Abstract
Plant-based overexpression of heterologous proteins has attracted much interest and development in recent years. To date, the most efficient vectors have been based on RNA virus-derived replicons. A system based on a disabled version of cowpea mosaic virus RNA-2 has been developed, which overcomes limitations on insert size and introduces biocontainment. This system involves positioning a gene of interest between the 5′ leader sequence and 3′ untranslated region (UTR) of RNA-2, thereby emulating a presumably stable mRNA for efficient translation. Thus far, the sequence of the 5′ UTR has been preserved to maintain the ability of the modified RNA-2 to be replicated by RNA-1. However, high-level expression may be achieved in the absence of RNA-1-derived replication functions using Agrobacterium-mediated transient transformation. To investigate those features of the 5′ UTR necessary for efficient expression, we have addressed the role of two AUG codons found within the 5′ leader sequence upstream of the main initiation start site. Deletion of an in-frame start codon upstream of the main translation initiation site led to a massive increase in foreign protein accumulation. By 6 d postinfiltration, a number of unrelated proteins, including a full-size IgG and a self-assembling virus-like particle, were expressed to >10% and 20% of total extractable protein, respectively. Thus, this system provides an ideal vehicle for high-level expression that does not rely on viral replication of transcripts.
The production of eukaryotic proteins for academic and industrial purposes can present significant challenges in terms of solubility and posttranslational modifications. For this reason, a number of eukaryotic protein production systems have been developed (Aricescu et al., 2006; Yin et al., 2007). Plants and plant cells possess many advantages over other eukaryotic expression hosts, such as high biomass, ease of scale-up, cost effectiveness, and low risk of contamination (Ma et al., 2003; Twyman et al., 2003). Although much work has been carried out using stably transformed plants, the significantly reduced development and production timelines make transient plant-based expression a particularly attractive option for the production of proteins of both commercial and academic interest.
To date, the most efficient means of achieving high-level transient expression of foreign proteins in plants has involved the use of vectors based on RNA plant viruses (Giritch et al., 2006; Lindbo, 2007), including the bipartite comovirus Cowpea mosaic virus (CPMV; Sainsbury et al., 2007). These systems take advantage of the ability of RNA viruses to replicate to high titers within infected cells. However, virus-directed replication of RNA has a number of undesirable features, including restrictions on the size of insert that can be accommodated without affecting replication and compromised fidelity of transcripts due to the lack of proofreading by RNA-dependent RNA polymerases (Ahlquist et al., 2005; Castro et al., 2005). In addition, vectors based on full-length viral replicons, which can move throughout a plant, suffer from problems of biocontainment.
To address the issue of biocontainment and to overcome the problem of insert size, we recently developed a system based on a disabled version of CPMV RNA-2 (delRNA-2; Cañizares et al., 2006; Sainsbury et al., 2008b). In this approach, the majority of the coding region of RNA-2 was replaced by a gene of interest. The sequence to be expressed was fused to the AUG at position 512 of RNA-2 because sequences upstream of this site had previously been shown to be essential for replication of RNA-2 by the RNA-1-encoded replication complex (Rohll et al., 1993). In addition, it was positioned immediately upstream of the 3′ untranslated region (UTR) to create a molecule that mimics RNA-2. Such constructs were shown to be capable of replication when agroinfiltrated into plants in the presence of RNA-1 and a suppressor of silencing and to direct the synthesis of substantial levels of heterologous proteins (Cañizares et al., 2006). Furthermore, it was demonstrated that the system was suitable for the production of heteromeric proteins, such as full-length antibodies (Sainsbury et al., 2008a).
Although the AUG at position 512 constitutes the major site of translation initiation on RNA-2 (Holness et al., 1989), the upstream sequence contains two additional AUGs at positions 115 and 161. Whereas the AUG at 115 is out of frame with that at 512 and has no known function (Wellink et al., 1993b), the AUG at position 161 is in-frame with AUG 512 and is functional as an initiation codon (Holness et al., 1989). Either deleting AUG 161 or disrupting its frame relationship with AUG 512 effectively eliminates RNA-2 replication (Holness et al., 1989; van Bokhoven et al., 1993). The need to preserve the frame relationship between AUG 161 and 512 to retain the replication ability of RNA-2-based constructs complicates the construction of vectors (Sainsbury et al., 2008b). However, whereas replication of the RNA-2-based constructs is essential to achieve high levels of expression when the mRNA is expressed from a transgene (Cañizares et al., 2006), it is less important with transient expression because large quantities of mRNA accumulate in agroinfiltrated tissue. This is particularly the case if a suppressor of silencing is coinfiltrated. We have therefore examined whether the upstream AUG codons can be eliminated without unduly compromising expression levels. Unexpectedly, the results obtained showed that expression can be greatly enhanced by eliminating the AUG at position 161. This observation has been used to design a simple and effective method for the production of high levels of proteins within plants.
