Abstract
Shade avoidance in plants involves rapid shoot elongation to grow toward the light. Cell wall-modifying mechanisms are vital regulatory points for control of these elongation responses. Two protein families involved in cell wall modification are expansins and xyloglucan endotransglucosylase/hydrolases. We used an alpine and a prairie ecotype of Stellaria longipes differing in their response to shade to study the regulation of cell wall extensibility in response to low red to far-red ratio (R/FR), an early neighbor detection signal, and dense canopy shade (green shade: low R/FR, blue, and total light intensity). Alpine plants were nonresponsive to low R/FR, while prairie plants elongated rapidly. These responses reflect adaptation to the dense vegetation of the prairie habitat, unlike the alpine plants, which almost never encounter shade. Under green shade, both ecotypes rapidly elongate, showing that alpine plants can react only to a deep shade treatment. Xyloglucan endotransglucosylase/hydrolase activity was strongly regulated by green shade and low blue light conditions but not by low R/FR. Expansin activity, expressed as acid-induced extension, correlated with growth responses to all light changes. Expansin genes cloned from the internodes of the two ecotypes showed differential regulation in response to the light manipulations. This regulation was ecotype and light signal specific and correlated with the growth responses. Our results imply that elongation responses to shade require the regulation of cell wall extensibility via the control of expansin gene expression. Ecotypic differences demonstrate how responses to environmental stimuli are differently regulated to survive a particular habitat.
Plants growing in dense stands often ensure survival by an escape syndrome known as shade avoidance. The imposition of shade by surrounding taller plants causes a change in the spectral composition of light reaching these plants. For plants that are beginning to get shaded, an early warning signal is the lowering of the red to far-red ratio (R/FR). The preferential absorption of red wavelengths by surrounding vegetation causes the reflected/transmitted light to be enriched in FR wavelengths and thus lowers the R/FR. Within a dense canopy, there are further spectral changes involving not just a lowering of R/FR but also a reduction of blue light and the total light intensity. These spectral changes are vital cues for plants to start elongation of their shoots and move their leaves upward. This in turn improves their chances of regaining access to sunlight and thus survival (Franklin and Whitelam, 2005; Vandenbussche et al., 2005; Franklin, 2008). In general, such functionally adaptive plasticity allows plants to survive despite widely varying environmental conditions and can also explain the success of certain species in colonizing diverse habitats.
This is clearly evident in the herbaceous perennial Stellaria longipes, which exhibits a circumpolar distribution and is able to inhabit diverse environmental niches such as alpine, prairie, montane, and sand dune, where, among other characteristics, light conditions vary drastically (Chinnappa et al., 2005). Two ecotypes of S. longipes, the alpine and the prairie, are of particular interest in the context of shade avoidance. The alpine ecotype inhabits an area of sparse vegetation and significant exposure to wind, and its dwarf phenotype is assumed to be advantageous in minimizing wind damage. The prairie ecotype, in contrast, grows among dense vegetation, where it is more prone to being shaded by surrounding plants. Consequently, its tall phenotype ensures a better chance of survival among the competing neighbors (Macdonald et al., 1988). Previous studies have shown that while the alpine ecotype is nonresponsive to low R/FR, an early shade signal, the prairie ecotype exhibits a typical shade avoidance response under similar conditions (Alokam et al., 2002). Differences within the same species in response to the same cue reflect the specialization of each ecotype to its specific habitat. However, so far, the underlying mechanisms are poorly studied. The prairie and alpine ecotypes, therefore, constitute an excellent system in which to study the molecular mechanisms underlying differential plasticity to similar environmental cues, in this case shading.
Shoot elongation, which is a primary feature of shade avoidance, mostly involves cellular expansion. This in turn requires an increased extensibility of the cell wall in response to an increased turgor pressure within the cells. Increased cell wall extensibility is made possible by certain proteins that act on the molecular framework of the cell wall and thus allow the walls to stretch out, a process termed “wall loosening” (Cosgrove, 2005). The most well-characterized groups of cell wall-loosening proteins are the expansins and the xyloglucan endotransglycosylase/hydrolases (XTHs).
Expansins are currently distinguished into four sequence-specific classes: Expansin A (EXPA), EXPB, Expansin-like A (EXLA), and EXLB (Kende et al., 2004). These proteins are proposed to act on the noncovalent interactions between cellulose and hemicelluloses in the cell wall in a nonenzymatic process that results in a more extensible cell wall and consequent cellular expansion (McQueen-Mason and Cosgrove, 1994; Cosgrove, 2000). Manipulation of the expression of expansins has confirmed the functionality of these proteins in plant growth and development (Cho and Cosgrove, 2000; Zenoni et al., 2004). Silencing of expansin genes results in shorter plants (Choi et al., 2003), while expansin overexpressors exhibit accelerated (Choi et al., 2003; Lee et al., 2003) or even abnormal (Rochange et al., 2001) growth responses. However, some studies also suggest that the correlation between expansin activity and growth does not always hold (Caderas et al., 2000; Reidy et al., 2001). Expansins are strongly regulated during plant responses to environmental stresses, such as drought (Jones and McQueen-Mason, 2004; Zhu et al., 2007), flooding (Cho and Kende, 1997; Colmer et al., 2004; Vreeburg et al., 2005), and response to pathogens (Ding et al., 2008; Fudali et al., 2008).
