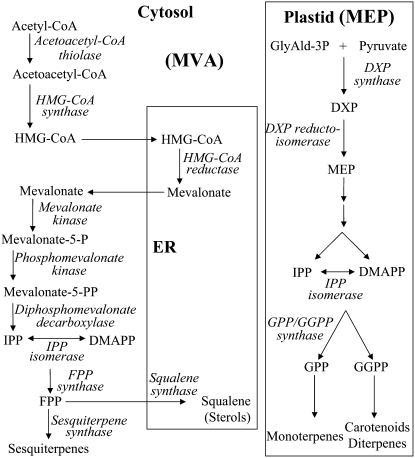
Isoprenoids, the largest family of natural products, play numerous vital roles in basic plant processes, including photosynthesis, growth and development, reproduction, plant defense, and adaptation to environmental conditions (Gershenzon and Kreis, 1999; Rodriguez-Concepcion and Boronat, 2002). All isoprenoids are synthesized via the condensation of the five-carbon universal isoprenoid precursors, isopentenyl diphosphate (IPP) and its allylic isomer dimethylallyl diphosphate (DMAPP; McGarvey and Croteau, 1995). In higher plants, two independent pathways located in separate intracellular compartments are involved in the biosynthesis of IPP and DMAPP (see current model for plant isoprenoid pathways compartmentalization in Fig. 1). In the mevalonic acid (MVA) pathway, which is referred to as the cytosolic pathway, biosynthesis of IPP/DMAPP starts from the condensation of acetyl-CoA (Qureshi and Porter, 1981; Newman and Chappell, 1999), whereas in plastids, IPP/DMAPP are formed from pyruvate and glyceraldehyde-3-P via the methylerythritol phosphate (MEP; or non-mevalonate) pathway (Lichtenthaler et al., 1997, 1999; Eisenreich et al., 1998, 2001; Rohmer, 1999). Initial research indicated that the MVA-pathway-derived pool of IPP/DMAPP serves as a precursor of farnesyl diphosphate (FPP; C15) and, ultimately, the sesquiterpenes, triterpenes, and sterols, whereas the plastidic MEP-pathway-derived pool of IPP/DMAPP provides the precursors of geranyl diphosphate (C10), geranylgeranyl diphosphate (C20), and, ultimately, the monoterpenes, diterpenes, and tetraterpenes. Although the subcellular compartmentalization of MVA and MEP pathways allows them to operate independently, metabolic cross talk between these two pathways was recently implied (Bick and Lange, 2003; Hemmerlin et al., 2003; Laule et al., 2003; Schuhr et al., 2003; Dudareva et al., 2005; Hampel et al., 2005).
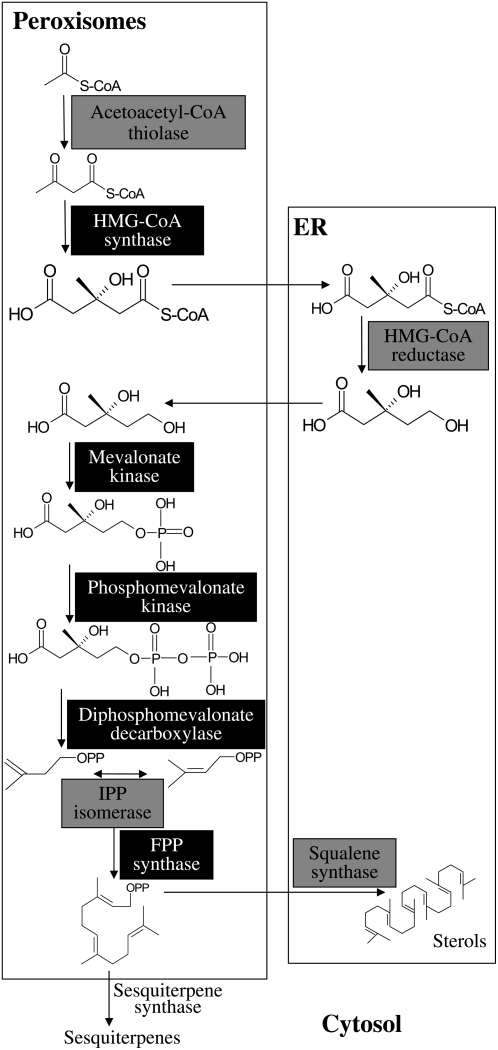
Figure 1.
Scheme of plant isoprenoid biosynthetic pathways. The MEP pathway is localized to the plastid; the MVA pathway is referred to as cytosolic and is localized partially in the ER compartment. We note that the mitochondrial branch of isoprenoid biosynthesis is not illustrated in the figure. GlyAld-3P, Glyceraldehyde-3-P; DXP, 1-deoxy-d-xylulose-5-P.
Regulation of plant isoprenoid biosynthetic pathways, the control of flux, and the channeling of intermediates are issues of major interest but still not well understood. Flux of the MVA pathway appears to be tightly regulated. Data obtained mainly from mammalian systems, but also from plant systems, suggest that 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) is a rate-limiting enzyme, which is posttranslationally regulated by feedback-inhibition mechanisms (Chappell et al., 1995; Gardner and Hampton, 1999). A major enigma, specific for plants, concerns the differential channeling of the MVA pathway flux between the two pathway branches, one leading to sesquiterpenes and the other to sterols. Several bodies of evidence support the notion that FPP, which is positioned at the branch point between sterol and sesquiterpenes, is available mainly for sterol biosynthesis (Hartmann, 2003), which is required in every cell, while its availability for sesquiterpene biosynthesis is limited to specific physiological conditions (i.e. pathogen attack) and/or to specific tissues (i.e. essential oil glands). (1) In healthy tobacco (Nicotiana tabacum) plants and tobacco-derived cell culture, sesquiterpenes, including the phytoalexin capsidiol, are almost undetectable, but are produced to significant levels only following exposure to pathogens or elicitation (Chappell and Nable, 1987; Vogeli and Chappell, 1988). (2) Constitutive overexpression in tobacco of a nonregulated form of HMGR (lacking the N-terminal membrane anchor) resulted in a 3- to 10-fold increase in total sterol levels but no increase in sesquiterpene levels (Chappell et al., 1995). (3) Constitutive overexpression of citrus valencene synthase (Sharon-Asa et al., 2003) in transgenic tobacco did not result in synthesis of the sesquiterpene valencene. However, treatment of these transgenic plants with elicitor resulted in valencene synthesis (S. Tal-Meshulam, A. Frydman, E. Bar, E. Lewinsohn, and Y. Eyal, unpublished data). (4) Constitutive overexpression of a germacrene A synthase in Arabidopsis (Arabidopsis thaliana) resulted in production of only trace amounts of germacrene A (Aharoni et al., 2003). (5) Constitutive overexpression of patchoulol synthase in tobacco results in negligible accumulation of the sesquiterpene patchoulol, compared to the high levels obtained when cotargeting the enzyme together with FPP synthase into plastids (Wu et al., 2006). These accumulating data support a mechanism that directs/channels the MVA pathway flux possibly through compartmentalization or metabolic channeling.
