Abstract
Trichome initiation in Arabidopsis (Arabidopsis thaliana) is controlled by the TRANSPARENT TESTA GLABRA1 (TTG1) network that consists of R2R3- and R1-type MYB-related transcription factors, basic helix-loop-helix (bHLH) proteins, and the WD40 protein TTG1. An experimental method was designed to investigate the molecular mechanisms by which jasmonates, cytokinins, and gibberellins modulate Arabidopsis leaf trichome formation. All three phytohormones provoked a seemingly common effect on cell patterning by promoting trichome initiation but caused strikingly distinct effects on cell and trichome maturation. The phytohormonal control was mediated by transcriptional regulation of the established TTG1 complex and depended on the R2R3-MYB factor GLABRA1. However, unsuspected degrees of functional specialization of the bHLH factors and a resultant differential molecular regulation of trichome initiation on leaf lamina and leaf margins were revealed. Trichome formation on leaf lamina relied entirely on GLABRA3 and ENHANCER OF GLABRA3. Conversely, TRANSPARENT TESTA8 (TT8) was particularly important for marginal trichome development. This hitherto unknown role for TT8 in trichome formation further underscored the functional redundancy between the three TTG1-dependent bHLH proteins.
Trichomes of Arabidopsis (Arabidopsis thaliana) are developmentally important because they are involved in temperature control, water regulation, and protection against insect herbivores and UV irradiation (Traw and Bergelson, 2003; Mauricio, 2005; Serna and Martin, 2006). Arabidopsis trichomes are single-celled, spiky hair-like structures that differentiate from individual protoderm cells into the developing epidermis of leaves, stems, and sepals. Each trichome is surrounded by eight to 12 rectangular epidermal cells at its base and reaches sizes from 200 to 300 μm (Marks, 1997). In contrast to root hair patterning, which is predictable with respect to the underlying cortex, the regular trichome patterning is an example of de novo pattern formation (Larkin et al., 1996, 1997; Hülskamp and Schnittger, 1998; Hülskamp et al., 1999; Schnittger et al., 1999).
Trichome patterns are generated by an activator/inhibitor system (Schellmann et al., 2002; Larkin et al., 2003; Pesch and Hülskamp, 2004), which has been characterized at the molecular level through mutant screens and yeast two-hybrid analysis (Y2H). The activator complex consists of a WD40 protein, a basic helix-loop-helix (bHLH) protein, and an R2R3-type MYB-related transcription factor (Serna and Martin, 2006; Schellmann et al., 2007). The WD40 protein is encoded by TRANSPARENT TESTA GLABRA1 (TTG1; Walker et al., 1999). For the other two elements of the activator complex, functional redundancy exists: the bHLH and R2R3-MYB factors can be encoded either by GLABRA3 (GL3) or ENHANCER OF GLABRA3 (EGL3) and GLABRA1 (GL1) or MYB23, respectively (Oppenheimer et al., 1991; Payne et al., 2000; Kirik et al., 2001, 2005; Zhang et al., 2003). TTG1 and GL1 or MYB23 physically interact with GL3 or EGL3, but not with each other (Payne et al., 2000; Zhang et al., 2003; Zimmermann et al., 2004). The activity of this TTG1-dependent complex can be repressed by single repeat (R1)-type MYB-related transcription factors that lack a transactivation domain and compete with the R2R3-MYB factors for binding to the bHLH protein (Esch et al., 2003; Zhang et al., 2003). To date, five homologous R1-MYB proteins have been identified as negative regulators of trichome initiation: TRIPTYCHON (TRY), CAPRICE (CPC), ENHANCER OF TRY AND CPC1 (ETC1), ETC2, and CAPRICE-LIKE MYB3 (Wada et al., 1997; Schellmann et al., 2002; Esch et al., 2004; Kirik et al., 2004a, 2004b; Tominaga et al., 2008). The active TTG1 complex stimulates expression of the inhibitors that move into the neighboring cells, where they repress the activators (Larkin et al., 1996, 2003; Schellmann et al., 2002; Pesch and Hülskamp, 2004).
Similar TTG1/bHLH/MYB complexes consisting of the same and/or other isoforms underlie root hair organogenesis, stomata patterning on hypocotyls, seed coat mucilage production, and control of phenylpropanoid biosynthesis (Zhang et al., 2003; Broun, 2005; Schellmann et al., 2007). For instance, the bHLH protein TRANSPARENT TESTA8 (TT8) and its close homologs GL3 and EGL3 control anthocyanin accumulation in leaves (Nesi et al., 2000; Heim et al., 2003; Baudry et al., 2004, 2006). However, in the case of anthocyanin biosynthesis, the corresponding MYB partner is not encoded by GL1 or MYB23 but by PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1), PAP2, MYB113, or MYB114 (Borevitz et al., 2000; Zhang et al., 2003; Gonzalez et al., 2008). Although the three bHLH proteins, GL3, EGL3, and TT8, often share common functions, as in leaf anthocyanin biosynthesis, they might also have specific roles; for example, TT8 is the only one that regulates seed coat pigment (proanthocyanidin) biosynthesis (Nesi et al., 2000; Zhang et al., 2003). Recently, the first R1-MYB, MYBL2, has been discovered that negatively regulates proanthocyanidin and anthocyanin biosynthesis in Arabidopsis (Dubos et al., 2008; Matsui et al., 2008) and that is expressed in various tissues, but not in those in which anthocyanins accumulated or TT8 was expressed (Matsui et al., 2008).
Previous research has pointed out that a number of phytohormones play an important role in trichome development. On Arabidopsis rosette leaves, trichome formation is negatively influenced by salicylic acid and positively influenced by GA3 and jasmonic acid (JA; Chien and Sussex, 1996; Telfer et al., 1997; Perazza et al., 1998; Traw and Bergelson, 2003). On Arabidopsis inflorescence organs, trichomes are positively influenced by GA3 and cytokinin (6-benzylaminopurine [BAP]; Greenboim-Wainberg et al., 2005; Gan et al., 2006, 2007). Knowledge of the molecular mechanisms by which phytohormones influence trichome formation is still limited. Pioneering reports have indicated that GAs promote trichome formation on Arabidopsis rosette leaves by up-regulating GL1 (Perazza et al., 1998) and that GAs and cytokinins might share C2H2 transcription factors to regulate Arabidopsis trichome initiation in late inflorescence organs (Gan et al., 2006, 2007). These C2H2 factors positively regulate the trichome initiation complex and act upstream of GL1. How jasmonates impinge on the trichome initiation machinery and whether phytohormones exert their effect through organ- or cell type-specific mechanisms remain to be revealed.
