Abstract
Mutations and copy number variation in the SNCA gene encoding the neuronal protein α-synuclein have been linked to familial Parkinson disease (Thomas, B., and Beal, M. F. (2007) Parkinson's disease. Hum. Mol. Genet. 16, R183–R194). The carboxyl terminus of α-synuclein can be phosphorylated at tyrosine 125 and serine 129, although only a small fraction of the protein is phosphorylated under normal conditions (Okochi, M., Walter, J., Koyama, A., Nakajo, S., Baba, M., Iwatsubo, T., Meijer, L., Kahle, P. J., and Haass, C. (2000) Constitutive phosphorylation of the Parkinson's disease associated α-synuclein. J. Biol. Chem. 275, 390–397). Under pathological conditions, such as in Parkinson disease, α-synuclein is a major component of Lewy bodies, a pathological hallmark of Parkinson disease, and is mostly phosphorylated at Ser-129 (Anderson, J. P., Walker, D. E., Goldstein, J. M., de Laat, R., Banducci, K., Caccavello, R. J., Barbour, R., Huang, J. P., Kling, K., Lee, M., Diep, L., Keim, P. S., Shen, X. F., Chataway, T., Schlossmacher, M. G., Seubert, P., Schenk, D., Sinha, S., Gai, W. P., and Chilcote, T. J. (2006) Phosphorylation of Ser-129 is the dominant pathological modification of α-synuclein in familial and sporadic Lewy body disease. J. Biol. Chem. 281, 29739–29752). Controversy exists over the extent to which phosphorylation of α-synuclein and/or the visible protein aggregation in Lewy bodies are steps in disease pathogenesis, are protective, or are neutral markers for the disease process. Here we used the combination of peptide pulldown assays and mass spectrometry to identify and compare protein-protein interactions of phosphorylated and non-phosphorylated α-synuclein. We showed that non-phosphorylated α-synuclein carboxyl terminus pulled down protein complexes that were highly enriched for mitochondrial electron transport proteins, whereas α-synuclein carboxyl terminus phosphorylated on either Ser-129 or Tyr-125 did not. Instead the set of proteins pulled down by phosphorylated α-synuclein was highly enriched in certain cytoskeletal proteins, in vesicular trafficking proteins, and in a small number of enzymes involved in protein serine phosphorylation. This targeted comparative proteomics approach for unbiased identification of protein-protein interactions suggests that there are functional consequences when α-synuclein is phosphorylated.
α-Synuclein is a 140-amino acid protein that is highly expressed in the brain and localizes at presynaptic terminals (1). The protein is associated with several neurodegenerative diseases, and mutations and copy number variants in the gene coding for α-synuclein have been linked to familial Parkinson disease (PD)1 (2, 44). The presence of large amounts of α-synuclein in the insoluble aggregates known as Lewy bodies, a pathological hallmark of all PD including sporadic disease, also suggests a connection between the protein and disease.
The protein is predominantly natively unfolded in solution but can bind to phospholipid membranes by adopting an amphipathic helical conformation in its amino-terminal 100 amino acids (3). The remainder of the protein, an acidic, hydrophilic 40-amino acid carboxyl-terminal tail, is thought unlikely to participate directly in membrane binding. A physiological role for vesicle binding by α-synuclein is suggested by the observation that in PC12 and chromaffin cells α-synuclein appears to be a negative regulator of synaptic vesicle exocytosis and neurotransmitter release (4). In addition, loss of α-synuclein in cell culture or in mice results in a significant decrease in the population of presynaptic vesicles in the resting or reserve pool (5–7). Further support for a role of α-synuclein in vesicular trafficking comes from modifier screens in Caenorhabditis elegans showing that the toxicity that arises from expression of α-synuclein may be modified by proteins involved in vesicle trafficking (8, 9).
The carboxyl-terminal portion of α-synuclein can be phosphorylated at tyrosine at position 125 (Tyr-125) and at serine at position 129 (Ser-129) by Src family kinases and various casein kinases, respectively (10–12). α-Synuclein phosphorylated at Ser-129 is the predominant form of α-synuclein in Lewy bodies but constitutes only a very small fraction of soluble α-synuclein in the neuron (13, 14). There is, however, controversy over the extent to which phosphorylation of α-synuclein at Ser-129 is important in the causation of PD. A phosphomimic form of α-synuclein with aspartic acid replacing serine at position 129 (S129D) was shown to enhance the toxicity of α-synuclein in a Drosophila model for PD (15). In contrast, overexpression of S129D α-synuclein using adeno-associated virus injected directly into rat brains appeared relatively non-toxic, whereas the non-phosphorylatable version, with alanine replacing serine 129, was highly neurotoxic (16). Thus, identification of protein-protein interactions of α-synuclein that depend on the phosphorylation state of α-synuclein at Ser-129 or Tyr-125 is likely to shed light on the normal physiological function of α-synuclein phosphorylation as well as identify potential pathways for Lewy body formation and α-synuclein toxicity.
We hypothesized that the hydrophilic tail of α-synuclein constitutes a domain that participates in phosphorylation state-dependent protein-protein interactions intrinsic to the normal function of α-synuclein. Mass spectrometric analysis of protein-protein interactions offers a uniquely unbiased tool for elucidating the components of protein complexes. We decided, therefore, to undertake a mass spectrometric analysis of synaptosomal protein complexes that could be pulled down in vitro with a peptide containing the hydrophilic domain comprising the carboxyl-terminal 40 amino acids of α-synuclein. We used a targeted mass spectrometry-based functional proteomics approach to identify qualitative and relative quantitative differences in protein-protein interactions of phosphorylated versus non-phosphorylated α-synuclein. We showed that a phosphorylated peptide containing the carboxyl-terminal 40 amino acids of α-synuclein interacted preferentially with cytoskeletal proteins, vesicular trafficking proteins involved in endocytosis, and enzymes involved in protein serine phosphorylation, whereas the non-phosphorylated peptide interacted preferentially with mitochondrial electron transport chain complexes. This suggests that phosphorylation likely has a profound effect on the function and/or localization of α-synuclein.
EXPERIMENTAL PROCEDURES
Peptide Pulldowns—
All animal work was done according to protocols reviewed and approved by the NHGRI Animal Care and Use Committee. Mouse brain synaptosomes were isolated as described previously (17) and frozen at −80 °C. For human brain extract fresh normal cortical tissue from human brain was obtained by surgical removal from a single epilepsy patient, embedded in optimum cutting temperature compound, snap frozen in dry ice/isopentane, and stored at −80 °C. The optimum cutting temperature compound was then removed from the tissue at −15 °C in a Leica CM1900 cryostat by surgical dissection, and human synaptosomes were isolated and stored at −80 °C as described previously (17). Protein was quantified using the BCA assay with BSA as a standard (Pierce). The protocol for surgical sample collection (02-N-0014, Research Study of Specimens Obtained during Epilepsy Surgery) was approved by the NINDS Intramural Institutional Review Board. For both preparations, synaptosomes were solubilized in a Triton buffer (∼1.5% Triton, 50 mm Tris-HCl (pH 7.4), 100 mm NaCl, 2 mm EGTA, 50 mm NaF, 0.5 mm sodium vanadate, 1× protease inhibitor mixture (Sigma-Aldrich)) resulting in final concentrations of 1% Triton and ∼5 mg/ml synaptosome protein and centrifuged at 16,000 × g for 10 min at 4 °C, and the supernatant was used for binding experiments.
Bait peptides consisted of amino-terminally tagged biotinylated peptides (Quality Controlled Biochemicals, Hopkinton, MA) (Table I). The peptides used were as follows: the non-phosphorylated carboxyl-terminal 40 amino acids of human α-synuclein (NP); carboxyl-terminal 40-amino acid peptides phosphorylated either at Ser-129 (pS129) or at Tyr-125 (pY125), respectively; and a scrambled control peptide with the same amino acid content as NP but with the residues in random order. Peptides were quantified by absorbance using a Beckman DU640 spectrophotometer (260 nm) and calculated using the molar extinction coefficient of each peptide or by the BCA assay with BSA as a standard (Pierce).
Table I.
Biotinylated peptides used for pulldown experiments
Phosphorylated residues in pS129 and pY125 are shown in bold.
| NP | GKNEEGAPQEGILEDMPVDPDNEAYEMPSEEGYQDYEPEA |
| pS129 | GKNEEGAPQEGILEDMPVDPDNEAYEMPSPO4EEGYQDYEPEA |
| pY125 | GKNEEGAPQEGILEDMPVDPDNEAYPO4EMPSEEGYQDYEPEA |
| Scrambled | AGEKPNEEYEGDAQPYQGEEGEILSEPDMEMPYVAEDPND |
For binding incubations, 400 μl of streptavidin-coated Dynabeads (Dynal, Carlsbad, CA) were incubated at 4 °C overnight in binding/wash buffer (50 mm Tris-HCl (pH 7.4), 100 mm NaCl, 2 mm EGTA, 50 mm NaF, 0.5 mm sodium vanadate, 1× protease inhibitor mixture (Sigma)) with ∼1.5 mg of solubilized mouse brain synaptosome protein and 4 nmol of biotinylated peptide. Final binding conditions contained ∼0.15% Triton and ∼800 mg/ml synaptosome protein. Samples were washed three times in binding/wash buffer, and proteins were eluted in SDS loading buffer (50 mm Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 0.1% bromphenol blue) by heating at 65 °C for 5 min. Binding incubations for each peptide were performed in duplicate and combined prior to elution. The eluted proteins were then heated at 95 °C for 5 min and separated by SDS-PAGE (4–20%) (Invitrogen), and the gel was stained with GelCode Blue (Pierce). All pulldowns and subsequent mass spectrometric analysis were repeated three times from three different pooled lysates.
In-gel Digestion—
Each gel lane was manually excised top to bottom into 40 ∼ 2-mm bands. In-gel tryptic digestion and peptide extraction followed a modified version of a standard protocol (18). Briefly individual gel bands were destained with 50% methanol in 100 mm ammonium bicarbonate, dehydrated with 50% acetonitrile followed by 100% acetonitrile, and dried. Samples were reduced and alkylated with 45 mm DTT at 60 °C followed by 100 mm iodoacetamide at room temperature. Bands were again dehydrated with 50% acetonitrile followed by 100% acetonitrile and then were rehydrated with 200 ng of sequencing grade trypsin (Promega, Madison, WI) in 25 mm ammonium bicarbonate. Digestion was allowed to proceed overnight at room temperature. Peptides were extracted from the gel pieces with 30% acetonitrile, 1% formic acid and sonicated. Extraction was repeated with 70% acetonitrile, 1% formic acid, and supernatants were pooled and dried prior to analysis.
Mass Spectrometry—
For one-dimensional LC-MS/MS samples were resuspended in 5% acetonitrile, 0.1% formic acid and injected into a series LC-VP HPLC system (Shimadzu, Columbia, MD) coupled to an ESI-LCQ Classic ion trap mass spectrometer (Thermo Electron, San Jose, CA). Samples were loaded onto a 75-μm PicoFrit BetaBasic C18 column (New Objectives, Woburn, MA). A linear separation gradient was developed from 10 to 60% B over 45 min (A, 5% ACN, 95% water, 0.1% formic acid; B, 80% ACN, 20% water, 0.1% formic acid). The chromatographic effluent was introduced at a flow rate of 400 nl/min (19). The LCQ was operated in positive ion mode, and spectra were acquired for 60 min in a data-dependent manner with the top three most intense ions in the MS survey scan selected for MS/MS by CID. Precursor ions selected three times were excluded for 60 s. Peak lists were extracted with Thermo Electron Excalibur 2.0 extract_msn utility without smoothing or signal-to-noise thresholding.
Informatics—
The resulting mass spectral peak lists were searched with the Mascot search engine (v.2.1.04; Matrix Sciences, London, UK) against the merged UniProtKB Swiss-Prot and TrEMBL protein sequence library (SPTremb_091706.fas). Search parameters were as follows: Rodentia species, trypsin specificity, one allowed missed cleavage, carbamidomethylation fixed modification, methionine oxidation variable modification, precursor ion mass tolerance of 2.0 Da, and fragment ion mass tolerance of ±0.8 Da.
