that were present in MMP-9-treated samples only (Fig. 7C). Peptide mapping suggests the 37-kDa fragment to be 59–398 (37,515 Da), the 33-kDa fragment to be either 59–367 (33, 947 Da) or 108–398 (32,355 Da), and the 12-kDa fragment to be 255–367 (12,100 Da) or 1–107 (11,700 Da; Fig. 7C). Evidence for the presence of the latter fragment was obtained by Q-TOF-MS analysis ([M + 3H]3+ 3901.13Da (data not shown)). Based on these results, MMP-9 appears to cleave PN-1 predominantly at the sites PHGI58↓A59 and I107↓V108, therefore preferentially before small aliphatic amino acids such as Ile, Gly, and Val, which is consistent with other studies that examined MMP-9 cleavage of non-collagenous substrates (53, 54). In particular, the cleavage site in the PN-1 sequence at position 58/59 corresponds to the major MMP-9-specific cleavage consensus sequence PXX↓Hy(S/T) (Hy is a hydrophobic amino acid) as described previously (54).
Fig. 6.
MMP-9-dependent degradation of PN-1 and LIF. A, recombinant PN-1 (40 ng) was incubated with 10 ng (lanes 2–4), 2 ng (lane 5), 5 ng (lane 6), or 10 ng (lane 7) of active MMP-9; 10 ng of pro-MMP-9 (lane 8), or 10 ng of active MMP-2 (lane 9) at 37 °C for various time periods as indicated. Lane 1 represents 40 ng of PN-1 only at 37 °C for 2 h. The cleaved products were resolved by 15% SDS-PAGE and immunoblotted with anti-PN-1 antibody. B, 30 ng of recombinant LIF was incubated with MMP-9 as described above and detected by anti-LIF antibody. Recombinant LIF has a molecular mass of 19.2 kDa in comparison with the glycosylated form in mammalian cells of ∼45 kDa.
DISCUSSION
In this study, we used a label-free quantitative proteomics approach to identify novel substrates for MMP-9 in cancer cells and confirmed changes in abundance levels and cleavage of some of them using biochemical assays and peptide mapping. Whereas mass spectrometry-based quantitation has been routinely used to measure changes in levels of small molecules, it has been less trivial to perform relative and absolute quantitation of protein levels in biological samples by MS (32). This was due in part to a low reliability and reproducibility of nano-LC as well as fluctuations in electrospray ionization that affect the MS acquisition quality (32). In addition, LC-MS analysis of complex biological samples (proteolytic digests) can give rise to ion suppression, which complicates the retrieval of quantitative information based on ion intensities (32). These shortcomings have been overcome by the introduction of isotope labels such as 15N labeling, stable isotope labeling with amino acids in cell culture (SILAC), ICAT, isotope-coded protein labels (ICPL), or iTRAQ that allow the mixing of samples prior to analysis to minimize variations during the data acquisition process (55). However, the data qualities derived from approaches that include introduction of isotopes depend on the yield of metabolic labeling or incorporation of thiol/amine-reactive chemical groups. In addition, the use of isotope-containing compounds can be costly for biological experiments (26). Therefore, a label-free approach remains attractive for such large scale comparisons of peptide and protein content.
Two recent technical developments contributed to major improvements that render this method applicable to such studies. First, UPLC, initially developed for more rapid separation solutions to reduce costs and increase throughput, has been introduced to nano-LC-based methods for proteomics (31). Elevated chromatography performance by nano-UPLC coupled to fast MS/MS significantly increases sensitivity and in-depth analysis of peptides and proteins (31, 33). Analysis by conventional nano-LC-MS as compared with nano-UPLC-MS has revealed the superior resolution power of the latter technology (43).2 Samples may contain several hundred proteins that result in more than 10,000 peptide species after proteolytic digestion. A common strategy to deconvolute and analyze a biological sample of this complexity was the use of multidimensional protein identification technology (MudPIT), which comprises two different chromatography steps (56). Such experiments are usually very time-consuming, require a significant amount of instrument time and complex data analysis, and are therefore feasible only in a laboratory that is equipped with a large infrastructure. In addition, multidimensional chromatography approaches are usually associated with sample loss (33, 57). In many cases, it represents a significant effort to prepare the amounts of biological material required to conduct such an analysis. Here we found that nano-UPLC may provide a significant advantage, namely that such complex biological samples can be analyzed by a smaller number of chromatography steps, resulting in a significant gain of time and effort to conduct such an experiment.