RESULTS
Removal of Upstream AUG Codons Greatly Improves GFP Expression Levels in a Transient Assay
To create a useful cloning vector, a derivative of the original delRNA-2 construct containing GFP (1-GFP; Cañizares et al., 2006), called pM81-FSC2, was created. This allows easy replacement of GFP by other sequences using unique NruI and XhoI restriction sites (Fig. 1A). Use of the NruI site allows the insert to be precisely aligned with AUG 512, where translation is expected to exclusively initiate (Cañizares et al., 2006). To examine the effect of the AUGs upstream of AUG 512 on GFP expression levels, mutations that removed one or both of AUG 115 and AUG 161 were introduced into pM81-FSC2 (Fig. 1B). Following transfer of the expression cassette into the binary vector pBINPLUS (van Engelen et al., 1995), the GFP constructs were agroinfiltrated into Nicotiana benthamiana leaves in the presence of the suppressor of silencing P19 (Voinnet et al., 2003) and the levels of GFP expression assessed.
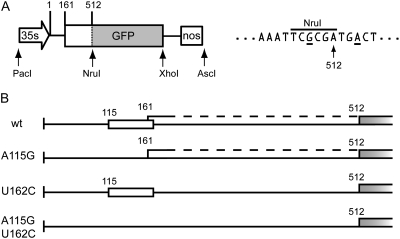
Figure 1.
Expression cassette used and mutations to the 5′ leader sequence of CPMV RNA-2. A, Schematic representation of the expression cassette from the cloning vector pM81-FSC2, with detail surrounding the AUG at position 512 showing the +4 and −3 Kozak consensus positions underlined. B, Schematic diagram of the mutations made to the 5′ leader (from nucleotides 1–512 in A). The positions of 5′ AUGs are indicated and the GFP sequence is shown as gray.
Examination of infiltrated tissue under UV light indicated that removal of AUG 115 alone resulted in a decrease in GFP expression to barely detectable levels (Fig. 2A, A115G). By contrast, removal of the in-frame AUG at 161 appeared to result in a dramatic increase in GFP expression levels (Fig. 2A, U162C). A similar enhancement was found when both AUGs at 115 and 161 were removed (Fig. 2A, A115G, U162C). Analysis of protein extracts by SDS-PAGE confirmed these findings and indicated that GFP accumulates to 20% to 25% of the extractable protein when AUG 161 is deleted, whether or not the AUG at 115 is present (Fig. 2B). This high-level expression was confirmed by analysis of the GFP fluorescence extracted from leaves by spectrofluorometry and corresponds to a level approaching 1.2 g/kg GFP of fresh-weight tissue (Fig. 2C). This represents at least a 10-fold increase in the levels obtained when the unmodified 5′ leader was used. The spectrofluorometric analysis also revealed that the maximum level of expression, 1.6 g/kg GFP of fresh-weight tissue, is obtained when both AUG 161 and AUG 115 are eliminated.
Figure 2.
Effect of the AUG 115 (A115G) and AUG 161 (U162C) mutations on GFP expression 5 d postinfiltration in N. benthamiana leaves. A, GFP fluorescence as seen under UV light. B, Coomassie-stained SDS-PAGE gel of approximately 25 μg of protein per lane extracted from infiltrated tissue. M, Marker with sizes indicated; C, control extract; wt, wild type; Std, 1 μg of recombinant GFP. C, GFP accumulation in infiltrated tissue measured by spectrofluorometry. Values represent averages from three extracts ± sd.