Pulldown assays were replicated with three different lysates resulting in three gels of four lanes each for MS/MS analysis. The tryptic digests of each of the 40 segments were analyzed by LC/MS/MS, resulting in ∼75,000 MS/MS spectra per lane when the files from all gel segments were concatenated. Only assigned probable peptide sequences with Mascot Ion Scores exceeding their Identity Scores were used throughout this study to produce minimal, parsimonious protein lists concatenated for each full gel lane. All results from a given pulldown experiment are the pooled data from triplicate gel lanes facilitated by the in-house software MassSieve (20) and DBParser 3.0 (21–23). DBParser 3.0 and MassSieve were also used for peptide and protein level parsimony comparisons across multiple experiments. Additionally individual peptides were culled prior to parsimony analysis if they were only observed once across all experiments.
Label-free relative quantification was determined by two methods. In the first method, differentiation was based on the total peptide “hits” or observations per protein (total number of independent spectra assigned by Mascot to peptides from a given protein and with Ion Scores greater than Identity). Each experiment was normalized relative to total peptide hits for a given bait (24, 25). The number of hits per peptide was distilled by MassSieve to generate the integral of the total number of observations of peptides from a given protein (20). Differences in the affinity of a bait peptide complex for a given protein were calculated as the ratio of normalized peptide hits of one α-synuclein form relative to another.
In the second quantification method, an integral of ion current intensities of the peptide parent ion was calculated by DBParser 3.0 from analysis of primary mass spectrometric survey scans. In brief, extracted ion chromatograms were generated from mass spectrometric raw data, and the integral areas were determined for peptides associated with Mascot peptide assignments identified with Ion Scores greater than Identity (21–23). Summed maximum ion intensities for all peptides mapped to respective proteins were normalized to the total assigned ion current for each experiment. For both methods corresponding gel migrations could also be used to validate peptide comparisons.
Enrichment of a particular protein in a pulldown with either the phosphorylated or non-phosphorylated bait peptide was assessed using the ratio of normalized peptide hits or normalized ion currents for the peptides of that protein obtained using either the phosphorylated or the non-phosphorylated peptide. Enrichment was arbitrarily defined as a 2-fold greater number of normalized peptide hits or normalized ion currents for the phosphorylated versus non-phosphorylated bait peptide or vice versa.
Western Analysis—
As an adjunct to mass spectrometric analysis, the gels containing synaptosomal proteins eluted from the washed streptavidin-coated beads were transferred to Hybond-P membrane (Amersham Biosciences), membranes were blocked in 5% nonfat milk, PBS with 0.1% Tween 20, and Western analysis was performed. Antibodies used were: core 1; core 2 complex III; and 39-kDa, 30-kDa (NDUFS3), and 17-kDa (NDUFB6) complex I (mouse monoclonal antibodies from Molecular Probes); adaptin β, clathrin heavy chain, and β-spectrin II (mouse monoclonal antibodies from BD Transduction Laboratories); and VDAC (rabbit polyclonal from Alexis Biochemicals). Proteins were then detected by ECL (Amersham Biosciences) and visualized by autoradiography.
RESULTS
Quantitative Analysis of Proteins Pulled Down with Different α-Synuclein Peptides—
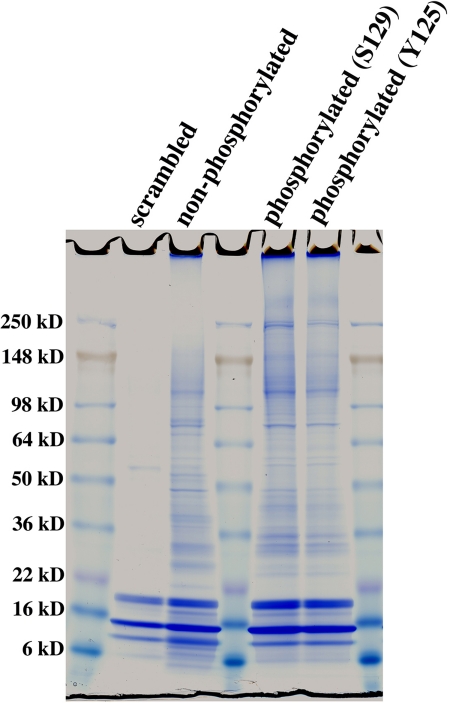
We sought to identify protein networks that interact with the carboxyl-terminal domain of α-synuclein in a phosphorylation-dependent manner. The experimental design was to pull down mouse brain synaptosomal proteins using biotinylated peptides representing the carboxyl-terminal portion (residues 101–140) of α-synuclein. The peptides were non-phosphorylated (NP), phosphorylated on serine 129 (pS129), or phosphorylated on tyrosine 125 (pY125). A biotinylated peptide with the same amino acids as the NP peptide but with the sequence scrambled was used as a control for nonspecific binding. The control lane (Fig. 1, Lane 2) shows very few visible bands other than contaminating streptavidin and BSA products, indicating minimal nonspecific binding. In contrast, many proteins were present in the NP (Fig. 1, Lane 3). The pattern was markedly different from that with the phosphorylated peptides pS129 and pY125 (Fig. 1, Lanes 5 and 6), which suggested a change in interaction proteins and justified pursuing the identification of these proteins.
Fig. 1.
Representative SDS-PAGE gel illustrating proteins binding to the carboxyl terminus of α-synuclein in a sequence- and phosphorylation-dependent manner. Pulldown experiments using biotinylated 40-amino acid peptides that were either scrambled, non-phosphorylated (NP), or phosphorylated on Ser-129 (pS129) or Tyr-125 (pY125) were performed as described under “Experimental Procedures.” Each lane is labeled with the peptide (scrambled, NP, pS129, and pY125) that was bound to the magnetic beads and used for each pulldown. Proteins bound to each peptide were eluted and heated at 95 °C for 5 min in Laemmli sample buffer, separated by SDS-PAGE (4–20%), and visualized by GelCode Blue staining. Lanes 2, 3, 5, and 6 are labeled with the peptide used for the pulldown. Protein markers are in Lanes 1, 4, and 7.
Pulldowns were prepared in three independent replicate experiments. Each group of experimental replicate data was parsed into a single merged experiment and yielded a total number of spectra identified, with a Mascot Ions Score greater than its Identity score, of ∼13,000 for NP, 15,900 pS129 spectra, 13,900 pY125 spectra, and 5300 for the control (supplemental Fig. 1).
Relative protein quantification determined using both peptide hits quantification and ion current intensity is shown in Tables II–V. We applied stringent thresholds to decide on the inclusion of proteins. First, comparisons were made across the three independent experimental replicates to establish reproducibility (supplemental Fig. 2). Only proteins identified in more than one gel and with greater than one peptide observation were included in the analysis; peptides observed only once across all experiments were discarded prior to analysis. Second, comparisons were based on proteins identified by 10 or more peptide observations. Third, to be considered specific, a protein had to have peptide hits greater than three times that of the scrambled control. Finally for affinity to be preferential, we required that there be 2 times the number of peptide hits depending on the phosphorylation status of the peptide. Ion current -fold changes were also calculated as an alternative to peptide hit quantification for confirmation of preferential affinity.
Table II.
Proteins with preferential affinity for the Ser-129 phosphorylated α-synuclein interaction network
To be considered specific a protein had to have peptide hits greater than 3 times that of the control. Comparisons are shown only for proteins identified by 10 or more peptide (Pep) hits. A protein was considered to show preferential enrichment if it had 2 times the number of peptide hits in the pS129 pulldown relative to the NP peptide. Ion current -fold changes are also shown as confirmation of peptide hit quantification. n/a, not applicable; Snare, soluble N-ethylmaleimide-sensitive factor attachment protein receptors; CNPase, 2′,3′-cyclic-nucleotide 3′-phosphodiesterase.
| UniProtKB/Swiss-Prot entry name | Primary accession number | Gene name | Pep hits
|
pS129 affinity
|
Unique peptides
|
Description | |||
|---|---|---|---|---|---|---|---|---|---|