Second, the development of MSE allows the collection of 5–10 times more precursor ions and fragmentation data as compared with DDA modes (35, 36, 43) because of a sequential low and high collision energy data acquisition cycle (Fig. 3). MSE-based data acquisition permits the collection of sufficient data points in low collision mode for the quantification of peak ion intensities and, at the same time, acquisition of fragmentation data in high collision mode for protein identification (Fig. 3). The successful analysis of this type of data was dependent on the development of specific software (PLGS) (36, 43). The assignment of precursor and its corresponding fragment ions is possible because of stringent settings for EMRT clusters (typically around 5 ppm and below ±0.25 min) even in complex mixtures (35, 36). Therefore, the mass accuracy achieved with this system reduces redundancy of multiple peptide assignments. In addition, when fragmentation data can be matched to peptide sequences that are observed in more than one protein that share similar probabilities, PLGS will exclude this data from further quantitative analyses to avoid potential errors in determining the relative abundance ratios of these proteins.
We noted that fragmentation spectra created via MSE were of lower quality than DDA-based MS/MS spectra because collision energies were adapted to mass and charge state in the latter method, whereas high collision energy ramping between 15 and 40 eV was used in MSE throughout the entire analysis. As a consequence, data-dependent acquisition appears to be more appropriate for the detection of post-translational modifications. However, the protein sequence coverage in DDA mode is generally lower as observed with MSE.2 The PLGS software takes into account some of the shortcomings of MSE data and gives more weight to the mass accuracy of the peptide precursor and fragment ions in addition to considering their retention times (EMRT clusters), whereas Mascot, at least the version used for this study, mainly considers the number of matched fragment ions, which favors data with high fragmentation quality obtained from data-dependent MS analysis experiments. Based on these reasons, the PLGS search engine is more optimized for the interpretation of MSE data. This is exemplified by the detection of the proteins neuropilin and LIF as well as their relative changes in abundance, which were confirmed by immunoblotting (Fig. 4). Interrogation of the MS data with Mascot failed to detect these proteins (Table I). Analysis of samples in triplicates permitted the improvement of the confidence level of assignments by PLGS based on multiple identifications in more than one replicate in some cases (Fig. 3D). PLGS analysis detected MMP-9-dependent changes in abundance levels of collagens II, IV, and XI, all of which are known to be MMP-9 substrates (1, 2, 4, 58), whereas Mascot searches did not assign confident scores for any of these proteins (Table I). The problem with proteomics data sets is that in the range below high confidence scores the false positive rate can increase considerably (supplemental Table S3), but these data cannot simply be discarded because they may contain biologically significant information. Future data curation and evaluation processes will certainly benefit from combining different search engines with complementary algorithms and selection criteria to further improve the accuracy of MS data interpretation.
Proteomics analysis of conditioned medium from PC-3ML cells after knockdown of MMP-9 revealed at least 100 proteins with variations in level. The variations detected were sometimes small (<2-fold) but were confirmed for substrates by Western blotting in several cases (Fig. 4). A possible reason for this is the use of a knockdown approach so that we would be comparing endogenous levels of MMP-9 expression without genetically engineered activation to avoid artifacts due to overexpression. The complexity of changing the expression of this one protease was surprisingly large. Several of the resultant changes were of proteases or their inhibitors that could lead to a cascade of effects. We cannot know how many of the affected proteins were altered in level as a result of direct action by MMP-9 or of indirect effects. Some of the proteins identified are known substrates of MMP-9, and we confirmed that several others could be cleaved by MMP-9 in vitro. Nonetheless this type of analysis may provide an initial snapshot and allow the construction of maps of protease interactions and networks.
The MMP-9 that we detected in the medium of the PC-3ML cells was 92 kDa, the proform, whereas the most active forms of MMP-9 are cleaved at the N terminus. However, MMP-9 has been shown to be active in its proform (16). In other cases MMP-9 activity has been detected at the cell surface without cleaved MMP-9 detection in the medium (59–61). Fridman and co-workers (62) reported cleaved cell surface MMP-9 that was a minority of the total MMP-9. We cannot be sure in this system whether activation by cleavage was required for the proteolysis we detected.