In addition to expansins, the XTHs are another group of proteins implicated in cell wall modification (Fry et al., 1992; Campbell and Braam, 1999). XTHs act on the xyloglucan tethers between the cellulose fibers in the cell wall, catalyzing either a transglucosylation or a hydrolytic reaction (Tabuchi et al., 1997, 2001; Kaku et al., 2002). Accordingly, most XTHs possess either one or both of these activities (Rose et al., 2002). XTH protein activity and transcript levels have also been correlated with various aspects of plant growth and development (Redgwell and Fry, 1993; Bourquin et al., 2002; Hyodo et al., 2003; Matsui et al., 2005; Van Sandt et al., 2007a) in which cellular expansion is required, and this functionality has been confirmed using transgenic approaches (Cho et al., 2006; Osato et al., 2006). Here too, there are instances where this correlation does not hold (Pritchard et al., 1993; Palmer and Davies, 1996). However, there is evidence suggesting that XTHs, via their effect on xyloglucan metabolism, are definitely involved in the cell expansion process (Takeda et al., 2002; Kaku et al., 2004). Expansins are generally considered primary wall-loosening agents because they can cause wall loosening in isolated cell walls, while XTHs are speculated to enhance or supplement their action and thus act as secondary wall-loosening agents (Cosgrove, 2005). However, a recent study suggests that XTHs can also cause wall loosening in isolated cell walls (Van Sandt et al., 2007b). Nevertheless, both expansin and XTH action on the cell wall network probably results in a cell wall that can stretch more easily in response to increased turgor pressure within the cell. Like expansins, XTHs are also regulated in response to environmental changes such as drought and salinity (Cho et al., 2006), wind (Antosiewicz et al., 1997), gravity (Zenko et al., 2004), and pathogen attack (Albert et al., 2004).
It is obvious, therefore, that the regulation of cell wall extensibility is important during plant adaptation to environmental changes. However, there is currently no such information relating to plant responses during shading or to changes in light quality. In this study, we use the aforementioned two ecotypes of S. longipes as a comparative system to study the molecular basis of plasticity and in doing so also investigate how changes in light quality can regulate cell wall extensibility and consequently growth in the functional context of shade avoidance. Our results present, to our knowledge, the first report on the regulation of expansins and XTHs in response to light signals that act as plant-plant interaction cues.
RESULTS
Shade Signals Induced Different Growth Responses in the Alpine and Prairie Ecotypes of S. longipes
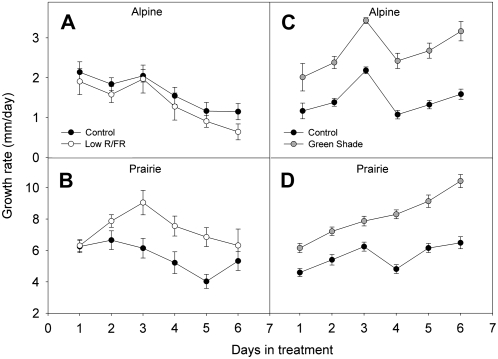
Plants of both ecotypes were grown under spectrally altered light conditions, and the increase in the length of the stem was measured every day for 1 week. We first subjected the plants to low R/FR conditions. This is an early neighbor detection signal, and our results confirmed previous studies of contrasting responses for the two ecotypes (Alokam et al., 2002). The alpine plants were not responsive to low R/FR and grew at rates similar to the plants grown under control light conditions (Figs. 1 and 2A). The prairie plants, in contrast, showed enhanced growth rates relative to controls in response to low R/FR (Fig. 2B). Growth rates became significantly (P < 0.05, Student's t test) higher relative to control plants by the 3rd d of treatment, and this difference was maintained until the 5th d of measurement. We then tested the responses of the two ecotypes to green shading, which mimics deep canopy shade, with combined reductions in blue, R/FR, and total light intensity. Surprisingly, we found that in response to this treatment, both ecotypes responded within 1 d with rapid internodal elongation and growth rates that were significantly higher than controls (Fig. 2, C and D). Also, the growth rates stayed significantly higher than those in controls until the last day of measurement. These results imply that the alpine ecotype can also detect changes in light quality. It must be noted that in the prairie ecotype, plants exposed to green shade are much taller than plants grown under low R/FR (Fig. 1). Furthermore, although the alpine ecotype did respond with higher growth rates relative to controls when subjected to green shade, the growth rates were much lower than those of prairie plants under similar conditions. In addition, in all cases of rapid internodal elongation, the strongest elongation growth was observed in the youngest internodes (Fig. 1).
Figure 1.
The alpine and prairie ecotypes of S. longipes. Shown are representative ramets from the alpine (left) and prairie (right) plants after 1 week in the indicated treatments. Low R/FR 0.25, PAR 140 μmol m−2 s−1; green shade, PAR 65 μmol m−2 s−1, R/FR 0.19, and blue light photon fluence rate of 2 μmol m−2 s−1. Control plants were grown in light with an unaltered spectral composition and a PAR of 140 μmol m−2 s−1. Each bar on the scale = 1 cm.
Figure 2.
The effects of canopy light signals on the growth rates of alpine and prairie plants. Ramet elongation rate was measured every day for 1 week. Alpine (A and C) and prairie (B and D) plants were grown under low R/FR (A and B; white circles) or green shade (C and D; gray circles) conditions. Controls (black circles) were grown under normal light conditions (spectral composition unaltered). Growth rates were calculated from length measurements of the total ramet height obtained using a digital caliper. Data points represent means of 30 to 35 ramets (mean ± se, n = 30–35). Experiments were carried out twice with similar results. Growth rates under green shading show statistically significant differences relative to controls, at all time points, in both ecotypes. Under low R/FR, only the growth rates for the prairie plants showed statistically significant differences relative to controls at days 3 to 5 (Student's t test, P < 0.05).