| pS129 | NP | Normalized Pep hits | Normalized ion current | pS129 | NP | ||||
| Signaling | |||||||||
| Q6PJ87_MOUSE | Q6PJ87 | Csnk1a1 | 24 | 0 | n/a | n/a | 7 | 0 | Casein kinase 1, α1 |
| MARK1_MOUSE | Q8VHJ5 | Mark1 | 13 | 0 | n/a | n/a | 4 | 0 | Serine/threonine-protein kinase MARK1 |
| Q5RJI5_MOUSE | Q5RJI5 | Brsk1 | 11 | 0 | n/a | n/a | 2 | 0 | BR serine/threonine kinase 1 |
| KCC2B_MOUSE | P28652 | Camk2b | 185 | 6 | 25.2 | 36.7 | 7 | 3 | CaM kinase II β chain |
| Q6PHZ2_MOUSE | Q6PHZ2 | Camk2d | 79 | 3 | 21.5 | 29.8 | 4 | 1 | CaM kinase II δ chain |
| Q80TN1_MOUSE | Q80TN1 | Camk2a | 168 | 13 | 10.6 | 25.8 | 7 | 4 | MKIAA0968 protein (fragment) |
| MARK2_MOUSE | Q05512 | Mark2 | 46 | 4 | 9.4 | 10 | 1 | Serine/threonine-protein kinase MARK2 | |
| PP1A_MOUSE | P62137 | Ppp1ca | 16 | 2 | 6.5 | 8.5 | 3 | 2 | Serine/threonine-protein phosphatase PP1-α catalytic |
| Cytoskeleton | |||||||||
| Q8C8R3_MOUSE | Q8C8R3 | Ank2 | 73 | 0 | n/a | n/a | 13 | 0 | Similar to Ankyrin-2 |
| Q6PCN2_MOUSE | Q6PCN2 | Ank2 | 48 | 0 | n/a | n/a | 11 | 0 | Ankyrin 2, brain |
| AINX_MOUSE | P46660 | Ina | 40 | 0 | n/a | n/a | 9 | 0 | α-Internexin |
| MYO5A_MOUSE | Q99104 | Myo5a | 28 | 0 | n/a | n/a | 6 | 0 | Myosin-5A |
| Q3UMG4_MOUSE | Q3UMG4 | Ina | 22 | 0 | n/a | n/a | 5 | 0 | Internexin neuronal intermediate filament protein |
| Q3THE2_MOUSE | Q3THE2 | Mylc2b | 22 | 0 | n/a | n/a | 5 | 0 | Myosin regulatory light chain |
| BCAS1_MOUSE | Q80YN3 | Bcas1 | 14 | 0 | n/a | n/a | 1 | 0 | Novel amplified in breast cancer 1 homolog |
| CTNB1_MOUSE | Q02248 | Ctnnb1 | 13 | 0 | n/a | n/a | 6 | 0 | β-Catenin |
| DYL1_MOUSE | P63168 | Dynll1 | 12 | 0 | n/a | n/a | 1 | 0 | Dynein light chain 1 |
| Q3TY37_MOUSE | Q3TY37 | Ctnna2 | 10 | 0 | n/a | n/a | 5 | 0 | Catenin α2 |
| SPTA2_MOUSE | P16546 | Spna2 | 256 | 4 | 52.3 | 298.1 | 33 | 3 | Spectrin α chain, brain |
| MYH10_MOUSE | Q61879 | Myh10 | 48 | 1 | 39.2 | 398.8 | 14 | 1 | Myosin-10 (myosin heavy chain, non-muscle IIb) |
| Q3V1V5_MOUSE | Q3V1V5 | Spna2 | 143 | 3 | 38.9 | 98.8 | 18 | 2 | Spectrin α chain, brain |
| NFL_MOUSE | P08551 | Nefl | 37 | 1 | 30.2 | 217.8 | 9 | 1 | Neurofilament triplet L protein |
| TRIM3_MOUSE | Q9R1R2 | Trim3 | 33 | 1 | 26.9 | 134.4 | 7 | 1 | Tripartite motif-containing protein 3 |
| CAZA2_MOUSE | P47754 | Capza2 | 34 | 2 | 13.9 | 112.3 | 6 | 1 | F-actin capping protein α-2 subunit |
| DCTN2_MOUSE | Q99KJ8 | Dctn2 | 16 | 1 | 13.1 | 47.8 | 5 | 1 | Dynactin subunit 2 |
| MLP3A_MOUSE | Q91VR7 | Map1lc3a | 16 | 1 | 13.1 | 10.8 | 2 | 1 | Microtubule-associated proteins 1A/1B light chain 3A |
| SPTB2_MOUSE | Q62261 | Spnb2 | 341 | 22 | 12.7 | 64.4 | 55 | 15 | Spectrin β chain, brain 1 |
| Q80UE4_MOUSE | Q80UE4 | Epb41l2 | 26 | 2 | 10.6 | 36.7 | 7 | 2 | Protein 4.1G |
| TMOD2_MOUSE | Q9JKK7 | Tmod2 | 11 | 1 | 9.0 | 40.8 | 5 | 1 | Tropomodulin-2 |
| E41L3_MOUSE | Q9WV92 | Epb41l3 | 75 | 8 | 7.7 | 17.3 | 11 | 5 | Band 4.1-like protein 3 |
| ACTB_MOUSE | P60710 | Actb | 182 | 21 | 7.1 | 31.1 | 15 | 5 | Actin, cytoplasmic 1 (β-actin) |
| Q9QZ83_MOUSE | Q9QZ83 | Actg1 | 173 | 21 | 6.7 | 30.5 | 13 | 5 | γ-Actin-like protein |
| Q7TSJ2_MOUSE | Q7TSJ2 | Mtap6 | 62 | 10 | 5.1 | 10.7 | 4 | 2 | Microtubule-associated protein 6 |
| MAP1B_MOUSE | P14873 | Mtap1b | 992 | 165 | 4.9 | 9.2 | 45 | 25 | Microtubule-associated protein 1B |
| TBB6_MOUSE | Q922F4 | Tubb6 | 51 | 9 | 4.6 | 15.3 | 5 | 4 | Tubulin β-6 chain |
| DYL2_MOUSE | Q9D0M5 | Dynll2 | 22 | 4 | 4.5 | 32.3 | 2 | 1 | Dynein light chain 2 |
| P25A_MOUSE | Q7TQD2 | Tppp | 11 | 2 | 4.5 | 8.6 | 2 | 1 | Tubulin polymerization-promoting protein (TPPP) |
| Q80Y54_MOUSE | Q80Y54 | Tubb4 | 100 | 29 | 2.8 | 15.3 | 12 | 10 | Tubulin β-4 |
| TBB5_MOUSE | P99024 | Tubb5 | 123 | 36 | 2.8 | 14.9 | 12 | 10 | Tubulin β-5 chain |
| TBB3_MOUSE | Q9ERD7 | Tubb3 | 99 | 29 | 2.8 | 11.4 | 8 | 7 | Tubulin β-3 chain |
| TBA4_MOUSE | P68368 | Tuba4 | 131 | 43 | 2.5 | 11.6 | 9 | 7 | Tubulin α-4 chain |
| ACTY_MOUSE | Q8R5C5 | Actr1b | 33 | 11 | 2.4 | 2.5 | 6 | 3 | β-Centractin |
| ACTZ_MOUSE | P61164 | Actr1a | 30 | 10 | 2.4 | 2.2 | 6 | 2 | α-Centractin |
| Endocytosis | |||||||||
| AP1B1_MOUSE | O35643 | Ap1b1 | 26 | 0 | n/a | n/a | 6 | 0 | AP-1 complex subunit β-1 |
| AP2M1_MOUSE | P84091 | Ap2m1 | 26 | 1 | 21.2 | 48.2 | 5 | 1 | AP-2 complex subunit μ-1 |
| AP2B1_MOUSE | Q9DBG3 | Ap2b1 | 82 | 8 | 8.4 | 20.3 | 15 | 4 | AP-2 complex subunit β-1 |
| AP2A1_MOUSE | P17426 | Ap2a1 | 89 | 15 | 4.8 | 25.5 | 14 | 4 | AP-2 complex subunit α-1 |
| CLH_MOUSE | Q68FD5 | Cltc | 105 | 25 | 3.4 | 8.5 | 26 | 13 | Clathrin heavy chain |
| 14-3-3 chaperones | |||||||||
| 1433E_MOUSE | P62259 | Ywhae | 24 | 0 | n/a | n/a | 4 | 0 | 14-3-3 protein ε |
| 1433Z_MOUSE | P63101 | Ywhaz | 26 | 1 | 21.2 | 103.8 | 5 | 1 | 14-3-3 protein ζ/δ |
| 1433G_MOUSE | P61982 | Ywhag | 23 | 2 | 9.4 | 28.5 | 4 | 1 | 14-3-3 protein γ |
| Neural adhesion molecule | |||||||||
| NTRI_MOUSE | Q99PJ0 | Hnt | 33 | 2 | 13.5 | 185.3 | 4 | 1 | Neurotrimin precursor |
| CNTN1_MOUSE | P12960 | Cntn1 | 78 | 16 | 4.0 | 13.2 | 18 | 7 | Contactin-1 precursor |
| Glutamate transport membrane | |||||||||
| EAA1_MOUSE | P56564 | Slc1a3 | 132 | 38 | 2.8 | 4.0 | 3 | 2 | Excitatory amino acid transporter 1 |
| Snare complex | |||||||||
| SNIP_MOUSE | Q9QWI6 | P140 | 15 | 5 | 2.4 | 17.4 | 4 | 2 | SNAP-25-interacting protein |
| Mitochondrial proteins | |||||||||
| Q8JZU2_MOUSE | Q8JZU2 | Slc25a1 | 13 | 2 | 5.3 | 4.6 | 3 | 1 | Solute carrier family 25 |
| GHC1_MOUSE | Q9D6M3 | Slc25a22 | 10 | 4 | 2.0 | 5.8 | 5 | 4 | Solute carrier family 25 member 22 |
| LACTB_MOUSE | Q9EP89 | Lactb | 72 | 0 | n/a | n/a | 9 | 0 | Serine β-lactamase-like protein LACTB |
| TFAM_MOUSE | P40630 | Tfam | 15 | 0 | n/a | n/a | 3 | 0 | Transcription factor A, mitochondrial precursor |
| ATPase | |||||||||
| VA0D_MOUSE | P51863 | Atp6v0d1 | 29 | 6 | 3.9 | 2.6 | 6 | 3 | Vacuolar ATP synthase subunit d |
| Q8CHX2_MOUSE | Q8CHX2 | Atp6v1a | 53 | 13 | 3.3 | 3.7 | 10 | 6 | Vacuolar ATP synthase catalytic subunit A |
| G proteins | |||||||||
| GBB1_MOUSE | P62874 | Gnb1 | 67 | 21 | 2.6 | 3.8 | 6 | 5 | Transducin β chain 1 |
| GBB2_MOUSE | P62880 | Gnb2 | 43 | 16 | 2.2 | 3.4 | 6 | 4 | Transducin β chain 2 |
| Ribosomal proteins | |||||||||
| RS7_MOUSE | P62082 | Rps7 | 69 | 26 | 2.2 | 6.6 | 8 | 6 | 40 S ribosomal protein S7 |
| RS10_MOUSE | P63325 | Rps10 | 11 | 4 | 2.2 | 2.2 | 2 | 2 | 40 S ribosomal protein S10 |
| Miscellaneous | |||||||||
| LSAMP_MOUSE | Q8BLK3 | Lsamp | 20 | 0 | n/a | n/a | 5 | 0 | Limbic system-associated membrane protein precursor |
| CALM_MOUSE | P62204 | Calm3 | 16 | 0 | n/a | n/a | 3 | 0 | CaM |
| Q6DFY2_MOUSE | Q6DFY2 | Opcml | 14 | 0 | n/a | n/a | 3 | 0 | Opioid-binding protein/cell adhesion molecule-like |
| ROA2_MOUSE | O88569 | Hnrpa2b1 | 10 | 0 | n/a | n/a | 1 | 0 | Heterogeneous nuclear ribonucleoproteins A2/B1 |
| H2A2B_MOUSE | Q64522 | Hist2h2ab | 11 | 1 | 9.0 | 40.0 | 3 | 1 | Histone H2A type 2-B |
| CN37_MOUSE | P16330 | Cnp1 | 74 | 7 | 8.6 | 12.3 | 7 | 4 | CNPase |
| CSKI1_MOUSE | Q6P9K8 | Caskin1 | 49 | 5 | 8.0 | 13.6 | 7 | 2 | CASK-interacting protein 1 |
| CENG1_MOUSE | Q3UHD9 | Centg1 | 155 | 17 | 7.4 | 21.6 | 14 | 5 | Centaurin-γ 1 |
| USMG5_MOUSE | Q78IK2 | Usmg5 | 10 | 2 | 4.1 | 21.4 | 2 | 1 | Up-regulated during skeletal muscle growth protein 5 |
| THY1_MOUSE | P01831 | Thy1 | 58 | 14 | 3.4 | 7.8 | 5 | 2 | Thy-1 membrane glycoprotein precursor |
| ALDOA_MOUSE | P05064 | Aldoa | 117 | 37 | 2.6 | 4.8 | 8 | 7 | Fructose-bisphosphate aldolase A |
| SYPH_MOUSE | Q62277 | Syp | 11 | 4 | 2.2 | 5.6 | 2 | 1 | Synaptophysin |
Table V.
Proteins with no detectable phosphorylation-dependent affinity
These proteins appeared equally in pulldowns with both forms of α-synuclein. Filter criteria were the same as in Table II. Pep, peptide; Snare, soluble N-ethylmaleimide-sensitive factor attachment protein receptors.