To study the molecular regulation of phytohormone-modulated trichome density on Arabidopsis rosette leaves, an experimental method was designed in which plants were first allowed to germinate on hormone-free medium before their transfer to hormone-supplemented medium. Here, we verified the trichome-stimulating potential of GA3, JA, and BAP. Whereas these hormones all increased trichome initiation, their effects on anthocyanin biosynthesis, trichome structure, and pavement cell formation differed distinctly. Detailed molecular analysis indicated that the positive effect of these hormones is mediated, at least in part, by the transcriptional regulation of the members of the TTG1/bHLH/MYB activator/inhibitor complex. Furthermore, our analysis pointed toward a differential regulation of marginal and laminal leaf trichome initiation and supported a hitherto unknown, but important, role for TT8 in trichome development.
RESULTS
Experimental Design to Study the Hormonal Influence on Trichome Formation
A robust experimental method was designed that allowed tight scoring the effects of the phytohormones JA, GA3, and BAP, whose positive involvement in the regulation of trichome formation had been suggested previously in the literature. Seeds of the Arabidopsis ecotype Columbia 0 (Col-0) were first germinated on control medium (K1) on which “normal” trichome initiation takes place. When the first four leaves were formed, seedlings were transferred to phytohormone-containing medium, on which they remained until the new leaves reached a size appropriate to explore the effect on trichome formation.
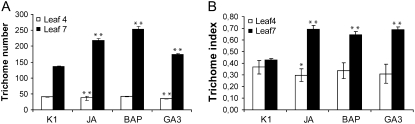
To determine at what stage trichome initiation is modulated by the phytohormones, we first compared the effect on the trichome numbers on leaves 3 and 4 with those on leaves 7 and 8. The former were already created before transfer and, thereby, supposed to have an established epidermal cell fate. Leaf 7 and consecutive leaves were formed upon or after the transfer (Larkin et al., 1996) and thus were amenable to phytohormone-mediated changes in cell fate development. A significant increase in the trichome number could be observed only from the seventh leaf on, whereas no effect, or a slight decrease, was seen on leaves 3 and 4 (Fig. 1A; Supplemental Fig. S1A; Supplemental Table S1), indicating that the experimental setup allows the control of trichome-promoting effects in a discrete developmental window. The effects of the three phytohormones were even more pronounced in ecotype C24 (Supplemental Fig. S1B), a glabrous derivative of Col-0 (Barth et al., 2002). For both ecotypes, the response was the strongest for BAP, with an increase of 47% compared with mock-treated plants in the Col-0 ecotype (Fig. 1A) and an increase from generally zero to approximately 70 to 80 trichomes per leaf in the C24 ecotype (data not shown).
Figure 1.
Influence of the phytohormones JA, BAP, and GA3 on trichome development in Arabidopsis Col-0 seedlings. Trichome number (A) and trichome index (B) of leaves 4 and 7 of Col-0 seedlings. Parameters to calculate the TI are listed in Supplemental Table S1. Error bars represent the se (n = 10 for A and n = 6 for B). Statistical significance was determined by Student's t test (** P < 0.01, * P < 0.05). Seedlings were mock treated (K1) or induced with JA (5 μM), BAP (1 μm), or GA3 (1 μm).
To verify whether the increase in trichome density was due to a direct effect on trichome initiation or to an indirect effect through pleiotropic effects on leaf development, we determined the trichome index (TI), which takes both the number of epidermal cells and the number of trichomes into account. The TI is calculated by the formula (numbers of trichomes/numbers of trichomes + numbers of pavement and guard cells) × 100 (Supplemental Table S1). The three hormones clearly increased the TI, confirming that BAP, GA3, and JA all can positively affect at least one aspect of leaf development (i.e. trichome formation; Fig. 1B; Supplemental Table S1). As for the trichome numbers, a marked effect on the TI was only observed from leaf 7 on.
JA, BAP, and GA3 Stimulate Trichome Initiation But Have Strikingly Distinct Effects on Cell Pattern Formation and Cell Maturation
As GAs and cytokinins are known to act antagonistically in leaf formation and meristem maintenance, the effects of the phytohormones on leaf and trichome development were studied in more detail to discern potentially divergent hormone specificities. The parameters assessed were leaf nuclear DNA content and pavement cell size (Fig. 2), trichome size and structure, and nuclear DNA content (Figs. 3 and 4). Leaf 7 was used throughout. Nuclear DNA content reflects the endoreduplication rate, a feature that is often correlated positively with cell size (Melaragno et al., 1993) and trichome branching (Melaragno et al., 1993; Hülskamp et al., 1994; Schnittger et al., 1998; Szymanski and Marks, 1998).
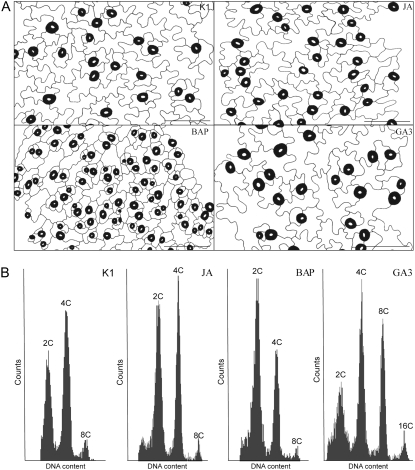
Figure 2.
Cell development in phytohormone-treated rosette leaves. Effects of the phytohormones on pavement cell sizes of the abaxial epidermis (A) and the DNA ploidy level distribution (B) in the seventh leaves of plants treated with K1 (mock), JA, BAP, and GA3 at 14 d after phytohormone treatment. Stomatal guard cells are colored in black. Bars = 100 μm.
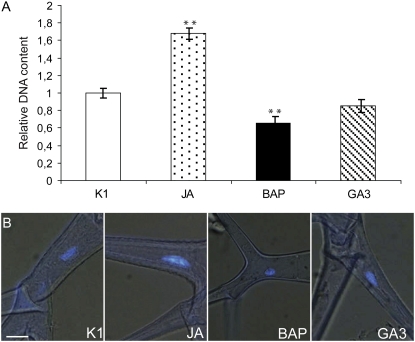
Figure 3.