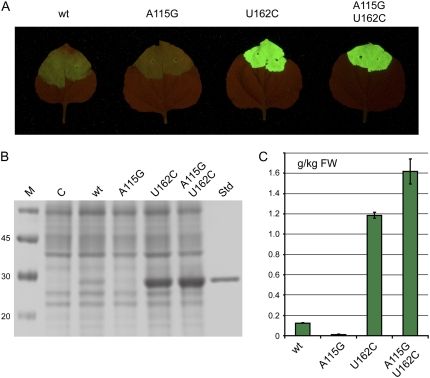
Increased Expression Levels Are Not Due to Increased mRNA Accumulation
To determine whether the increase in protein expression observed after removal of AUG 161 is due to increased levels of mRNA as a result of the mutations in the mutated 5′ leaders, quantitative reverse transcription (RT)-PCR was performed on RNA extracted from leaf tissue infiltrated with the various constructs. The levels of GFP-specific mRNA did not vary significantly with the nature of the 5′ leader sequence used. This lack of variation was found whether or not a construct expressing P19 was coinfiltrated (Fig. 3). These results indicate that the enhanced levels of protein expression found when AUG 161 is deleted are not due to enhanced levels of mRNA accumulation, but rather to the mRNA molecules being hypertranslated relative to the wild-type leader. For this reason, we refer to the RNA-2 leader lacking AUG 161 as the hypertranslatable (HT) leader.
Figure 3.
Analysis of RNA accumulation from the wild-type (wt) and mutant 5′ leader sequences either in the absence or presence of P19. Values indicate the average proportion of transcripts relative to the wild-type leader from three extracts ± se.
The HT Leader Is a General Enhancer of Protein Expression in Plants
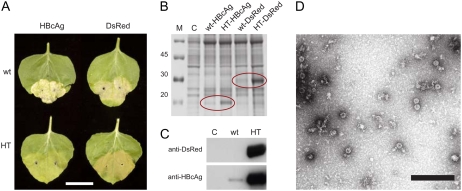
To examine whether the HT leader is generally effective at increasing expression of heterologous proteins, the Discosoma red fluorescent protein (DsRed) and the Hepatitis B core antigen (HBcAg) were each inserted downstream of either the wild-type or the HT 5′ leader. When infiltrated into N. benthamiana leaves, the HT-based constructs appeared to cause less necrosis in the infiltrated patches than the wild-type equivalent (Fig. 4A). Furthermore, infiltration with the DsRed construct gives a reddish hue to the infiltrated leaf patches, suggesting high levels of accumulation of this protein. SDS-PAGE of proteins extracted from infiltrated patches showed that, for both proteins, elimination of AUG 161 led to a substantial increase in accumulation over that obtained with the wild-type leader (Fig. 4B). In each case, the identity of the expressed proteins was confirmed by western blotting (Fig. 4C). DsRed appears to accumulate to similar levels to that of GFP (approximately 25% of the extractable protein; compare Figs. 2B and 4B). The HBcAg accumulates to approximately 1 g/kg of fresh-weight tissue as determined by ELISA using anti-HBcAg antibodies (E. Thuenemann, personal communication), which corresponds to around 20% of the extractable protein. To confirm that the expressed HBcAg was assembly competent, samples of leaf tissue were extracted in 3 volumes of Tris-NaCl buffer and subject to buffer exchange into Tris-EDTA, without concentration, using a 100-kD molecular cutoff column. Transmission electron microscopy of this crude preparation showed the presence of many virus-like particles, which we estimate comprise about 50% of the remaining total protein in the sample (Fig. 4D).
Figure 4.
Expression of DsRed and HBcAg cloned downstream of wild-type (wt) and HT (U162C) 5′ leaders. A, Leaves 6 d postinfiltration expressing HBcAg and DsRed downstream of either the wild-type or the HT 5′ leader. Scale bar, 2.5 cm. B, Coomassie-stained SDS-PAGE gel of approximately 25 μg of protein per lane extracted from tissue infiltrated with constructs as indicated. M, Marker with sizes indicated; C, control extract. C, Immunological detection of DsRed (top) or HBcAg (bottom) expression using either the wild-type or the HT 5′ leader. D, Transmission electron microscopy of crude extracts of HT-HBcAg following buffer exchange and negative staining with 2% uranyl acetate. Scale bar, 250 nm.
