Abstract
Plant cells exhibit a high rate of mitochondrial DNA (mtDNA) recombination. This implies that before cytokinesis, the different mitochondrial compartments must fuse to allow for mtDNA intermixing. When and how the conditions for mtDNA intermixing are established are largely unknown. We have investigated the cell cycle-dependent changes in mitochondrial architecture in different Arabidopsis (Arabidopsis thaliana) cell types using confocal microscopy, conventional, and three-dimensional electron microscopy techniques. Whereas mitochondria of cells from most plant organs are always small and dispersed, shoot apical and leaf primordial meristematic cells contain small, discrete mitochondria in the cell periphery and one large, mitochondrial mass in the perinuclear region. Serial thin-section reconstructions of high-pressure-frozen shoot apical meristem cells demonstrate that during G1 through S phase, the large, central mitochondrion has a tentaculate morphology and wraps around one nuclear pole. In G2, both types of mitochondria double their volume, and the large mitochondrion extends around the nucleus to establish a second sheet-like domain at the opposite nuclear pole. During mitosis, approximately 60% of the smaller mitochondria fuse with the large mitochondrion, whose volume increases to 80% of the total mitochondrial volume, and reorganizes into a cage-like structure encompassing first the mitotic spindle and then the entire cytokinetic apparatus. During cytokinesis, the cage-like mitochondrion divides into two independent tentacular mitochondria from which new, small mitochondria arise by fission. These cell cycle-dependent changes in mitochondrial architecture explain how these meristematic cells can achieve a high rate of mtDNA recombination and ensure the even partitioning of mitochondria between daughter cells.
Mitochondria are the principal source of ATP energy in eukaryotic cells. Although they are often portrayed as static, oval or rod-shaped organelles that sometimes exhibit a branched configuration, studies of living cells carried out over the past 30 years have demonstrated that they are among the most plastic organelles of cells in terms of form and distribution (Calvayrac et al., 1972; Osafune et al., 1972, 1975a, 1975b, 1975c; Hoffmann and Avers, 1973; Bereiter-Hahn, 1990; Bereiter-Hahn and Voth, 1994; Hermann and Shaw, 1998; Yaffe, 1999b; Logan and Leaver, 2000; Logan, 2006a, 2006b; Zadworny et al., 2007). Furthermore, changes in their architecture and their ability to translocate rapidly throughout the cytoplasm appear to be of critical importance for executing their cellular functions. For example, it has long been known that mitochondria congregate around cellular areas with high energy requirements (Bakeeva et al., 1978; Bawa and Werner, 1988; Bereiter-Hahn, 1990; Bereiter-Hahn and Voth, 1994; Logan, 2006a). In addition, in both mammalian and plant cells, they constantly undergo fission, fusion, and branching changes while sliding to different cellular locations (Bereiter-Hahn, 1990; Bereiter-Hahn and Voth, 1994; Logan and Leaver, 2000; Arimura et al., 2004; Logan, 2006a, 2006b).
Serial thin-section reconstruction of mitochondria of the unicellular algae Chlorella (Atkinson et al., 1974) and Chlamydomonas reinhardtii (Blank and Arnold, 1981) and analysis of the three-dimensional (3D) architecture of thin sectioned and GFP-tagged mitochondria in yeast (Calvayrac et al., 1972; Osafune et al., 1975a, 1975b, 1975c; Hermann and Shaw, 1998; Yaffe, 1999b, 2003) have shown that in each of these cell types, the mitochondria are joined into a single, reticulate structure. Extended tubules and networks have also been observed in the chondriome of different fungal and algal species (Floyd et al., 1972; Pickett-Heaps, 1974; Howard, 1981; Zadworny et al., 2007). In contrast, in mammalian cells, where large numbers of mitochondria can be resolved by light microscopy techniques, their architecture can vary from small (1–2 μm in diameter) spheres/ovals to 10-μm-long, sausage-like, and sometimes branched organelles whose longitudinal axis tends to parallel the orientation of the radial microtubules (MTs; Bereiter-Hahn, 1990). However, in specialized cell types such as muscle fibers (Bakeeva et al., 1978) and COS-7 cells (Yaffe, 1999b), some of the mitochondria have been shown to possess a more network/reticulate type of organization. All of these changes in mitochondrial shape and the accompanying fusion and fission events can occur within minutes (Bereiter-Hahn and Voth, 1994). Higher plant mitochondria are generally portrayed as being oval or sausage like, with only a few studies reporting the existence of branched mitochondria that undergo fusion, fission, and amoeba-like changes over short periods of time (Logan and Leaver, 2000; Logan et al., 2003; Arimura et al., 2004; Foissner, 2004; Logan, 2006a). To account for the high rate of recombination of the plant mitochondrial DNA (mtDNA; Lonsdale et al., 1988; Gillham, 1994), the “discontinuous whole” hypothesis postulates that the individual, small mitochondria must transiently fuse to transfer their mtDNA into a common physical space (Logan, 2006a, 2006b).
The relationship between mitochondrial propagation and the cell cycle has also been investigated for many years. New mitochondria do not arise de novo; they derive from preexisting ones (Bereiter-Hahn, 1990; Yaffe, 1999b; Logan, 2006a). It is generally assumed that the doubling of the mitochondrial volume and numbers occurs during the S and G2 stages of the cell cycle, as evidenced by the abundance of dumbbell-shaped mitochondrial profiles in electron micrographs during these stages of the cell cycle (Bereiter-Hahn, 1990). For many years, segregation of the mitochondria into the two daughter cells has been assumed to be a stochastic process. However, in yeast cells, the transfer of some of the reticulate mitochondrial domains to the growing buds has been shown to require intact cytoskeletal systems and GTPase-mediated activities (Hermann and Shaw, 1998; Hermann et al., 1998; Shepard and Yaffe, 1999; Fekkes et al., 2000; Boldogh and Pon, 2006). Although the information about mitochondrial propagation in plants is more limited, there are several reports on the coupling of the mitochondrial cycle to the cell cycle (Arimura et al., 2004; Sheahan et al., 2005; Zottini et al., 2006).
In this study, we have analyzed the changes in shape, number, and distribution of the mitochondrial population in different cell types of Arabidopsis (Arabidopsis thaliana) by means of conventional transmission electron microscopy (TEM), confocal laser scanning microscopy (CLSM) imaging, and 3D TEM modeling. Whereas in most cell types the shape of the mitochondria corresponds to the accepted pattern of oval or sausage-like organelles, the chondriome of shoot apical meristem (SAM) and leaf primordium (LP) meristematic cells is defined by the existence of a large mitochondrial mass that coexists with smaller mitochondria and undergoes changes in shape and size during the cell cycle. To obtain a more precise view of these mitochondrial masses, we have reconstructed entire cryofixed SAM cells at representative stages of the cell cycle by serial thin-section electron microscopy (EM) imaging and 3D modeling. These reconstructions confirm that Arabidopsis SAM and LP meristematic cells contain a large, network-forming mitochondrion that surrounds the nucleus and interacts with a set of smaller, cortical mitochondria in quantifiable ways. During mitosis and cytokinesis, most of the mitochondria fuse into a large, coherent, cage-like mitochondrion that encompasses the entire spindle/phragmoplast cytoskeletal arrays and possesses sheet-like domains at the spindle poles. During interphase, about half of the cage-like mitochondrion fragments into smaller, physically discrete mitochondria, while the other half adopts a tentaculate architecture, with its central, sheet-like domain forming a perinuclear cap. Based on the information contained in the 3D reconstructions, we have developed a model of how the architectural changes of the large mitochondrion relate to cell cycle-dependent functions of these meristematic cells.
RESULTS
During the course of a series of studies designed to elucidate the structural basis of plant cell division (Seguí-Simarro et al., 2004, 2007; Seguí-Simarro and Staehelin, 2006a), we have analyzed thousands of electron micrographs of Arabidopsis SAM, LP, and root meristem cells preserved by high-pressure-freezing/freeze-substitution techniques. One of the unexpected findings was a striking dichotomy in mitochondrial architecture. As illustrated in Figure 1, some of the cross-sectioned mitochondria exhibited a classical round, oval, or dumbbell- or rod-like configuration. In the shoot apex (Supplemental Fig. S1A), this pattern was consistently observed in differentiated cells of the cotyledons and the apical region of the LP (Supplemental Fig. S1, B and C). However, in SAM and LP meristematic (undifferentiated) cells, we found in many instances lobed mitochondria with narrow, elongated domains resembling the matrixules described by Logan (2006b) and Scott et al. (2007) together with mitochondria with even more complex cross-sectional profiles (Fig. 2, A and B). To gain a better understanding of the actual 3D morphology of these complex mitochondria, we analyzed randomly chosen sets of serial EM sections of SAM cells (Fig. 2C). To our surprise, these random sets of serial thin sections contained many more large and branched mitochondria than anticipated. They also showed that these mitochondria always possessed at least one sheet-like domain that was partly wrapped around the nucleus (Fig. 2A; Supplemental Fig. S1, D and E) and other domains that partly encompassed organelles and other cytoplasmic structures. These persistent spatial relationship patterns suggested that the large, branched mitochondria might serve specialized functional needs of these proliferating cells. To address this question, we examined the morphology of mitochondria in SAM and LP cells as well as in other cell types, not derived from the SAM, by means of confocal, conventional (two-dimensional), and 3D TEM techniques.