XTH Activity in Internodes of Alpine and Prairie Plants Correlated with Observed Growth Trends But Not under Low R/FR Conditions
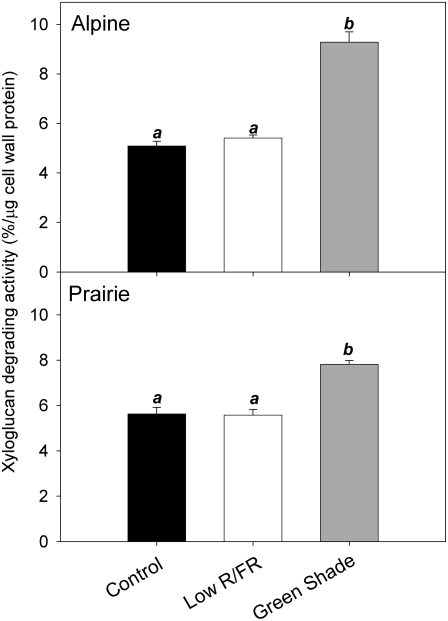
We investigated the activity of XTHs in order to relate growth responses to the regulation of cell wall extensibility and cellular expansion. Internodes from alpine and prairie plants that had been grown under low R/FR and green shade for 3 d were used as the source of a crude enzyme extract that was assayed for xyloglucan-degrading activity. This is a measure of both transglycolytic and hydrolytic activities of XTHs. In both ecotypes, XTH activity in the internodes of control and low-R/FR-treated plants was similar, suggesting a lack of correlation between XTH activity and elongation growth under low R/FR (Fig. 3). These results show that XTH activity is not responsive to changes in R/FR. However, XTH activity showed a significant increase in the internodes of plants in green shade relative to controls. Under these conditions, there was an increase in XTH activity relative to controls of approximately 30% and 45% in the prairie and alpine ecotypes, respectively (Fig. 3).
Figure 3.
Xyloglucan-degrading activity in response to canopy light signals. Shown is xyloglucan-degrading activity measured in the top internodes of alpine (top) and prairie (bottom) plants grown for 3 d under low R/FR (white bars) and green shade (gray bars) conditions. Controls (black bars) refer to data from plants grown under normal light conditions (unaltered spectral composition). Data points represent means ± se (n = 3); each biological replicate consisted of internodes pooled from different ramets from different pots. Different letters above each bar indicate statistically significant differences (P < 0.05, Tukey's b test). Experiments were repeated twice with similar results.
Growth Responses and XTH Activity in Response to Blue Light Depletion
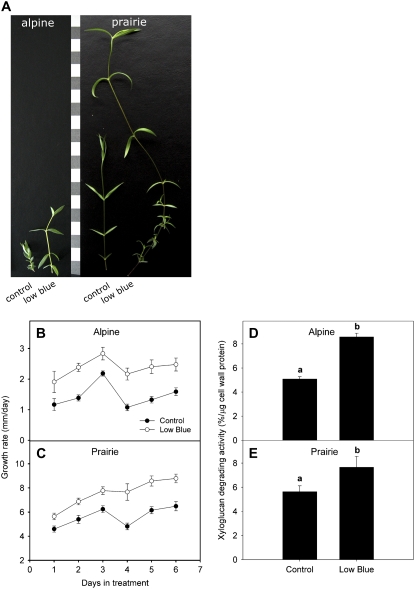
Since XTH activity was regulated only under conditions of green shade and not in response to low R/FR, we wanted to investigate whether this response was due to a reduction in blue light, which is the other spectral change in green shade. We measured the growth rates of plants from both ecotypes under low blue light conditions. The growth rates in both alpine and prairie plants were significantly higher than in controls (Fig. 4, A–C). Furthermore, the growth rates under low blue light were very similar to the growth rates under green shade conditions. We next measured XTH activity in the internodes of plants grown in low blue light and found that there was a significant increase in activity relative to plants grown under control conditions in both ecotypes (Fig. 4, D and E). This suggests that XTH activity is under the control of blue light receptors.
Figure 4.
Effects of low blue light conditions on the growth rates and xyloglucan-degrading activity in the alpine and prairie ecotypes. A, Representative ramets from the alpine and prairie plants after 1 week in the indicated treatments. Control plants were grown in light with an unaltered spectral composition. Each bar on the scale = 1 cm. B and C, Ramet elongation rate for alpine (B) and prairie (C) plants grown under low blue light (white circles) and control (black circles) conditions for 1 week. Data points represent means ± se (n = 30–35). Ramet length measurements were made using a digital caliper. Growth rate differences between control and low blue light treatments were statistically significant at all time points (P < 0.05, Student's t test). D and E, Xyloglucan-degrading activity measured in the top internodes of alpine (D) and prairie (E) plants grown for 3 d under low blue light conditions. Controls refer to data from plants grown under normal light conditions (unaltered spectral composition). Data points represent means ± se (n = 3); each biological replicate consisted of internodes pooled from different ramets from different pots. Different letters above each bar indicate statistically significant differences (P < 0.05, Tukey's b test). Experiments were repeated twice with similar results. [See online article for color version of this figure.]