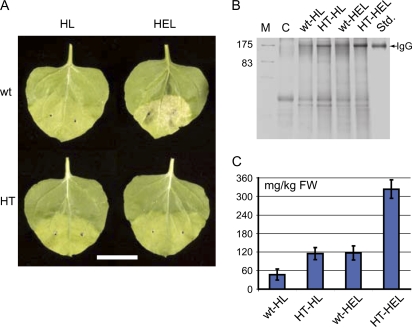
One of the advantages of CPMV expression systems over those based on other viruses is their ability to simultaneously express multiple polypeptides in the same plant cell (Sainsbury et al., 2008a). To test whether this ability is retained when the HT leader is used, the heavy (H) and light (L) chains of the human anti-HIV antibody 2G12 (Buchacher et al., 1994) were inserted downstream of either the wild-type or the HT 5′ leader. In both cases, the immunoglobulin chains retained their native leader peptides and two forms of the H chain were constructed, with (HE) and without (H) an endoplasmic reticulum (ER) retention motif. To obtain expression of full-size antibody, a combination of the L and either of the H chain constructs was coinfiltrated with P19 into N. benthamiana leaves (Fig. 5A). Analysis of total protein extracts by SDS-PAGE on nonreducing gels again suggested that higher levels of assembled antibody accumulation were obtained when the HT leader was used (Fig. 5B). The benefit of the HT over the wild-type leader was seen regardless of ER retention, although overall levels were higher when the antibody was retained. Quantification of IgG levels showed that the ER-retained 2G12 expressed using the HT leader (HT-HEL) accumulated to a yield in excess of 325 mg/kg of fresh-weight tissue (Fig. 5C). Furthermore, we have found that purified 2G12 produced in this way is comparable to 2G12 from other plant-based systems or from the mammalian cell-produced control in terms of antigen binding and virus neutralization in vitro (M. Sack and F. Sainsbury, unpublished data).
Figure 5.
Expression of the human anti-HIV IgG using wild-type (wt) and HT leaders. A, Leaves 6-d postinfiltration expressing 2G12, with (HEL) or without (HL) ER retention of the heavy chain, downstream of either the wild-type or the HT 5′ leader. Scale bar, 2.5 cm. B, Coomassie-stained nonreducing SDS-PAGE gel of approximately 12.5 μg of protein per lane extracted from tissue infiltrated with constructs as indicated. M, Marker with sizes indicated; C, control extract; Std, 1 μg of human IgG. C, 2G12 accumulation in extracts of 2G12 and HT-2G12 infiltrated tissue measured by SPR using a protein-A-coated surface. Values represent averages from three extracts ± sd.
DISCUSSION
The results presented here represent the highest reported level of plant-based protein production without the use of viral replication. We report the creation of an expression system based on a version of CPMV RNA-2 that is hypertranslatable relative to the wild-type version. By the removal of an upstream AUG that appears to inhibit translation, the system allows a variety of proteins to be produced to levels similar to that from state-of-the-art viral vectors in a matter of days, and without concomitant shortcomings of viral replication of transcripts. A recent study (Lindbo, 2007) showed 100-fold better expression for a single protein, GFP, from a tobacco mosaic virus (TMV)-based vector than when P19 was coinfiltrated with a cauliflower mosaic virus 35S promoter-driven construct. The HT constructs used in this study produced GFP levels in the same order of magnitude as the highest achieved with the TMV vector used in that study.
A significant disadvantage of vectors based on monopartite viruses, such as TMV, is their inability to coexpress multiple proteins. This limitation can be overcome by using vectors based on two different viruses that exist synergistically in nature, such as TMV and Potato virus X (Pruss et al., 1997). Using this noncompeting viral vector approach, Giritch et al. (2006) expressed the separate H and L chains of a tumor-specific IgG in TMV and potato virus X-based vectors. Depending on the vector-IgG combination used, yields of assembled antibody of 0.2 to 0.5 g/kg fresh-weight tissue were reported. In the case of the CPMV-HT system, levels of assembled 2G12 in excess of 0.3 g/kg fresh-weight tissue were obtained, a level comparable with the virus-based system. However, the viral vector-based system involved the coinfiltration of six Agrobacterium cultures took 10 d to reach maximum expression, and resulted in the production of infectious virus particles from the potato virus X construct used. In contrast, the HT expression required the coinfiltration of only three cultures, an incubation of only 6 d, and is fully biocontained, with no infectious virus being produced. Furthermore, the noncompeting viral vector approach is likely to be limited to the coexpression of only two proteins, unless additional noncompeting viruses can be found. In contrast, there is no obvious limit on the number of CPMV RNA-2-based constructs that can be coinfiltrated, raising the possibility of the production of multichain complexes.