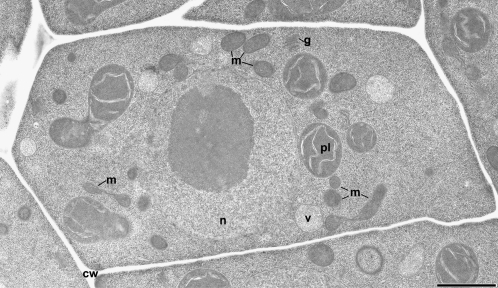
Figure 1.
Thin-section electron micrograph of an Arabidopsis LP cell at interphase. Most of the organelles—mitochondria (m), plastids (pl), Golgi (g), and vacuoles (v)—are positioned close to the central nucleus (n). The mitochondrial profiles vary from oval to dumbbell shaped. cw, Cell wall. Bar = 2 μm.
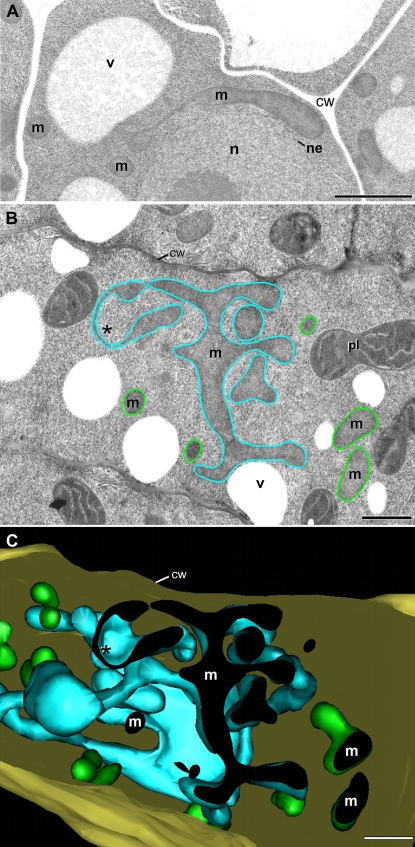
Figure 2.
Complex network-like mitochondrial morphologies in Arabidopsis SAM cells. A, Thin-section electron micrograph of a large mitochondrion (m) that is partly wrapped around the nucleus (n) but does not make contact with the nuclear envelope (ne; see also Supplemental Fig. S1C). This central sheet-like domain connects to more tubular, peripheral domains, as shown in B and C. B, Tangential thin section through the central region of a large mitochondrion with tubular extensions (blue outline) that is surrounded by smaller, individual mitochondria (green outlines). C, 3D model of the large (blue) and smaller (green) mitochondria seen in B reconstructed from serial thin-section images. Asterisks in B and C indicate a region of the network where a thin mitochondrial sheet forms a pocket-shaped subdomain. cw, Cell wall; v, vacuole. Bars = 2 μm.
Mitochondria of Living SAM and LP Meristematic Cells Display a Network-Like Configuration
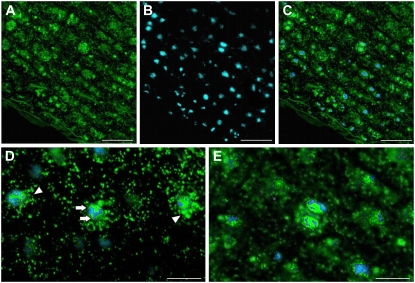
To determine if the morphology of the large, complex mitochondria of SAM and LP meristematic cells changed in a cell cycle-dependent manner, we analyzed their architecture and distribution in living cells (Fig. 3). Excised SAM and LP tissues were incubated first in Mitotracker, a mitochondria-specific stain (Fig. 3A, green signal), and then in 4′,6-diamidino-2-phenylindole (DAPI), a DNA-specific fluorescent stain (Fig. 3B, blue signal), prior to viewing by CLSM. Images of the confocal sections were recorded in both green and blue channels and then merged (Fig. 3C). As seen in Figure 3, D and E, some small, rounded mitochondria could be resolved in the periphery of the cells, but most of the staining was associated with the mitochondrial mass around the nucleus. In the majority of the smaller interphasic cells (Fig. 3D), the network-forming mass stained more intensely at one pole of the nucleus than at the other (arrowheads), whereas in larger cells the distribution around the nucleus appeared more balanced (arrows). Since the size of meristematic cells increases as they progress through the cell cycle, the different distributions of mitochondria in differently sized cells suggested a cell cycle-dependent control of the mitochondrial networks. Thus, in the larger, dividing cells (Fig. 3E), the number of small, round mitochondria in the cell periphery appeared to be reduced and the size of the large, network-forming mitochondrial masses increased compared with the interphase cells.
Figure 3.
Confocal micrographs of mitochondria and nuclei in Arabidopsis LP meristematic cells. A to C, Overview images of a very young LP stained with Mitotracker (green; A) and DAPI (blue; B). The merged image is shown in C. D, higher magnification micrograph of three interphasic SAM cells in which a differential distribution of the mitochondrial signal can be observed. In the two lateral cells, most of the signal is concentrated in a mass at one pole of the nucleus (arrowheads), whereas in the central cell, large mitochondrial masses are seen on both sides of the nucleus (arrows). Smaller, round, individual mitochondria are seen in the periphery of the cells. E, Dividing cell in which the large mitochondrial masses seem to encompass the reforming nuclei. Fewer individual mitochondria can be discerned in the cell periphery. Bars = 25 μm in A to C and 10 μm in D and E.
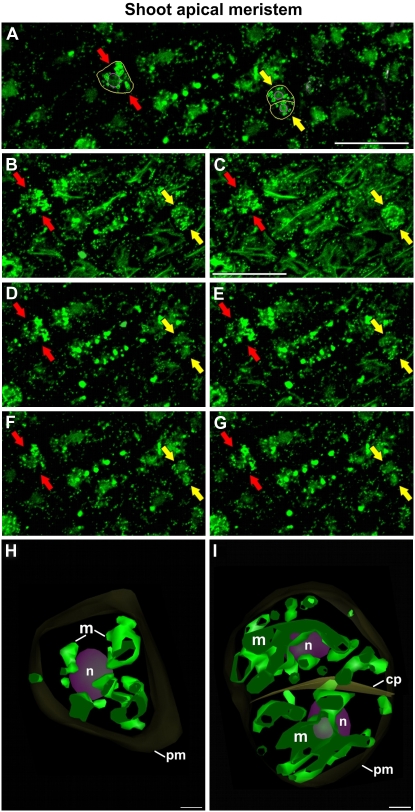
To obtain more detailed information on the organization of the mitochondria in living cells, we produced 3D models of the fluorescing mitochondria of both SAM and LP meristematic cells from serial CLSM sections of both interphasic cells (red arrows in Figs. 4, A–G, and 5, A–G) and dividing cells (yellow arrows in Figs. 4, A–G, and 5, A′–G′). In all of these cell types, large mitochondrial masses with variable morphologies as well as smaller, individual mitochondria could be discerned. Typically, the large mitochondrial masses were concentrated around the nuclei (Figs. 4, H and I, and 5, H and H′) and the smaller, individual mitochondria were concentrated in the cell cortex. Quantitative analyses of these models also suggested that both the size of the large mitochondrial mass and the number of individual mitochondria differed between small and large interphasic cells and dividing cells (Fig. 4, compare H and I). Our CLSM data were consistent with the idea that the chondriome of SAM and LP meristematic cells was composed of two types of mitochondria: small, discrete mitochondria in the cell periphery and a larger mitochondrial mass in the cell center whose size increased during mitosis and cytokinesis.
Figure 4.
Confocal micrographs of mitochondria in interphase and dividing SAM cells. A, Overview of a SAM region with a highlighted interphase cell (red arrows, yellow outline) and a dividing cell (yellow arrows, yellow outline). B to G, Consecutive, 0.5-μm-thick, confocal sections of the highlighted cells. H, 3D model of the interphase cell. I, 3D model of the dividing cell. The plasma membranes (pm) and the cell plate (cp) are shown in dark yellow, the nuclei (n) are shown in purple, and the mitochondria (m) are shown in green. Bars = 25 μm in A to G and 1 μm in H and I.
Figure 5.