Compartmentalization of MVA pathway enzymes has been extensively studied in mammalian systems, although some debate regarding this issue persists. Compelling evidence locates many of the mammalian MVA pathway enzymes to peroxisomes (for review, see Kovacs et al., 2002), organelles that have been recently shown to be involved in various metabolic pathways in both mammals and plants. The first MVA pathway enzyme to be localized to peroxisomes was rat (Rattus norvegicus) HMGR (Keller et al., 1985). This, in addition to previous endoplasmic reticulum (ER) localization of HMGR (Goldfarb, 1972), demonstrates dual localization. Acetoacetyl-CoA thiolase, which is involved but not unique to the MVA pathway, was identified as a peroxisomal protein in mammals (Thompson and Krisans, 1990; Olivier et al., 2000). Immunofluorescence was used to demonstrate that overexpressed hamster (Mesocricetus auratus) IPP isomerase fused to a hemagglutinin epitope tag was targeted to peroxisomes (Paton et al., 1997). Subcellular fractionation studies and immunoelectronmicroscopy show that mammalian mevalonate kinase is a peroxisomal protein (Stamellos et al., 1992; Biardi et al., 1994) and immunofluorescence microscopy was used to demonstrate that FPP synthase is localized to mammalian peroxisomes (Kovacs et al., 2007). Phosphomevalonate kinase was also identified as a peroxisomal protein by monitoring the location of the gene product fused to the GFP (Olivier et al., 1999). These results have been challenged by data supporting the possibility that the biosynthetic reactions leading to mammalian sterols (e.g. cholesterol) occur mostly in the cytosol (Hogenboom et al., 2004; Wanders and Waterham, 2006). Localization of the rate-limiting enzyme, HMGR, and the first committed enzyme of sterol biosynthesis, squalene synthase, to the ER are in general agreement.
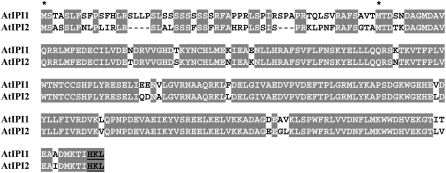
In plants, only fragmented experimental data exists regarding the localization of the MVA pathway enzymes. The rate-limiting enzyme, HMGR, has been localized to the ER as well as to unidentified spherical structures (Campos and Boronat, 1995; Leivar et al., 2005), thus mostly conforming to the picture seen in mammals (Goldfarb, 1972). Squalene synthase was shown to be targeted to the ER by a C-terminal hydrophobic trans-membrane domain (Busquets et al., 2008), also conforming to the data from mammalian systems (Stamellos et al., 1993). Acetoacetyl-CoA thiolase, an enzyme involved but not unique to the MVA pathway, was recently identified as a peroxisomal protein by a proteomic approach (Reumann et al., 2007), again conforming to the data from mammalian systems (Thompson and Krisans, 1990). In contrast, localization studies performed up to now on plant IPP isomerases (IPIs), which catalyze the conversion between the isoprenoid building blocks IPP and DMAPP, do not conform to the peroxisomal localization seen in mammalian systems. The emerging picture suggests that Arabidopsis and other plant species contain two IPI genes (Campbell et al., 1998), which are transcribed each as long and short isoforms (see Fig. 2). The long transcripts contain the entire open reading frame (ORF) starting from the first Met codon, and their protein products appear to be targeted to organelles (IPI1-long to plastids and IPI2-long to mitochondria). The short transcripts contain ORFs that start at the second Met codon, and their respective protein products are suggested to be cytoplasmic (Nakamura et al., 2001; Okada et al., 2008; Phillips et al., 2008). However, all localization studies of the IPI gene products were based on C-terminal fusions of IPI sequences to GFP, which may mask C-terminal targeting signals (Tian et al., 2004).
Figure 2.
Alignment of Arabidopsis IPP isomerase deduced amino acid sequences. AtIPI1 and AtIPI2 represent Arabidopsis IPP isomerase 1 and IPP isomerase 2, respectively. Identical amino acids are gray shadowed. Asterisks denote the first Met amino acids of the ORFs of long and short IPI transcripts, previously described by Phillips et al. (2008) and prevalent in the EST databases. Putative PTS1 peroxisomal-targeting motif {HKL} appears in bold black and is underlined.
In this study we present evidence that previous localizations of IPI versions to the cytosol likely resulted from mislocalization due to GFP fusion design. Internal GFP fusions of Arabidopsis IPI enzymes confirm that the long versions encoded by the full IPI gene ORFs are targeted to chloroplasts/mitochondria, and demonstrate that the short versions that start from the second Met codon of the gene ORF are localized to peroxisomes. A bioinformatic survey of Arabidopsis MVA pathway enzyme sequences implies that additional enzymes of this pathway may be localized to peroxisomes, suggesting that part of the MVA pathway is compartmentalized in the peroxisomes. The results offer a mechanistic explanation for substrate channeling in the MVA pathway.