The selection of substrates by MMP-9 is not a simple matter of recognition of a linear amino acid sequence. The triple helical domains of collagens tend to be resistant to MMP-9 (63). A detailed examination of the susceptible peptides in collagen VI is consistent with this pattern (Fig. 5). The cleavage sites sequenced in protease nexin-1 do not clearly conform to the identified MMP-9 cleavage sites in collagenous substrates, although the site at position 59 has proline in the P3 position as has been identified for MMP-9 cleavage of triple helical substrates (54). The exact components that allow MMP-9 to recognize substrates are not yet clear. Some of the sites in PN-1 have similarities to the sequences termed Group II constructed from recognition of cleavage sites in phage libraries. Group II consists of Gly-Leu↓(Lys/Arg). The ultimate determination of the sites to be utilized by MMP-9 awaits confirmation by mutation analysis.
Taken together, UPLC-MSE provides a label-free quantitative proteomics approach that was used to discover candidate MMP-9 substrates. Subsequent biochemical analysis and identification of cleavage patterns suggested that some of these proteins are likely to be MMP-9 substrates. This method proved to be robust and allowed the identification of peptide differences of relatively small magnitude, such as 2-fold or lower. Our results provide the framework to examine the functional role of these substrates in cancer metastasis.
Acknowledgments
We thank Dr. Holger Kramer for expert assistance with the sample analysis by tandem mass spectrometry and members of the group for insightful discussion. We are grateful to Dr. Ghislain Opdenakker for the kind comments on the manuscript. We also acknowledge the services of the Computational Biology Research Group at the University of Oxford in this project.
Footnotes
Published, MCP Papers in Press, July 2, 2008, DOI 10.1074/mcp.M800095-MCP200
The abbreviations used are: MMP, matrix metalloproteinase; PN-1, protease nexin-1; TIMP, tissue inhibitor of metalloproteinases; UPLC, ultraperformance LC; MSE, high/low collision energy MS; MudPIT, multidimensional protein identification technology; APP, amyloid-β precursor protein; LIF, leukemia inhibitory factor; PCDGF, PC cell-derived growth factor; iTRAQ, isobaric tags for relative and absolute quantitation; DMEM, Dulbecco's modified Eagle's medium; PMA, phorbol 12-myristate 13-acetate; shRNA, short hairpin RNA; PLGS, ProteinLynx Global Server; PC, prostate cancer; KD, knockdown; DDA, data-dependent acquisition; EMRT, exact mass and retention time; siRNA, small interfering RNA; C, control; ICAT, isotope-coded affinity tags; ICPL, isotope-coded protein labels.
D. Xu, N. Suenaga, M. J. Edelmann, R. Fridman, R. J. Muschel, and B. M. Kessler, unpublished results.
This work was supported, in whole or in part, by National Institutes of Health grants (to D. X. and R. J. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Reponen, P., Sahlberg, C., Munaut, C., Thesleff, I., and Tryggvason, K. ( 1994) High expression of 92-kD type IV collagenase (gelatinase B) in the osteoclast lineage during mouse development. J. Cell Biol. 124, 1091–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olson, M. W., Toth, M., Gervasi, D. C., Sado, Y., Ninomiya, Y., and Fridman, R. ( 1998) High affinity binding of latent matrix metalloproteinase-9 to the α2(IV) chain of collagen IV. J. Biol. Chem. 273, 10672–10681 [DOI] [PubMed] [Google Scholar]
- 3.Allan, J. A., Docherty, A. J., Barker, P. J., Huskisson, N. S., Reynolds, J. J., and Murphy, G. ( 1995) Binding of gelatinases A and B to type-I collagen and other matrix components. Biochem. J. 309, 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steffensen, B., Wallon, U. M., and Overall, C. M. ( 1995) Extracellular matrix binding properties of recombinant fibronectin type II-like modules of human 72-kDa gelatinase/type IV collagenase. High affinity binding to native type I collagen but not native type IV collagen. J. Biol. Chem. 270, 11555–11566 [DOI] [PubMed] [Google Scholar]