Confocal micrographs of mitochondria in interphasic and dividing LP meristematic cells. A and A′, Overview of LP regions where interphase (A, red arrows) and dividing (A′, yellow arrows) cells have been outlined in yellow. B to G and B′ to G′, Consecutive, 0.5-μm-thick, confocal sections of the interphase (B–G) and the dividing (B′–G′) cells were recorded and mounted into a stack for partial cell reconstruction and modeling. H, 3D model of the interphasic cell marked by red arrows in A to G. H′, 3D model of the dividing cell marked by yellow arrows in A′ to G′. For both cells, the plasma membrane (pm) is modeled in dark yellow, nuclei (n) are modeled in purple, and mitochondria (m) are modeled in green. Bars = 25 μm in A to G and A′ to G′ and 1 μm in H and H′.
A Large Perinuclear Mitochondrial Mass Is Not Observed in Cells outside the Shoot Apical Region
To determine whether the mitochondrial architecture described above for SAM and LP meristematic cells is typical of all Arabidopsis cell types, we reexamined our extensive collection of EM images from different Arabidopsis cell types, including root tip, stem, mature leaf, meiocyte, microspore, pollen, endosperm, endothelial, and embryo cells (Kiss et al., 1990; Staehelin et al., 1990; Otegui and Staehelin, 2000, 2004; Otegui et al., 2002, 2005, 2006; Seguí-Simarro et al., 2004, 2007; Austin et al., 2005, 2006; Seguí-Simarro and Staehelin, 2006a; Borsics et al., 2007; Christopher et al., 2007; J.M. Seguí-Simarro and L.A. Staehelin, unpublished data). After an exhaustive viewing of thousands of micrographs, we could not find any evidence in any of these cell types for the presence of mitochondria with a complex 3D architecture resembling the structures seen in TEM micrographs of SAM and LP meristematic cells. Instead, all of the micrographs showed mitochondrial profiles reflecting the conventional and generally accepted round, oval, or sausage-like morphology. Nevertheless, to confirm this notion, we also examined by CLSM cells in two of these tissue types, stem cells (Fig. 6) and root tip cells (Supplemental Fig. S2) stained with Mitotracker and DAPI. As expected, none of the cells in these tissues displayed mitochondrial structures that resembled the large, nucleus-associated mitochondrial masses of SAM cells. Instead, all of the mitochondria of these cells produced fluorescence signals that were both small and discrete (Fig. 6, A–G; Supplemental Fig. S2, A–G). When selected cells of these tissues were modeled, an oval or sausage-like morphology was evident for mitochondria of both stem cells (Fig. 6H) and root cells (Supplemental Fig. S2H). Thus, modeling of the stem and root cell image stacks demonstrated that the mitochondria of these cell types differ in their morphology, their distribution, and their numbers from those observed in SAM and LP meristematic cells. Most notably, these cells lacked a nucleus-associated mitochondrial mass.
Figure 6.
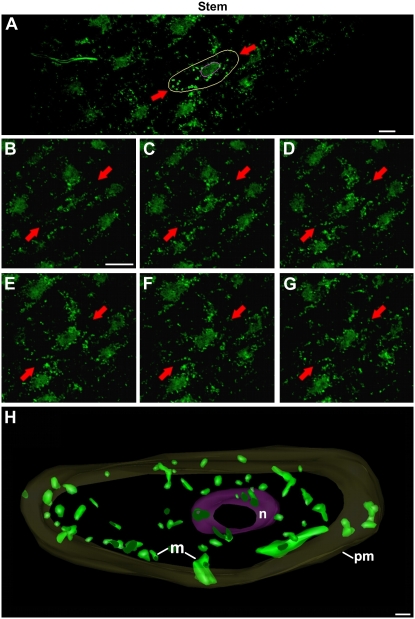
Confocal micrographs of mitochondria of interphase stem cells. A, Overview of a stem region in which an interphase cell (red arrows) has been outlined in yellow. B to G, Consecutive, 0.5-μm-thick, confocal sections of the highlighted cell. H, 3D model of the cell marked by red arrows in A to G. The plasma membrane (pm) is modeled in dark yellow, the nucleus (n) is modeled in purple, and the mitochondria (m) are modeled in green. Bars = 10 μm in A to G and 1 μm in H.
Serial Thin-Section Analysis of Whole Cells Demonstrates That the Large Mitochondrion of SAM Cells Undergoes Characteristic Architectural Changes during the Cell Cycle
To overcome the spatial resolution limitations of fluorescence microscopy techniques, and in order to obtain higher resolution data of mitochondrial architecture during the cell cycle, we produced electron micrographs of serial thin sections of six entire cryofixed SAM cells (approximately 130 sections per cell) at representative stages of the cell cycle, traced the mitochondria, nuclei and plasma membranes, and finally generated 3D models of the cells using the IMOD software program. Each cell was chosen according to well-defined morphological criteria (see Supplemental Table S1 and “Materials and Methods” for details) to represent a specific stage in the cell cycle (G1, G1-S, G2, prometaphase, early telophase, and late telophase). Arabidopsis SAM and LP meristematic cells are characterized, like other meristematic cells, by a small size (a major diameter of approximately 8 μm in G1 cells and up to approximately 12 μm in G2/dividing cells), a polyhedral shape, a nearly spherical large nucleus (from approximately 4 μm in diameter in G1 nuclei to approximately 5 μm in diameter for G2 nuclei), high organellar density, limited vacuolation, and a dense cytoplasm (Fig. 1). All of these features, typical of highly active, proliferating cells, make them perfect candidates to apply the above-mentioned criteria for cell cycle staging (Supplemental Table S1). Indeed, this combination of cell staging according to morphological criteria and 3D modeling of entire serial sectioned cells has previously proven useful to analyze quantitative, morphological, and distributional changes in organelles such as the Golgi apparatus, multivesicular bodies, and vacuoles in Arabidopsis SAM cells (Seguí-Simarro and Staehelin, 2006a).
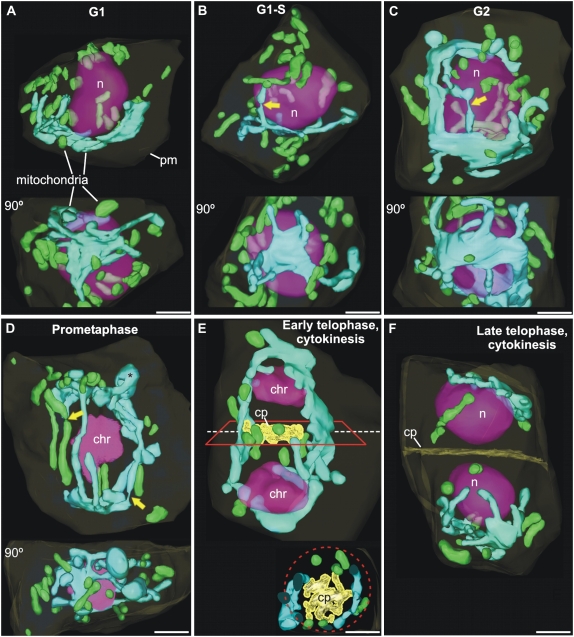
The large type of mitochondrion observed in randomly chosen serial EM sections and in CLSM stacks of SAM and LP meristematic cells was consistently found in all of the fully reconstructed cells (Fig. 7), and always possessed at least one sheet-like subdomain that extended over part of the interphasic nucleus. Figure 7 documents the changes in mitochondrial architecture during G1, G1-S, G2, prometaphase, as well as early and late telophase stages of the cell cycle of SAM cells. In Figure 7, A to D, each of the reconstructed cells is shown in two perpendicular orientations to demonstrate both the general distribution of the two mitochondrial types and the persistence of the sheet-like nuclear cap domain throughout the cell cycle. In Figure 7E, the longitudinal view is supplemented by a cross-sectional view at the level of the forming cell plate.
Figure 7.
EM-based, 3D models of mitochondria in Arabidopsis SAM cells at different stages of the cell cycle. The plasma membrane (pm) is outlined in faint yellow, the cell plate (cp) is shown in yellow, the nuclei (n) and chromosomes (chr) are depicted in purple, and the mitochondria (m) are colored in either green (individual mitochondria) or turquoise (tentaculate/cage-like mitochondria). A and B, Throughout the G1 and S phases, the cells exhibit a large, single tentaculate mitochondrion closely apposed to the nuclear envelope as well as numerous, smaller individual mitochondria in the cell periphery. The top, side view of the tentaculate mitochondrion in A is complemented by the bottom, face-on view, rotated 90°, where its central, sheet-like domain is more evident. The yellow arrows in B to D mark narrow regions of the mitochondria, which might correspond to fissioning sites. C, Growth of the G2 cells is matched by the expansion of the large mitochondrion, which develops a clamp-like morphology as it grows around the nucleus and develops a second nucleus-capping domain at the opposite nuclear pole. Abundant small, individual mitochondria are present in the cell periphery. D, The large mitochondrion in this prometaphase cell exhibits a cage-like structure that surrounds the entire mitotic apparatus. The reduction in the number of small mitochondria is matched by a corresponding increase in the size of the large, cage-like mitochondrion. At the poles of the cage-like mitochondrion, pocket-like domains are occasionally observed (asterisk). E, During early telophase, elements of the cage-like mitochondrion and individual mitochondria redistribute to form a belt that surrounds the growing cell plate (see the cross-sectional view of this belt and of the cell plate in the inset). F, During late telophase, the cell plate divides the cell into two and the cage-like mitochondrion becomes separated into two independent tentaculate mitochondria, similar to those of G1 cells. Bars = 2 μm.