Nuclear content of phytohormone-treated trichomes. A, DNA content of DAPI-stained nuclei in phytohormone-treated trichomes relative to that of mock-treated (K1) trichomes. Parameters to calculate the trichome DNA content are listed in Supplemental Table S2. Error bars represent se (n = 10). Statistical significance was determined by Student's t test (** P < 0.01). B, Microscopic analysis of the nuclear area of leaf 7 trichomes at 14 d after phytohormone treatment stained with DAPI. Bar = 10 μm.
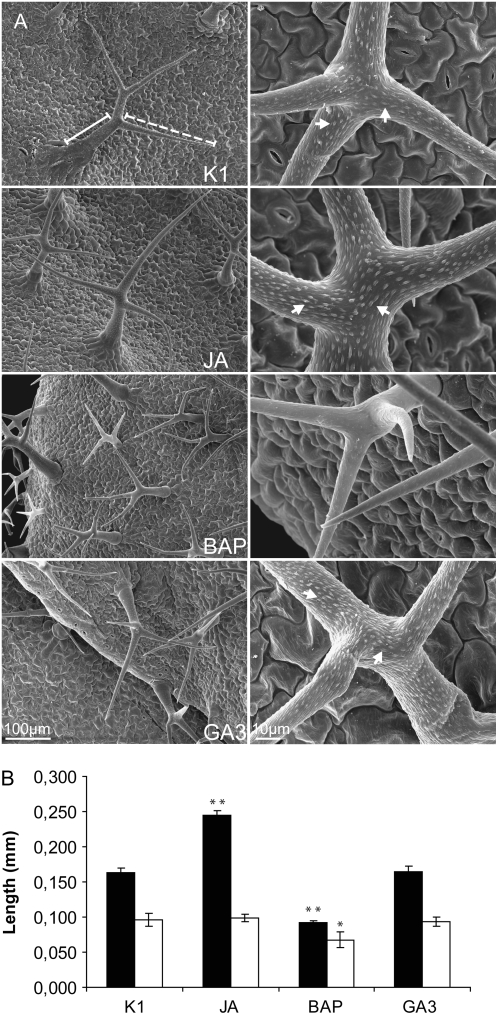
Figure 4.
Trichome structure of phytohormone-treated rosette leaves. A, Scanning electron micrographs of trichomes treated with JA, BAP, and GA3 14 d after treatment. Solid and dashed white lines indicate trichome stalk and trichome branch, respectively, and white arrows indicate the trichome papillae. B, Trichome stalk (white bars) and branch (black bars) length. Error bars represent se (n = 9). Statistical significance was determined by Student's t test (** P < 0.01, * P < 0.05).
Three general trends were discernible. First, BAP stimulated cell division and differentiation but prevented or delayed maturation of cells, independently of their developmental fate. The influence of BAP was particularly reflected by an increase in the number of cells and trichomes per leaf (Figs. 1 and 2; Supplemental Table S1) and a decrease in nuclear ploidy levels or DNA content, both in leaves (Fig. 2) and trichomes (Fig. 3). Endoreduplication rates correlated positively with cell size, as mirrored by the small leaf pavement cells (Fig. 2) and short trichomes (Fig. 4) of BAP-treated leaves. The inhibitory effect of BAP on trichome maturation was also apparent from the absence of surface papillae (Fig. 4A), the formation of which is correlated with the cell wall changes that occur normally during trichome maturation (Rerie et al., 1994). Notably, neither BAP nor any of the other phytohormonal treatments affected significantly the trichome branch number (data not shown). The curved and shortened branch phenotypes of BAP trichomes were reminiscent of those observed after pharmacological disruption of long actin cables (Szymanski et al., 1999) or overproduction of the actin-depolymerizing factor and key regulator of F-actin organization, ADF1 (Dong et al., 2001).
Second, the influence of GA3 and JAs on endoreduplication and cell maturation depended seemingly on the developmental fate, contrary to cytokinins. For instance, pavement cells of GA3-treated leaves were larger than those of the control (Fig. 2; Supplemental Table S1), an event accompanied by an additional round of endoreduplication (Fig. 2). In contrast, no significant differences in leaf cell number and size or in leaf nuclear ploidy levels were observed in JA-treated leaves when compared with mock-treated leaves (Fig. 2; Supplemental Table S1). Conversely, JA-treated trichomes had higher DNA contents and an enlarged nuclear area, whereas trichomes of GA3-treated and mock-treated plants did not significantly differ for any of the scored parameters (Fig. 3). Accordingly, trichome outgrowth was clearly stimulated in JA-treated plants, as reflected by the increase in trichome branch size (Fig. 4).
Third, the three phytohormones distinctly affected cell pattern formation and maturation depending on the epidermal cell fate. Remarkably, the stomatal index (Supplemental Table S1) was only affected by GA3, further underscoring the cell type-specific effect of the phytohormones on developmental pattern formation. Furthermore, GA3, JA, and BAP differentially regulated trichome maturation, despite the shared promotion of the trichome formation.
JA, BAP, and GA3 Stimulate Trichome Initiation through Transcriptional Regulation of the TTG1 Network
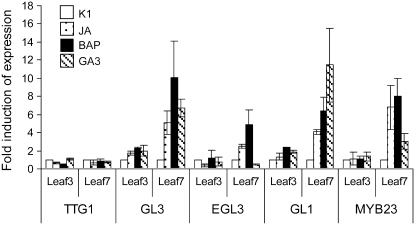
To characterize the molecular mechanisms behind the phytohormone-mediated control of trichome formation, we evaluated the transcriptional regulation of the genes coding for known components of the trichome initiation machinery by quantitative (q)PCR. Previously it was reported that the promotion of trichome formation by GAs in rosette leaves was accompanied by up-regulation of GL1 (Perazza et al., 1998) and that GL1 is cytokinin and GA inducible in inflorescence organs (Gan et al., 2007). Here, the expression levels of the known positive trichome regulator genes TTG1, GL3, EGL3, GL1, and MYB23 were compared on (pooled) leaves 3 and 4 and leaves 7 and 8. The expression levels differed only in leaves 7 and 8 after the various treatments (Fig. 5). As this observation was consistent with the discrete developmental window in which trichome pattern formation could be promoted (Fig. 1; Supplemental Table S1), we focused on the seventh and eighth leaves for all further expression analyses. TTG1 expression did not significantly differ at any of the developmental stages or treatments. In contrast, the R2R3-MYB genes (GL1 and MYB23) and the bHLH genes (GL3 and EGL3) were transcriptionally induced by the three phytohormones (Fig. 5). For the known negative regulators of trichome development, the expression of the R1-MYB genes TRY, ETC1, and ETC2 was reduced, whereas that of CPC was stimulated (Supplemental Fig. S2). Although the relation to the presumed different functions of the individual members of the R1-MYB family remains unclear, experimental evidence hints at a more important role for TRY in short-range inhibition and for CPC and, particularly, ETC1 in long-range inhibition (Schellmann et al., 2002; Kirik et al., 2004a). In conclusion, the phytohormone-mediated increased trichome initiation can, at least partly, be accounted for by the modulation of gene expression for trichome-inducing genes.