- 5.Hotary, K., Li, X. Y., Allen, E., Stevens, S. L., and Weiss, S. J. ( 2006) A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev. 20, 2673–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Page-McCaw, A., Ewald, A. J., and Werb, Z. ( 2007) Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 8, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egeblad, M., and Werb, Z. ( 2002) New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 8.Cauwe, B., Van den Steen, P. E., and Opdenakker, G. ( 2007) The biochemical, biological, and pathological kaleidoscope of cell surface substrates processed by matrix metalloproteinases. Crit. Rev. Biochem. Mol. Biol. 42, 113–185 [DOI] [PubMed] [Google Scholar]
- 9.Kaplan, R. N., Riba, R. D., Zacharoulis, S., Bramley, A. H., Vincent, L., Costa, C., MacDonald, D. D., Jin, D. K., Shido, K., Kerns, S. A., Zhu, Z., Hicklin, D., Wu, Y., Port, J. L., Altorki, N., Port, E. R., Ruggero, D., Shmelkov, S. V., Jensen, K. K., Rafii, S., and Lyden, D. ( 2005) VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 438, 820–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiratsuka, S., Watanabe, A., Aburatani, H., and Maru, Y. ( 2006) Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat. Cell Biol. 8, 1369–1375 [DOI] [PubMed] [Google Scholar]
- 11.Bjorklund, M., and Koivunen, E. ( 2005) Gelatinase-mediated migration and invasion of cancer cells. Biochim. Biophys. Acta 1755, 37–69 [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Otin, C., and Matrisian, L. M. ( 2007) Emerging roles of proteases in tumour suppression. Nat. Rev. Cancer 7, 800–808 [DOI] [PubMed] [Google Scholar]
- 13.Bendrik, C., Robertson, J., Gauldie, J., and Dabrosin, C. ( 2008) Gene transfer of matrix metalloproteinase-9 induces tumor regression of breast cancer in vivo. Cancer Res. 68, 3405–3412 [DOI] [PubMed] [Google Scholar]
- 14.Van Wart, H. E., and Birkedal-Hansen, H. ( 1990) The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc. Natl. Acad. Sci. U. S. A. 87, 5578–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stocker, W., Grams, F., Baumann, U., Reinemer, P., Gomis-Ruth, F. X., McKay, D. B., and Bode, W. ( 1995) The metzincins—topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidases. Protein Sci. 4, 823–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bannikov, G. A., Karelina, T. V., Collier, I. E., Marmer, B. L., and Goldberg, G. I. ( 2002) Substrate binding of gelatinase B induces its enzymatic activity in the presence of intact propeptide. J. Biol. Chem. 277, 16022–16027 [DOI] [PubMed] [Google Scholar]
- 17.Rosenblum, G., Meroueh, S., Toth, M., Fisher, J. F., Fridman, R., Mobashery, S., and Sagi, I. ( 2007) Molecular structures and dynamics of the stepwise activation mechanism of a matrix metalloproteinase zymogen: challenging the cysteine switch dogma. J. Am. Chem. Soc. 129, 13566–13574 [DOI] [PubMed] [Google Scholar]
- 18.Van den Steen, P. E., Dubois, B., Nelissen, I., Rudd, P. M., Dwek, R. A., and Opdenakker, G. ( 2002) Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9). Crit. Rev. Biochem. Mol. Biol. 37, 375–536 [DOI] [PubMed] [Google Scholar]
- 19.Rudd, P. M., Mattu, T. S., Masure, S., Bratt, T., Van den Steen, P. E., Wormald, M. R., Kuster, B., Harvey, D. J., Borregaard, N., Van Damme, J., Dwek, R. A., and Opdenakker, G. ( 1999) Glycosylation of natural human neutrophil gelatinase B and neutrophil gelatinase B-associated lipocalin. Biochemistry 38, 13937–13950 [DOI] [PubMed] [Google Scholar]