During the G1 and G1-S interphase stages (Fig. 7, A and B), many individual round (0.4–0.6 μm in diameter) and tubular (0.4–0.6 μm in section diameter and highly variable length) mitochondria are seen dispersed throughout the peripheral cytoplasm. This contrasts with the distribution of the large mitochondrion (highly pleomorphic and up to 7 μm of maximal length) that is centered on one pole of the nucleus. Typically, this latter type of mitochondrion possesses tentacle-like tubular extensions that originate from the margins of the sheet-like central domain. Few individual mitochondria are seen in the vicinity of the tentaculate mitochondrion. As documented in Supplemental Figure S1, D and E, the nucleus-capping tentaculate mitochondrion comes fairly close to the nuclear envelope but the two membranes never touch, as evidenced by the presence of ribosomes between them. During G2, the first major 3D architectural change of the tentaculate mitochondrion is observed (Fig. 7C). Besides increasing in size, this mitochondrion acquires a clamp-like morphology by forming a second sheet-like nuclear cap domain opposite the first one. This duplication of the nuclear cap domain sets the stage for conversion of the large mitochondrion into a cage-like organelle that encompasses the spindle during mitosis and the reforming daughter nuclei and phragmoplast during cytokinesis (Fig. 7, D and E).
During mitosis (Fig. 7D), the two sheet-like mitochondrial domains that bracket the spindle are connected to each other through large, tubular mitochondrial elements. Careful analysis of these tubular bridging elements shows that while some form continuous links between the two sheet-like pole domains, others exhibit distinct breaks and/or highly constricted domains (Fig. 7D, arrows). These structural features are consistent with the idea that the tubular bridging elements are unstable mitochondrial domains that undergo cycles of fission and fusion, while the overall cage-like conformation of the large mitochondrion around the spindle is maintained. After formation of the phragmoplast and initiation of cell plate growth, a number of individual mitochondria as well as tubular domains of the cage-like mitochondrion assemble in the plane of division around the margins of the forming cell plate in a belt-like conformation (Fig. 7E). As the cell plate expands, small, individual mitochondria begin to appear again in the cortical region of the cell, while the large mitochondrion is converted back to the tentaculate morphology of the G1-stage interphasic cells by severing of the tubular bridging elements of the cage-like mitochondrion (Fig. 7F).
Prior to Mitosis, Most of the Individual SAM Mitochondria Fuse to the Cage-Like Mitochondrion, Which Constitutes Approximately 80% of the Mitochondrial Volume during Cell Division
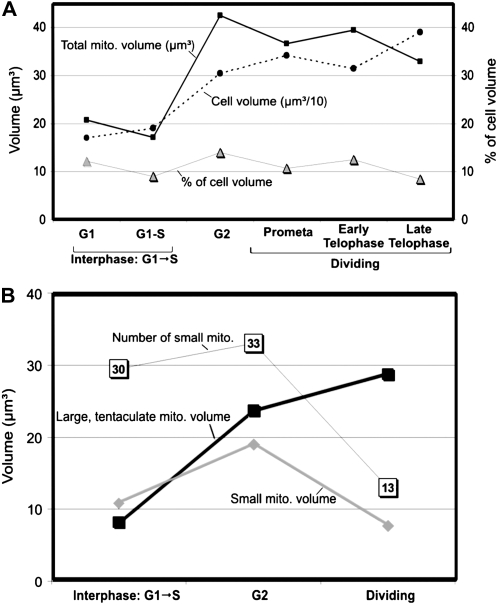
To further characterize the two types of mitochondria of these meristematic cells, we have determined their volume, surface area, number, and contribution to the total cell volume during the cell cycle. To obtain accurate quantitative data, these calculations were made exclusively over the serial sectioned, reconstructed, and modeled cells. The volume occupied by mitochondria was found to fluctuate slightly from 8% to 12% of total cell volume (Fig. 8A), indicating that the mitochondrial volume expands in parallel with the cell volume during the cell cycle. Doubling of the total mitochondrial volume and of the total surface area of the outer envelope membrane (data not shown) occurs during G2. Thereafter, they remain relatively constant during mitosis and cytokinesis (Fig. 8A). The observed doubling of the total mitochondrial volume and of the surface area during G2 occurs in parallel in the individual mitochondria and in the perinuclear tentaculate mitochondrion (Fig. 8B) and is consistent with the observed morphological changes (Fig. 7). This suggests that the increase in volume is not only due to a simple change in mitochondrial shape, such as swelling, but reflects mitochondrial growth related to the duplication of these organelles.
Figure 8.
Quantitative and volumetric analysis of the two types of mitochondria in SAM cells. A, Cell cycle-dependent changes in total mitochondrial volume (black squares) and cell volume (black circles). Note that the cell volumes are divided by 10 to fit them into the chart. The percentage of cell volume occupied by mitochondria (gray triangles) remains relatively constant during the cell cycle. B, Comparison of the volume occupied by the large, tentacular mitochondrion (black squares), the volume occupied by small mitochondria (gray triangles), and the number of small individual mitochondria (white squares) during the G1/S, G2, and dividing cell stages of the cell cycle. Note that the doubling of the total mitochondrial volume during G2 (see Fig. 7A) is due to the doubling of volume of the tentaculate mitochondrion (black squares) and of the individual mitochondria (gray diamonds), while the number of small mitochondria (white squares) remains relatively unchanged. Between G2 and the onset of cell division, the volume loss of small mitochondria is paralleled by a remarkable decrease in the number of small mitochondria, which seem to fuse with the large, tentaculate mitochondrion, as deduced from the increase in volume of the large mitochondrion.
Quantitative comparisons of the volumes of the two types of mitochondria during the cell cycle have also yielded functionally relevant insights. Thus, whereas the tentaculate mitochondrion contributes from 42% to 44% of the total mitochondrial volume in interphase (G1, G1-S, and G2) cells, it contributes approximately 80% of the total mitochondrial volume during mitosis and cytokinesis (i.e. when it assumes a cage-like organization). Conversely, the volume percent of the small, dispersed mitochondria reaches a maximum during interphase (56%–58%) and drops to a low of approximately 20% during mitosis. These volumetric changes suggest that the dramatic enlargement of the tentaculate/cage-like mitochondrion shortly before the onset of mitosis involves the fusion of a significant number of small, individual mitochondria with the large mitochondrion. The putative fusion of small mitochondria with the large, cage-like mitochondrion prior to mitosis is also reflected in an approximately 60% reduction in the number of small, individual mitochondria as the cells progress from interphase to mitosis and cytokinesis (Fig. 8B).
Taken together, these data indicate that during G2, both the tentaculate mitochondrion and the individual mitochondria double their volume, but without a net increase in the number of individual mitochondria. Instead, the enlarged, individual G2 mitochondria fuse to the cage-like mitochondrion prior to mitosis, and only after cytokinesis and fission of the cage-like mitochondrion are new, small mitochondria released by the fissioning of the tentaculate mitochondrion. This reestablishes the nearly equal distribution of volume between the individual mitochondria and the tentaculate mitochondrion, as observed in G1 cells.
DISCUSSION
In Arabidopsis, Only SAM and LP Meristematic Cells Contain Two Types of Mitochondria That Undergo Changes during the Cell Cycle
The main finding of this study is the discovery that in SAM and LP meristematic cells of Arabidopsis the chondriome consists of two structurally distinct types of mitochondria that undergo cell cycle-dependent changes in shape, size, and distribution, whereas in all other cell types (approximately 10) studied by us, only one mitochondrial morphotype could be identified by TEM. This finding is based (1) on the reexamination of thousands of random thin-section electron micrographs of many different, high-pressure-frozen cells produced over a 15-year period in the Staehelin laboratory; (2) on serial CLSM image studies of SAM, LP, and root meristematic cells as well as stem and other root and shoot tip cells; and (3) on 3D reconstructions of six entire SAM cells based on serial thin-section micrographs.