| UniProtKB/Swiss-Prot entry name | Primary accession number | Gene name | Pep hits
|
pS129 affinity
|
Unique peptides
|
Description | |||
|---|---|---|---|---|---|---|---|---|---|
| pS129 | NP | Normalized Pep hits | Normalized ion current | pS129 | NP | ||||
| Microtubule-based vesicle mobility | |||||||||
| MAP2_MOUSE | P20357 | Mtap2 | 152 | 85 | 1.5 | 4.3 | 22 | 14 | Microtubule-associated protein 2 |
| Q3UHB7_MOUSE | Q3UHB7 | Map1a | 723 | 497 | 1.2 | 1.9 | 35 | 33 | Microtubule-associated protein 1 A |
| MAP1A_MOUSE | Q9QYR6 | Map1a | 625 | 437 | 1.2 | 2.2 | 20 | 19 | Microtubule-associated protein 1A |
| Q7TPD4_MOUSE | Q7TPD4 | Mtap4 | 53 | 50 | 0.9 | 1.3 | 6 | 7 | Microtubule-associated protein 4 |
| Q3UIS2_MOUSE | Q3UIS2 | Mtap4 | 115 | 146 | 0.6 | 1.0 | 9 | 11 | Microtubule-associated protein 4 |
| Q8CIL3_MOUSE | Q8CIL3 | Mtap7d1 | 17 | 23 | 0.6 | 0.6 | 2 | 3 | Arginine/proline-rich coiled-coil 1 |
| Mitochondrial carrier membrane proteins | |||||||||
| MPCP_MOUSE | Q8VEM8 | Slc25a3 | 28 | 12 | 1.9 | 1.7 | 7 | 5 | Solute carrier family 25 member 3 |
| MTCH2_MOUSE | Q791V5 | Mtch2 | 15 | 17 | 0.7 | 1.3 | 3 | 3 | Mitochondrial carrier homolog 2 |
| CMC1_MOUSE | Q8BH59 | Slc25a12 | 176 | 206 | 0.7 | 1.4 | 16 | 19 | Solute carrier family 25 member 12 |
| M2OM_MOUSE | Q9CR62 | Slc25a11 | 77 | 95 | 0.7 | 0.5 | 7 | 7 | Solute carrier family 25 member 11 |
| CMC2_MOUSE | Q9QXX4 | Slc25a13 | 10 | 14 | 0.6 | 8.3 | 2 | 3 | Solute carrier family 25 member 13 |
| Heat shock proteins | |||||||||
| GRP75_MOUSE | P38647 | Hspa9a | 27 | 12 | 1.8 | 2.5 | 9 | 6 | GRP 75 (mortalin) |
| GRP78_MOUSE | P20029 | Hspa5 | 20 | 9 | 1.8 | 2.2 | 5 | 1 | GRP 78 (BiP) |
| Mitochondrial membrane proteins | |||||||||
| TOM22_MOUSE | Q9CPQ3 | Tomm22 | 18 | 12 | 1.2 | 0.8 | 4 | 4 | Translocase of outer membrane 22 kDa |
| VDAC1_MOUSE | Q60932 | Vdac1 | 393 | 333 | 1.0 | 0.9 | 17 | 17 | Voltage-dependent anion-selective channel protein 1 |
| VDAC2_MOUSE | Q60930 | Vdac2 | 170 | 269 | 0.5 | 0.8 | 8 | 8 | Voltage-dependent anion-selective channel protein 2 |
| Q5EBQ0_MOUSE | Q5EBQ0 | Vdac3 | 180 | 288 | 0.5 | 0.7 | 8 | 8 | Voltage-dependent anion channel 3 |
| Voltage-gated potassium channel | |||||||||
| KCAB2_MOUSE | P62482 | Kcnab2 | 185 | 142 | 1.1 | 1.6 | 12 | 10 | Voltage-gated potassium channel subunit β-2 |
| KCAB1_MOUSE | P63143 | Kcnab1 | 35 | 28 | 1.0 | 3.3 | 3 | 3 | Voltage-gated potassium channel subunit β-1 |
| ATP synthase | |||||||||
| ATP5H_MOUSE | Q9DCX2 | Atp5h | 32 | 13 | 2.0 | 3.1 | 6 | 4 | ATP synthase D chain |
| ATPG_MOUSE | Q91VR2 | Atp5c1 | 137 | 58 | 1.9 | 2.3 | 4 | 4 | ATP synthase γ chain |
| AT5F1_MOUSE | Q9CQQ7 | Atp5f1 | 27 | 16 | 1.4 | 2.1 | 4 | 3 | ATP synthase B chain |
| ATPase | |||||||||
| VATB2_MOUSE | P62814 | Atp6v1b2 | 43 | 22 | 1.6 | 2.5 | 10 | 9 | Vacuolar proton pump B isoform 2 |
| VPP1_MOUSE | Q9Z1G4 | Atp6v0a1 | 27 | 15 | 1.5 | 2.3 | 9 | 6 | Vacuolar proton translocating ATPase 116-kDa subunit a isoform 1 |
| G proteins | |||||||||
| GNAO1_MOUSE | P18872 | Gnao1 | 55 | 32 | 1.4 | 2.9 | 7 | 4 | Guanine nucleotide-binding protein Go subunit α1 |
| Membrane proteins | |||||||||
| MGST3_MOUSE | Q9CPU4 | Mgst3 | 37 | 35 | 0.9 | 1.1 | 3 | 2 | Microsomal glutathione S-transferase 3 |
| IMMT_MOUSE | Q8CAQ8 | Immt | 103 | 158 | 0.5 | 10 | 12 | Mitochondrial inner membrane protein (mitofilin) | |
| Snare complex | |||||||||
| STX1A_MOUSE | O35526 | Stx1a | 30 | 34 | 0.7 | 1.1 | 6 | 7 | Syntaxin-1A (neuron-specific antigen HPC-1) |
| Glutamate transport, membrane | |||||||||
| EAA2_MOUSE | P43006 | Slc1a2 | 49 | 42 | 1.0 | 0.9 | 7 | 4 | Sodium-dependent glutamate/aspartate transporter 2 |
| Electron transport chain | |||||||||
| NDUA2_MOUSE | Q9CQ75 | Ndufa2 | 8 | 11 | 0.6 | 0.2 | 1 | 1 | NADH-ubiquinone oxidoreductase B8 subunit |
| CX6B1_MOUSE | P56391 | Cox6b1 | 11 | 17 | 0.5 | 0.1 | 3 | 3 | Cytochrome c oxidase subunit VIb isoform 1 |
| Miscellaneous | |||||||||
| CD47_MOUSE | Q61735 | Cd47 | 18 | 11 | 1.3 | 0.8 | 3 | 2 | Integrin-associated protein |
| Q91VC6_MOUSE | Q91VC6 | Glul | 18 | 11 | 1.3 | 3.9 | 4 | 4 | Glutamine synthetase |
| SYT2_MOUSE | P46097 | Syt2 | 45 | 30 | 1.2 | 2.1 | 5 | 3 | Synaptotagmin-2 |
| Q2KHL7_MOUSE | Q2KHL7 | Icam5 | 58 | 47 | 1.0 | 2.2 | 8 | 8 | Intercellular adhesion molecule 5, telencephalin |
| PHB_MOUSE | P67778 | Phb | 36 | 30 | 1.0 | 0.9 | 8 | 8 | Prohibitin |
| CHCH3_MOUSE | Q9CRB9 | Chchd3 | 21 | 25 | 0.7 | 0.4 | 3 | 4 | Coiled-coil-helix-coiled-coil-helix domain-containing protein 3 |
| Q6GQU1_MOUSE | Q6GQU1 | Hk1 | 97 | 131 | 0.6 | 1.0 | 16 | 17 | Hk1 protein |
| PHB2_MOUSE | O35129 | Phb2 | 11 | 15 | 0.6 | 1.4 | 5 | 5 | Prohibitin-2 |
| PDIP3_MOUSE | Q8BG81 | Poldip3 | 6 | 10 | 0.5 | 0.7 | 3 | 3 | Polymerase δ-interacting protein 3 |
There were 85 proteins that showed very significant enrichment (affinity or -fold change) in pulldowns performed with pS129 and pY125 versus NP (Table II). These proteins fell into a number of discrete functional groupings. Thirty-nine of these were cytoskeletal proteins, including microtubule-associated protein 1B (MAP1B) previously reported to interact with the carboxyl terminus of α-synuclein (26). Our new finding is the significant enrichment for interaction with MAP1B when α-synuclein was phosphorylated. We also made the novel observation of significant enrichment for a number of presynaptic cytoskeletal elements, particularly the non-erythrocyte αII and βII spectrins (fodrins) along with the known spectrin-binding proteins ankyrin and band 4.1B (27, 28). We also saw significant enrichment for β- and γ-actin (29, 30), non-muscle myosins, and one species of neurofilament. These proteins are present in the presynaptic portion of neurons and form a dense presynaptic cytoskeletal structure known as the presynaptic particle web (31). A second functional group contained six proteins involved in synaptic vesicle endocytosis (32). In particular, we saw the heavy chain of clathrin and the α-1, β-1, and μ subunits of the adaptor protein complex 2 (AP-2) enriched with pS129 and pY125 versus NP. A third group of nine proteins enriched in pulldowns with phosphorylated α-synuclein peptide were subunits of enzymes involved in serine/threonine phosphorylation and signaling such as three subunits of calmodulin (CaM) kinase II, the CaM kinase family member MARK2, casein kinase 1, and a serine/threonine-protein phosphatase PP1A. Interestingly casein kinase 2, which is considered to be the kinase primarily responsible for Ser-129 phosphorylation of α-synuclein, was not seen (33). Finally three members of the 14-3-3 family of protein chaperones were enriched in pulldowns using phosphorylated peptides. The interaction between 14-3-3 proteins and α-synuclein has been reported previously, although a preference for phosphorylated α-synuclein was not previously known (34).
When we compared the enrichment for proteins pulled down by pS129 and pY125 versus NP, greater enrichment was seen, in nearly every case, with pS129 as compared with the pY125 peptide (Table III). The one exception to this trend was the α subunit of casein kinase 1.
Table III.
Protein interactions with a phosphorylation site preference
Preferential enrichment with structural proteins was usually seen with either Ser-129 or Tyr-125 phosphorylation over the NP, although the affinity for the Ser-129 phosphorylated peptide was generally greater than for peptides phosphorylated at Tyr-125. Exceptions are several kinases that were not enriched in pulldowns with the peptide phosphorylated at Tyr-125 and the first protein in the table, casein kinase 1, α1, for which peptide phosphorylated at Tyr-125 showed greater enrichment. CNPase, 2′,3′-cyclic-nucleotide 3′-phosphodiesterase; Pep, peptide.
| UniProtKB/Swiss-Prot entry name | Primary accession number | Gene name | Pep hits
|
pS129 affinity
|
Unique peptides
|
Description | |||
|---|---|---|---|---|---|---|---|---|---|
| pS129 | pY125 | Normalized Pep hits | Normalized ion current | pS129 | pY125 | ||||
| pY125 affinity > pS129 affinity | |||||||||
| Q6PJ87_MOUSE | Q6PJ87 | Csnk1a1 | 24 | 43 | 0.5 | 0.5 | 7 | 7 | Casein kinase 1, α1 |
| pS129 affinity > pY125 affinity | |||||||||
| Signaling | |||||||||
| Q6PHZ2_MOUSE | Q6PHZ2 | Camk2d | 79 | 24 | 2.9 | 1.8 | 4 | 4 | CaM kinase II δ chain |
| MARK2_MOUSE | Q05512 | Mark2 | 46 | 16 | 2.5 | n/a | 10 | 7 | Serine/threonine-protein kinase MARK2 |
| Q5RJI5_MOUSE | Q5RJI5 | Brsk1 | 11 | 4 | 2.4 | 3.4 | 2 | 2 | BR serine/threonine kinase 1 |
| PP1A_MOUSE | P62137 | Ppp1ca | 16 | 3 | 4.7 | 4.5 | 3 | 2 | Serine/threonine-protein phosphatase PP1-α catalytic |
| Cytoskeletal | |||||||||
| Q3V1V5_MOUSE | Q3V1V5 | Spna2 | 143 | 62 | 2.0 | 1.7 | 18 | 15 | Spectrin α chain, brain |
| MYH10_MOUSE | Q61879 | Myh10 | 48 | 13 | 3.2 | 4.0 | 14 | 8 | Myosin-10 (myosin heavy chain, non-muscle IIb) |
| AINX_MOUSE | P46660 | Ina | 40 | 13 | 2.7 | 4.4 | 9 | 7 | α-Internexin |
| ACTZ_MOUSE | P61164 | Actr1a | 30 | 11 | 2.4 | 5.4 | 6 | 2 | α-Centractin |
| Q3THE2_MOUSE | Q3THE2 | Mylc2b | 22 | 7 | 2.8 | 3.0 | 5 | 3 | Myosin regulatory light chain |
| Q3UMG4_MOUSE | Q3UMG4 | Ina | 22 | 7 | 2.8 | 4.0 | 5 | 3 | Internexin neuronal intermediate filament protein |
| BCAS1_MOUSE | Q80YN3 | Bcas1 | 14 | 1 | 12.3 | 8.8 | 1 | 1 | Novel amplified in breast cancer 1 homolog |
| TRIM3_MOUSE | Q9R1R2 | Trim3 | 33 | 5 | 5.8 | 3.3 | 7 | 4 | Tripartite motif-containing protein 3 |
| Q3TY37_MOUSE | Q3TY37 | Ctnna2 | 10 | 4 | 2.2 | 4.4 | 5 | 3 | Catenin α2 |
| Mitochondria-related | |||||||||
| Q8JZU2_MOUSE | Q8JZU2 | Slc25a1 | 13 | 5 | 2.3 | 2.2 | 3 | 2 | Solute carrier family 25 |
| LACTB_MOUSE | Q9EP89 | Lactb | 72 | 2 | 31.6 | 138.4 | 9 | 1 | Serine β-lactamase-like protein LACTB |
| TFAM_MOUSE | P40630 | Tfam | 15 | 5 | 2.6 | 3.9 | 3 | 2 | Transcription factor A, mitochondrial precursor |
| Miscellaneous | |||||||||
| CN37_MOUSE | P16330 | Cnp1 | 74 | 28 | 2.3 | 5.8 | 7 | 6 | CNPase |
| RS10_MOUSE | P63325 | Rps10 | 11 | 3 | 3.2 | 2.8 | 2 | 2 | 40 S ribosomal protein S10 |
| H2A2B_MOUSE | Q64522 | Hist2h2ab | 11 | 4 | 2.4 | 3.4 | 3 | 2 | Histone H2A type 2-B |
A second class of proteins included those that were preferentially pulled down by NP as compared with pS129 and pY125 (Table IV). The striking finding was that this class of proteins consisted nearly entirely of subunits of the mitochondrial electron transport chain. Of complex I proteins, we observed three of the seven mitochondrially encoded peptides and 28 of 39 nuclearly encoded proteins. We also observed seven of the 11 complex III proteins and eight of 13 complex IV proteins but no complex II proteins. Complexes I, III, and IV, but not complex II, together form a supercomplex, the respirasome (35).