Acid-Induced Extension of Internodes of Both Alpine and Prairie Ecotypes Correlated with Growth under Different Shade Signals
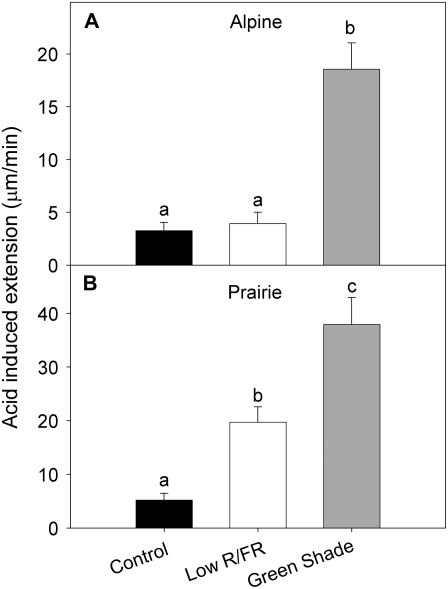
Expansins are considered primary mediators of cell wall loosening, which aid cellular expansion (Cosgrove, 2005). Acid-induced extension (AIE) is a reflection of the in planta expansin activity. Unlike the trends seen in XTH activity, AIE values corresponded well with low-R/FR-induced growth trends (Fig. 5). Under low R/FR, the AIE of the internodes from prairie plants was significantly higher than in controls (Fig. 5B). Alpine internodes from plants in low R/FR had similar AIE values as controls (Fig. 5A). Plants from both ecotypes grown in green shade also showed much higher AIE relative to controls (Fig. 5). This correlation of AIE values with growth suggests a role for expansins in shade-induced elongation in S. longipes ecotypes.
Figure 5.
The effects of canopy light signals on the AIE of internodes from alpine and prairie plants. AIE of the topmost internodes of ramets from alpine (A) and prairie (B) plants grown under low R/FR (white bars) and green shade (gray bars) growth conditions for 3 d. Control plants (black bars) were grown under light with an unaltered spectral composition. AIE was measured using a constant-load extensometer with a pulling weight of 20 g and is calculated as the difference in the slopes of lines fitted through 10-min intervals before and after the bending point observed due to a change in pH from 6.8 to 4.5. Data points represent means ± se (n = 8–10). Each biological replicate consisted of the topmost internode from different ramets from different pots. Different letters above each bar indicate statistically significant differences (P < 0.05, Tukey's b test). Experiments were repeated twice with similar results.
Expansins in the Internodes of Alpine and Prairie Ecotypes of S. longipes
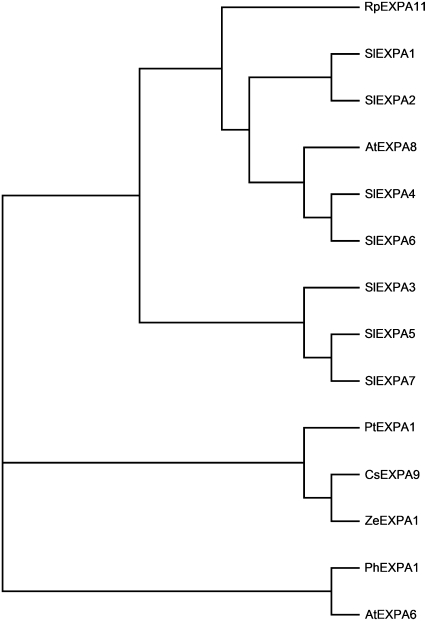
In order to identify expansin genes required for shade-induced growth responses, we cloned expansins from the internodes of both ecotypes. In total, 12 expansin (GenBank accession nos. EU84703–EU84721) sequences were identified in both ecotypes. Of these, seven were chosen for expression analysis, since we were able to design gene-specific primers for these. Amino acid sequence alignment of these α-expansins from S. longipes revealed high homology among these genes. The similarity ranged from 71.7% (SlEXPA6 and SlEXPA7, SlEXPA5 and SlEXPA6) to 100% (SlEXPA5 and SlEXPA7, SlEXPA1 and SlEXPA2). Phylogenetic analysis of these seven α-expansins using α-expansin sequences with the highest similarity from GenBank showed that S. longipes α-expansins grouped into two clades (Fig. 6). Out of these, SlEXPA1, SlEXPA2, SlEXPA4, and SlEXPA6 fell into a clade with α-expansins from Arabidopsis (Arabidopsis thaliana) and Rumex palustris. The other three sequences formed their own branch and were separate from all of the other sequences from GenBank that were found to be highly similar to these sequences. These seven α-expansin sequences were further used to evaluate transcript abundance in the internodes of alpine and prairie plants in response to low R/FR and green shade.
Figure 6.
Phylogenetic analysis of α-expansin proteins. The deduced amino acid sequences of S. longipes α-expansins were aligned with highly similar amino acid sequences from the GenBank database using ClustalX software. This alignment was then used to generate a phylogenetic tree using TreeView software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). The following are the sequences used along with their accession numbers in parentheses: Stellaria longipes, SlEXPA1 to SlEXPA7 (EU840703–EU840714, EU840720, and EU840721); Arabidopsis thaliana, AtEXPA6 (NP_180461), AtEXPA8 (O22874); Cucumis sativus, CsEXPA9 (AAL31480); Petunia hybrida, PhEXPA1 (AAR82849); Populus tremula, PtEXPA1 (AAR09168); Rumex palustris, RpEXPA11 (AAM22625); and Zinnia elegans, ZeEXPA1 (AAF35900).