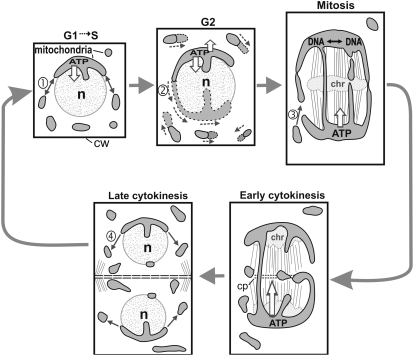
The two types of mitochondria of SAM and LP meristematic cells are termed small, round/tubular and large, tentaculate/cage-like mitochondria. The population of conventional, small, round or tubular mitochondria is located in the cell periphery. In contrast, the single, very large, tentaculate/cage-like mitochondrion is seen to be closely apposed to the nucleus and to occupy the cell center. The most notable cell cycle-dependent changes in mitochondrial organization are all associated with this latter mitochondrial type (Figs. 7 and 8), which nearly doubles its relative volume as the cells progress from interphase to mitosis. By integrating structural and quantitative data, we have been able to identify the growth, fission and fusion, and morphological remodeling activities that bring about the architectural changes during the cell cycle. A schematic summary of these events is presented in Figure 9.
Figure 9.
Model of the cell cycle-dependent changes in mitochondrial architecture in Arabidopsis SAM and LP meristematic cells and their potential functional roles. This model postulates that the different configurations of the tentaculate/cage-like mitochondrion help funnel ATP energy to cell structures that are of critical importance for the proliferative functions of the meristematic cells. In parallel, they provide a structural framework to explain how mtDNA intermixing for mtDNA recombination can occur in these cell types through the creation of the giant, cage-like mitochondrion. The numbered arrows refer to the most likely fusion, fission, and mitochondrial growth events that may occur at each stage, namely fusion/fission equilibrium at G1 (1), net mitochondrial growth at G2 (2), fusion during mitosis (3), and fission during cytokinesis (4). See text for further details. chr, Chromosomes; cp, cell plate; cw, cell wall; n, nucleus.
The Volume Percent of the Large Tentaculate/Cage-Like Mitochondrion of SAM Cells Varies from Approximately 40% during Interphase to Approximately 80% Prior to Mitosis
The mitochondrial cycle of Arabidopsis SAM cells is defined by three events: mitochondrial volume doubling during G1/S to G2, changes in the number and volumetric ratios of the small, peripheral mitochondria to the large central mitochondrion, and the dramatic changes in architecture of the large mitochondrion prior to mitosis and after cytokinesis. During G2, the mass of the mitochondria doubles (Fig. 9, arrow 2) at a rate that appears to correspond to the rate of doubling of the cytoplasmic volume, as evidenced by the constancy of the percentage of cell volume (approximately 10%) occupied by mitochondria during the entire cell cycle.
Mitochondrial enlargement during G2 appears to be a common feature of cells of many multicellular organisms (Bereiter-Hahn, 1990; Kennady et al., 2004); thus, our data on mitochondrial growth fit into this general pattern. As the nuclear envelope breaks down and the cells enter mitosis, both the number and the volume percent of the small, individual mitochondria population decreases to approximately 20% and the volume percent of the cage-like large mitochondrion increases to approximately 80%, which most likely reflects a change in the balance of fission and fusion events between the small mitochondria and the large mitochondrion (Fig. 9, arrow 3). During late cytokinesis, this shift in net volume from the small mitochondria to the large mitochondrion is reversed. Thus, after the large mitochondrion has divided into two, the balance of fission and fusion events reverts to an increase in fission events (Fig. 9, arrow 4), leading to both a significantly increased (approximately three times) number of small, individual mitochondria per cell and an increase (approximately three times) in their percent volume. This new balance between the two mitochondria types is maintained throughout the G1 to S stages of the cell cycle (Fig. 9, arrow 1).
Reticular Mitochondria Have Not Been Observed in Unperturbed Wild-Type Higher Plant Meristematic Cells to Date
For over 30 years, reticular mitochondria have been reported as a characteristic feature of unicellular organisms such as trypanosomes, yeast, fungi, Chlamydomonas, Chlorella, and Euglena (Floyd et al., 1972; Osafune et al., 1972; Hoffmann and Avers, 1973; Atkinson et al., 1974; Pickett-Heaps, 1974; Howard, 1981; Bereiter-Hahn, 1990; Hermann and Shaw, 1998; Yaffe, 1999a, 1999b; Jakobs, 2006; Hoog et al., 2007). In contrast, the mitochondria of animal cells are typically discrete round or sausage-like organelles (for review, see Bereiter-Hahn, 1990; Bereiter-Hahn and Voth, 1994). Only in certain types of differentiated mammalian cells have reticulate mitochondria been observed (Bakeeva et al., 1978; Bawa and Werner, 1988; Bereiter-Hahn, 1990; Smirnova et al., 1998; Yaffe, 1999b). Similarly, higher plant mitochondria have been typically portrayed as round to sausage-like organelles (Olyslaegers and Verbelen, 1998; Logan and Leaver, 2000; Nebenführ et al., 2000; Arimura and Tsutsumi, 2002; Van Gestel and Verbelen, 2002; Logan et al., 2003; Arimura et al., 2004; Logan, 2006a). Although some studies report on the presence of reticulate mitochondria in unperturbed eggs (Kuroiwa et al., 1996, 2002) and vascular cell types (Gamalei and Pakhomova, 1981), most of the documented examples of reticular or disc-shaped mitochondria come from mitochondrial mutants (Arimura and Tsutsumi, 2002; Logan et al., 2003) or from cells subjected to experimental perturbations such as low oxygen pressure (Bereiter-Hahn, 1990; Van Gestel and Verbelen, 2002; Logan, 2006a), prolonged cell culturing (Rohr, 1978), or protoplasting (Sheahan et al., 2004, 2005). It may be argued that the exposure of the root, stem, and shoot tissues to Suc (a cryoprotectant) in the growth and freezing medium prior to high-pressure freezing might have produced the “reticulate” (tentaculate/cage-like) mitochondrial architecture observed in this study. This seems unlikely, since plants grown for the CLSM studies, which were never exposed to 150 mm Suc, displayed the same mitochondrial configuration seen in the high-pressure-frozen samples (compare Figs. 2, 4, 5, and 7). Furthermore, in all of our different studies, tentaculate, cage-like mitochondria were seen exclusively in SAM and LP meristematic cells.
The presence of a large tentaculate/cage-like mitochondrion in SAM and LP meristematic cells makes these cell types unique. This begs the question of why these cells contain mitochondria with this type of morphology. For reasons detailed below, we postulate that the proliferative function of SAM cells and the fact that they are the precursors of, among others, the germ cell lines can explain the presence of a reticulate mitochondrion in these cells.
The Tentaculate/Cage-Like Mitochondrial Architecture Provides a Means for Efficient Delivery of ATP to Cell Proliferation-Related Activities
Mitochondrial shape, number, and distribution have been shown to be affected by the developmental state and the physiological status of a cell (Bereiter-Hahn, 1990; Stickens and Verbelen, 1996). From this perspective, the cell cycle-dependent changes in the spatial organization of the tentaculate/cage-like mitochondrion of SAM and LP cells can be viewed as a mechanism for becoming more energy efficient in their primary function, cell proliferation. As shown in Figure 8, over 40% of the total mitochondrial volume is associated with the tentaculate mitochondrion that wraps itself around one pole of the nucleus (Fig. 7; Supplemental Fig. S1, D and E). This spatial relationship between the nucleus and mitochondria would appear to be optimal for funneling large quantities of ATP into the nucleus to support the reassembly of the interphasic nucleus in G1, the replication of the nuclear genome in S (Fig. 9, G1-S), and the high rate of transcription in the G2 phase (Fig. 9, G2). Similarly, the transformation of the enlarged tentacular mitochondrion into a cage-like mitochondrion around the forming spindle at the onset of mitosis would ensure an even supply of energy to the entire spindle apparatus to support activities such as MT growth, kinesin-based MT and chromosome movements, and MT bundling (Fig. 9, mitosis; Asada et al., 1997; Liu and Lee, 2001).
Upon separation of the chromosomes, the largest energy sink becomes the phragmoplast and the forming cell plate, where in addition to the MT-related events, the assembly of the Golgi-derived vesicles into a cell plate and the subsequent stages of cell plate maturation involve a multitude of biochemical and structural activities (Verma, 2001; Seguí-Simarro et al., 2004, 2007). These activities might also be expedited by the formation of the mitochondrial belt (Figs. 7E and 9, early cytokinesis), similar to the one reported by Sheahan et al. (2004). However, considering the fact that the root apical meristem cells lack a tentaculate/cage-like mitochondrion, meristematic cells per se do not require such a unique type of mitochondrion to meet their energy needs; thus, the tentaculate/cage-like mitochondria are, from a strictly energetic point of view, not essential for proliferating plant cells. Instead, as elaborated in the following section, the major role of the tentaculate/cage-like mitochondrial system is likely related to the need for mixing of mtDNA in determinate SAM-derived cell lines.