Table IV.
Proteins with preferential affinity for the non-phosphorylated α-synuclein interaction network
Criteria were the same as in Table II. A protein was considered to show preferential affinity if it had 2 times the number of peptide (Pep) hits in the non-phosphorylated pulldown relative to the phosphorylated. n/a, not applicable.
| UniProtKB/Swiss-Prot entry name | Primary accession number | Gene name | Pep hits
|
NP affinity
|
Unique peptides
|
Description | |||
|---|---|---|---|---|---|---|---|---|---|
| NP | pS129 | Normalized Pep hits | Normalized ion current | NP | pS129 | ||||
| Complex I | |||||||||
| NDUAB_MOUSE | Q9D8B4 | Ndufa11 | 28 | 0 | n/a | n/a | 3 | 0 | NADH-ubiquinone oxidoreductase subunit B14.7 |
| NU1M_MOUSE | P03888 | ND1 | 22 | 0 | n/a | n/a | 1 | 0 | NADH-ubiquinone oxidoreductase chain 1 |
| NIPM_MOUSE | Q99LY9 | Ndufs5 | 14 | 0 | n/a | n/a | 2 | 0 | NADH-ubiquinone oxidoreductase 15-kDa subunit |
| NU5M_MOUSE | P03921 | ND5 | 47 | 2 | 28.8 | 22.9 | 3 | 2 | NADH-ubiquinone oxidoreductase chain 5 |
| NUBM_MOUSE | Q91YT0 | Ndufv1 | 164 | 14 | 14.3 | 28.1 | 11 | 5 | NADH-ubiquinone oxidoreductase 51-kDa subunit |
| NDUBB_MOUSE | O09111 | Ndufb11 | 35 | 3 | 14.3 | 12.1 | 5 | 2 | NADH-ubiquinone oxidoreductase ESSS subunit |
| NUYM_MOUSE | Q9CXZ1 | Ndufs4 | 11 | 1 | 13.5 | 10.0 | 3 | 1 | NADH-ubiquinone oxidoreductase 18-kDa subunit |
| NDUB3_MOUSE | Q9CQZ6 | Ndufb3 | 20 | 2 | 12.2 | 4.9 | 2 | 1 | NADH-ubiquinone oxidoreductase B12 subunit |
| NUCM_MOUSE | Q91WD5 | Ndufs2 | 167 | 26 | 7.9 | 14.3 | 14 | 8 | NADH-ubiquinone oxidoreductase 49-kDa subunit |
| NDUB8_MOUSE | Q9D6J5 | Ndufb8 | 12 | 2 | 7.3 | 6.9 | 3 | 1 | NADH-ubiquinone oxidoreductase ASHI subunit |
| NUIM_MOUSE | Q8K3J1 | Ndufs8 | 71 | 12 | 7.2 | 23.8 | 6 | 3 | NADH-ubiquinone oxidoreductase 23-kDa subunit |
| NDUB6_MOUSE | Q3UIU2 | Ndufb6 | 23 | 4 | 7.0 | 8.3 | 2 | 2 | NADH-ubiquinone oxidoreductase B17 subunit |
| NDUB7_MOUSE | Q9CR61 | Ndufb7 | 97 | 17 | 7.0 | 11.5 | 9 | 4 | NADH-ubiquinone oxidoreductase B18 subunit |
| NDUAA_MOUSE | Q99LC3 | Ndufa10 | 74 | 13 | 7.0 | 18.9 | 12 | 6 | NADH-ubiquinone oxidoreductase 42-kDa subunit |
| NUAM_MOUSE | Q91VD9 | Ndufs1 | 491 | 89 | 6.8 | 17.1 | 28 | 15 | NADH-ubiquinone oxidoreductase 75-kDa subunit |
| NUKM_MOUSE | Q9DC70 | Ndufs7 | 69 | 13 | 6.5 | 33.9 | 6 | 3 | NADH-ubiquinone oxidoreductase 20-kDa subunit |
| NU2M_MOUSE | P03893 | Mtnd2 | 15 | 3 | 6.1 | 8.0 | 2 | 1 | NADH-ubiquinone oxidoreductase chain 2 |
| N4BM_MOUSE | Q9CQ54 | Ndufc2 | 18 | 4 | 5.5 | 15.0 | 2 | 1 | NADH-ubiquinone oxidoreductase subunit B14.5b |
| NUHM_MOUSE | Q9D6J6 | Ndufv2 | 95 | 22 | 5.3 | 13.3 | 9 | 7 | NADH-ubiquinone oxidoreductase 24-kDa subunit |
| NDUB9_MOUSE | Q9CQJ8 | Ndufb9 | 73 | 17 | 5.3 | 13.5 | 3 | 2 | NADH-ubiquinone oxidoreductase B22 subunit |
| NDUA6_MOUSE | Q9CQZ5 | Ndufa6 | 29 | 7 | 5.1 | 15.2 | 3 | 1 | NADH-ubiquinone oxidoreductase B14 subunit |
| NDUB5_MOUSE | Q9CQH3 | Ndufb5 | 21 | 6 | 4.3 | 15.0 | 3 | 2 | NADH-ubiquinone oxidoreductase SGDH subunit |
| Q6GTD3_MOUSE | Q6GTD3 | Ndufa9 | 87 | 25 | 4.3 | 7.5 | 9 | 6 | NADH dehydrogenase (ubiquinone) 1α subcomplex, 9 |
| NU4M_MOUSE | P03911 | ND4 | 24 | 7 | 4.2 | 7.8 | 4 | 3 | NADH-ubiquinone oxidoreductase chain 4 |
| NDUBA_MOUSE | Q9DCS9 | Ndufb10 | 131 | 39 | 4.1 | 11.7 | 7 | 6 | NADH-ubiquinone oxidoreductase PDSW subunit |
| NDUA5_MOUSE | Q9CPP6 | Ndufa5 | 17 | 6 | 3.5 | 5.4 | 3 | 2 | NADH-ubiquinone oxidoreductase 13-kDa B subunit |
| NDUAC_MOUSE | Q7TMF3 | Ndufa12 | 16 | 6 | 3.3 | 10.0 | 4 | 2 | NADH-ubiquinone oxidoreductase subunit B17.2 |
| NUGM_MOUSE | Q9DCT2 | Ndufs3 | 113 | 43 | 3.2 | 10.7 | 10 | 7 | NADH-ubiquinone oxidoreductase 30-kDa subunit |
| NDUAD_MOUSE | Q9ERS2 | Ndufa13 | 65 | 26 | 3.1 | 13.8 | 5 | 5 | NADH-ubiquinone oxidoreductase B16.6 subunit |
| NDUA8_MOUSE | Q9DCJ5 | Ndufa8 | 40 | 18 | 2.7 | 10.8 | 5 | 3 | NADH-ubiquinone oxidoreductase 19-kDa subunit |
| NDUB4_MOUSE | Q9CQC7 | Ndufb4 | 21 | 11 | 2.3 | 9.1 | 2 | 2 | NADH-ubiquinone oxidoreductase B15 subunit |
| Complex III | |||||||||
| UCR6_MOUSE | Q9D855 | Uqcrb | 18 | 2 | 11.0 | 30.6 | 3 | 1 | Ubiquinol-cytochrome c reductase complex 14-kDa protein |
| UQCR1_MOUSE | Q9CZ13 | Uqcrc1 | 289 | 62 | 5.7 | 14.5 | 14 | 9 | Ubiquinol-cytochrome c reductase complex core protein I |
| CY1_MOUSE | Q9D0M3 | Cyc1 | 168 | 40 | 5.1 | 14.5 | 7 | 5 | Cytochrome c1, heme protein, mitochondrial precursor |
| UCRI_MOUSE | Q9CR68 | Uqcrfs1 | 118 | 36 | 4.0 | 9.3 | 8 | 5 | Ubiquinol-cytochrome c reductase iron-sulfur subunit |
| UQCR2_MOUSE | Q9DB77 | Uqcrc2 | 215 | 66 | 4.0 | 9.4 | 15 | 13 | Ubiquinol-cytochrome c reductase complex core protein 2 |
| UCR10_MOUSE | Q8R1I1 | Uqcr10 | 19 | 7 | 3.3 | 3.6 | 2 | 1 | Ubiquinol-cytochrome c reductase complex 7.2-kDa protein |
| UCRQ_MOUSE | Q9CQ69 | Uqcrq | 16 | 7 | 2.8 | 8.4 | 1 | 1 | Ubiquinol-cytochrome c reductase complex 9.5-kDa protein |
| Complex IV | |||||||||
| COX6C_MOUSE | Q9CPQ1 | Cox6c | 20 | 2 | 12.2 | 44.6 | 2 | 1 | Cytochrome c oxidase polypeptide Vic |
| Q9MD68_MOUSE | Q9MD68 | mt-Co1 | 19 | 3 | 7.8 | 8.0 | 1 | 1 | Cytochrome oxidase subunit 1 |
| COX7R_MOUSE | Q61387 | Cox7a2l | 15 | 3 | 6.1 | 7.3 | 2 | 1 | Cytochrome c oxidase subunit VIIa-related protein |
| CX7A2_MOUSE | P48771 | Cox7a2 | 57 | 12 | 5.8 | 13.1 | 2 | 2 | Cytochrome c oxidase subunit VIIa-L |
| COX5B_MOUSE | P19536 | Cox5b | 15 | 4 | 4.6 | 12.5 | 1 | 1 | Cytochrome c oxidase polypeptide Vb |
| COX2_MOUSE | P00405 | COX2 | 192 | 73 | 3.2 | 9.3 | 4 | 4 | Cytochrome c oxidase subunit 2 |
| COX5A_MOUSE | P12787 | Cox5a | 28 | 13 | 2.6 | 2.8 | 3 | 2 | Cytochrome c oxidase polypeptide Va |
| CX6A1_MOUSE | P43024 | Cox6a1 | 21 | 11 | 2.3 | 1.6 | 1 | 1 | Cytochrome c oxidase polypeptide Via |
| Mitochondrial membrane proteins | |||||||||
| Q3U5Y8_MOUSE | Q3U5Y8 | Fam82c | 18 | 0 | n/a | n/a | 4 | 0 | Protein FAM82C |
| GLPK_MOUSE | Q64516 | Gyk | 10 | 2 | 6.1 | 4.0 | 2 | 1 | Glycerol kinase |
| Synaptic vesicle proteins | |||||||||
| VAPA_MOUSE | Q9WV55 | Vapa | 81 | 17 | 5.8 | 21.2 | 6 | 3 | Vesicle-associated membrane protein-associated protein A |
| VAPB_MOUSE | Q9QY76 | Vapb | 49 | 11 | 5.5 | 13.5 | 3 | 3 | Vesicle-associated membrane protein-associated protein B |
| Mitochondrial transport membrane proteins | |||||||||
| Q3TWD3_MOUSE | Q3TWD3 | Samm50 | 25 | 8 | 3.8 | 3.0 | 10 | 6 | SAM50-like protein CGI-51 homolog |
| Q3TBZ2_MOUSE | Q3TBZ2 | Tomm40 | 16 | 8 | 2.4 | 1.2 | 4 | 3 | Translocase of outer mitochondrial membrane 40 homolog |
| Miscellaneous | |||||||||
| STXB1_MOUSE | O08599 | Stxbp1 | 111 | 50 | 2.7 | 5.3 | 17 | 14 | Syntaxin-binding protein 1 (Unc-18-1) |
A third major class were proteins pulled down specifically by the carboxyl-terminal portion of α-synuclein as compared with the scrambled control peptide, but the affinity was independent of the phosphorylation status of the bait peptide (Table V). The proteins in this group included microtubule-associated proteins other than MAP1B, i.e. MAP1A, MAP2, and MAP4, as well as outer mitochondrial membrane transporters from the solute carrier 25 family and from the porin family of voltage-dependent anion channels. Although no enrichment was seen here, peptides that showed no quantitative differences may require a more targeted approach, such as multiple reaction monitoring analysis, to provide increased accuracy.