SUBCELLULAR LOCALIZATION OF IPI ISOFORMS
We examined the predicted subcellular localization of the long and short isoforms of Arabidopsis IPI1 and IPI2 by bioinformatic tools (Table I). While IPI-long versions (starting from the first Met codon) are predicted by several algorithms to be localized to plastids and/or mitochondria, IPI-short versions (starting from the second Met codon) are predicted to be localized to peroxisomes by a C-terminal peroxisomal targeting signal (peroxisomal targeting signal 1 [PTS1]; Elgersma et al., 1996; Emanuelsson et al., 2003; see Fig. 2). This peroxisomal prediction is limited to two algorithms, since most motif-based localization predictors do not cover the peroxisomal option (Table I). Thus, bioinformatic targeting predictions of IPI-long isoforms conform to the previous experimental localizations to chloroplasts and/or mitochondria, while bioinformatic targeting predictions of the IPI-short isoforms to peroxisomes are in disagreement with the previous experimental localization to the cytosol (Nakamura et al., 2001; Okada et al., 2008; Phillips et al., 2008). This discrepancy between the bioinformatic prediction and the experimental localizations of the short IPI isoforms prompted us to investigate the localization of Arabidopsis IPI gene products in greater depth. We note that all previous localization experiments were conducted using C-terminal fusions to GFP, which would mask putative C-terminal PTS1 peroxisomal targeting sequences. Thus, it is possible that the cytosolic localizations obtained in previous experiments are mislocalizations due to the GFP fusion design.
Table I.
Bioinformatic predictions for the subcellular localization of IPI protein versions
NR, Nonrelevant due to lack of software possibilities to allow for prediction of either peroxisome or plastid/mitochondria targeting.
| Program | IPI1-Long Prediction | IPI1-Short Prediction | IPI2-Long Prediction | IPI2-Short Prediction |
|---|---|---|---|---|
| iPSORT | Plastid | NR | Plastid | NR |
| MultiLoc | Plastid | NR (other) | Plastid | NR (other) |
| Predotar | Plastid | NR (other) | Mitochondria | NR (other) |
| TargetP | Plastid | NR (other) | Plastid/mitochondria | NR (other) |
| PTS1 predictor | NR | Peroxisome | NR | Peroxisome |
| PeroxiP | NR | Peroxisome | NR | Peroxisome |
To obtain GFP-fusion-based localizations without masking N- and C-terminal potential targeting signals, we designed internal GFP fusions within Arabidopsis IPI1 versions (Fig. 3). The internal GFP fusion approach follows recent work for fluorescent tagging of full-length proteins in Arabidopsis (Tian et al., 2004). Considerations regarding the location of the internal GFP fusions include: (1) the default criteria of about 10 amino acids upstream from the C terminus, unless specific protein features suggest otherwise (Tian et al., 2004); (2) a protein region that is distant from the catalytic site and that is not likely to affect the native folding (i.e. based on the three-dimensional structure of human [Homo sapiens] IPI PDB no. 2icj); and (3) a protein region that is hydrophilic. Chimeric internal fusion genes, IPI1-long:GFP and IPI1-short:GFP, were generated by insertion of sequences encoding GFP within the IPI1 ORF (Fig. 3). Chimeric genes were transiently expressed under control of the cauliflower mosaic virus (CaMV) 35S promoter in tobacco mesophyll protoplasts. Confocal laser scanning microscopy was used to determine the localization pattern of the expressed proteins in correlation to localization standards.
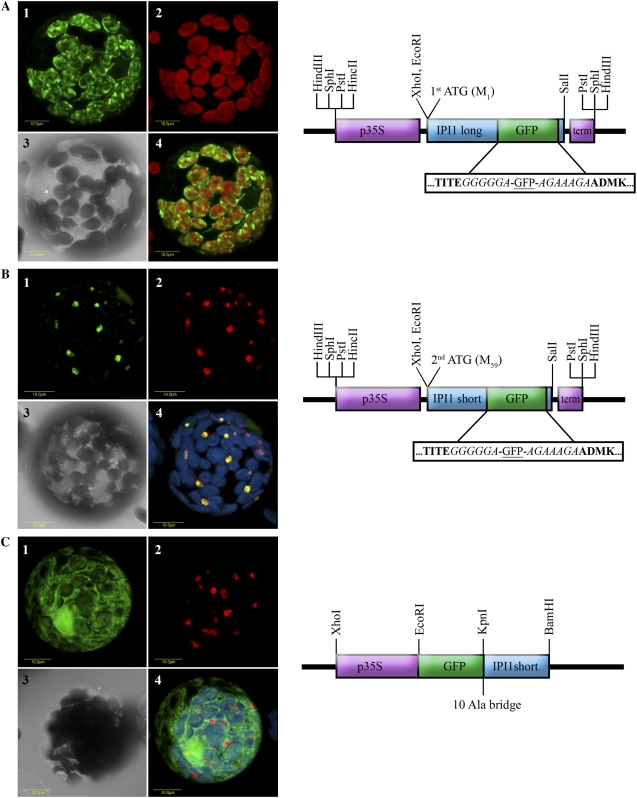
Figure 3.