The Cell Cycle-Dependent Changes in Tentaculate/Cage-Like Mitochondrion Architecture Provide a Structural Framework for Efficient Mixing and Recombination of mtDNA
The mtDNA of flowering plants exhibits a higher frequency of intraorganelle recombination than the mtDNA of mammalian cells (Lonsdale et al., 1988; Gillham, 1994). Indeed, the angiosperm mitochondrial genome has evolved to become recombinationally active, a condition that promotes extensive genomic rearrangements (Gray et al., 1999). A prerequisite for intraorganelle recombination is mitochondrial fusion, which allows for the genomes (nucleoids) from different mitochondria to intermix. To reconcile these facts with the finding that plant mitochondria tend to be small, structurally independent units, it has been proposed that plant mitochondria are “discontinuously interconnected” by means of transient fusion and fission events, as defined by the “discontinuous whole” hypothesis (Logan, 2006a, 2006b).
However, not all plants use the same strategy to produce this discontinuous interconnected state. For example, in nondividing, highly vacuolated cells such as onion (Allium cepa) epidermal cells, mtDNA intermixing between the small, grain-shaped mitochondria appears to be achieved by means of a high rate of fusion and fission events (Arimura et al., 2004). Nevertheless, this mechanism does not guarantee an equal distribution of the mtDNA after fission. In tobacco (Nicotiana tabacum) mesophyll protoplasts induced to fuse, the fusion process triggers the formation of a large reticular mitochondrion from smaller mitochondria to homogenize the mtDNA of the two cells (Sheahan et al., 2005). In cultured Medicago truncatula cells, the mitochondrial cycle is defined by the presence of punctate mitochondria at the onset of the log phase, the formation of more reticulate mitochondria during cell growth, and the accumulation of mitochondria around the cytokinetic apparatus during the division phase (Zottini et al., 2006).
The mitochondrial cycle reported here for SAM cells constitutes another variation on this theme. As discussed above, the organization of the SAM cell chondriome is optimized for the delivery of ATP energy to the structures involved in cell proliferation. Equally important, however, is the fact that the tentaculate/cage-like architecture also provides an efficient means for the intermixing of mtDNA and for the equal partitioning of the intermixed DNA to the two daughter cells. Thus, as the tentaculate mitochondrion is converted to a cage-like mitochondrion during the G2 stage of the cell cycle by growth and the fusion of smaller mitochondria (Fig. 9), its volume increases from approximately 40% to approximately 80% of the total mitochondrial volume. Thus, during mitosis and early cytokinesis, approximately 80% of the mtDNA is in the same mitochondrial compartment and available for recombination. Furthermore, since the remaining individual mitochondria are in a dynamic fusion/fission equilibrium with the cage-like mitochondrion, virtually 100% of the mtDNA of each meristematic cell can intermix and potentially participate in recombination events during this stage of the cell cycle.
The Tentaculate/Cage-Like Mitochondrion Provides a Means for Preventing Speciation
Since SAM cells are the precursors of all of the aerial parts of plants, their unique mitochondrial cycle has the potential to affect the inheritance of mtDNA in all of the aerial cell types, including germ lines. In particular, we hypothesize that the cell cycle-dependent changes in the tentaculate/cage-like mitochondria are expressed in those cell lines destined to originate, sooner or later, germ lines. This provides a means for passing homogeneous, recombined mtDNA to the next generation of plants, thereby avoiding the accumulation of undesirable mtDNA mutations. This capability would be characteristic of vegetative development, being expressed in SAM cells as well as in cells directly derived from them, including the LP cells. However, once the proliferative phase of the LP cells stops and differentiation to a mature leaf occurs, such a mechanism is no longer required. This would explain why the complex mitochondrial morphologies described in this study have not been observed in cells of differentiated organs, including mature leaves. This would also apply to any other SAM-derived differentiated cell, tissue, or organ, including ovules and also embryos, since after meiosis and gamete formation there would be no need for this feature until a new seedling started a new cycle of vegetative development., Finally, this hypothesis provides a rationale for why reticulate (tentaculate/cage-like) mitochondria have not been observed in other meristematic tissues such as root meristems, since they do not give rise to future germ cell lines.
Generating 3D Models of Organelles from Serial Thin-Section Micrographs Is Less Time Consuming Than in the Past and Opens Up New Future Possibilities
Confocal microscopy has become a mainstay of modern cell biology research and continues to produce exquisite insights into the structural organization of living cells. However, as demonstrated in this study, the spatial resolution of the CLSM micrographs is often insufficient to allow for an unambiguous interpretation of the cellular structures that produce the fluorescence signals. We have overcome this limitation by producing 3D mitochondrial models based on serial thin sections of high-pressure-frozen tissues. This approach yields 3D models with an approximately 100-fold increase in resolution in the x/y axes and an approximately 4-fold increase in resolution in the z axis (Staehelin and Kang, 2008). Prior to the advent of computer-assisted reconstruction and modeling with programs such as those included in the IMOD package (Kremer et al., 1996), the generation of 3D physical models of organelles from serial thin sections typically involved many months of labor, which limited the interest in and the use of this type of experimental approach. With current desktop computers, alignment of the thin-section images, correction for section distortions, and the tracing of individual organelles can be achieved in weeks, and quantitative data from the 3D models can be gotten in minutes. In recent years, we have shown that the production of 3D models from serial thin-section micrographs is a viable option for visualizing the 3D organization of cellular structures that cannot be adequately resolved by means of confocal microscopy (Seguí-Simarro and Staehelin, 2006a; this study). We believe that the use of this approach has the potential to provide novel insights on the 3D architecture of mitochondria in other cell types in which perinuclear congregations of mitochondria have been reported with the use of CLSM.
MATERIALS AND METHODS
CLSM and Reconstruction
Seeds of Arabidopsis (Arabidopsis thaliana Landsberg erecta) were grown on 0.8% (w/v) agar plates with Murashige and Skoog medium for 5 d at 24°C (16-h photoperiod). SAMs, LP, stems, and root tips were excised, incubated in 2 μm Mitotracker Green FM (Molecular Probes), diluted in 0.4 m mannitol, washed several times in phosphate-buffered saline, stained with DAPI, and mounted with Mowiol. We analyzed a minimum of 20 cells of each type, corresponding to at least five different plants, grown at different times.
Images of the specimens were collected with a Leica TCS SP2 AOBS confocal laser scanning microscope with 20× HC PL APO CS (0.70 numerical aperture), 40× HCX PL APO CS (1.25–0.75 numerical aperture), and 63× HCX PL APO (1.4–1.60 numerical aperture) oil-immersion optics. Laser lines at 351 and 490 nm for excitation of DAPI and MitoTracker were provided by a UV laser and an argon laser, respectively. The x and y resolution of the CCD chip was 4,096 pixels with a dynamic range of 12 bits per channel, while the resolution of the z axis control (galvanometric stage) was 40 nm. Z-series images were obtained through the collection of serial, confocal sections at 0.5-μm intervals. For partial cell confocal section reconstructions, up to 11 confocal sections were converted into stacks of serial images, aligned, and modeled with the IMOD software package (Kremer et al., 1996). To adjust the reconstructions to the actual cell volume, a correction factor was applied only to the z axis, calculated by dividing the Z-interval length of each virtual section (0.5 μm) by the pixel size in the confocal micrographs.
Processing of EM Samples by High-Pressure Freezing and Freeze Substitution
Seeds of Arabidopsis (Landsberg erecta) were grown and acclimated to increasing Suc concentrations as described previously (Seguí-Simarro et al., 2004). Shoot apices were excised, transferred to aluminum sample holders, cryoprotected with 150 mm Suc, frozen in a Baltec HPM 010 high-pressure freezer (Technotrade), and transferred to liquid nitrogen. The samples were freeze substituted in 4% OsO4 in anhydrous acetone at −80°C for 5 d, followed by slow warming to room temperature over 2 d. After rinsing in several acetone washes, they were removed from the holders, incubated in propylene oxide for 30 min, rinsed again in acetone, and infiltrated with increasing concentrations of Epon resin (Ted Pella) in acetone, according to the following schedule: 4 h in 5% resin, 4 h in 10% resin, 12 h in 25% resin, and 24 h in 50%, 75%, and 100% resin. Polymerization was carried out at 60°C for 2 d under vacuum.