Western Blot Confirmation of Quantitative Mass Spectroscopy—
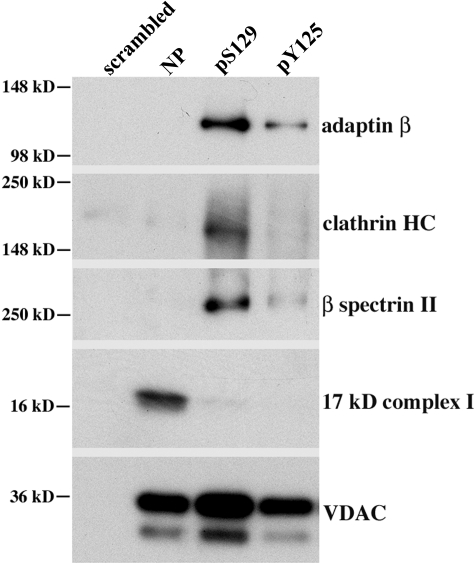
We used Western blotting of the mixture of proteins pulled down by phosphorylated and non-phosphorylated peptides as an independent method of assessing the validity of the mass spectrometry results. We chose a subset of five proteins for which robust antibodies were available for Western analysis of brain proteins pulled down by the various peptides. As shown in Fig. 2, we confirmed that all five of the identified proteins showed a similar pattern of enrichment in complexes pulled down by pS129 and pY125 peptides or the NP peptide as we found with the mass spectrometric analysis.
Fig. 2.
Composite of Western analyses confirming phosphorylation-dependent binding of selected mouse proteins. Pulldown experiments were performed as in Fig. 1. Each lane is labeled with the peptide (scrambled, NP, pS129, and pY125) that was bound to the magnetic beads and used for each pulldown. Eluted proteins bound to each peptide were separated by SDS-PAGE (4–20%), and Western analysis was performed as described under “Experimental Procedures” with various antibodies as indicated to the right of the Western blots. HC, heavy chain.
Physiological Relevance for Human Brain—
Because these studies were all carried out using human α-synuclein peptides to pull down mouse synaptosomal proteins, we wanted to make sure that the marked differences in affinity of synaptosomal proteins based on the phosphorylation state of the bait peptide was conserved with human synaptosomal proteins. We carried out a Western blot analysis of proteins pulled down by the bait peptides using human brain extracts and found very similar findings with human and mouse brain extracts (Fig. 3).
Fig. 3.
Composite of Western analyses confirming phosphorylation-dependent binding of selected human proteins. Human brain proteins bind the carboxyl terminus of α-synuclein in a sequence- and phosphorylation-dependent manner. A composite of Western blots of total human synaptosomal protein pulled down by magnetic beads is shown. Each lane is labeled with the peptide bound to the magnetic beads that was used for each pulldown (scrambled, NP, and pS129). Pulldown experiments were performed as in Fig. 1. Eluted proteins bound to each peptide were separated by SDS-PAGE (4–20%), and Western analysis was performed with various antibodies as indicated to the right of the Western blots as described under “Experimental Procedures.” HC, heavy chain.
Finally we asked whether the protein interactions seen with the carboxyl-terminal domain of α-synuclein would be replicated when the domain was part of the full-length protein. Full-length α-synuclein at 3-fold molar excess to NP was able to partially compete the pulldown of three different electron transport chain proteins as well as the mitochondrial VDAC; 10-fold molar excess completely blocked the pulldown (Fig. 4). Because recombinant α-synuclein containing amino acids 1–124 (lacking the last 15 amino acids) failed to compete even when present at 10-fold molar excess, the competition seen with the full-length protein was not a nonspecific effect of adding an excess of recombinant protein or peptide. Although we do not know the minimum extent of the interacting domain, interruption between residues 124 and 125 destroyed the interaction because neither the truncated protein containing residues 1–124 nor a peptide containing the terminal 16 amino acids, including the Ser-129 residue, was able to compete (supplemental Fig. 3). Finally full-length mouse α-synuclein and full-length human β-synuclein were able to compete with biotinylated NP peptide in the pulldown, whereas human γ-synuclein, another α-synuclein paralog, was not (supplemental Fig. 3). Mouse α-synuclein and human β-synuclein are highly homologous to human α-synuclein at their carboxyl termini and undergo phosphorylation on serine (36, 37). In contrast, γ-synuclein has very little homology to α-synuclein at its carboxyl terminus, and, therefore, it is not surprising that it was not able to compete with the NP peptide in our pulldown experiments (37).
Fig. 4.
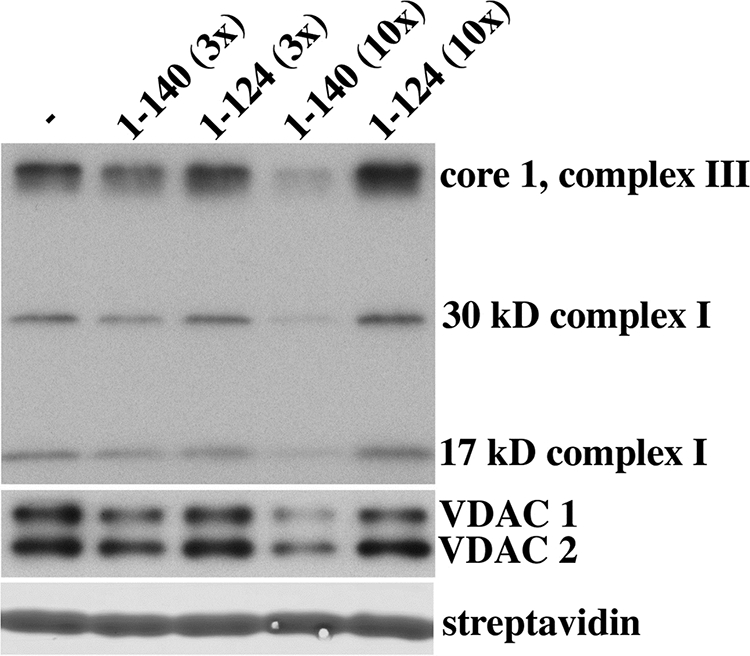
Competition with the NP peptide pulldown by full-length recombinant human α-synuclein. Pulldown experiments using biotinylated NP peptide were performed in the presence or absence of a 3× or 10× molar excess of recombinant full-length human α-synuclein (residues 1–140) or a control protein consisting of truncated α-synuclein lacking the terminal 16 amino acids (residues 1–124). Eluted proteins bound to the biotinylated peptide were separated by SDS-PAGE (4–20%), and Western analysis was performed using specific antibodies. The eluted protein mixture was stained with GelCode Blue to identify the streptavidin, which served as a gel loading control.
DISCUSSION
Our targeted functional proteomics approach provides a broad and unbiased look at the differences in protein networks associated with phosphorylation of α-synuclein. Label-free relative quantification was determined by two methods: the number of independent assignments or hits of a given peptide (more sensitive to minor components) and the ion current intensity (more accurate for major components). Both quantification methods are widely used for initial profiling in high throughput comparative proteomics studies (38, 39). Tables II–IV provide strong evidence to support a change in the role of α-synuclein upon phosphorylation.
Phosphorylated α-Synuclein Affinity—
A striking and biologically intriguing observation is the enrichment for cytoskeletal proteins seen with phosphorylated versus non-phosphorylated α-synuclein carboxyl-terminal peptide. Structural proteins are frequent contaminants in pulldown assays and proteomics studies. However, we used very strict cutoffs for inclusion and for consideration as relevant preferential interactions. Many more structural proteins did not make our threshold for inclusion and can be found in the supplemental material. Thus, the proteins shown in Tables II–V are shown with confidence.
The interaction of MAP1B with the carboxyl-terminal 45 amino acids of α-synuclein has been observed previously, although an increased affinity with phosphorylation was not previously recognized (26). A striking and novel observation in the pulldown using phosphorylated peptide was the prominence of non-erythrocyte αII and βII spectrins (fodrins) and the spectrin-interacting proteins ankyrin and band 4.1B (27, 28). In addition, we saw two isoforms of cytoplasmic actin, β- and γ-actin (29, 30); non-muscle myosins; and one species of neurofilament. These proteins are present in the presynaptic portion of neurons and form what has been referred to as the presynaptic particle web involved in stabilization of the synapse (31, 40). This result suggests that phosphorylation could promote tethering of α-synuclein to the synaptic cytoskeleton, thereby also holding synaptic vesicles bound to the amino-terminal amphipathic helix of α-synuclein in place as well.
In Lewy bodies, α-synuclein is predominantly phosphorylated at Ser-129 (7). Lewy bodies have been shown to contain cytoskeletal proteins, including MAP1B, spectrin, cytoplasmic actin, neurofilament L, and tau (41). The data presented here suggest that the pathway to the production of Lewy bodies involves the interaction between phosphorylated α-synuclein and cytoskeletal elements.
Another interesting group of proteins interacting with phosphorylated α-synuclein are enzymes and signaling proteins involved in serine/threonine phosphorylation. Previous work has indicated that casein kinase 2 is the major kinase responsible for phosphorylation of α-synuclein at Ser-129 with much less activity demonstrated by casein kinase 1 and CaM kinase (12, 33). However, the increased affinity we saw for casein kinase 1, CaM kinase, and MARK2 with the phosphorylated peptide suggests either that these kinases are recognizing the product of phosphorylation rather than substrate or that they are not direct interactors with α-synuclein but are part of a complex that recognizes phosphorylated α-synuclein preferentially. It is worth noting, however, that the affinity of casein kinase 1 for phosphorylated α-synuclein is greater when the phosphorylation is at Tyr-125 rather than at Ser-129, raising the interesting possibility that Tyr-125 phosphorylation may be involved in a cooperative manner with serine phosphorylation at Ser-129. The tyrosine at position 125 can be phosphorylated by Src family kinases in cell culture, but we lack conclusive evidence that physiological phosphorylation at that site occurs in the brain (2, 3).
Clathrin heavy chain and subunits of AP-2 and AP-1 adapter complexes involved in clathrin-mediated endocytosis are also enriched among the proteins preferentially pulled down by phosphorylated α-synuclein over non-phosphorylated α-synuclein. AP-2 interacts with clathrin and is involved in endocytosis of synaptic vesicles destined to enter the recycling pool in the presynaptic region (42). If the interaction between the clathrin·AP-2 complex and α-synuclein is direct, this finding is of interest because recent data show that α-synuclein may be required for the genesis and/or maintenance of the “reserve” or “resting” pools of presynaptic vesicles (5–7). Proteins involved in vesicle trafficking were also identified as modifiers of α-synuclein toxicity in screens of C. elegans (8, 43). Thus, phosphorylation of α-synuclein may be important for its involvement in synaptic vesicle endocytosis.
There remains a long list of proteins showing a substantial preference for phosphorylated α-synuclein such as limbic system-associated membrane protein precursor, serine β-lactamase-like protein, and opioid-binding protein/cell adhesion molecule-like. Although it is impossible to group all of these proteins into a coherent interaction network, our hope is that many of these proteins will serve as impetus for further experiments in the field.