- 20.Mattu, T. S., Royle, L., Langridge, J., Wormald, M. R., Van den Steen, P. E., Van Damme, J., Opdenakker, G., Harvey, D. J., Dwek, R. A., and Rudd, P. M. ( 2000) O-Glycan analysis of natural human neutrophil gelatinase B using a combination of normal phase-HPLC and online tandem mass spectrometry: implications for the domain organization of the enzyme. Biochemistry 39, 15695–15704 [DOI] [PubMed] [Google Scholar]
- 21.Van den Steen, P. E., Opdenakker, G., Wormald, M. R., Dwek, R. A., and Rudd, P. M. ( 2001) Matrix remodelling enzymes, the protease cascade and glycosylation. Biochim. Biophys. Acta 1528, 61–73 [DOI] [PubMed] [Google Scholar]
- 22.Yu, Q., and Stamenkovic, I. ( 1999) Localization of matrix metalloproteinase 9 to the cell surface provides a mechanism for CD44-mediated tumor invasion. Genes Dev. 13, 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van den Steen, P. E., Van Aelst, I., Hvidberg, V., Piccard, H., Fiten, P., Jacobsen, C., Moestrup, S. K., Fry, S., Royle, L., Wormald, M. R., Wallis, R., Rudd, P. M., Dwek, R. A., and Opdenakker, G. ( 2006) The hemopexin and O-glycosylated domains tune gelatinase B/MMP-9 bioavailability via inhibition and binding to cargo receptors. J. Biol. Chem. 281, 18626–18637 [DOI] [PubMed] [Google Scholar]
- 24.Matthews, D. J., and Wells, J. A. ( 1993) Substrate phage: selection of protease substrates by monovalent phage display. Science 260, 1113–1117 [DOI] [PubMed] [Google Scholar]
- 25.Turk, B. E., Huang, L. L., Piro, E. T., and Cantley, L. C. ( 2001) Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat. Biotechnol. 19, 661–667 [DOI] [PubMed] [Google Scholar]
- 26.Schilling, O., and Overall, C. M. ( 2007) Proteomic discovery of protease substrates. Curr. Opin. Chem. Biol. 11, 36–45 [DOI] [PubMed] [Google Scholar]
- 27.Overall, C. M., and Blobel, C. P. ( 2007) In search of partners: linking extracellular proteases to substrates. Nat. Rev. Mol. Cell Biol. 8, 245–257 [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Otin, C., and Overall, C. M. ( 2002) Protease degradomics: a new challenge for proteomics. Nat. Rev. Mol. Cell Biol. 3, 509–519 [DOI] [PubMed] [Google Scholar]
- 29.Tam, E. M., Morrison, C. J., Wu, Y. I., Stack, M. S., and Overall, C. M. ( 2004) Membrane protease proteomics: isotope-coded affinity tag MS identification of undescribed MT1-matrix metalloproteinase substrates. Proc. Natl. Acad. Sci. U. S. A. 101, 6917–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dean, R. A., and Overall, C. M. ( 2007) Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol. Cell. Proteomics 6, 611–623 [DOI] [PubMed] [Google Scholar]
- 31.Plumb, R., Castro-Perez, J., Granger, J., Beattie, I., Joncour, K., and Wright, A. ( 2004) Ultra-performance liquid chromatography coupled to quadrupole-orthogonal time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 18, 2331–2337 [DOI] [PubMed] [Google Scholar]
- 32.Qian, W. J., Jacobs, J. M., Liu, T., Camp, D. G., II, and Smith, R. D. ( 2006) Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol. Cell. Proteomics 5, 1727–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Motoyama, A., Venable, J. D., Ruse, C. I., and Yates, J. R., III ( 2006) Automated ultra-high-pressure multidimensional protein identification technology (UHP-MudPIT) for improved peptide identification of proteomic samples. Anal. Chem. 78, 5109–5118 [DOI] [PubMed] [Google Scholar]
- 34.Batycka, M., Inglis, N. F., Cook, K., Adam, A., Fraser-Pitt, D., Smith, D. G., Main, L., Lubben, A., and Kessler, B. M. ( 2006) Ultra-fast tandem mass spectrometry scanning combined with monolithic column liquid chromatography increases throughput in proteomic analysis. Rapid Commun. Mass Spectrom. 20, 2074–2080 [DOI] [PubMed] [Google Scholar]