EM and Serial Section Reconstruction
Ribbons of Epon (approximately 80–100 nm thick) serials sections were collected and mounted on formvar-coated copper slot grids and stained with uranyl acetate and lead citrate (Seguí-Simarro et al., 2004). Meristematic cells in G1, S, G2, mitosis, and cytokinesis were selected from the sections according to previously described morphological criteria (Seguí-Simarro and Staehelin, 2006a, 2006b; Supplemental Table S1): nuclear to cytoplasmic surface area ratio, which progressively decreases from G1 to G2 (Seguí-Simarro and Staehelin, 2006a); nuclear size, which increases as the nuclear genome replicates at the S phase (Jovtchev et al., 2006); architecture of the nucleolus and its different subdomains (dense fibrillar component, fibrillar centers, granular component, and nucleolar vacuoles), which change during the cell cycle (Risueño and Medina, 1986; Risueño et al., 1988); and cell wall thickness, which progressively increases after the new cell wall is formed (Risueño et al., 1968; Seguí-Simarro and Staehelin, 2006a). A prometaphase cell with a discontinuous nuclear envelope and condensed chromosomes was also selected. The cytokinetic stages of early telophase and late telophase were selected according to the differential architecture of the developing cell plate (Seguí-Simarro et al., 2004, 2007). For whole cell serial section reconstructions, single digital micrographs were converted into stacks of serial images and aligned with the IMOD software package (Kremer et al., 1996). To adjust the reconstructions to the actual cell volume, a correction factor was applied only to the z axis, calculated by dividing the section physical thickness (100 nm) by the pixel size in the digital micrographs.
Modeling and 3D Analysis of Serial EM Section Reconstructions
Serial EM section reconstructions were displayed and modeled with the 3DMOD program of the IMOD software package, as described previously (Seguí-Simarro et al., 2004). Since two morphologically different mitochondrial subpopulations were observed, they were modeled as different objects. This enabled us to independently acquire numerical data from each subpopulation. Once a model was completed, it was meshed and numerically analyzed using the imodmesh and imodinfo programs from the IMOD software package. Total volumes and surface areas of organelles were calculated with the imodinfo program, and the numerical data were processed in a spreadsheet.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Mitochondrial architecture in different cell types of the shoot apical region, as seen in thin-section electron micrographs.
Supplemental Figure S2. Mitochondria of interphasic root cells.
Supplemental Table S1. The different morphological criteria used to identify Arabidopsis SAM cells at different stages of the cell cycle.
Supplementary Material
Acknowledgments
Thanks are due to David Mastronarde and the other members of the Boulder Laboratory for Three-Dimensional Electron Microscopy of Cells (grant no. RR00592) and to Thomas Giddings for their technical help.
This work was supported by the National Institutes of Health (grant no. GM 61306 to L.A.S.) and the Ministerio de Educación y Ciencia (grant no. AGL2006–06678 to J.M.S.-S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: José M. Seguí-Simarro (seguisim@btc.upv.es).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Arimura S, Tsutsumi N (2002) A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc Natl Acad Sci USA 99 5727–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura S, Yamamoto J, Aida GP, Nakazono M, Tsutsumi N (2004) Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc Natl Acad Sci USA 101 7805–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada T, Kuriyama R, Shibaoka H (1997) TKRP125, a kinesin-related protein involved in the centrosome- independent organization of the cytokinetic apparatus in tobacco BY-2 cells. J Cell Sci 110 179–189 [DOI] [PubMed] [Google Scholar]
- Atkinson AW, John PCL, Gunning BES (1974) The growth and division of the single mitochondrion and other organelles during cell cycle of Chlorella, studied by quantitative stereology and three-dimensional reconstruction. Protoplasma 81 77–109 [DOI] [PubMed] [Google Scholar]
- Austin JR II, Frost E, Vidi PA, Kessler F, Staehelin LA (2006) Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 18 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin JR, Seguí-Simarro JM, Staehelin LA (2005) Quantitative analysis of changes in spatial distribution and plus-end geometry of microtubules involved in plant-cell cytokinesis. J Cell Sci 118 3895–3903 [DOI] [PubMed] [Google Scholar]
- Bakeeva LE, Chentsov YS, Skulachev VP (1978) Mitochondrial framework (reticulum mitochondrial) in rat diaphragm muscle. Biochim Biophys Acta 501 349–369 [DOI] [PubMed] [Google Scholar]
- Bawa SR, Werner G (1988) Mitochondrial changes in spermatogenesis of the pseudoscorpion, Diplotemnus sp. J Ultrastruct Mol Struct Res 98 281–293 [Google Scholar]
- Bereiter-Hahn J (1990) Behaviour of mitochondria in the living cell. Int Rev Cytol 122 1–63 [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J, Voth M (1994) Dynamics of mitochondria in living cells: shape changes, dislocations, fusion and fission of mitochondria. Microsc Res Tech 27 198–219 [DOI] [PubMed] [Google Scholar]
- Blank R, Arnold CG (1981) Structural-changes of mitochondria in Chlamydomonas reinhardtii after chloramphenicol treatment. Eur J Cell Biol 24 244–251 [PubMed] [Google Scholar]
- Boldogh IR, Pon LA (2006) Interactions of mitochondria with the actin cytoskeleton. Biochim Biophys Acta 1763 450–462 [DOI] [PubMed] [Google Scholar]
- Borsics T, Webb D, Andeme-Ondzighi C, Staehelin LA, Christopher DA (2007) The cyclic nucleotide-gated calmodulin-binding channel AtCNGC10 localizes to the plasma membrane and influences numerous growth responses and starch accumulation in Arabidopsis thaliana. Planta 225 563–573 [DOI] [PubMed] [Google Scholar]
- Calvayrac R, Butow RA, Lefort-Tran M (1972) Cyclic replication of DNA and changes in mitochondrial morphology during cell cycle of Euglena gracilis (Z). Exp Cell Res 71 422–432 [DOI] [PubMed] [Google Scholar]
- Christopher DA, Borsics T, Yuen CYL, Ullmer W, Andeme-Ondzighi C, Andres MA, Kang BH, Staehelin LA (2007) The cyclic nucleotide gated cation channel AtCNGC10 traffics from the ER via Golgi vesicles to the plasma membrane of Arabidopsis root and leaf cells. BMC Plant Biol 7 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekkes P, Shepard KA, Yaffe MP (2000) Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J Cell Biol 151 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd GL, Stewart KD, Mattox KR (1972) Cellular organization, mitosis, and cytokinesis in ulotrichalean alga, Klebsormidium. J Phycol 8 176–184 [Google Scholar]
- Foissner I (2004) Microfilaments and microtubules control the shape, motility, and subcellular distribution of cortical mitochondria in characean internodal cells. Protoplasma 224 145–157 [DOI] [PubMed] [Google Scholar]
- Gamalei YV, Pakhomova MV (1981) The structure of companion cells in the leaf phloem: results of the 3-dimensional cell reconstruction using serial sections. Tsitologiya 23 117–129 [Google Scholar]
- Gillham NW (1994) Organelle Genes and Genomes. Oxford University Press, New York
- Gray MW, Burger G, Lang BF (1999) Mitochondrial evolution. Science 283 1476–1481 [DOI] [PubMed] [Google Scholar]
- Hermann GJ, Shaw JM (1998) Mitochondrial dynamics in yeast. Annu Rev Cell Dev Biol 14 265–303 [DOI] [PubMed] [Google Scholar]
- Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM (1998) Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol 143 359–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HP, Avers CJ (1973) Mitochondrion of yeast: ultrastructural evidence for one giant branched organelle per cell. Science 181 749–751 [DOI] [PubMed] [Google Scholar]
- Hoog JL, Schwartz C, Noon AT, O'Toole ET, Mastronarde DN, McIntosh JR, Antony C (2007) Organization of interphase microtubules in fission yeast analyzed by electron tomography. Dev Cell 12 349–361 [DOI] [PubMed] [Google Scholar]
- Howard RJ (1981) Ultrastructural analysis of hyphal tip cell growth in fungi: spitzenkorper, cytoskeleton and endomembranes after freeze-substitution. J Cell Sci 48 89–103 [DOI] [PubMed] [Google Scholar]
- Jakobs S (2006) High resolution imaging of live mitochondria. Biochim Biophys Acta 1763 561–575 [DOI] [PubMed] [Google Scholar]
- Jovtchev G, Schubert V, Meister A, Barow M, Schubert I (2006) Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet Genome Res 114 77–82 [DOI] [PubMed] [Google Scholar]