The question arises as to why deletion of AUG 161 enhances expression from AUG 512. Although translation does occur from AUG 161 on wild-type CPMV RNA-2, the massive increase in expression resulting from the removal of AUG 161 suggests that the presence of AUG 161 is inhibitory to overall translation. A possible mechanism for this is that the majority of ribosomes that do not initiate at AUG 161 are unable to proceed to the downstream AUG 512. If this is the case, it suggests a possible function for the short open reading frame (ORF), which begins at AUG 115 and overlaps AUG 161, in bypassing this start codon. Initiation is known to occur at AUG 115 in vitro (Wellink et al., 1993b) and a possible bypassing of AUG 161 would potentially permit efficient translation at AUG 512 following reinitiation. This hypothesis is supported by the observed reduction in expression from AUG 512 when AUG 115 is removed and AUG 161 is retained (Fig. 2). Examples of short ORFs regulating expression from the main ORF following reinitiation have been described for eukaryotic as well as virus genes (Morris and Geballe, 2000; Ryabova et al., 2006). Thus, removal of the AUG at 161 appears to free translation from inhibition imposed by the presence of this start codon. A possible reason for a deliberate reduction in translation is to allow 3′ to 5′ movement of the RNA-1-encoded replicase on at least some transcripts, which would require RNA relatively free of ribosomes (Gamarnik and Andino, 1998).
An unexpected benefit of the removal of AUG 161 was that the increase in foreign protein production was accompanied by a reduction in the amount of tissue necrosis previously observed with some constructs (Figs. 4 and 5). Although an N-terminal fusion protein that would theoretically be produced by initiation at AUG 161 has not been detected (Cañizares et al., 2006), the N-terminal sequence of such a fusion may direct the polypeptide to the nucleus (Wellink et al., 1993a) with a potential toxic effect. Any such effect would therefore be alleviated by the prevention of initiation at AUG 161.
CONCLUSION
The results reported here show that it is possible to express very high levels of foreign proteins in plants without viral replication through the use of a modified version of the CPMV RNA-2 5′ leader. CPMV-HT provides a quick, easy, and inexpensive eukaryotic expression system that will prove very useful for the production of large quantities of recombinant proteins. Expression levels are similar to the highest reported so far from systems relying on viral replication. In addition to the biological advantages over viral vectors, such as the absence of RNA-dependent RNA polymerases and restrictions on insert size, the use of CPMV-HT does not require a license for work with plant pathogens. Therefore, this system presents an extremely useful and accessible tool in the fields of plant biology and biotechnology.
MATERIALS AND METHODS
Plasmid Constructs
A combination of oligonucleotide insertion and site-directed mutagenesis on pM81-FSC1 (Sainsbury et al., 2008b) resulted in the production of pM81-FSC2 (Fig. 1), which allows cloning with NruI and either XhoI or StuI. The terminal adenine of the NruI site lies at position 512, thereby allowing preservation of the AUG at this position. The modifications altered nucleotides immediately 5′ to the AUG at 512; however, a good context was maintained. The GFP sequence was amplified by PCR from pBinP-NS-1 (Liu et al., 2005) with primers that incorporated a 5′ NruI site and a 3′ XhoI site. The resulting NruI/XhoI fragment was inserted into similarly digested pM81-FSC2 to give pM81-FSC2-GFP. Complementary pairs of oligonucleotides were used in the site-directed mutagenesis of pM81-FSC2-GFP (Quickchange; Stratagene). Oligos to remove the AUG at 115 were A115G-F, 5′-CTTGTCTTTCTTGCGTGAGCGATCTTCAACG-3′ and A115G-R, 5′-CGTTGAAGATCGCTCACGCAAGAAAGACAAG-3′. Oligos to remove the AUG at 161, while maintaining the sequence of the putative uORF were U162C-F, 5′-GGCACCAGTACAACGTTTTCTTTCACTGAAGCG-3′ and U162C-R, 5′-CGCTTCAGTGAAAGAAAACGTTGTACTGGTGCC-3′. The mutant nucleotide is underlined in bold. The double mutation was made by applying the mutagenesis of AUG 161 to the AUG 115 mutant. The pM81-FSC2-derived plasmids were digested with AscI and PacI and the fragment containing the expression cassette including the foreign sequences transferred to the similarly digested binary vector, pBINPLUS (van Engelen et al., 1995).