Localization of Arabidopsis IPI1 versions by transient expression of IPI1-GFP fusions in tobacco protoplasts. A, The full-length ORF of IPI1 encoded by the long transcript (starting from the first Met codon) was fused internally with GFP as schematically shown and as described in the methods. The construct was transiently transformed into tobacco protoplasts, which were visualized using a laser scanning confocal microscope. A representative protoplast is visually presented as follows: section 1, green fluorescence corresponds to GFP; section 2, red fluorescence corresponds to chlorophyll; section 3, bright-field image; section 4, confocal image recorded simultaneously for green and red fluorescence (i.e. GFP and chlorophyll fluorescence overlaid). B, The ORF of IPI1 encoded by the short transcript (starting from the second Met codon) was fused internally with GFP as schematically shown and as described in the methods. The construct was transiently transformed into tobacco protoplasts, which were visualized using a laser scanning confocal microscope. A representative protoplast is visually presented as follows: section 1, green fluorescence corresponds to GFP; section 2, red fluorescence corresponds to peroxisome marker Cherry-PTS1 (Avisar et al., 2008); section 3, bright-field image; section 4, confocal image recorded simultaneously for GFP (green fluorescence), Cherry-PTS1 (red fluorescence), and chlorophyll fluorescence (coded blue, overlaid). C, The ORF of IPI1 encoded by the short transcript (starting from the second Met codon) was fused N terminally to GFP as schematically shown and as described in the methods. The construct was transiently transformed into tobacco protoplasts, which were visualized using a laser scanning confocal microscope. A representative protoplast is visually presented as follows: section 1, green fluorescence corresponds to GFP; section 2, red fluorescence corresponds to peroxisome marker Cherry-PTS1 (Avisar et al., 2008); section 3, bright-field image; section 4, confocal image recorded simultaneously for GFP (green fluorescence), Cherry-PTS1 (red fluorescence), and chlorophyll fluorescence (coded blue, overlaid). Schematic representation of the constructs are as follows: p35S (purple box) designates CaMV 35S promoter; term (purple box) designates CaMV 35S terminator; GFP (green box) designates the ORF of the green fluorescent protein; IPI1 long and IPI1 short designate the ORF of Arabidopsis IPI1 starting from the first or second Met codon, respectively (blue box). Boxed sequence blowup shows amino acid residues flanking the GFP sequence; residues in bold are Arabidopsis IPI1 sequences; residues in italic are protein spacer sequences. 10 Ala bridge designates a spacer of 10 Ala residues between the GFP and IPI1 sequences.
Figure 3A demonstrates the localization of IPI1-long:GFP to plastids in a representative cell. The green fluorescence of GFP (Fig. 3A, section 1) and the red fluorescence of chlorophyll (section 2) entirely overlap (section 4). This result is in agreement with the bioinformatic predictions of the protein encoded by the full-length ORF (IPI1-long) to plastids (Table I) and with previous experimental localizations (Okada et al., 2008; Phillips et al., 2008). We note that the green fluorescence is also evident in stromules, which are plastid-associated stroma-filled tubules (Fig. 3A, section 1). We also note occasional colocalization of IPI1-long:GFP to chloroplasts and mitochondria (data not shown), suggesting that IPI1-long may be dual localized (chloroplast/mitochondria) albeit at much higher efficiency to the chloroplast.
Figure 3B demonstrates the localization of IPI1-short:GFP to peroxisomes in a representative cell. The green fluorescence of GFP (Fig. 3B, section 1) and the red fluorescence of the peroxisomal marker Cherry-PTS1 (Avisar et al., 2008; section 2) entirely overlap (section 4). This result, documenting localization of a dedicated plant MVA pathway enzyme to peroxisomes, is in agreement with bioinformatic predictions for the protein encoded by the ORF starting with the second Met codon (IPI1-short; Table I), and also with extensive results demonstrating the localization of the mammalian IPI to peroxisomes (Paton et al., 1997; Kovacs et al., 2007).
Figure 3C shows the apparent mislocalization of IPI1-short to the cytosol when fused N terminally to GFP. We note that GFP fluorescence is also seen in the nucleus, which is likely the result of the well-known diffusion of relatively small GFP fusions through the nuclear pore complex (Kohler, 1998). Thus, it appears that the C-terminal PTS1 signal is insufficient to target IPI1-short to peroxisomes on its own, and that essential N-terminal signals are masked when constructing N-terminal fusions.
Recent extensive work suggests that the function of the two Arabidopsis IPIs, IPI1 and IPI2, is partly redundant, since single homozygous mutants of each had no major morphological or chemical difference from wild-type plants, while the double mutant was severely growth impaired or lethal (Okada et al., 2008; Phillips et al., 2008). Indeed the organ-specific expression profile of the two genes is similar, although expression levels are somewhat different (Okada et al., 2008; Phillips et al., 2008). Thus, it was of much interest to determine whether the long and short isoforms of the two genes overlap also at the level of subcellular localization. We therefore designed internal GFP fusions within Arabidopsis IPI2 long and short versions (IPI2-long:GFP and IPI2-short:GFP), similar to the design for IPI1 versions. Chimeric IPI2-GFP genes were transiently expressed in tobacco mesophyll protoplasts and localization of the products was studied by confocal microscopy, in correlation to localization standards (Fig. 4).
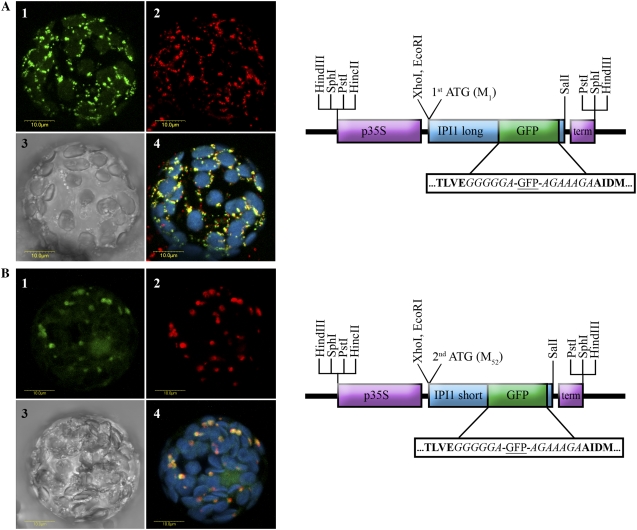
Figure 4.