Figure 5.
Expression of trichome initiation genes in phytohormone-treated rosette leaves. Comparison of TTG1, GL3, EGL3, GL1, and MYB23 expression between the third and fourth leaves (pooled) and the seventh and eighth leaves (pooled) at 5 d after phytohormone treatment. Numbers on the ordinate give the fold induction compared with that in the mock treatment (K1). Error bars represent se (n = 9).
Phytohormone-Mediated Trichome Initiation Is Not Consistently Linked with Anthocyanin Biosynthesis
Because trichome initiation and leaf anthocyanin production are regulated by the same bHLH proteins (GL3 and EGL3) in a TTG1-dependent manner, we verified whether the increase in trichome number consistently correlated with an increase in anthocyanin accumulation. Interestingly, whereas JA and BAP both increased anthocyanin production, GA3 did not (Supplemental Fig. S3A), indicating that different phytohormones can specifically impinge on particular components of the TTG1-dependent complexes, despite the partly shared molecular machinery. Correspondingly, JA and BAP, but not GA3, induced expression of the bHLH (TT8) and R2R3-MYB (PAP1 and PAP2) factors dedicated to the regulation of anthocyanin biosynthesis (Supplemental Fig. S3B).
Formation of Marginal and Laminal Trichomes Is Steered by Different TTG1-Dependent Complexes
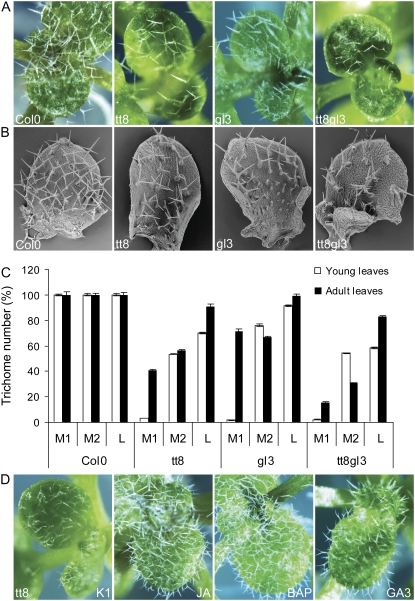
Since the phytohormone-mediated increase in trichome initiation correlated with transcriptional up-regulation of the known activator components of the trichome initiation network, we assessed whether this hormonal effect depended exclusively on these components. Therefore, we analyzed the trichome-promoting effects of the three phytohormones on several glabrous Arabidopsis mutants deficient in TTG1 network components. Surprisingly, the phytohormones were found to impinge differently on the trichome initiation machinery depending on the leaf zone, namely lamina versus margin. None of the phytohormones could fully restore the trichome formation on the leaf lamina in any of the ttg1, gl3egl3, and gl1 mutants (Fig. 6). Sporadically, small trichome-primordium-like structures were formed on the leaf lamina of ttg1 mutants (Fig. 6B), indicating that the phytohormone-mediated induction of laminal trichome initiation depends exclusively on the known WD40, bHLH, and MYB elements. However, trichome formation was induced at the leaf margin in the ttg1 and gl3egl3 mutants, but not in the gl1 mutant. Because simultaneous overexpression of GL3 and EGL3 can suppress the ttg1 trichome defect (Zhang et al., 2003), the increased expression of GL3 and EGL3 upon phytohormone treatment (Fig. 5) might explain the observation in the ttg1 mutant. In contrast, the marginal trichome induction in the gl3egl3 mutant clearly pointed toward the existence of distinct control pathways for the laminal and marginal leaf trichomes. These two epidermal differentiation programs might share the WD40 and R2R3-MYB elements of the TTG1-dependent complex, but apparently they involved different bHLH factors.
Figure 6.
Distinct trichome formation in the leaf margin and lamina. Influence of phytohormones on trichome development in the glabrous mutants ttg1, gl3egl3, gl1, and gl3egl3tt8, as visualized by light microscopy (A) and scanning electron microscopy (B). Marginal trichomes and laminal trichome primordia are indicated by yellow and white arrowheads, respectively.
TT8 Plays an Important Role in Marginal Trichome Development
A potential candidate for the induction of marginal trichomes in the gl3egl3 double mutant was TT8, which hitherto was not linked with trichome initiation. Analysis of the gl3egl3tt8 triple mutant showed that the phytohormone-mediated induction of marginal trichomes in the gl3egl3 background was completely abolished in the triple mutant (Fig. 6). Hence, TT8 might be involved in the establishment of marginal trichomes, at least in certain genetic backgrounds and under certain conditions, such as phytohormone elicitation. To verify whether TT8 is involved also in marginal trichome initiation in the presence of GL3 and EGL3 and in “normal” growth medium, we checked the marginal trichome development in tt8 single mutants grown in the absence of exogenously added hormones. No trichomes could be detected at the margin of young developing tt8 leaves (Fig. 7, A–C). To assess whether marginal trichome development depended exclusively on TT8, we also verified the marginal trichome development on gl3 seedling leaves (Fig. 7, A–C). Young developing gl3 leaves lacked marginal trichomes, a phenotype further enhanced in the tt8gl3 double mutant. Hence, both TT8 and GL3 seem to be involved in and required for marginal trichome induction.
Figure 7.