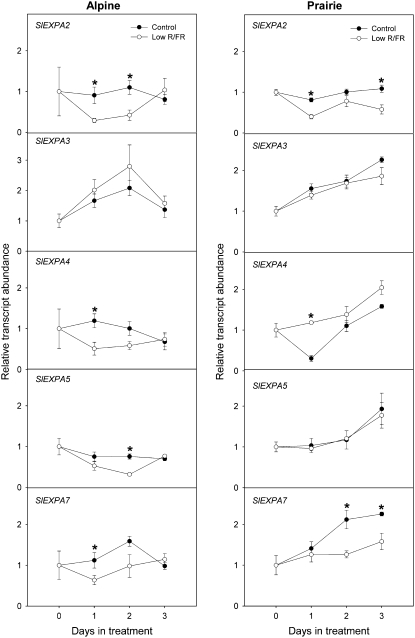
Shade Signals Differentially Regulated Expansin Gene Expression in Both Ecotypes
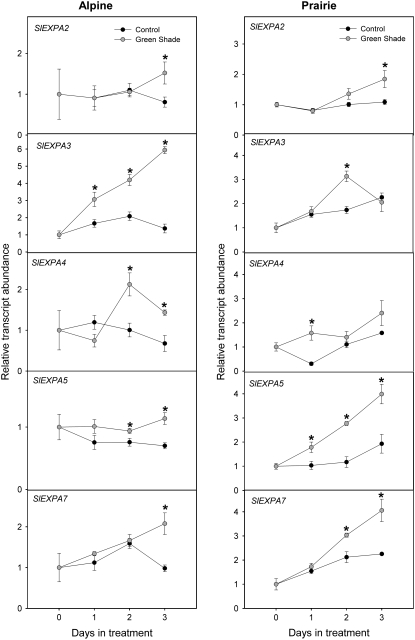
Figure 7 shows the transcript profiles for five α-expansin genes in the internodes of plants from the two ecotypes grown under low R/FR conditions. Of the seven genes selected for gene expression studies, SlEXPA1 and SlEXPA6 did not show any detectable expression at the time points and tissues tested (data not shown). In the alpine ecotype, none of the five genes examined showed any significant up-regulation relative to controls under low R/FR. Instead, there was a down-regulation of SlEXPA2, SlEXPA4, SlEXPA5, and SlEXPA7 in the internodes of low-R/FR-grown alpine plants at day 1 or day 2 of treatment. For SlEXPA3, however, there was no significant difference between control and low R/FR values. A large variation is seen in the transcript profiles for these same genes for the prairie ecotype. Under low-R/FR treatment, SlEXPA4 transcript abundance in prairie internodes was almost 4-fold higher than in controls by day 1. The low-R/FR treatment caused SlEXPA2 and SlEXPA7 transcripts to be down-regulated, while the abundance of SlEXPA3 and SlEXPA5 was maintained similar to that in controls. Figure 8 shows the transcript profiles for the same five α-expansins in alpine and prairie internodes upon a green shade treatment, which resulted in dramatic internodal elongation in both ecotypes. In the alpine internodes, green shade caused a massive up-regulation of SlEXPA3. Expression was doubled by day 1, and there was an approximately 6-fold increase in SlEXPA3 expression relative to controls by day 3. Although on the 1st d of treatment none of the other expansins showed any up-regulation, by day 3 green shade caused a significant increase in the relative abundance of SlEXPA2, SlEXPA4, SlEXPA5, and SlEXPA7. In the prairie internodes, expression of another expansin, SlEXPA5, was enhanced dramatically in response to green shade. SlEXPA5 expression doubled already by day 1 of treatment, and the up-regulation was maintained until day 3, when there was an approximately 2.5-fold difference relative to controls. SlEXPA4 transcript abundance was approximately 5-fold higher at day 1 relative to controls. However, this difference leveled off at day 2. Two other expansins, SlEXPA2 and SlEXPA7, also showed high relative transcript abundance from day 2 onward. SlEXPA3 was up-regulated only slowly and transiently and only at day 2 showed an increase in expression levels relative to controls. In conclusion, there is good correlation between the regulation of expansin gene expression and the growth responses to shade signals.
Figure 7.
Differential regulation of S. longipes α-expansins in response to low R/FR. Relative transcript abundance of S. longipes α-expansins expressed in the internodes of alpine and prairie plants exposed to low R/FR (white circles) and control (unaltered spectral composition; black circles) light conditions for 3 d. Values were measured using real-time RT-PCR with 18S as an internal standard (means ± se; n = 3–4). Statistically significant differences are indicated by asterisks (Student's t test, P < 0.05).
Figure 8.
Differential regulation of S. longipes α-expansins in response to green shading. Relative transcript abundance of S. longipes α-expansins expressed in the internodes of alpine and prairie plants exposed to green shade (gray circles) and control (unaltered spectral composition; black circles) light conditions for 3 d. Values were measured using real-time RT-PCR with 18S as an internal standard (means ± se; n = 3–4). Statistically significant differences are indicated by asterisks (Student's t test, P < 0.05).