Localization of Arabidopsis IPI2 versions by transient expression of IPI2-GFP fusions in tobacco protoplasts. A, The full-length ORF of IPI2 encoded by the long transcript (starting from the first Met codon) was fused internally with GFP as schematically shown and as described in the methods. The construct was transiently transformed into tobacco protoplasts, which were visualized using a laser scanning confocal microscope. A representative protoplast is visually presented as follows: section 1, green fluorescence corresponds to GFP; section 2, red fluorescence corresponds to the mitochondria marker MitoTracker (Invitrogen); section 3, bright-field image; section 4, confocal image recorded simultaneously for GFP (green fluorescence), MitoTracker (red fluorescence), and chlorophyll fluorescence (coded blue, overlaid). B, The ORF of IPI2 encoded by the short transcript (starting from the second Met codon) was fused internally with GFP as schematically shown and as described in the methods. The construct was transiently transformed into tobacco protoplasts, which were visualized using a laser scanning confocal microscope. A representative protoplast is visually presented as follows: section 1, green fluorescence corresponds to GFP; section 2, red fluorescence corresponds to peroxisome marker Cherry-PTS1 (Avisar et al., 2008); section 3, bright-field image; section 4, confocal image recorded simultaneously for GFP (green fluorescence), Cherry-PTS1 (red fluorescence), and chlorophyll fluorescence (coded blue, overlaid). Schematic representation of the constructs are as follows: p35S (purple box) designates CaMV 35S promoter; term (purple box) designates CaMV 35S terminator; GFP (green box) designates the ORF of the green fluorescent protein; IPI2 long and IPI2 short designate the ORF of Arabidopsis IPI2 starting from the first or second Met codon, respectively (blue box). Boxed sequence blowup shows amino acid residues flanking the GFP sequence; residues in bold are Arabidopsis IPI2 sequences; residues in italic are protein spacer sequences.
Figure 4A demonstrates the localization of IPI2-long:GFP to mitochondria in a representative cell, in correlation to the mitochondrial marker MitoTracker (Invitrogen). The green fluorescence of GFP (Fig. 4A, section 1) and the red fluorescence of the MitoTracker (section 2) entirely overlap (section 4). We note that the green fluorescence signal is also apparent weakly in plastids in many of the cells (data not shown), suggesting that IPI2-long may be dual localized (mitochondria/chloroplast) albeit at much higher efficiency to the mitochondria. These results are in agreement with the bioinformatic predictions of the protein encoded by the full-length ORF (IPI2-long) to mitochondria and plastids (Table I) and with previous experimental localizations of IPI2-long to mitochondria (Okada et al., 2008; Phillips et al., 2008).
Figure 4B demonstrates the localization of IPI2-short:GFP to peroxisomes in a representative cell. The green fluorescence of GFP (Fig. 4B, section 1) and the red fluorescence of the peroxisomal marker Cherry-PTS1 (Avisar et al., 2008, section 2) entirely overlap (section 4). This result is in agreement with bioinformatic predictions for the protein encoded by the ORF starting with the second Met codon (IPI2-short; Table I), and also with extensive results demonstrating the localization of the mammalian IPI to peroxisomes (Paton et al., 1997; Kovacs et al., 2007). Thus, our data shows that the short versions of IPI are both localized exclusively to peroxisomes. The overlap in expression and subcellular localization of the short versions explains why the single gene mutants are not impaired in sterol biosynthesis (Phillips et al., 2008). However, the overlap in subcellular localization of the long IPI versions is only partial. Our data suggests that both Arabidopsis long versions are dual localized to plastids and mitochondria, albeit at different efficiencies. While IPI1 is predominantly plastidic, IPI2 is predominantly mitochondrial. Nevertheless, it appears that this partial overlap is sufficient for complementation of function evident in the single IPI gene mutant plants (Okada et al., 2008; Phillips et al., 2008).
BIOINFORMATIC EVIDENCE FOR PEROXISOMAL LOCALIZATION OF ADDITIONAL PLANT MVA PATHWAY ENZYMES
The peroxisomal localization of Arabidopsis IPI1-short and IPI2-short versions motivated us to seek evidence for the potential localization of additional plant MVA pathway enzymes to peroxisomes. Deduced amino acid sequences of the Arabidopsis MVA pathway enzymes were analyzed for the existence of consensus signals for PTS and their similarity to the previously described consensus sequences of their respective human MVA pathway enzymes (Kovacs et al., 2007). The analysis was done based on the currently characterized classes of peptide signals for peroxisomal targeting. The first characterized PTS (PTS1) is a tripeptide with the consensus sequence (S/A/C)(K/H/R)(L/M) found at the extreme C terminus of many peroxisomal proteins (Gould et al., 1987; Elgersma et al., 1996). The second PTS (PTS2) is a nine-amino-acid consensus sequence (R/K)(L/V/I)(X5)(H/Q)(L/A) that can be found at variable distances from the N terminus (Swinkels et al., 1991; Gietl et al., 1994; Tsukamoto et al., 1994). Although these consensus sequences have been defined, there is evidence that the targeting information may consist of a structural or charge-based motif (Flynn et al., 1998). Moreover, there are continuous additions/modifications to the consensus sequences for PTS1 and PTS2 motifs and it was recently demonstrated that functional PTS2 can be located at an internal position of the protein (Reumann et al., 2007).
We identified PTS1-like C-terminal motifs in the deduced amino acid sequences of Arabidopsis acetoacetyl-CoA thiolase, which was recently shown by proteomic analysis to reside in the peroxisome (Reumann et al., 2007), and of Arabidopsis IPIs (Table II). In both cases, the corresponding mammalian MVA pathway enzymes contain a PTS1 motif (Table II). PTS2-like motifs were found in Arabidopsis 3-hydroxy-3-methylglutaryl (HMG)-CoA synthase, mevalonate kinase, and mevalonate diphosphate decarboxylase (Table II). The corresponding mammalian MVA pathway enzymes contain similar PTS2 motifs at similar positions relative to their respective N terminus. We note that mammalian IPIs present a special case by containing both PTS1 and PTS2 motifs (Table II). It is likely that Arabidopsis IPI similarly requires both C-and N-terminal signals for peroxisomal targeting, since both C-terminal and N-terminal GFP fusions to Arabidopsis IPI-short were apparently mislocalized to the cytosol (Okada et al., 2008; Fig. 3C). However, we do not detect an obvious PTS2 consensus sequence in the Arabidopsis IPI deduced amino acid sequence. We note that FPP synthase bears no clear PTS motifs in Arabidopsis as well as in mammals, but was localized to peroxisomes in mammals (Krisans et al., 1994; Kovacs et al., 2007). Thus, the comparison of deduced amino acid sequences of MVA pathway enzymes reveals high conservation of PTS motifs between mammals and plants.