Development of marginal trichomes. A and B, Comparison of wild-type plants with tt8, gl3, and tt8gl3 mutants for the developing leaf stage by light microscopy (A) and scanning electron microscopy (B). C, Quantification of marginal trichomes on developing and mature leaves (from 1-week-old and 4-week-old seedlings, respectively) of single and double bHLH mutants. In both stages, three zones were arbitrarily defined based on the distance from the visible leaf margin. From the leaf margin to the center, these zones are M1 (margin 1), the outer 5% (calculated on the total leaf surface) that includes the visible margin; M2 (margin 2), the 5% inward layer adjacent to M1; and L (lamina), the central region. Trichome numbers in leaves of the Col-0 wild type for each zone were set at 100% and corresponded to 12 (M1), 15 (M2), and 33 (L) in developing leaves and to 49 (M1), 32 (M2), and 73 (L) in adult leaves. Error bars represent se (n = 15). D, Trichome development in the tt8 mutant after transfer from mock (K1) to phytohormone-containing medium.
To determine whether TT8 and GL3 are essential for trichome development on margins or rather control its timing, single and double mutants were scored for (marginal) trichome numbers on young developing leaves and adult leaves (of 1-week-old and 4-week-old seedlings, respectively). This quantitative analysis indicated that marginal trichome formation was almost completely impaired in developing leaves of tt8 and gl3 single and double mutants. Laminal trichome formation still occurred, albeit slightly reduced, both in tt8 and gl3 mutants (Fig. 7B). In adult leaves, marginal trichome formation was still decreased, most clearly in tt8gl3 but also in the single tt8 and, to a lesser extent, gl3 mutants (16%, 41%, and 72% of the Col-0 level, respectively), indicating that both TT8 and GL3 are essential for trichome development on margins, independently of the developmental stage. Possibly, EGL3 also might participate, because marginal trichome development was still observed on (adult) tt8gl3 leaves. Moreover, when the tt8 and gl3 single and double mutants were subjected to phytohormone treatment, marginal trichome development was restored, further supporting the functional redundancy of TT8, GL3, and, probably, EGL3 (Fig. 7D).
Spatial Expression of GL3, EGL3, and TT8 Depends on Phytohormone Treatment and the Presence of TTG1
GL3 has been postulated to act in a negative autoregulatory loop (Morohashi et al., 2007), whereas TT8 expression in seeds is regulated by a positive feedback loop involving TT8 itself and GL3 and EGL3 (Baudry et al., 2006). Therefore, the marginal trichome phenotypes in the tt8 and gl3 mutants might possibly be ascribed to a general decrease in bHLH factor expression. However, qPCR analysis showed that EGL3, GL3, and TT8 expression was not considerably altered, either in the gl3 and tt8 mutants or in the ttg1 and gl3egl3 mutants (Supplemental Fig. S4). Thus, the presence of both GL3 and TT8 seems to be required for marginal trichome initiation.
Nonetheless, the altered transcriptional regulation of bHLH genes could be responsible for the marginal trichome phenotypes in the single bHLH mutants, because the conditional mutual presence was overcome by phytohormonal treatment in both the gl3 and tt8 mutants (Fig. 7D) and GL3 expression was clearly induced (Fig. 5). However, TT8 expression was only induced markedly by BAP, and not by JA or GA3, suggesting that marginal trichome formation in the gl3 mutant might not be restored by the simple transcriptional up-regulation of TT8 expression (Supplemental Fig. S3B).
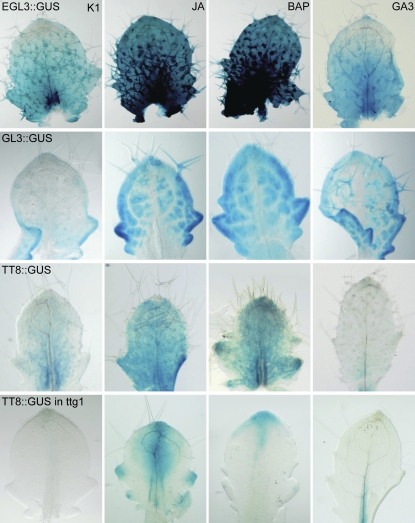
Therefore, we visualized promoter activities and spatial expression of trichome developmental genes by histochemical analysis of transgenic plants with promoter-GUS fusions in the absence or presence of the three phytohormones. The GUS activity patterns in all promoter-GUS lines tested were affected markedly by the phytohormones (Fig. 8). The effects observed were all consistent with the qPCR data (Fig. 5; Supplemental Fig. S3B). In the ProGL3:GUS line, the strongest GUS staining was detected at the leaf margins and further increased after phytohormonal treatment without substantial alteration of the overall spatial expression pattern; hence, it was in agreement with the above-described role of GL3 in marginal trichome initiation. In contrast, the expression pattern of ProEGL3:GUS was also strongly pronounced in the lower half of the leaf lamina in addition to the leaf margin. This activity was further enhanced by JA and BAP but not by GA3.
Figure 8.
In situ expression patterns of the trichome initiation machinery in the presence of phytohormones. Histochemical studies on the effect of plant hormones on Pro:GUS activity for different trichome developmental genes. Leaf 7 or 8 was stained for GUS activity 5 d after phytohormone treatment.
In mock-treated ProTT8:GUS plants, we could not detect TT8 promoter activity in leaf margins but we detected a weak blue staining in the lamina. JA and BAP clearly induced promoter activity in the leaf lamina, whereas GA3 treatment reduced GUS staining dramatically. Intriguingly, even in the presence of JA and BAP, the TT8 promoter activity remained lower in the leaf margin than in the lamina. Notably, the ProTT8:GUS expression pattern was clearly affected by the absence of TTG1. In contrast to the situation in the wild-type background, ProTT8:GUS staining could not be detected anymore in ttg1 mutant leaves under mock conditions. Furthermore, ProTT8:GUS staining was now visible at the margins of ttg1 leaves after treatment with JA and BAP (Fig. 8), which is consistent with the restoration of marginal trichome initiation in this mutant background (Fig. 6). We did not notice any differences in the spatial expression of ProTT8:GUS in the gl3egl3 mutant background when compared with that of the wild type (data not shown), indicating that in leaves, TT8 expression is not regulated by GL3 or EGL3. Our findings indicate that for each of the three trichome-regulating bHLH factors, spatial gene expression is divergent in leaf margin and lamina, depends on the presence of phytohormones and of TTG1, and is in accordance with the respective functions in trichome initiation.
DISCUSSION
Multiple genetic and environmental factors affect trichome density, a trait that is of great ecological importance for many plants, in particular in defense against herbivores (Traw and Bergelson, 2003; Mauricio, 2005; Serna and Martin, 2006). Here, we investigated how the phytohormones GA3, JA, and BAP stimulate Arabidopsis trichome formation and how it is integrated into other cellular processes.