DISCUSSION
Shade Avoidance and Cell Wall Extensibility
One of the primary objectives of this study was to examine cell wall-modifying proteins as downstream targets in shade-induced elongation responses and to correlate the regulation of these genes with the magnitude of the growth response. Two candidate protein families that are implicated in increasing cell wall extensibility needed during rapid cellular expansion leading to shoot elongation are the expansins and the XTHs. The control of cell wall extensibility requires the expression of these proteins at the right time and place, and this is a vital regulatory point during growth (Cosgrove, 2005). The activity and gene expression for both of these proteins have been found to correlate in many instances with elongation growth in response to a variety of environmental stimuli (Rose et al., 2002; Cosgrove, 2005). However, there are no studies on the regulation of cell wall extensibility via the control of the expression and/or activity of these proteins in response to shade signals. Therefore, we studied the effect of early neighbor signals (low R/FR) and deep canopy shade on the activity of expansins and XTHs. We used a system consisting of two ecotypes of S. longipes that behave differently in response to these canopy signals. The alpine ecotype with a constitutive dwarf phenotype showed no change in growth rates upon exposure to low R/FR, while the tall prairie ecotype grew still taller under low R/FR conditions (Fig. 2, A and B). These responses are consistent with the requirements needed to survive in the respective habitats of the two ecotypes. The prairie plants frequently encounter shade from neighboring plants, but the alpine plants do not. However, we also found that a combination of shade signals was able to induce rapid internodal elongation in both ecotypes (Fig. 2, C and D). This seems to imply that while the prairie plants can detect impending shade, the alpine plants are able to respond only when complete shading has already occurred. In order to see whether these growth responses could be correlated with the regulation of cell wall-modifying proteins, we measured AIE as a measure of expansin activity as well as XTH activity in the internodes of the two ecotypes.
Regulation of Expansin and XTH Activity in Response to Canopy Signals
We found that although XTH activity correlated well with growth under green shade conditions in both ecotypes, the correlation did not hold for low R/FR responses (Fig. 3). Even the prairie plants, which respond to low R/FR with enhanced internodal elongation, showed no measurable increase in XTH activity levels in their youngest internodes compared with plants grown under normal light conditions (Fig. 3, bottom). XTH activity, therefore, does not appear to be regulated by R/FR. However, it could be argued that there is a spatial regulation of XTH activity (Wu et al., 2005). It is possible that within the internode, the most rapidly growing part has higher XTH activity than the more mature parts. Since we used whole internodes, this difference, if present, might be diluted. Nevertheless, we also found that internodes from plants grown in green shade did have an increased XTH activity (Fig. 3). This confirms the robustness of the methods used. Furthermore, this is also not the first instance in which the correlation between growth and XTH activity does not hold (Pritchard et al., 1993; Palmer and Davies, 1996). Since green shading involves a lowering of both R/FR and blue light, and low R/FR had no apparent effect on XTH activity, we hypothesized that the green shading effect might be through the low blue light component. We first proceeded to record growth responses for both ecotypes under low blue light conditions (Fig. 4, A–C) and found that in both cases, growth rates were higher relative to controls and around the same magnitude as in green shade. Also, there was a significant increase in XTH activity in response to the depletion of blue light in the internodes of both alpine and prairie plants (Fig. 4, D and E). This in turn points toward a blue light photoreceptor-mediated regulation for the XTHs and opens up exciting new research questions. For instance, it would be interesting to know which particular blue light photoreceptors regulate XTH activity in S. longipes and whether low blue light-induced shade avoidance responses reported in other species such as tobacco (Nicotiana tabacum; Pierik et al., 2004) and cucumber (Cucumis sativus; Ballaré et al., 1991) also involve XTH regulation. Unlike S. longipes, in Arabidopsis seedlings XTH genes are regulated by both R/FR ratios and blue light, as suggested by microarray analyses. While XTH22 (At5g57560) and XTH32 (At2g36870) were suppressed by cryptochrome 1 in blue light (Folta et al., 2003), another study found that XTH15 (At4g14130) was strongly regulated by changes in light quality as well as dark/light transitions (Ma et al., 2001). In S. longipes, XTHs might be required for elongation responses during shade avoidance, but perhaps only when there is complete canopy closure, which results in blue light-depleted growth conditions. This up-regulation in XTH activity might help amplify the action of other wall-loosening agents that are already active in the cell wall, such as expansins, in order to result in an acceleration of cellular expansion and consequent growth.
Accordingly, AIE of alpine and prairie internodes was used as a measure of expansin activity. Unlike XTH activity, we found expansin activity to correlate well with growth responses (Fig. 5). Under low R/FR, alpine internodes had similar AIE values as controls, while prairie internodes showed an increase in AIE compared with control values. During green shade conditions, internodes from both ecotypes had much higher AIE levels than controls, reflecting their higher growth rates under these conditions.
We cloned members of the expansin gene family from S. longipes in order to identify specific expansins that are regulated in response to different light signals as well as to find possible ecotypic differences in the regulation of these genes in response to similar light cues. Our focus was on expansins since, first, unlike the XTHs, we found a good correlation between expansin activity (as reflected by AIE values) and growth responses to the light manipulations used. Second, expansins are considered the primary mediators of cell wall loosening (Cosgrove, 2000), and most reports show a strong correlation between expansins and growth. Furthermore, reports on the regulation of expansins in response to changes in light conditions are scant. A study with tomato (Solanum lycopersicum) hypocotyls failed to find any correlation between expansin gene expression and growth, even though expansin transcript accumulation showed a typical phytochrome response (Caderas et al., 2000). A microarray analysis on Arabidopsis seedlings found two expansins (N37536 and R29778) to be regulated by changes in light quality as well as light/dark transitions (Ma et al., 2001). Another study using genome-wide expression profiling with light quality manipulations found expansin regulation in Arabidopsis seedlings and cotyledons, but the regulation did not always positively correlate with elongation responses (Jiao et al., 2005).