Table II.
Sequences of putative PTS motifs in MVA pathway enzymes
At, A. thaliana; Hs, H. sapiens; *, no consensus PTS1 motif; **, no consensus PTS2 motif.
| Protein Name | PTS1 Motif | Organism | Accession No. |
|---|---|---|---|
| Acetoacetyl-CoA thiolase | SAL | At | NM_124146 |
| QKL | Hs | NM_000019 | |
| Phosphomevalonate kinase | * | At | NM_102927 |
| SRL | Hs | BC007694 | |
| IPP isomerase | HKL | At (IPI1) | NM_121649 |
| HKL | At (IPI2) | NM_111146 | |
| YRM | Hs | AF271720 | |
| PTS2 Motif
|
|||
| HMG-CoA synthase | SIKTFLMQL | At | NM_117251 |
| SVKTNLMQL | Hs | BC000297 | |
| Mevalonate kinase | KIILAGEHA | At | NM_180753 |
| KVILHGEHA | Hs | M88468 | |
| Mevalonate-PP decarboxylase | SVTLDPDHL | At | NM_129427 |
| SVTLHQDQL | Hs | U49260 | |
| IPP isomerase | ** | At (IPI1) | NM_121649 |
| ** | At (IPI2) | NM_111146 | |
| HLDKQQVQL | Hs | AF271720 | |
| Consensus PTS1 | (S/C/A)(K/R/H)(L/M) | ||
| Consensus PTS2 | (R/K)(L/V/I)X5(H/Q)(L/A) |
We present evidence for compartmentalization of part of the plant MVA pathway to the peroxisomes: (1) The enzymes IPI1 and IPI2 (short isoforms) from Arabidopsis were experimentally localized to the peroxisome (this work); (2) most of the Arabidopsis MVA pathway enzymes were found to contain PTS-like motifs, which are very similar in sequence and position to their mammalian counterparts (this work); (3) a strong body of evidence locates most of the mammalian MVA pathway enzymes to peroxisomes (for review, see Kovacs et al., 2002, 2007); (4) one plant MVA pathway-related enzyme (acetoacetyl-CoA thiolase) was detected in a proteomic study of Arabidopsis peroxisomes (Reumann et al., 2007). Other enzymes were likely not identified by this proteomic study due to low constitutive expression of the enzymes in the healthy plant material (i.e. plants not exposed to pathogens or elicitors) that was used. Similarly, most of the mammalian MVA pathway enzymes were not detected in peroxisomal proteomic studies. Detection of the first two mammalian MVA pathway peroxisomal enzymes (HMG-CoA synthase and HMG-CoA lyase) in a proteomic study was only recently described (Mi et al., 2007). (5) We note that previous molecular/biochemical findings regarding the plant MVA pathway do not preclude compartmentalization to the peroxisome. Recent elegant feeding experiments using radiolabeled mevalonate show that sterols are derived solely from MVA-pathway-derived IPP/DMAPP (Phillips et al., 2008). However, since mevalonate feeding at the cellular level is not compartment specific, this experimental setup could not distinguish between a cytosolic versus peroxisomal location of the MVA pathway.
Based on the results and the arguments raised above we suggest a new model for compartmentalization of the plant MVA pathway in three cellular compartments: cytosol, ER, and peroxisome (Fig. 5). The model resembles that proposed for mammals, but includes the sesquiterpene biosynthesis branch absent from mammalian organisms. Beyond constituting a new perspective on the plant MVA pathway, the model provides a mechanistic explanation for channeling of substrates toward sterol biosynthesis (but not sesquiterpenes) in healthy plants via compartmentalization. According to the proposed model, FPP is produced in the peroxisome and is usually destined for sterol biosynthesis via squalene synthase, located in the ER (Busquets et al., 2008), similar to mammals. Therefore, in most tissues of healthy plants, the vast majority of FPP synthesized is not available for catalysis by sesquiterpene synthases that are located in the cytoplasm. However, exposure of plants to pathogens (or elicitors) results in enhanced flux of the MVA pathway (Vogeli and Chappell, 1988), and apparently the FPP surplus is directed to the cytoplasm, where it becomes available for catalysis by sesquiterpene synthases.
Figure 5.
Scheme of suggested plant MVA pathway, partly compartmentalized in the peroxisomes. Enzymes highlighted by light-gray background were experimentally localized in both mammals and plants (including this work); enzymes highlighted by black background were experimentally localized only in mammals. The putatively cytosolic sesquiterpene synthases are indicated for reference purposes.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_121649 and NM_111146 (the Arabidopsis IPI1 and IPI2 genes, respectively).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Materials and Methods S1. A supplemental Materials and Methods section.
Acknowledgments
We thank Prof. Valerian Dolja, Dr. Dror Avisar, and Dr. Einat Sadot for generous contributions of plasmids; Dr. Oren Ostersetzer for advice; Dr. Moria Zik and Dr. Naomi Ori for providing Arabidopsis cDNA; Arnon Brand at www.bio-graphica.com for graphic solutions; and Tamar Azoulay for helpful discussions.
This work was supported by a grant from the Chief Scientist of the Israeli Ministry of Agriculture and a grant from the Israel Science Foundation (grant no. 492/08).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yoram Eyal (eyalab@volcani.agri.gov.il).
The online version of this article contains Web-only data.