DsRed (CLONTECH), HBcAg (Mechtcheriakova et al., 2006), and the H and L chains of 2G12 (Buchacher et al., 1994) were initially cloned into pM81-FSC1 via BspHI/StuI sites. For expression with the wild-type leader, PacI/AscI fragments were transferred into similarly digested pBINPLUS. For expression with the modified leaders, DraIII/AscI fragments containing the gene of interest, the 3′ UTR, and the nos terminator were transferred into a similarly digested FSC2-GFP-U162C expression cassette within pBINPLUS.
Agroinfiltration
Binary plasmid constructs were maintained in Agrobacterium tumefaciens strain LBA4404 and agroinfiltration into Nicotiana benthamiana was carried out as follows. Cultures grown to stable phase in Luria-Bertani medium supplemented with the appropriate antibiotics were pelleted by centrifugation at 2,000g and resuspended in MMA (10 mm MES, pH 5.6, 10 mm MgCl2, 100 μm acetosyringone) to an OD600 of 1.2. After 2- to 4-h incubations at room temperature, CPMV-based expression constructs were coinfiltrated at a 1:1 ratio with pBIN61-P19 (Voinnet et al., 2003) and a mix of pBIN61-P19 and pBINPLUS was infiltrated as a control.
Protein Extractions and Electrophoresis
For the extraction of GFP, DsRed, and HBcAg infiltrated leaf tissue was homogenized in 3 volumes of protein extraction buffer (50 mm Tris-HCl, pH 7.25, 150 mm NaCl, 2 mm EDTA, 0.1% [v/v], Triton X-100). For the extraction of 2G12, infiltrated leaf tissue was homogenized in 3 volumes of phosphate-buffered saline with 5 mm EDTA, 3 mm β-mercaptoethanol, 0.05% Triton X-100). Lysates were clarified by centrifugation and protein concentrations determined by the Bradford assay. The protein concentrations of extracts were consistently 2 to 2.5 mg/mL. Approximately 20 μg of GFP, DsRed, and HBcAg extracts were separated on 12% NuPage gels (Invitrogen) under reducing conditions and approximately 12.5 μg of 2G12 protein extract was separated by Tris-Gly SDS-PAGE under nonreducing conditions. For western blotting, separated extracts were transferred to nitrocellulose membranes and probed with Living Colors DsRed monoclonal antibody (CLONTECH) or rabbit anti-HBcAg (AbD Serotec). Anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibodies were used as appropriate (Amersham Biosciences). Signals were generated by chemiluminescence and captured on Hyperfilm (Amersham Biosciences).
Plants and Photography
N. benthamiana plants were grown from November to March in greenhouses maintained at 23°C to 25°C with 16 h of supplementary light per day. Infiltrated leaves were photographed with a Nikon D1x digital camera under visible light or, for the detection of GFP, under UV illumination from a Blak-Ray B-100AP UV lamp (Blak-Ray).
GFP Assay
GFP fluorescence measurements were made using a protocol modified from Richards et al. (2003). Soluble protein extracts were diluted in 0.1 m Na2CO3 and loaded in triplicate onto a fluorescently neutral black 96-well plate (Costar). Recombinant GFP from CLONTECH is the same variant of GFP as was used in this study and was, therefore, used to generate standard curves in a control plant extract at the same dilution as samples. Excitation (wavelength of 395 nm) and emission (509 nm) maxima were matched to CLONTECH's GFP and read using a SPECTRAmax spectrofluorometer (Molecular Devices).