Non-phosphorylated α-Synuclein Affinity—
The pulldown using the NP peptide was enriched for a very different set of proteins than was found with either the pS129 or pY125 peptides (Table IV). The most striking finding is that the NP peptide pulled down a large number of subunits of electron transport chain proteins with much greater affinity than did either of the phosphorylated peptides. Table IV includes 31 of the 46 subunits of complex I of the electron transport chain as well as many complex III and complex IV proteins but not complex II proteins. Complex I is an L-shaped multimer with one hydrophobic arm that is inside the inner mitochondrial membrane and a more hydrophilic arm that is outside the membrane (45, 46). The 31 subunits of complex I listed in Table IV are not from any known precursor subcomplex of complex I and include proteins that seed complex I assembly, such as the mitochondrially encoded proteins, those that enter the complex somewhat later but prior to incorporation of the B17.2L chaperone, and proteins that come into the complex late and are located in the hydrophilic arm, such as NDUFS4 and NDUFS6 (45). A previous proteomics analysis of purified complex I showed that many of the complex III and IV proteins identified here co-purify as contaminants (47), reflecting the existence of a supercomplex consisting of complexes I, III, and IV referred to as the respirasome (35). Thus, we propose that the enrichment for electron transport chain proteins pulled down with the non-phosphorylated carboxyl terminus of α-synuclein is consistent with a preferential interaction of α-synuclein with one or more components of the respirasome complex.
There are a number of limitations to the study reported here. The first is that some of the proteins pulled down in a phosphorylation-dependent manner are likely false positives. Some are likely technical false positives due to nonspecific interactions with biotinylated peptide and the streptavidin beads. Based on the results with scrambled peptide, we believe this class of false positives is rather small. A larger class of false positives is made up of biological false positives. These proteins are likely to be in protein complexes in which only a small number of proteins are actually interacting, whereas the rest are pulled down through their association in the complex. Identifying which proteins are directly interacting will require substantial additional experiments to sort through these lists and test them for direct interaction. We have initiated such experiments using the yeast two-hybrid system to test for direct interactions and have already found domains in some of the cytoskeletal proteins in Table II that can interact directly with the carboxyl-terminal portion of α-synuclein.2 A second limitation is that the experiments were performed with only a portion of α-synuclein. Although the competition experiment demonstrated that the COOH terminus when part of the full-length protein was able to specifically compete with a representative sample of the interactions, it still remains true that these experiments do not capture all the changes in conformation and subsequent interactions of the full-length protein conferred by phosphorylation. Finally the results described here are all derived from in vitro experiments, and the biological significance of many of these interactions remains an open question. Additional studies need to be performed in relevant living cells, such as neurons, in which the α-synuclein is either constitutively phosphorylated or is unphosphorylatable. The results reported here are only the first steps that should prove useful for generating novel hypotheses to be tested in more biologically complex and authentic systems.
There are a number of published proteomics studies describing interactions of α-synuclein with cellular proteins. These studies have primarily involved treating cells or whole animals with mitochondrial poisons, such as rotenone or 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine, and then performing a quantitative proteomics analysis of the changes in the identity and quantity of proteins found to interact with α-synuclein on an affinity column (48–50). To the best of our knowledge, ours is the first proteomics study to evaluate phosphorylation-dependent α-synuclein interactions. Fewer than a dozen proteins were found to overlap between these published studies and the research presented here. This may not be quite so surprising given that the previously published studies were designed to address different questions than the question being asked here. The published studies were looking for changes in proteins that interact with α-synuclein after toxic stress, whereas this study was designed to identify proteins with differential binding affinity for phosphorylated versus non-phosphorylated α-synuclein. None of the studies on proteins interacting with α-synuclein after exposure to toxins addressed the question of the phosphorylation status of α-synuclein in the treated versus untreated samples of cells or brain tissues. If there were little in the way of significant changes in the phosphorylation status of α-synuclein following these treatments, then we would not expect that the changes seen in those studies should overlap substantially with our data. If the bulk of α-synuclein remained non-phosphorylated after treatment with these toxins, an interaction with mitochondrial proteins would be missed because mitochondria were specifically excluded in two of these studies, whereas in the third there was substantial mitochondrial damage and cell loss.
Complex I dysfunction has long been hypothesized as an important component of the pathogenesis of PD (51–54). Li et al. (55) proposed that α-synuclein co-purifies with a mitochondrial fraction from brain and reported an immunogold electron micrograph showing a small number of gold particles marking the outer mitochondrial membrane using antibody against α-synuclein. In published results, we also demonstrated that α-synuclein, when expressed at low to moderate levels in stably transfected neuroblastoma cells, could translocate onto mitochondrial membranes following oxidative stress or reduced intracellular pH (56). This membrane interaction was likely through binding to cardiolipin, an acidic phospholipid for which the lipid binding amino-terminal portion of α-synuclein has high affinity, and no evidence was found for α-synuclein actually entering the mitochondria. The electron transport chain complexes are located within the inner mitochondrial membrane, however, and it is therefore puzzling to find a predominance of electron transport chain proteins pulled down by non-phosphorylated α-synuclein in the current study.
In contrast to our results (56), Devi et al. (57) and Parihar et al. (58) demonstrated that in cells that overexpressed α-synuclein the protein can enter mitochondria and interfere with mitochondrial function. Devi et al. (57) and Li et al. (55) went on to propose that the large number of lysines present in the repeated motifs in the amino terminus of the protein can serve as a cryptic mitochondrial targeting sequence that allows α-synuclein to enter mitochondria. One can hypothesize that with high levels of expression of non-phosphorylated α-synuclein the protein is untethered to cytoskeletal elements and is free to enter mitochondria where it can interfere with complex I of the electron transport chain. If perhaps the capacity to phosphorylate α-synuclein is limiting, overexpression of the protein or mutations that prolong its half-life would increase the levels of non-phosphorylated protein disproportionately and, therefore, cause more mitochondrial inhibition. These results are consistent with the results of Gorbatyuk et al. (16) and support a model that places non-phosphorylated α-synuclein in the pathway leading to mitochondrial dysfunction and the development of PD. The enzymes involved in phosphorylating and dephosphorylating α-synuclein might, therefore, be potential therapeutic targets in PD.
Acknowledgments
We thank Douglas Slotta, Sara Yang, and Anthony J. Makusky for help with data processing; Jeffrey Kowalak and Joanne Connolly for preliminary observations on this project; and Dr. Yien-Ming Kuo and Dr. Valerie Drews for help in preparing and critically reading the manuscript. We thank Nelson Cole for providing some recombinant synucleins and helping us to make others.
Footnotes
Published, MCP Papers in Press, July 9, 2008, DOI 10.1074/mcp.M800116-MCP200
The abbreviations used are: PD, Parkinson disease; NP, peptide consisting of carboxyl-terminal 40 amino acids of α-synuclein; pS129, peptide consisting of carboxyl-terminal 40 amino acids of α-synuclein that is phosphorylated at serine position 129; pY125, peptide consisting of carboxyl-terminal 40 amino acids of α-synuclein that is phosphorylated at tyrosine position 125; VDAC, voltage-dependent anion-selective channel; MAP, microtubule-associated protein; AP-2, adaptor protein complex 2; CaM, calmodulin.
V. Drews and R. L. Nussbaum, unpublished data.
This work was supported, in whole or in part, by National Institutes of Health Grant Z01 MH000279 from the intramural programs of the National Institute of Mental Health and by the NHGRI. This work was also supported by the Sandler Family Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.George, J. M. ( 2002) The synucleins. Genome Biol. 3, REVIEWS3002 [DOI] [PMC free article] [PubMed]
- 2.Beal, M. F. ( 2001) Experimental models of Parkinson's disease. Nat. Rev. Neurosci. 2, 325–334 [DOI] [PubMed] [Google Scholar]
- 3.Bennett, M. C. ( 2005) The role of α-synuclein in neurodegenerative diseases. Pharmacol. Ther. 105, 311–331 [DOI] [PubMed] [Google Scholar]
- 4.Larsen, K. E., Schmitz, Y., Troyer, M. D., Mosharov, E., Dietrich, P., Quazi, A. Z., Savalle, M., Nemani, V., Chaudhry, F. A., Edwards, R. H., Stefanis, L., and Sulzer, D. ( 2006) α-Synuclein overexpression in PC12 and chromaffin cells impairs catecholamine release by interfering with a late step in exocytosis. J. Neurosci. 26, 11915–11922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy, D. D., Rueter, S. M., Trojanowski, J. Q., and Lee, V. M. ( 2000) Synucleins are developmentally expressed, and α-synuclein regulates the size of the presynaptic vesicular pool in primary hippocampal neurons. J. Neurosci. 20, 3214–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabin, D. E., Shimazu, K., Murphy, D., Cole, N. B., Gottschalk, W., McIlwain, K. L., Orrison, B., Chen, A., Ellis, C. E., Paylor, R., Lu, B., and Nussbaum, R. L. ( 2002) Synaptic vesicle depletion correlates with attenuated synaptic responses to prolonged repetitive stimulation in mice lacking α-synuclein. J. Neurosci. 22, 8797–8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gureviciene, I., Gurevicius, K., and Tanila, H. ( 2007) Role of α-synuclein in synaptic glutamate release. Neurobiol. Dis. 28, 83–89 [DOI] [PubMed] [Google Scholar]
- 8.Cooper, A. A., Gitler, A. D., Cashikar, A., Haynes, C. M., Hill, K. J., Bhullar, B., Liu, K., Xu, K., Strathearn, K. E., Liu, F., Cao, S., Caldwell, K. A., Caldwell, G. A., Marsischky, G., Kolodner, R. D., Labaer, J., Rochet, J. C., Bonini, N. M., and Lindquist, S. ( 2006) α-Synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science 313, 324–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuwahara, T., Koyama, A., Koyama, S., Yoshina, S., Ren, C. H., Kato, T., Mitani, S., and Iwatsubo, T. ( 2008) A systematic RNAi screen reveals involvement of endocytic pathway in neuronal dysfunction in α-synuclein transgenic C. elegans. Hum. Mol. Genet., in press [DOI] [PubMed]
- 10.Ellis, C. E., Schwartzberg, P. L., Grider, T. L., Fink, D. W., and Nussbaum, R. L. ( 2001) α-Synuclein is phosphorylated by members of the Src family of protein-tyrosine kinases. J. Biol. Chem. 276, 3879–3884 [DOI] [PubMed] [Google Scholar]