References
- Aharoni A, Giri AP, Deuerlein S, Griepink F, de Kogel WJ, Verstappen FWA, Verhoeven HA, Jongsma MA, Schwab W, Bouwmeester HJ (2003) Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell 15 2866–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avisar D, Prokhnevsky AI, Makarova KS, Koonin EV, Dolja VV (2008) Myosin XI-K is required for rapid trafficking of golgi stacks, peroxisomes and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol 146 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biardi L, Sreedhar A, Zokaei A, Vartak NB, Bozeat RL, Shackelford JE, Keller GA, Krisans SK (1994) Mevalonate kinase is predominantly localized in peroxisomes and is defective in patients with peroxisome deficiency disorders. J Biol Chem 269 1197–1205 [PubMed] [Google Scholar]
- Bick JA, Lange BM (2003) Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Arch Biochem Biophys 415 146–154 [DOI] [PubMed] [Google Scholar]
- Busquets A, Keim V, Closa M, del Arco A, Boronat A, Arro M, Ferrer A (2008) Arabidopsis thaliana contains a single gene encoding squalene synthase. Plant Mol Biol 67 25–36 [DOI] [PubMed] [Google Scholar]
- Campbell M, Hahn FM, Poulter CD, Leustek T (1998) Analysis of the isopentenyl diphosphate isomerase gene family from Arabidopsis thaliana. Plant Mol Biol 36 323–328 [DOI] [PubMed] [Google Scholar]
- Campos N, Boronat A (1995) Targeting and topology in the membrane of plant 3-hydroxy-3 methylglutaryl coenzyme A reductase. Plant Cell 7 2163–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J, Nable R (1987) Induction of sesquiterpenoid biosynthesis in tobacco cell suspension cultures by fungal elicitor. Plant Physiol 85 469–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J, Wolf F, Proulx J, Cuellar R, Saunders C (1995) Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme A reductase a rate-limiting step for isoprenoid biosynthesis in plants? Plant Physiol 109 1337–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Andersson S, Orlova I, Gatto N, Reichelt M, Rhodes D, Boland W, Gershenzon J (2005) The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc Natl Acad Sci USA 102 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenreich W, Rohdich F, Bacher A (2001) Deoxyxylulose phosphate pathway to terpenoids. Trends Plant Sci 6 78–84 [DOI] [PubMed] [Google Scholar]
- Eisenreich W, Schwarz M, Cartayrade A, Arigoni D, Zenk MH, Bacher A (1998) The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem Biol 5 R221–R233 [DOI] [PubMed] [Google Scholar]
- Elgersma Y, Vos A, van den Berg M, van Rosermund CWT, van ser Sluijs P, Distel B, Tabak HF (1996) Analysis of the carboxyl-terminal peroxisomal targeting signal 1 in a homologous context in Saccharomyces cerevisiae. J Biol Chem 271 26375–26382 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Elofsson A, von Heijne G, Cristobal S (2003) In silico prediction of the peroxisomal proteome in fungi, plants and animals. J Mol Biol 330 443–456 [DOI] [PubMed] [Google Scholar]
- Flynn CR, Mullen RT, Trelease RN (1998) Mutational analyses of a type 2 peroxisomal targeting signal that is capable of directing oligomeric protein import into tobacco BY-2 glyoxysomes. Plant J 16 709–720 [DOI] [PubMed] [Google Scholar]
- Gardner RG, Hampton RY (1999) A highly conserved signal controls degradation of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase in eukaryotes. J Biol Chem 274 31671–31678 [DOI] [PubMed] [Google Scholar]
- Gershenzon J, Kreis W (1999) Biosynthesis of monoterpenes, sesquiterpenes, diterpenes, sterols, cardiac glycosides and steroid saponins. In M Wink, ed, Biochemistry of Plant Secondary Metabolism, Annual Plant Reviews, Vol 2. Sheffield Academic Press, Sheffield, UK, pp 222–229
- Gietl C, Faber KN, van der Klei IJ, Veenhuis M (1994) Mutational analysis of the N-terminal topogenic signal of watermelon glyoxysomal malate dehydrogenase using the heterologous host Hansenula polymorpha. Proc Natl Acad Sci USA 91 3151–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb S (1972) Submicrosomal localization of hepatic 3-hydroxy-3-methylglutaryl coenzyme a (HMG-CoA) reductase. FEBS Lett 24 153–155 [DOI] [PubMed] [Google Scholar]
- Gould SJ, Keller GA, Subramani S (1987) Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J Cell Biol 105 2923–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel D, Mosandl A, Wust M (2005) Biosynthesis of mono- and sesquiterpenes in carrot roots and leaves (Daucus carota L.): metabolic cross talk of cytosolic mevalonate and plastidial methylerythritol phosphate pathways. Phytochemistry 66 305–311 [DOI] [PubMed] [Google Scholar]
- Hartmann MA (2003) Sterol metabolism and function in higher plants. In G Daum, ed, Lipid Metabolism and Membrane Biogenesis. Springer, Heidelberg, pp 183–211
- Hemmerlin A, Hoeffler JF, Meyer O, Tritsch D, Kagan IA, Grosdemange C, Rohmer M, Bach TJ (2003) Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright-yellow-2 cells. J Biol Chem 278 26666–26676 [DOI] [PubMed] [Google Scholar]
- Hogenboom S, Tuyp JJM, Espeel M, Koster J, Wanders RJA, Waterham HR (2004) Phodphomevalonate kinase is a cytosolic protein in humans. J Lipid Res 45 697–705 [DOI] [PubMed] [Google Scholar]
- Keller GA, Barton MC, Shapiro DJ, Singer SJ (1985) 3-Hydroxy-3-methylglutaryl-coenzyme A reductase is present in peroxisomes in normal rat liver cells. Proc Natl Acad Sci USA 82 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler RH (1998) GFP for in vivo imaging of subcellular structures in plant cells. Trends Plant Sci 3 317–320 [Google Scholar]
- Kovacs WJ, Olivier LM, Krisans SK (2002) Central role of peroxisomes in isoprenoid biosynthesis. Prog Lipid Res 41 369–391 [DOI] [PubMed] [Google Scholar]
- Kovacs WJ, Tape KN, Shackelford JE, Duan X, Kasumov T, Kelleher JK, Brunengraber H, Krisans SK (2007) Localization of the pre-squalene segment of the isoprenoid biosynthetic pathway in mammalian peroxisomes. Histochem Cell Biol 127 273–290 [DOI] [PubMed] [Google Scholar]