- Kennady PK, Ormerod MG, Singh S, Pande G (2004) Variation of mitochondrial size during the cell cycle: a multiparameter flow cytometric and microscopic study. Cytometry A 62A: 97–108 [DOI] [PubMed] [Google Scholar]
- Kiss JZ, Giddings TH Jr, Staehelin LA, Sack FD (1990) Comparison of the ultrastructure of conventionally fixed and high pressure frozen/freeze substituted root tips of Nicotiana and Arabidopsis. Protoplasma 157 64–74 [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116 71–76 [DOI] [PubMed] [Google Scholar]
- Kuroiwa H, Nishimura Y, Higashiyama T, Kuroiwa T (2002) Pelargonium embryogenesis: cytological investigations of organelles in early embryogenesis from the egg to the two-celled embryo. Sex Plant Reprod 15 1–12 [Google Scholar]
- Kuroiwa H, Ohta T, Kuroiwa T (1996) Studies on the development and three-dimensional reconstruction of giant mitochondria and their nuclei in egg cells of Pelargonium zonale Ait. Protoplasma 192 235–244 [Google Scholar]
- Liu B, Lee YRJ (2001) Kinesin-related proteins in plant cytokinesis. J Plant Growth Regul 20 141–150 [Google Scholar]
- Logan DC (2006. a) The mitochondrial compartment. J Exp Bot 57 1225–1243 [DOI] [PubMed] [Google Scholar]
- Logan DC (2006. b) Plant mitochondrial dynamics. Biochim Biophys Acta 1763 430–441 [DOI] [PubMed] [Google Scholar]
- Logan DC, Leaver CJ (2000) Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J Exp Bot 51 865–871 [PubMed] [Google Scholar]
- Logan DC, Scott I, Tobin AK (2003) The genetic control of plant mitochondrial morphology and dynamics. Plant J 36 500–509 [DOI] [PubMed] [Google Scholar]
- Lonsdale DM, Brears T, Hodge TP, Melville SE, Rottmann WH (1988) The plant mitochondrial genome: homologous recombination as a mechanism for generating heterogeneity. Philos Trans R Soc Lond B Biol Sci 319 149–163 [Google Scholar]
- Nebenführ A, Frohlick JA, Staehelin LA (2000) Redistribution of Golgi stacks and other organelles during mitosis and cytokinesis in plant cells. Plant Physiol 124 135–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olyslaegers G, Verbelen JP (1998) Improved staining of F-actin and co-localization of mitochondria in plant cells. J Microsc 192 73–77 [Google Scholar]
- Osafune T, Mihara S, Hase E, Ohkuro I (1975. a) Formation and division of giant mitochondria during cell-cycle of Euglena gracilis Z in synchronous culture. 1. Some characteristics of changes in morphology of mitochondria and oxygen-uptake activity of cells. Plant Cell Physiol 16 313–326 [Google Scholar]
- Osafune T, Mihara S, Hase E, Ohkuro I (1975. b) Formation and division of giant mitochondria during cell-cycle of Euglena gracilis Z in synchronous culture. 2. Modes of division of giant mitochondria. J Electron Microsc (Tokyo) 24 33–39 [PubMed] [Google Scholar]
- Osafune T, Mihara S, Hase E, Ohkuro I (1975. c) Formation and division of giant mitochondria during cell-cycle of Euglena gracilis Z in synchronous culture. 3. 3-Dimensional structures of mitochondria after division of giant forms. J Electron Microsc (Tokyo) 24 283–286 [PubMed] [Google Scholar]
- Osafune T, Ohkuro I, Hase E, Mihara S (1972) Electron microscope studies on vegetative cellular life-cycle of Chlamydomonas reinhardtii dangeard in synchronous culture. 1. Some characteristics of changes in subcellular structures during cell cycle, especially in formation of giant mitochondria. Plant Cell Physiol 13 211–217 [Google Scholar]
- Otegui MS, Capp R, Staehelin LA (2002) Developing seeds of Arabidopsis store different minerals in two types of vacuoles and in the endoplasmic reticulum. Plant Cell 14 1311–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Herder R, Schulze J, Jung R, Staehelin LA (2006) The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18 2567–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Noh YS, Martinez DE, Vila Petroff MG, Staehelin LA, Amasino RM, Guiamet JJ (2005) Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J 41 831–844 [DOI] [PubMed] [Google Scholar]
- Otegui MS, Staehelin LA (2000) Syncytial-type cell plates: a novel kind of cell plate involved in endosperm cellularization of Arabidopsis. Plant Cell 12 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Staehelin LA (2004) Electron tomographic analysis of post-meiotic cytokinesis during pollen development in Arabidopsis thaliana. Planta 218 501–515 [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps JD (1974) Cell division in Stichococcus. Br Phycol J 9 63–73 [Google Scholar]
- Risueño MC, Giménez M, López-Saez F (1968) Development on the middle lammela in rib meristem cells. Experientia 24 514 [Google Scholar]
- Risueño MC, Medina FJ (1986) The Nucleolar Structure in Plant Cells. Servicio Editorial de la Universidad del País Vasco, Leioa, Vizcaya, Spain
- Risueño MC, Testillano PS, Sánchez-Pina MA (1988) Variations of nucleolar ultrastructure in relation to transcriptional activity during G1, S, G2 of microspore interphase. In M Cresti, P Gori, E Paccini, eds, Sexual Plant Reproduction. Springer-Verlag, Berlin/Heidelberg, pp 9–14
- Rohr R (1978) Existence of a mitochondrial reticulum in haploid tissue-culture of a vascular plant. Biol Cell 33 89–94 [Google Scholar]
- Scott I, Sparkes IA, Logan DC (2007) The missing link: inter-organellar connections in mitochondria and peroxisomes? Trends Plant Sci 12 380–381 [DOI] [PubMed] [Google Scholar]
- Seguí-Simarro JM, Austin JR, White EA, Staehelin LA (2004) Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell 16 836–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguí-Simarro JM, Otegui MS, Austin JR, Staehelin LA (2007) Plant cytokinesis: insights gained from electron tomography studies. In DPS Verma, Z Hong, eds, Cell Division Control in Plants, Vol 9. Springer, Berlin/Heidelberg, pp 251–287
- Seguí-Simarro JM, Staehelin LA (2006. a) Cell cycle-dependent changes in Golgi stacks, vacuoles, clathrin-coated vesicles and multivesicular bodies in meristematic cells of Arabidopsis thaliana: a quantitative and spatial analysis. Planta 223 223–236 [DOI] [PubMed] [Google Scholar]
- Seguí-Simarro JM, Staehelin LA (2006. b) Mechanisms of cytokinesis in flowering plants: new pieces for an old puzzle. In JA Teixeira da Silva, ed, Floriculture, Ornamental and Plant Biotechnology: Advances and Topical Issues. Global Science Books, London, pp 185–196
- Sheahan MB, McCurdy DW, Rose RJ (2005) Mitochondria as a connected population: ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J 44 744–755 [DOI] [PubMed] [Google Scholar]
- Sheahan MB, Rose RJ, McCurdy DW (2004) Organelle inheritance in plant cell division: the actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J 37 379–390 [DOI] [PubMed] [Google Scholar]
- Shepard KA, Yaffe MP (1999) The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J Cell Biol 144 711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM (1998) A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol 143 351–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin LA, Giddings TH Jr, Kiss JZ, Sack FD (1990) Macromolecular differentiation of Golgi stacks in root tips of Arabidopsis and Nicotiana seedlings as visualized in high pressure frozen and freeze-substituted samples. Protoplasma 157 75–91 [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Kang BH (2008) Nanoscale architecture of endoplasmic reticulum export sites and of Golgi membranes as determined by electron tomography. Plant Physiol Biochem 147 1454–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickens D, Verbelen JP (1996) Spatial structure of mitochondria and ER denotes changes in cell physiology of cultured tobacco protoplasts. Plant J 9 85–92 [Google Scholar]
- Van Gestel K, Verbelen JP (2002) Giant mitochondria are a response to low oxygen pressure in cells of tobacco (Nicotiana tabacum L.). J Exp Bot 53 1215–1218 [DOI] [PubMed] [Google Scholar]
- Verma DPS (2001) Cytokinesis and building of the cell plate in plants. Annu Rev Plant Physiol Plant Mol Biol 52 751–784 [DOI] [PubMed] [Google Scholar]
- Yaffe MP (1999. a) Dynamic mitochondria. Nat Cell Biol 1 E149–E150 [DOI] [PubMed] [Google Scholar]
- Yaffe MP (1999. b) The machinery of mitochondrial inheritance and behavior. Science 283 1493–1497 [DOI] [PubMed] [Google Scholar]
- Yaffe MP (2003) The cutting edge of mitochondrial fusion. Nat Cell Biol 5 497–499 [DOI] [PubMed] [Google Scholar]
- Zadworny M, Tuszynska S, Samardakiewicz S, Werner A (2007) Effects of mutual interaction of Laccaria laccata with Trichoderma harzianum and T. virens on the morphology of microtubules and mitochondria. Protoplasma 232 45–53 [DOI] [PubMed] [Google Scholar]
- Zottini M, Barizza E, Bastianelli F, Carimi F, Lo Schiavo F (2006) Growth and senescence of Medicago truncatula cultured cells are associated with characteristic mitochondrial morphology. New Phytol 172 239–247 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.