- 35.Silva, J. C., Denny, R., Dorschel, C., Gorenstein, M. V., Li, G. Z., Richardson, K., Wall, D., and Geromanos, S. J. ( 2006) Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: a sweet tale. Mol. Cell. Proteomics 5, 589–607 [DOI] [PubMed] [Google Scholar]
- 36.Silva, J. C., Gorenstein, M. V., Li, G. Z., Vissers, J. P., and Geromanos, S. J. ( 2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 37.Wessel, D., and Flugge, U. I. ( 1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 38.Kinter, M., and Sherman, N. E. ( 2000) Protein Sequencing and Identification Using Tandem Mass Spectrometry, pp. 147–164 Wiley Interscience, New York
- 39.Lakka, S. S., Gondi, C. S., Yanamandra, N., Olivero, W. C., Dinh, D. H., Gujrati, M., and Rao, J. S. ( 2004) Inhibition of cathepsin B and MMP-9 gene expression in glioblastoma cell line via RNA interference reduces tumor cell invasion, tumor growth and angiogenesis. Oncogene 23, 4681–4689 [DOI] [PubMed] [Google Scholar]
- 40.Taylor, G. K., and Goodlett, D. R. ( 2005) Rules governing protein identification by mass spectrometry. Rapid Commun. Mass Spectrom. 19, 3420. [DOI] [PubMed] [Google Scholar]
- 41.Jiang, Y., and Muschel, R. J. ( 2002) Regulation of matrix metalloproteinase-9 (MMP-9) by translational efficiency in murine prostate carcinoma cells. Cancer Res. 62, 1910–1914 [PubMed] [Google Scholar]
- 42.Foda, H. D., George, S., Conner, C., Drews, M., Tompkins, D. C., and Zucker, S. ( 1996) Activation of human umbilical vein endothelial cell progelatinase A by phorbol myristate acetate: a protein kinase C-dependent mechanism involving a membrane-type matrix metalloproteinase. Lab. Investig. 74, 538–545 [PubMed] [Google Scholar]
- 43.Plumb, R. S., Johnson, K. A., Rainville, P., Smith, B. W., Wilson, I. D., Castro-Perez, J. M., and Nicholson, J. K. ( 2006) UPLC/MS(E); a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun. Mass Spectrom. 20, 1989–1994 [DOI] [PubMed] [Google Scholar]
- 44.Seftor, R. E., Seftor, E. A., Stetler-Stevenson, W. G., and Hendrix, M. J. ( 1993) The 72 kDa type IV collagenase is modulated via differential expression of αvβ3 and α5β1 integrins during human melanoma cell invasion. Cancer Res. 53, 3411–3415 [PubMed] [Google Scholar]
- 45.Ochieng, J., Fridman, R., Nangia-Makker, P., Kleiner, D. E., Liotta, L. A., Stetler-Stevenson, W. G., and Raz, A. ( 1994) Galectin-3 is a novel substrate for human matrix metalloproteinases-2 and -9. Biochemistry 33, 14109–14114 [DOI] [PubMed] [Google Scholar]
- 46.Weidemann, A., Konig, G., Bunke, D., Fischer, P., Salbaum, J. M., Masters, C. L., and Beyreuther, K. ( 1989) Identification, biogenesis, and localization of precursors of Alzheimer's disease A4 amyloid protein. Cell 57, 115–126 [DOI] [PubMed] [Google Scholar]
- 47.Miyazaki, K., Hasegawa, M., Funahashi, K., and Umeda, M. ( 1993) A metalloproteinase inhibitor domain in Alzheimer amyloid protein precursor. Nature 362, 839–841 [DOI] [PubMed] [Google Scholar]
- 48.Miyazaki, K., Funahashi, K., Umeda, M., and Nakano, A. ( 1994) Gelatinase A and APP. Nature 368, 695–696 [DOI] [PubMed] [Google Scholar]
- 49.LePage, R. N., Fosang, A. J., Fuller, S. J., Murphy, G., Evin, G., Beyreuther, K., Masters, C. L., and Small, D. H. ( 1995) Gelatinase A possesses a β-secretase-like activity in cleaving the amyloid protein precursor of Alzheimer's disease. FEBS Lett. 377, 267–270 [DOI] [PubMed] [Google Scholar]
- 50.Soker, S., Takashima, S., Miao, H. Q., Neufeld, G., and Klagsbrun, M. ( 1998) Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell 92, 735–745 [DOI] [PubMed] [Google Scholar]