Different Phytohormones Determine the Frequency of Trichome Initiation through Distinct Cellular Events
Using a robust trichome-promoting experimental setup, we showed that exogenous application of the phytohormones GA3, JA, and BAP all stimulated trichome formation in a seemingly similar way when scored merely by leaf trichome numbers and density. However, despite this common effect, the three hormones have strikingly distinct or even antagonistic effects on various other cellular and developmental processes. For instance, cytokinins stimulated leaf cell division and blocked pavement cell maturation, whereas GAs promoted the latter event. In spite of their positive effect on trichome initiation, cytokinins clearly blocked trichome maturation, whereas JAs promoted it. Thus, different cell types can adjust their response to a particular hormone to the context of the developmental programs they are committed to. This finding is in agreement with recent observations that cytokinin functions seem to be diverse and context dependent and that different cytokinin threshold levels are required for cell proliferation, elongation, and differentiation (Müller and Sheen, 2007) or that JAs can transcriptionally activate different metabolic and cellular responses in Arabidopsis in a context-dependent manner (L. Pauwels and A. Goossens, unpublished data).
In general, our observations correspond well with the established physiological roles of GAs, cytokinins, and JAs in plant development and defense. GAs promote the juvenile-to-adult growth transition, and this vegetative phase change influences trichome production (Telfer et al., 1997; Perazza et al., 1998). Cytokinins keep leaves in a division state, as reflected by the increased expression of the cyclin D3-encoding gene CYCD3 (L. Maes and A. Goossens, unpublished data), a crucial driver in cytokinin-induced cell division (Riou-Khamlichi et al., 1999). CYCD3 transcript levels were not affected in leaves treated with JA or GA3 (L. Maes and A. Goossens, unpublished data). Maintenance of leaves in such a prolonged division state prevents full maturation and, eventually, might promote trichome initiation (Larkin et al., 1996). JAs are well-known defense hormones that are synthesized upon wounding by herbivores and that activate various stress responses. Considering the role of trichomes in defense against herbivores, it is plausible to assume that JAs might be capable of affecting cellular differentiation or other developmental programs when this is appropriate for the plant's survival.
Functional Specialization of the TTG1-Dependent bHLH Factors in Trichome Initiation
Arabidopsis trichome formation depends on the activity of the TTG1 transcriptional activator/repressor complex, in which the bHLH proteins are postulated to share largely overlapping functions and the MYB proteins to provide the specificity for the downstream effects. However, hitherto, functional redundancy between the bHLH proteins TT8, GL3, and EGL3 was suggested to be incomplete, because TT8 was presumably involved only in seed coat mucilage production and in phenylpropanoid biosynthesis in leaves and seeds (Zhang et al., 2003; Broun, 2005). Accordingly, no trichome phenotype was reported for the TT8 overexpressors, and complementation of tt8 was less complete with Pro-35S:GL3 than with Pro-35S:EGL3 (Nesi et al., 2000; Broun, 2005). Furthermore, the combined TT8/GL1 or TT8/MYB23 overexpression could not transactivate anthocyanin/phenylpropanoid biosynthetic gene expression in protoplasts, in contrast to the TT8/PAP1 and TT8/PAP2 combinations (Zimmermann et al., 2004). This functional specificity might possibly be due to structural features that prevent interaction between certain MYB and bHLH proteins (Broun, 2005). Our results attenuate this latter hypothesis and reinforce the assumption of overlapping functions, because we clearly demonstrate the involvement of TT8 in trichome formation. We propose that, in a particular molecular and developmental context, TT8 and the trichome-stimulating R2R3-MYB factors GL1 and/or MYB23 might interact. Whereas no such interaction between TT8 and GL1 or MYB23 was implied by Y2H analysis based on growth on selective medium, the semiquantitative β-galactosidase-based Y2H assay hinted at such a possibility, although definitely much weaker, for instance, than that between EGL3 and GL1 or MYB23 (Zimmermann et al., 2004).
Crucial factors in the control of TTG1-dependent pathways are the existence of autoregulatory loops and the degree of overlap in expression of MYB- and bHLH-encoding genes. For instance, GL3 is recruited to its own promoter with a decreased GL3 expression as a result, implying the presence of a GL3-negative autoregulatory loop (Morohashi et al., 2007). Homologous MYB and bHLH proteins can also bind simultaneously to the TT8 promoter (e.g. TT8/PAP1, EGL3/PAP1, and GL3/PAP1); hence, TT8 might be involved in the control of its own transcription, and a broad set of MYB/bHLH complexes might participate in the TT8 activation (Baudry et al., 2006). This hypothesis correlates with the ProTT8:GUS patterns in seeds of wild-type and tt8, egl3, and ttg1 mutant plants. In developing seeds and cotyledons, the TT8 promoter activity is restricted to specific cell types, and this spatial expression pattern is altered in the different mutants (Baudry et al., 2006). Accordingly, our study showed that the TT8 promoter activity, as well as that of GL3 and EGL3, in rosette leaves was restricted to particular zones and depended on the presence of TTG1. Furthermore, BAP, JA, and GA3 influenced the spatial expression pattern of TT8 as well as those of GL3 and EGL3. Thus, the divergence in transcriptional control allowed the functional specialization of largely equivalent bHLH proteins, in turn enabling plants to respond similarly to distinct developmental, molecular, or hormonal signals. Likewise, the study of the GIS clade of C2H2 factors has revealed that functional specialization within a transcription factor family can facilitate and integrate the regional influence of phytohormones to regulate plant cell differentiation (Gan et al., 2007).
MATERIALS AND METHODS
Plant Material and Maintenance
Mutant and wild-type Arabidopsis (Arabidopsis thaliana) seeds were incubated on Murashige and Skoog germination medium on horizontally oriented square plates (Greiner Labortechnik). After the first four leaves had been formed, seedlings were transferred to Murashige and Skoog medium without (K1) or with 5 μm JA (Sigma-Aldrich), 1 μm GA3 (Sigma-Aldrich), or 1 μm BAP (Sigma-Aldrich). Plants were grown under continuous light (110 μE m−2 s−1 photosynthetically active radiation supplied by cool-white fluorescent tungsten tubes [Osram]) at 22°C.