- Krisans SK, Ericsson J, Edwards PA, Keller GA (1994) Farnesyl-diphosphate synthase is localized in peroxisomes. J Biol Chem 269 14165–14169 [PubMed] [Google Scholar]
- Laule O, Furholz A, Chang HS, Zhu T, Wang X, Heifetz PB, Gruissem W, Lange BM (2003) Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 100 6866–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Gonzales VM, Castel S, Trelease RN, Lopez-Iglesias C, Arro M, Boronat A, Campos N, Ferrer A, Fernandez-Busquets X (2005) Subcellular localization of Arabidopsis 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Plant Physiol 137 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK (1999) The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 50 47–66 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Schwender J, Disch A, Rohmer M (1997) Biosynthesis of isoprenoids in higher plant chloroplast proceeds via a mevalonate-independent pathway. FEBS Lett 400 271–274 [DOI] [PubMed] [Google Scholar]
- McGarvey DJ, Croteau R (1995) Terpenoid metabolism. Plant Cell 7 1015–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi J, Kirchner E, Cristobal S (2007) Quantitative proteomic comparison of mouse peroxisomes from liver and kidney. Proteomics 7 1916–1928 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Shimada H, Masuda T, Ohta H, Takamiya K (2001) Two distinct isopentenyl diphosphate isomerase in cytosol and plastid are differentially induced by environmental stresses in tobacco. FEBS Lett 506 61–64 [DOI] [PubMed] [Google Scholar]
- Newman JD, Chappell J (1999) Isoprenoid biosynthesis in plants: carbon partitioning within the cytoplasmic pathway. Crit Rev Biochem Mol Biol 34 95–106 [DOI] [PubMed] [Google Scholar]
- Okada K, Kasahara H, Yamaguchi S, Kawaide H, Kamiya Y, Nojiri H, Yamane H (2008) Genetic evidence for the role of isopentenyl diphosphate isomerase in the mevalonate pathway and plant development in Arabidopsis. Plant Cell Physiol 49 604–616 [DOI] [PubMed]
- Olivier LM, Chambliss KL, Gibson KM, Krisans SK (1999) Characterization of phosphomevalonate kinase: chromosomal localization, regulation, and subcellular targeting. J Lipid Res 40 672–679 [PubMed] [Google Scholar]
- Olivier LM, Kovacs W, Masuda K, Keller GA, Krisans SK (2000) Identification of peroxisomal targeting signals in cholesterol biosynthetic enzymes: AA-CoA thiolase, HMG-CoA synthase, MPPD, and FPP synthase. J Lipid Res 41 1921–1935 [PubMed] [Google Scholar]
- Paton VG, Shackelford JE, Krisans SK (1997) Cloning and subcellular localization of hamster and rat isopentenyl diphosphate dimethylallyl diphosphate isomerase: a PTS1 motif targets the enzyme to peroxisomes. J Biol Chem 272 18945–18950 [DOI] [PubMed] [Google Scholar]
- Phillips MA, D'Auria JC, Gershenzon J, Pichersky E (2008) The Arabidopsis thaliana type I isopentenyl diphosphate isomerases are targeted to multiple subcellular compartments and have overlapping functions in isoprenoid biosynthesis. Plant Cell 20 677–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi N, Porter W (1981) Conversion of acetyl-coenzyme A to isopentenyl pyrophosphate. In JW Porter, SL Spurgeon, eds, Biosynthesis of Isoprenoid Compounds, Vol 1. John Wiley & Sons, New York, pp 47–94
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Luder F, Weckwerth W, Jahn O (2007) Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M, Boronat A (2002) Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids: a metabolic milestone achieved through genomics. Plant Physiol 130 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer M (1999) The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat Prod Rep 16 565–574 [DOI] [PubMed] [Google Scholar]
- Schuhr CA, Radykewicz T, Sagner S, Latzel C, Zenk MH, Arigoni D, Bacher A, Rohdich F, Eisenreich W (2003) Quantitative assessment of crosstalk between the two isoprenoid biosynthesis pathways in plants by NMR spectroscopy. Phytochem Rev 2 3–16 [Google Scholar]
- Sharon-Asa L, Shalit M, Frydman A, Bar E, Holland D, Or E, Lavi U, Lewinsohn E, Eyal Y (2003) Citrus fruit flavor and aroma biosynthesis: isolation, functional characterization, and developmental regulation of Cstps1, a key gene in the production of the sesquiterpene aroma compound valencene. Plant J 36 664–674 [DOI] [PubMed] [Google Scholar]
- Stamellos KD, Shackelford JE, Shechter I, Jiang G, Conrad D, Keller GA, Krisans SK (1993) Subcellular localization of squalene synthase in rat hepatic cells. J Biol Chem 268 12818–12824 [PubMed] [Google Scholar]
- Stamellos KD, Shackelford JE, Tanaka RD, Krisans SK (1992) Mevalonate kinase is localized in rat liver peroxisomes. J Biol Chem 267 5560–5568 [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bondar AG, Rachubinski RA, Subramani S (1991) A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J 10 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Krisans SK (1990) Rat liver peroxisomes catalyze the initial step in cholesterol synthesis: the condensation of acetyl-CoA units into acetoacetyl-CoA. J Biol Chem 265 5731–5735 [PubMed] [Google Scholar]
- Tian GW, Mohanty A, Chary N, Li S, Paap B, Drakakaki G, Kopac CD, Li J, Ehrhardt D, Jackson D, et al (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Hata S, Yokota S, Mirura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T, Osumi T (1994) Characterization of the signal peptide at the amino terminus of the rat peroxisomal 3-ketoacyl-CoA thiolase precursor. J Biol Chem 269 6001–6010 [PubMed] [Google Scholar]
- Vogeli U, Chappell J (1988) Induction of sesquiterpene cyclase and suppression of squalene synthetase activities in plant cell cultures treated with fungal elicitor. Plant Physiol 88 1291–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders RJA, Waterham HR (2006) Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem 75 295–332 [DOI] [PubMed] [Google Scholar]
- Wu S, Schalk M, Clark A, Miles RB, Coates R, Chappell J (2006) Redirection of cytosolic or plastidic isoprenoid precursors elevates terpene production in plants. Nat Biotechnol 24 1441–1447 [DOI] [PubMed] [Google Scholar]