- 51.Gagnon, M. L., Bielenberg, D. R., Gechtman, Z., Miao, H. Q., Takashima, S., Soker, S., and Klagsbrun, M. ( 2000) Identification of a natural soluble neuropilin-1 that binds vascular endothelial growth factor: in vivo expression and antitumor activity. Proc. Natl. Acad. Sci. U. S. A. 97, 2573–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iyengar, P., Espina, V., Williams, T. W., Lin, Y., Berry, D., Jelicks, L. A., Lee, H., Temple, K., Graves, R., Pollard, J., Chopra, N., Russell, R. G., Sasisekharan, R., Trock, B. J., Lippman, M., Calvert, V. S., Petricoin, E. F., III, Liotta, L., Dadachova, E., Pestell, R. G., Lisanti, M. P., Bonaldo, P., and Scherer, P. E. ( 2005) Adipocyte-derived collagen VI affects early mammary tumor progression in vivo, demonstrating a critical interaction in the tumor/stroma microenvironment. J. Clin. Investig. 115, 1163–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minond, D., Lauer-Fields, J. L., Nagase, H., and Fields, G. B. ( 2004) Matrix metalloproteinase triple-helical peptidase activities are differentially regulated by substrate stability. Biochemistry 43, 11474–11481 [DOI] [PubMed] [Google Scholar]
- 54.Kridel, S. J., Sawai, H., Ratnikov, B. I., Chen, E. I., Li, W., Godzik, A., Strongin, A. Y., and Smith, J. W. ( 2002) A unique substrate binding mode discriminates membrane type-1 matrix metalloproteinase from other matrix metalloproteinases. J. Biol. Chem. 277, 23788–23793 [DOI] [PubMed] [Google Scholar]
- 55.Cravatt, B. F., Simon, G. M., and Yates, J. R., III ( 2007) The biological impact of mass-spectrometry-based proteomics. Nature 450, 991–1000 [DOI] [PubMed] [Google Scholar]
- 56.Washburn, M. P., Wolters, D., and Yates, J. R., III ( 2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 57.Delahunty, C. M., and Yates, J. R., III ( 2007) MudPIT: multidimensional protein identification technology. BioTechniques 43, 563, 565, 567 passim [PubMed] [Google Scholar]
- 58.Yu, L. P., Jr., Smith, G. N., Jr., Hasty, K. A., and Brandt, K. D. ( 1991) Doxycycline inhibits type XI collagenolytic activity of extracts from human osteoarthritic cartilage and of gelatinase. J. Rheumatol. 18, 1450–1452 [PubMed] [Google Scholar]
- 59.Partridge, C. A., Phillips, P. G., Niedbala, M. J., and Jeffrey, J. J. ( 1997) Localization and activation of type IV collagenase/gelatinase at endothelial focal contacts. Am. J. Physiol. 272, L813–L822 [DOI] [PubMed] [Google Scholar]
- 60.Ginestra, A., Monea, S., Seghezzi, G., Dolo, V., Nagase, H., Mignatti, P., and Vittorelli, M. L. ( 1997) Urokinase plasminogen activator and gelatinases are associated with membrane vesicles shed by human HT1080 fibrosarcoma cells. J. Biol. Chem. 272, 17216–17222 [DOI] [PubMed] [Google Scholar]
- 61.Mazzieri, R., Masiero, L., Zanetta, L., Monea, S., Onisto, M., Garbisa, S., and Mignatti, P. ( 1997) Control of type IV collagenase activity by components of the urokinase-plasmin system: a regulatory mechanism with cell-bound reactants. EMBO J. 16, 2319–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olson, M. W., Bernardo, M. M., Pietila, M., Gervasi, D. C., Toth, M., Kotra, L. P., Massova, I., Mobashery, S., and Fridman, R. ( 2000) Characterization of the monomeric and dimeric forms of latent and active matrix metalloproteinase-9. Differential rates for activation by stromelysin 1. J. Biol. Chem. 275, 2661–2668 [DOI] [PubMed] [Google Scholar]
- 63.Bode, W., Reinemer, P., Huber, R., Kleine, T., Schnierer, S., and Tschesche, H. ( 1994) The X-ray crystal structure of the catalytic domain of human neutrophil collagenase inhibited by a substrate analogue reveals the essentials for catalysis and specificity. EMBO J. 13, 1263–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morodomi, T., Ogata, Y., Sasaguri, Y., Morimatsu, M., and Nagase, H. ( 1992) Purification and characterization of matrix metalloproteinase 9 from U937 monocytic leukaemia and HT1080 fibrosarcoma cells. Biochem. J. 285, 603–611 [DOI] [PMC free article] [PubMed] [Google Scholar]