- 11.Nakamura, T., Yamashita, H., Takahashi, T., and Nakamura, S. ( 2001) Activated Fyn phosphorylates α-synuclein at tyrosine residue 125. Biochem. Biophys. Res. Commun. 280, 1085–1092 [DOI] [PubMed] [Google Scholar]
- 12.Okochi, M., Walter, J., Koyama, A., Nakajo, S., Baba, M., Iwatsubo, T., Meijer, L., Kahle, P. J., and Haass, C. ( 2000) Constitutive phosphorylation of the Parkinson's disease associated α-synuclein. J. Biol. Chem. 275, 390–397 [DOI] [PubMed] [Google Scholar]
- 13.Anderson, J. P., Walker, D. E., Goldstein, J. M., de Laat, R., Banducci, K., Caccavello, R. J., Barbour, R., Huang, J. P., Kling, K., Lee, M., Diep, L., Keim, P. S., Shen, X. F., Chataway, T., Schlossmacher, M. G., Seubert, P., Schenk, D., Sinha, S., Gai, W. P., and Chilcote, T. J. ( 2006) Phosphorylation of Ser-129 is the dominant pathological modification of α-synuclein in familial and sporadic Lewy body disease. J. Biol. Chem. 281, 29739–29752 [DOI] [PubMed] [Google Scholar]
- 14.Fujiwara, H., Hasegawa, M., Dohmae, N., Kawashima, A., Masliah, E., Goldberg, M. S., Shen, J., Takio, K., and Iwatsubo, T. ( 2002) α-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 4, 160–164 [DOI] [PubMed] [Google Scholar]
- 15.Mattei, M. G., Mattei, J. F., Vidal, I., and Giraud, F. ( 1981) Expression in lymphocyte and fibroblast culture of the fragile-X chromosome: a new technical approach. Hum. Genet. 59, 166–169 [DOI] [PubMed] [Google Scholar]
- 16.Gorbatyuk, O. S., Li, S., Sullivan, L. F., Chen, W., Kondrikova, G., Manfredsson, F. P., Mandel, R. J., and Muzyczka, N. ( 2008) The phosphorylation state of Ser-129 in human α-synuclein determines neurodegeneration in a rat model of Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 105, 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellis, C. E., Murphy, E. J., Mitchell, D. C., Golovko, M. Y., Scaglia, F., Barcelo-Coblijn, G. C., and Nussbaum, R. L. ( 2005) Mitochondrial lipid abnormality and electron transport chain impairment in mice lacking α-synuclein. Mol. Cell. Biol. 25, 10190–10201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. ( 1996) Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 19.Masuda, J., Maynard, D. A., Nishimura, M., Uedac, T., Kowalak, J. A., and Markey, S. P. ( 2005) Fully automated micro- and nanoscale one- or two-dimensional high-performance liquid chromatography system for liquid chromatography-mass spectrometry compatible with non-volatile salts for ion exchange chromatography. J. Chromatogr. A 1063, 57–69 [DOI] [PubMed] [Google Scholar]
- 20.Slotta, D. J., McFarland, M. A., Makusky, S. J., and Markey, S. P. ( 2007) in the 55th ASMS Conference on Mass Spectrometry and Allied Topics, Indianapolis, June 3–7, 2007, Abstract 157, American Society for Mass Spectrometry, Santa Fe, NM
- 21.Yang, X. Y., Dondeti, V., Dezube, R., Maynard, D. M., Geer, L. Y., Epstein, J., Chen, X. F., Markey, S. P., and Kowalak, J. A. ( 2004) DBParser: Web-based software for shotgun proteomic data analyses. J. Proteome Res. 3, 1002–1008 [DOI] [PubMed] [Google Scholar]
- 22.Dosemeci, A., Makusky, A. J., Jankowska-Stephens, E., Yang, X., Slotta, D. J., and Markey, S. P. ( 2007) Composition of the synaptic PSD-95 complex. Mol. Cell. Proteomics 6, 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McFarland, M. A., Yang, X., Geer, L. Y., Makusky, A. J., Kowalak, J. A., Nussbaum, R. L., and Markey, S. P. ( 2006) in the 54th ASMS Conference on Mass Spectrometry and Allied Topics, Seattle, May 28–June 1, 2006, Abstract 584, American Society for Mass Spectrometry, Santa Fe, NM
- 24.Florens, L., Carozza, M. J., Swanson, S. K., Fournier, M., Coleman, M. K., Workman, J. L., and Washburn, M. P. ( 2006) Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paoletti, A. C., Parmely, T. J., Tomomori-Sato, C., Sato, S., Zhu, D. X., Conaway, R. C., Conaway, J. W., Florens, L., and Washburn, M. P. ( 2006) Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl. Acad. Sci. U. S. A. 103, 18928–18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen, P. H., Islam, K., Kenney, J., Nielsen, M. S., Power, J., and Gai, W. P. ( 2000) Microtubule-associated protein 1B is a component of cortical Lewy bodies and binds α-synuclein filaments. J. Biol. Chem. 275, 21500–21507 [DOI] [PubMed] [Google Scholar]
- 27.Bennett, V., Baines, A. J., and Davis, J. Q. ( 1985) Ankyrin and synapsin: spectrin-binding proteins associated with brain membranes. J. Cell. Biochem. 29, 157–169 [DOI] [PubMed] [Google Scholar]
- 28.Bennett, V., Baines, A. J., and Davis, J. ( 1986) Purification of brain analogs of red blood cell membrane skeletal proteins: ankyrin, protein 4.1 (synapsin), spectrin, and spectrin subunits. Methods Enzymol. 134, 55–69 [DOI] [PubMed] [Google Scholar]
- 29.Vandekerckhove, J., and Weber, K. ( 1978) Mammalian cytoplasmic actins are the products of at least two genes and differ in primary structure in at least 25 identified positions from skeletal muscle actins. Proc. Natl. Acad. Sci. U. S. A. 75, 1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrels, J. I., and Gibson, W. ( 1976) Identification and characterization of multiple forms of actin. Cell 9, 793–805 [DOI] [PubMed] [Google Scholar]
- 31.Phillips, G. R., Huang, J. K., Wang, Y., Tanaka, H., Shapiro, L., Zhang, W., Shan, W. S., Arndt, K., Frank, M., Gordon, R. E., Gawinowicz, M. A., Zhao, Y., and Colman, D. R. ( 2001) The presynaptic particle web: ultrastructure, composition, dissolution, and reconstitution. Neuron 32, 63–77 [DOI] [PubMed] [Google Scholar]
- 32.Voglmaier, S. M., and Edwards, R. H. ( 2007) Do different endocytic pathways make different synaptic vesicles? Curr. Opin. Neurobiol. 17, 374–380 [DOI] [PubMed] [Google Scholar]
- 33.Ishii, A., Nonaka, T., Taniguchi, S., Saito, T., Arai, T., Mann, D., Iwatsubo, T., Hisanaga, S., Goedert, M., and Hasegawa, M. ( 2007) Casein kinase 2 is the major enzyme in brain that phosphorylates Ser129 of human α-synuclein: implication for α-synucleinopathies. FEBS Lett. 581, 4711–4717 [DOI] [PubMed] [Google Scholar]
- 34.Ostrerova, N., Petrucelli, L., Farrer, M., Mehta, N., Choi, P., Hardy, J., and Wolozin, B. ( 1999) α-Synuclein shares physical and functional homology with 14-3-3 proteins. J. Neurosci. 19, 5782–5791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schagger, H., de Coo, R., Bauer, M. F., Hofmann, S., Godinot, C., and Brandt, U. ( 2004) Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J. Biol. Chem. 279, 36349–36353 [DOI] [PubMed] [Google Scholar]
- 36.Nakajo, S., Tsukada, K., Omata, K., Nakamura, Y., and Nakaya, K. ( 1993) A new brain-specific 14-kDa protein is a phosphoprotein. Its complete amino acid sequence and evidence for phosphorylation. Eur. J. Biochem. 217, 1057–1063 [DOI] [PubMed] [Google Scholar]
- 37.Lavedan, C. ( 1998) The synuclein family. Genome Res. 8, 871–880 [DOI] [PubMed] [Google Scholar]
- 38.Mueller, L., Brusniak, M., Mani, D., and Aebersold, R. ( 2008) An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J. Proteome Res. 1, 51–61 [DOI] [PubMed] [Google Scholar]
- 39.Ong, S. E., and Mann, M. ( 2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 40.Pielage, J., Fetter, R. D., and Davis, G. W. ( 2005) Presynaptic spectrin is essential for synapse stabilization. Curr. Biol. 15, 918–928 [DOI] [PubMed] [Google Scholar]
- 41.Leverenz, J. B., Umar, I., Wang, Q., Montine, T. J., McMillan, P. J., Tsuang, D. W., Jin, J. H., Pan, C., Shin, J., Zhu, D., and Zhang, J. ( 2007) Proteomic identification of novel proteins in cortical Lewy bodies. Brain Pathol. 17, 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knoll, J. H., Chudley, A. E., and Gerrard, J. W. ( 1984) Fragile-X X-linked mental retardation II. Frequency and replication pattern of fragile (X)(q28) in heterozygotes. Am. J. Hum. Genet. 36, 640–645 [PMC free article] [PubMed] [Google Scholar]
- 43.van Ham, T. J., Thijssen, K. L., Breitling, R., Hofstra, R. M., Plasterk, R. H., and Nollen, E. A. ( 2008) C. elegans model identifies genetic modifiers of α-synuclein inclusion formation during aging. PLoS Genet. 4, e1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas, B., and Beal, M. F. ( 2007) Parkinson's disease. Hum. Mol. Genet. 16, R183–R194 [DOI] [PubMed] [Google Scholar]
- 45.Lazarou, M., McKenzie, M., Ohtake, A., Thorburn, D. R., and Ryan, M. T. ( 2007) Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol. Cell. Biol. 27, 4228–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenzie, M., Lazarou, M., Thorburn, D. R., and Ryan, M. T. ( 2007) Analysis of mitochondrial subunit assembly into respiratory chain complexes using Blue Native polyacrylamide gel electrophoresis. Anal. Biochem. 364, 128–137 [DOI] [PubMed] [Google Scholar]
- 47.Pocsfalvi, G., Cuccurullo, M., Schlosser, G., Cacace, G., Siciliano, R. A., Mazzeo, M. F., Scacco, S., Cocco, T., Gnoni, A., Malorni, A., and Papa, S. ( 2006) Shotgun proteomics for the characterization of subunit composition of mitochondrial complex I. Biochim. Biophys. Acta 1757, 1438–1450 [DOI] [PubMed] [Google Scholar]
- 48.Zhou, Y., Gu, G., Goodlett, D. R., Zhang, T., Pan, C., Montine, T. J., Montine, K. S., Aebersold, R. H., and Zhang, J. ( 2004) Analysis of α-synuclein-associated proteins by quantitative proteomics. J. Biol. Chem. 279, 39155–39164 [DOI] [PubMed] [Google Scholar]
- 49.Jin, J., Li, G. J., Davis, J., Zhu, D., Wang, Y., Pan, C., and Zhang, J. ( 2007) Identification of novel proteins associated with both α-synuclein and DJ-1. Mol. Cell. Proteomics 6, 845–859 [DOI] [PubMed] [Google Scholar]
- 50.Jin, J. H., Meredith, G. E., Chen, L., Zhou, Y., Xu, J., Shie, F. S., Lockhart, P., and Zhang, J. ( 2005) Quantitative proteomic analysis of mitochondrial proteins: relevance to Lewy body formation and Parkinson's disease. Mol. Brain Res. 134, 119–138 [DOI] [PubMed] [Google Scholar]
- 51.Greenamyre, J. T., Sherer, T. B., Betarbet, R., and Panov, A. V. ( 2001) Complex I and Parkinson's disease. IUBMB Life 52, 135–141 [DOI] [PubMed] [Google Scholar]
- 52.Hanagasi, H. A., Ayribas, D., Baysal, K., and Emre, M. ( 2005) Mitochondrial complex I, II/III, and IV activities in familial and sporadic Parkinson's disease. Int. J. Neurosci. 115, 479–493 [DOI] [PubMed] [Google Scholar]
- 53.Song, D. D., Shults, C. W., Sisk, A., Rockenstein, E., and Masliah, E. ( 2004) Enhanced substantia nigra mitochondrial pathology in human α-synuclein transgenic mice after treatment with MPTP. Exp. Neurol. 186, 158–172 [DOI] [PubMed] [Google Scholar]
- 54.Fornai, F., Schluter, O. M., Lenzi, P., Gesi, M., Ruffoli, R., Ferrucci, M., Lazzeri, G., Busceti, C. L., Pontarelli, F., Battaglia, G., Pellegrini, A., Nicoletti, F., Ruggieri, S., Paparelli, A., and Sudhof, T. C. ( 2005) Parkinson-like syndrome induced by continuous MPTP infusion: convergent roles of the ubiquitin-proteasome system and α-synuclein. Proc. Natl. Acad. Sci. U. S. A. 102, 3413–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li, W. W., Yang, R., Guo, J. C., Ren, H. M., Zha, X. L., Cheng, J. S., and Cai, D. F. ( 2007) Localization of α-synuclein to mitochondria within midbrain of mice. Neuroreport 18, 1543–1546 [DOI] [PubMed] [Google Scholar]
- 56.Cole, N. B., Dieuliis, D., Leo, P., Mitchell, D. C., and Nussbaum, R. L. ( 2008) Mitochondrial translocation of α-synuclein is promoted by intracellular acidification. Exp. Cell Res. 314, 2076–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Devi, L., Raghavendran, V., Prabhu, B. M., Avadhani, N. G., and Anandatheerthavarada, H. K. ( 2008) Mitochondrial import and accumulation of α-synuclein impair complex I in human dopaminergic neuronal cultures and Parkinson disease brain. J. Biol. Chem. 283, 9089–9100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parihar, M. S., Parihar, A., Fujita, M., Hashimoto, M., and Ghafourifar, P. ( 2008) Mitochondrial association of α-synuclein causes oxidative stress. CMLS Cell. Mol. Life Sci. 65, 1272–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]