Measurement of Epidermal Cell Number and Trichome Density
For microscopy, leaves were harvested, cleared overnight in methanol, and subsequently stored in lactic acid (90%). Total leaf area was determined with the public domain image-analysis program ImageJ (version 1.37; http://rsb.info.nih.gov/ij/) from digitized images taken with a CCD camera installed on a binocular (Stemi SV11; Zeiss). To measure cell density, leaves were mounted on a slide and observed with a microscope fitted with differential interference contrast optics (Leica). Epidermal cell number and area were determined with ImageJ from scanned drawing-tube images of outlines of at least 100 cells of the abaxial epidermis located 25% and 75% from the distance between the tip and the base of the leaf and halfway between the midrib and the leaf margin. The following parameters were obtained with ImageJ: the areas of all individual cells in the drawing, the total number of cells, and the number of guard cells. From these data, we calculated the average cell area and estimated the total number of cells per leaf by dividing the leaf area by the average cell area (averaged between the apical and basal positions). Finally, the number of trichomes per whole leaf was counted.
Histochemical GUS Staining
GUS activity was assayed by immersing seedlings in a staining solution consisting of 50 mm sodium phosphate buffer (pH 7.0), 0.1% Triton X-100, 10 μL mL−1 dimethylformamide, and 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl glucuronide at 37°. To limit the diffusion of the blue staining, 5 mm K3Fe(CN)6 and 5 mm K4Fe(CN)6 were added. After staining for 24 h, tissues were cleared in absolute ethanol for 1 h and in 70% ethanol for 2 h to remove the chlorophyll. Seedlings were mounted in lactic acid (90%) and visualized on a binocular (Stemi SV11; Zeiss).
Flow Cytometry Analysis
Leaf blades from three plants were pooled and chopped with a razor blade in 300 μL of 45 mm MgCl2, 30 mm sodium citrate, 20 mm MOPS (pH 7.0), and 1% Triton X-100 (Galbraith et al., 1991). To the supernatant, 1 μL of 4′,6-diamidino-2-phenylindole (DAPI) from a stock of 1 mg mL−1 was added that had been filtered over a 30-μm mesh. The nuclei were analyzed with the CyFlow cytometer and Flomax software (Partec).
DAPI Staining and Measurements of Nuclear DNA Quantity
Leaves were fixed in a solution of three-quarters 95% ethanol and one-quarter glacial acetic acid for 2 h at room temperature and stored in 70% ethanol at 4°C. Fixed tissue was soaked first in water and then in 0.5 m EDTA (pH 8.0). Trichomes from leaves from six plants were removed from the abaxial epidermis with a pair of fine forceps, pooled, and mounted on a glass slide. A drop of DAPI at a concentration of 0.005 mg mL−1 in McIlvaines's buffer (pH 4.1; 60 mL of 0.1 mol L−1 citric acid + 40 mL of 0.2 mol mL−1 Na2HPO4) was put on the isolated trichomes. The trichomes were covered with a drop of Vectashield medium (Vector Laboratories) and observed with a 20× objective and a 40× objective on an Axioskop microscope equipped with an Axiocam CCD camera (Zeiss). Images were obtained with the Axiovision software and analyzed in gray-scale with ImageJ. The nuclear area was manually circumscribed, and the size and fluorescence intensity were determined. The relative DNA content is reflected by the integrated density, which is the product of the area and the average fluorescence (Boudolf et al., 2004).
Scanning Electron Microscopy
Leaves were harvested and fixed overnight at 4°C in a solution of 4% (v/v) paraformaldehyde, 1% (v/v) glutaraldehyde (25% solution), and 0.02 m sodium phosphate buffer (pH 7.0). After fixation, samples were rinsed three times in sodium phosphate buffer and postfixed in 1% (v/v) OsO4 (in the same buffer) for 2 h. After dehydration through a graded ethanol series (30%, 50%, 70%, 95%, and 100% three times, 15 min for each step), specimens were dried with CO2 by the critical point dryer CPD 030 (Bal-Tec). The samples were coated with a gold layer with a JFC-1200 sputter coater (JEOL), mounted on scanning electron microscope stubs with double-sided sticky carbon tape, and examined with a scanning electron microscope (JSM-5600 LV; JEOL) under an accelerating voltage of 5 kV.
Anthocyanin Measurement
Pigments were extracted by incubating pooled whole seedlings (n = 3) for 48 h at 4°C in acidic (1% HCl [w/v]) methanol. The absorbance of the extracts, clarified by filtration, was measured at 530 nm (absorption peak of anthocyanin) and 657 nm (absorption peak of chlorophyll in acidic methanol) with a Life Science UV/VIS spectrophotometer (Beckman). The formula A530 − 0.33 A657 was used to compensate for the contribution of chlorophyll and its degradation products to the absorption at 530 nm (Mancinelli and Schwartz, 1984).
qPCR
RNA was extracted with Plant Reagent (Invitrogen) from pooled leaves of 20 plants. Poly(dT) cDNA was prepared from 1 μg of total RNA with SuperScriptII reverse transcriptase (Invitrogen) and quantified on an iCycler apparatus (Bio-Rad) with the qPCR core kit for SYBR Green I (Eurogentec). PCR was carried out on 96-well optical reaction plates heated for 10 min at 95°C to activate hot-start Taq DNA polymerase, followed by 50 cycles of denaturation for 60 s at 95°C and annealing extension for 60 s at 58°C. Targets were quantified with specific primer pairs designed with the Beacon Designer 4.0 (Premier Biosoft International) or Probe library (http://www.probelibrary.com/). All PCRs were done in triplicate. Expression levels were normalized to those of the ACTIN2 control gene.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Influence of the phytohormones JA, BAP, and GA3 on trichome development in Arabidopsis Col-0 and C24 seedlings.
Supplemental Figure S2. Expression of R1-MYB genes in phytohormone-treated rosette leaves.
Supplemental Figure S3. Phytohormonal elicitation of leaf anthocyanin biosynthesis.
Supplemental Figure S4. Expression of bHLH genes in single bHLH mutants.
Supplemental Table S1. Overview of the effects of phytohormone treatment on Col-0 leaf parameters.
Supplemental Table S2. Trichome nuclear area parameters.