Expansins usually exist as large multigene families, and we found the same to be true in S. longipes. We limited our cloning to RNA extracted from internodes in order to improve our chances of finding all possible expansins expressed in this organ. In the alpine ecotype (Fig. 7), in response to low R/FR, none of the five α-expansins showed any significant up-regulation. Furthermore, four of these five were actually down-regulated by low R/FR relative to controls. In the prairie ecotype, in response to low R/FR, SlEXPA4 transcript levels were 4-fold higher relative to controls by the end of the 1st d of treatment. This preceded the increase in growth rates seen under low R/FR (Fig. 2B). The regulation of expansin gene expression, therefore, could account for the different growth responses to low R/FR conditions in both the alpine and prairie plants. Green shade conditions led to an induction of α-expansin gene expression in the internodes of both ecotypes (Fig. 8). However, in each ecotype, different α-expansin genes were up-regulated. In the alpine ecotype, SlEXPA3 showed the most dramatic increase, which was visible within 1 d of treatment, and increased up to 6-fold relative to controls by day 3. All other expansins that we could study were up-regulated as well at various time points upon green shade exposure. In the prairie ecotype, SlEXPA5 and SlEXPA7 showed the strongest up-regulation in green shade relative to controls. Here also, all of the other expansins were up-regulated as well. Furthermore, there was no down-regulation of any of the other genes, as seen in response to low R/FR. These results implicate cell wall extensibility as an important regulatory point during light-mediated elongation responses. This is further corroborated by numerous microarray-based expression profiling studies, which have often found genes associated with cell wall modification to be regulated by R/FR ratios and during seedling deetiolation in Arabidopsis (Ma et al., 2001; Devlin et al., 2003; Salter et al., 2003). Although these studies used seedlings, which have a very different developmental program compared with the mature plants used in this study, taken together with our results, it is evident that cell wall extensibility is a major control point during light-induced elongation responses.
Our results also demonstrate a possible reason for the existence of expansins as large gene families. Although multigene families can imply either redundancy or specialization, our results suggest that the latter is more important. This study clearly implies that specific expansins are required upon exposure of the plant to different light signals. In response to the same light signal, there is again a difference between ecotypes in the identity of the expansin expressed. Previous studies with the alpine and prairie ecotypes of S. longipes implicated the hormones ethylene and gibberellin in the observed differential responses to shade (Kurepin et al., 2006a, 2006b). Phytohormones have a good potential to regulate the amount of plasticity in response to environmental cues. The two ecotypes of S. longipes clearly show different levels of, and sensitivities to, these hormones (Kurepin et al., 2006a, 2006b). Cell wall-modifying proteins like expansins and XTHs may very well be the downstream targets of these hormones, allowing control of the timing and magnitude of the elongation response. It is possible that differential regulation of particular expansins by these hormones is responsible for the differential response in the two ecotypes to the same environmental cue.
Different members of a multigene family are not only regulated differently in response to hormones and environmental stimuli, they are also expressed only in certain tissues and organs. In rice (Oryza sativa), for example, expansins are differentially regulated by developmental, hormonal, and environmental signals (Cho and Kende, 1997). In R. palustris, in response to flooding, ethylene caused the up-regulation of one expansin gene out of 13 (Vreeburg et al., 2005). Specific expansins are also up-regulated during fruit ripening (Rose et al., 1997), abscission (Belfield et al., 2005), root hair development (Cho and Cosgrove, 2002), and drought (Jones and McQueen-Mason, 2004) responses, to cite a few examples.
Ecotypic Specificity of Shade Avoidance
Shade avoidance is a good example of adaptive plasticity in plants in response to their environment. The speed and magnitude with which plants are able to sense and respond to their ambient environment is key to their survival. This is demonstrated well in the alpine and prairie ecotypes of S. longipes. Each of these ecotypes has specialized itself in sensing, responding to, and adapting to the changes in its specific habitat, and this is best exhibited by their growth responses to changes in low R/FR. It is surprising, therefore, that the alpine plants, which never encounter shade, still retain their ability to increase their growth rates when exposed to a combination of shade signals. It is possible that the response observed is primarily a vestige of etiolation responses (i.e. mainly mediated by a depletion of blue light). However, it could also imply that the alpine plants simply do not respond to shade or invest resources to deal with this stress until the threat is severe, as would happen in cases of complete canopy closure. An insensitivity to low R/FR would prevent a situation in which shade avoidance phenotypes, which are sensitive to wind damage, develop at the earliest detection of shade. In contrast, the prairie ecotype has probably fine-tuned its shade-sensing mechanism and is able to respond to low R/FR conditions that occur when a plant is beginning to get shaded (Ballaré et al., 1990) and can then accelerate responses upon sensing other canopy signals.
Our results also provide support for the concept of multigene families being the molecular basis of phenotypic plasticity (Smith, 1990). The presence of large multigene families like expansins could provide species like S. longipes with the flexibility to respond appropriately to the ever-changing environment as well as to inhabit diverse habitats successfully.