Quantitative PCR
RNA extractions were performed using Ambion's RNAqueous kit with the plant RNA isolation aid (Ambion) according to the manufacturer's instructions. RNA concentration and quality was determined using a NanoDrop spectrophotometer (NanoDrop Technologies). cDNA was synthesized using the ProtoScript first-strand cDNA synthesis kit (New England BioLabs). RT quantification of target transcripts relative to actin transcripts was revealed by quantitative real-time PCR as measured by a Chromo 4 continuous fluorescence detector coupled to a PTC-200 peltier thermal cycler (MJ research) using SYBR Green JumpStart Taq ready mix (Sigma). Target transcripts were detected with the primers GFP-F, 5′-CTTGACTTCAGCACGTGTCTTGTAGTTCCC-3′ and GFP-R, 5′-AGAGGGTGAAGGTGATGCAACATACGG-3′; and actin transcripts were detected with the primers NbActin-F, 5′-CAGAAAGAGGCTACTCTTTTACCACCACGG-3′ and NbActin-R, 5′-GTGGTTTCATGAATGCCAGCAGCTTCC-3′. The amplification threshold was set and Ct values were calculated by OpticonMONITOR and Microsoft Excel. Triplicate leaf extracts representing infiltrated tissue from six plants were assayed and relative abundance of GFP RNA was calculated by dividing 0.5Ct-GFP by 0.5Ct-actin.
2G12 Measurements
Antibody concentrations were measured by surface plasmon resonance as described previously using a BIACORE 2000 (Biacore; GE Healthcare; Rademacher et al., 2008). 2G12 accumulation was measured from triplicate leaf extracts representing infiltrated tissue from six plants.
Transmission Electron Microscopy
Extraction buffer was exchanged for TE (10 mm Tris-HCl, pH 7.5, 1 mm EDTA) using a 100-kD molecular mass cutoff column and eluted in the same volume as the initial sample loaded onto the column. Droplets were placed onto carbon-coated electron microscopy grids and left to settle for 60 s. After drawing off excess liquid, grids were negatively stained by placing them upside down onto droplets of 2% uranyl acetate, then washed three times on droplets of water. Imaging was performed using a JEOL 1200 transmission electron microscope at 80 kV.
Acknowledgments
We would like to thank Markus Sack for help with 2G12 measurements and Kim Findlay for assistance with electron microscopy.
This work was supported in part by the European Union FP6 “PharmaPlanta” project, by funding from a Marie Curie Early Stage Training Fellowship (grant no. MEST–CT–2004–504273 to F.S.), and by the Trustees of the John Innes Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: George P. Lomonossoff (george.lomonossoff@bbsrc.ac.uk).
References
- Ahlquist P, Schwartz M, Chen JB, Kushner D, Hao LH, Dye BT (2005) Viral and host determinants of RNA virus vector replication and expression. Vaccine 23 1784–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aricescu AR, Assenberg R, Bill RM, Busso D, Chang VT, Davis SJ, Dubrovsky A, Gustafsson L, Hedfalk K, Heinemann U, et al (2006) Eukaryotic expression: developments for structural proteomics. Acta Crystallogr D Biol Crystallogr 62 1114–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, et al (1994) Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses 10 359–369 [DOI] [PubMed] [Google Scholar]
- Cañizares MC, Liu L, Perrin Y, Tsakiris E, Lomonossoff GP (2006) A bipartite system for the constitutive and inducible expression of high levels of foreign proteins in plants. Plant Biotechnol J 4 183–193 [DOI] [PubMed] [Google Scholar]
- Castro C, Arnold JJ, Cameron CE (2005) Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective. Virus Res 107 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R (1998) Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev 12 2293–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giritch A, Marillonnet S, Engler C, van Eldik G, Botterman J, Klimyuk V, Gleba Y (2006) Rapid high-yield expression of full-size IgG antibodies in plants coinfected with noncompeting viral vectors. Proc Natl Acad Sci USA 103 14701–14706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holness CL, Lomonossoff GP, Evans D, Maule AJ (1989) Identification of the initiation codons for translation of cowpea mosaic-virus middle component RNA using site-directed mutagenesis of an infectious cDNA clone. Virology 172 311–320 [DOI] [PubMed] [Google Scholar]