Supplementary Material
Acknowledgments
We thank John Larkin for providing the ProGL1:GUS line and the ttg1, gl3egl3, and gl1 mutants; Alan Lloyd for the ProGL2:GUS, ProGL3:GUS, and ProEGL3:GUS lines; Martin Hülskamp for the gl1myb23 mutant; Loïc Lepiniec for the ProTT8:GUS line and the tt8 and gl3egl3tt8 mutants; Renaat Dasseville for excellent technical microscope assistance; Gerrit Beemster and Tom Beeckman for helpful discussion and critical reading of the manuscript; and Martine De Cock for help in preparing it.
This work was supported by grants from Ghent University (project no. VARL9104) and the Vlaamse Interuniversitaire Raad (grant no. VLADOC–B/09269/02).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Alain Goossens (algoo@psb.ugent.be).
The online version of this article contains Web-only data.
References
- Barth S, Melchinger AE, Lübberstedt T (2002) Genetic diversity in Arabidopsis thaliana L. Heynh. investigated by cleaved amplified polymorphic sequence (CAPS) and inter-simple sequence repeat (ISSR) markers. Mol Ecol 11 495–505 [DOI] [PubMed] [Google Scholar]
- Baudry A, Caboche M, Lepiniec L (2006) TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J 46 768–779 [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39 366–380 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Barrôco R, de Almeida Engler J, Verkest A, Beeckman T, Naudts M, Inzé D, De Veylder L (2004) B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol 8 272–279 [DOI] [PubMed] [Google Scholar]
- Chien JC, Sussex IM (1996) Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol 111 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Xia GX, Hong Y, Ramachandran S, Kost B, Chua NH (2001) ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55 940–953 [DOI] [PubMed] [Google Scholar]
- Esch JJ, Chen M, Sanders M, Hillestad M, Ndkium S, Idelkope B, Neizer J, Marks MD (2003) A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130 5885–5894 [DOI] [PubMed] [Google Scholar]
- Esch JJ, Chen MA, Hillestad M, Marks MD (2004) Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J 40 860–869 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Knapp S (1991) Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol 96 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Kumimoto R, Liu C, Ratcliffe O, Yu H, Broun P (2006) GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell 18 1383–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Liu C, Yu H, Broun P (2007) Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8, and GIS2 in the regulation of epidermal cell fate. Development 134 2073–2081 [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53 814–827 [DOI] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20 735–747 [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Miséra S, Jürgens G (1994) Genetic dissection of trichome cell development in Arabidopsis. Cell 76 555–566 [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Schnittger A (1998) Spatial regulation of trichome formation in Arabidopsis thaliana. Semin Cell Dev Biol 9 213–220 [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Schnittger A, Folkers U (1999) Pattern formation and cell differentiation: trichomes in Arabidopsis as a genetic model system. Int Rev Cytol 186 147–178 [DOI] [PubMed] [Google Scholar]
- Kirik V, Lee MM, Wester K, Herrmann U, Zheng Z, Oppenheimer D, Schiefelbein J, Hülskamp M (2005) Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132 1477–1485 [DOI] [PubMed] [Google Scholar]
- Kirik V, Schnittger A, Radchuk V, Adler K, Hülskamp M, Bäumlein H (2001) Ectopic expression of the Arabidopsis AtMYB23 gene induces differentiation of trichome cells. Dev Biol 235 366–377 [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Hülskamp M, Schiefelbein J (2004. a) The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol 268 506–513 [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Wester K, Schiefelbein J, Hülskamp M (2004. b) ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol Biol 55 389–398 [DOI] [PubMed] [Google Scholar]
- Larkin JC, Brown ML, Schiefelbein J (2003) How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu Rev Plant Biol 54 403–430 [DOI] [PubMed] [Google Scholar]
- Larkin JC, Marks MD, Nadeau J, Sack F (1997) Epidermal cell fate and patterning in leaves. Plant Cell 9 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin JC, Young N, Prigge M, Marks MD (1996) The control of trichome spacing and number in Arabidopsis. Development 122 997–1005 [DOI] [PubMed] [Google Scholar]
- Mancinelli AL, Schwartz OM (1984) The photoregulation of anthocyanin synthesis. IX. The photosensitivity of the response in dark and light-grown tomato seedlings. Plant Cell Physiol 25 93–105 [Google Scholar]
- Marks MD (1997) Molecular genetic analysis of trichome development in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol 48 137–163 [DOI] [PubMed] [Google Scholar]
- Matsui K, Umemura Y, Ohme-Takagi M (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55 954–967 [DOI] [PubMed] [Google Scholar]
- Mauricio R (2005) Ontogenetics of QTL: the genetic architecture of trichome density over time in Arabidopsis thaliana. Genetica 123 75–85 [DOI] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW (1993) Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Zhao M, Yang M, Read B, Lloyd A, Lamb R, Grotewold E (2007) Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol 145 736–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Sheen J (2007) Advances in cytokinin signaling. Science 318 68–69 [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67 483–493 [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Vachon G, Herzog M (1998) Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant Physiol 117 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M, Hülskamp M (2004) Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr Opin Genet Dev 14 422–427 [DOI] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD (1994) The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8 1388–1399 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JAH (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283 1541–1544 [DOI] [PubMed] [Google Scholar]
- Schellmann S, Hülskamp M, Uhrig J (2007) Epidermal pattern formation in the root and shoot of Arabidopsis. Biochem Soc Trans 35 146–148 [DOI] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J 21 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A, Folkers U, Schwab B, Jürgens G, Hülskamp M (1999) Generation of spacing pattern: the role of TRIPTYCHON in trichomes patterning in Arabidopsis. Plant Cell 11 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A, Jürgens G, Hülskamp M (1998) Tissue layer and organ specificity of trichome formation are regulated by GLABRA1 and TRIPTYCHON in Arabidopsis. Development 125 2283–2289 [DOI] [PubMed] [Google Scholar]
- Serna L, Martin C (2006) Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci 11 274–280 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Marks MD (1998) GLABROUS1 overexpression and TRIPTYCHON alter the cell cycle and trichome cell fate in Arabidopsis. Plant Cell 10 2047–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski DB, Marks MD, Wick SM (1999) Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11 2331–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124 645–654 [DOI] [PubMed] [Google Scholar]
- Tominaga R, Iwata M, Sano R, Inoue K, Okada K, Wada T (2008) Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135 1335–1345 [DOI] [PubMed] [Google Scholar]
- Traw MB, Bergelson J (2003) Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol 133 1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K (1997) Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277 1113–1116 [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11 1337–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40 22–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.