- Lindbo JA (2007) TRBO: a high-efficiency tobacco mosaic virus RNA-Based overexpression vector. Plant Physiol 145 1232–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cañizares MC, Monger W, Perrin Y, Tsakiris E, Porta C, Shariat N, Nicholson L, Lomonossoff GP (2005) Cowpea mosaic virus-based systems for the production of antigens and antibodies in plants. Vaccine 23 1788–1792 [DOI] [PubMed] [Google Scholar]
- Ma JKC, Drake PMW, Christou P (2003) The production of recombinant pharmaceutical proteins in plants. Nat Rev Genet 4 794–805 [DOI] [PubMed] [Google Scholar]
- Mechtcheriakova IA, Eldarov MA, Nicholson L, Shanks M, Skryabin KG, Lomonossoff GP (2006) The use of viral vectors to produce hepatitis B virus core particles in plants. J Virol Methods 131 10–15 [DOI] [PubMed] [Google Scholar]
- Morris DR, Geballe AP (2000) Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 20 8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss G, Ge X, Shi XM, Carrington JC, Vance V (1997) Plant viral synergism: the potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 9 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher T, Sack M, Arcalis E, Stadlmann J, Balzer S, Altmann F, Quendler H, Stiegler G, Kunert R, Fischer R, et al (2008) Recombinant antibody 2G12 produced in maize endosperm efficiently neutralizes HIV-1 and contains predominantly single-GlcNAc N-glycans. Plant Biotechnol J 6 189–201 [DOI] [PubMed] [Google Scholar]
- Richards HA, Halfhill MD, Millwood RJ, Stewart CN Jr (2003) Quantitative GFP fluorescence as an indicator of recombinant protein synthesis in transgenic plants. Plant Cell Rep 22 117–121 [DOI] [PubMed] [Google Scholar]
- Rohll JB, Holness CL, Lomonossoff GP, Maule AJ (1993) 3′-Terminal nucleotide sequences important for the accumulation of cowpea mosaic virus M-RNA. Virology 193 672–679 [DOI] [PubMed] [Google Scholar]
- Ryabova LA, Pooggin MM, Hohn T (2006) Translation reinitiation and leaky scanning in plant viruses. Virus Res 119 52–62 [DOI] [PubMed] [Google Scholar]
- Sainsbury F, Cañizares MC, Lomonossoff GP (2007) Cowpea mosaic virus-based expression vectors. In K Hefferon, ed, Virus Expression Vectors. Transworld Research Network, Kerala, India, pp 339–555
- Sainsbury F, Lavoie PO, D'Aoust MA, Vezina LP, Lomonossoff GP (2008. a) Expression of multiple proteins using full-length and deleted versions of cowpea mosaic virus RNA-2. Plant Biotechnol J 6 82–92 [DOI] [PubMed] [Google Scholar]
- Sainsbury F, Liu L, Lomonossoff GP (2008. b) Cowpea mosaic virus-based expression of antigens and antibodies in plants. In L Faye, V Gomord, eds, Methods in Biotechnology: Recombinant Pharmaceutical Proteins from Plants. Humana Press, NY (in press)
- Twyman RM, Stoger E, Schillberg S, Christou P, Fischer R (2003) Molecular farming in plants: host systems and expression technology. Trends Biotechnol 21 570–578 [DOI] [PubMed] [Google Scholar]
- van Bokhoven H, Le Gall O, Kasteel D, Verver J, Wellink J, van Kammen AB (1993) Cis- and trans-acting elements in cowpea mosaic virus RNA replication. Virology 195 377–386 [DOI] [PubMed] [Google Scholar]
- van Engelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res 4 288–290 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33 949–956 [DOI] [PubMed] [Google Scholar]
- Wellink J, van Lent JWM, Verver J, Sijen T, Goldbach RW, van Kammen AB (1993. a) The cowpea mosaic virus M RNA-encoded 48-kilodalton protein is responsible for induction of tubular structures in protoplasts. J Virol 67 3660–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellink J, Verver J, van Kammen A (1993. b) Mutational analysis of AUG codons of Cowpea Mosaic Virus-M RNA. Biochimie 75 741–747 [DOI] [PubMed] [Google Scholar]
- Yin J, Li G, Ren X, Herrler G (2007) Select what you need: a comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. J Biotechnol 127 335–347 [DOI] [PubMed] [Google Scholar]