MATERIALS AND METHODS
Plant Material
The two ecotypes of Stellaria longipes were originally collected from the Chain Lakes (prairie; elevation, 1,310 m) and the summit of the Plateau Mountain (alpine; elevation, 2,453 m) in southern Alberta, Canada. Plants were clonally propagated and were potted in 220-mL plastic pots in a mixture of 2:1 (v/v) potting soil and sand with 2.8 g of MgOCaO (Magkal; 17% MgO; Vitasol) per liter of potting mixture. Prior to potting the ramets, each pot received approximately 60 mL of nutrient solution containing 7.5 m (NH4)2SO4, 15 mm KH2PO4, 15 mm KNO3, 3.3 μm MnSO4, 1.8 μm ZnSO4, 0.32 μm CuSO4, 43 μm H3BO3, 0.53 μm Na2MoO4, and 86 μm Fe-EDTA. Freshly potted ramets were allowed to establish for 2 weeks in a climate-controlled growth room (16-h photoperiod, 200 μmol m−2 s−1 photosynthetically active radiation [PAR; Philips Master HPI 400 W], 8 h of dark), after which they were transferred to a short-day (8-h photoperiod), cold (8°C day, 5°C night) growth chamber for at least 60 d to simulate the winter cycle.
Light Treatments
Plants that had been in the cold for 60 d were first transferred to a climate-controlled growth chamber (16-h photoperiod, 200 μmol m−2 s−1 PAR [Philips Master HPI 400 W], 8 h of dark) for 1 week, after which the plants were put under specific light treatments. Light quality manipulations took place in a white light background (Philips Master HPI-T Plus 400 W and Philips Plus line Pro 150 W). The R/FR was reduced from 1.2 to 0.25 by supplemental far-red light (730-nm light-emitting diode; Shinto Electronics [http://www.shinkohelecs.com]). Blue light photon fluence rates (400–500 nm) were reduced from 26 to less than 1 μmol m−2 s−1 using two layers of Lee 010 medium yellow filter (Lee Filters; http://www.leefilters.com). PAR for these light treatments was maintained at 140 μmol m−2 s−1. Green shading mimicking light conditions in a dense canopy was achieved using two layers of Lee 122 Fern Green, which reduced the PAR to 65 μmol m−2 s−1, the R/FR to 0.19, and the blue light photon fluence rate to 2 μmol m−2 s−1. Wherever mentioned, “controls” refers to data from plants grown in light conditions with an unaltered spectral composition and PAR of 140 μmol m−2 s−1. All light treatments were started at approximately 10 am each time the experiments were performed.
Measurement of Plant Growth
In order to measure stem elongation, ramet lengths were measured using a digital caliper every day at the same time to calculate daily growth increments. Care was taken to choose ramets that had similar starting lengths. For each light treatment, a total of at least 30 to 35 ramets were measured. Measurements were made for three independent trials.
AIE
AIE was measured using a custom-built constant-load extensometer, modified from the design of Cosgrove (1989), with a pulling weight of 20 g. This acid-induced extensibility of native cell walls reflects the expansin content of the cell wall as well as its susceptibility to expansin action (Cosgrove, 1996). The topmost internodes from plants of both S. longipes ecotypes grown for 3 d under different light conditions were harvested for measurements and immediately frozen in liquid nitrogen. These internodes were then thawed, abraded, and pressed, and 10-mm segments were clamped in the extensometer cuvette. The internodes were first bathed in 160 μL of a 50 mm HEPES (pH 6.8) buffer for 30 min, after which the buffer was replaced with a 50 mm sodium acetate (pH 4.5) buffer for another 30 min. AIE was measured as the difference in the slopes of lines fitted through 10-min intervals before and after the observed bending point obtained upon the change in pH of the buffer.
Measurement of Xyloglucan-Degrading Activity
The top internodes from plants of both ecotypes growing under different light conditions were harvested on the 3rd d after the start of the light treatment. Harvested material was immediately frozen in liquid nitrogen and stored at −80°C until they were used. Enzyme extracts were prepared as described (Soga et al., 1999). Briefly, frozen internodes were homogenized in ice-cold sodium phosphate buffer (10 mm, pH 7). The homogenate was centrifuged and the supernatant was discarded. The remaining cell wall pellet was washed twice with sodium phosphate buffer (10 mm, pH 7), after which the wall pellet was resuspended in sodium phosphate buffer (10 mm, pH 6) containing 1 m NaCl. The walls were then allowed to extract in this buffer for 24 h at 4°C before centrifugation and removal of the supernatant. This supernatant was then used as a crude enzyme extract to measure xyloglucan-degrading activity. Xyloglucan-degrading activity was measured as described by Sulova et al. (1995). The reaction mixture contained 15 μL of the enzyme extract, 0.4 mg mL−1 xyloglucan (Megazyme International), and 0.2 mg mL−1 xyloglucan oligosaccharides (XGOs; Megazyme International) in 0.2 mL of 0.1 m sodium phosphate buffer, pH 6. The mixture was incubated for 1.5 h at 37°C. The reaction was terminated by adding 0.1 mL of 1 n HCl, following which the remaining xyloglucan was quantitated via the iodine staining method. Color development was allowed to proceed for 1 h in the dark, after which the absorbance of the samples was read at 620 nm against a blank that contained no xyloglucan or XGOs. The activity measured is designated xyloglucan-degrading activity, which includes the transglycosylating activity and hydrolytic activity of XTHs as well as the hydrolytic activity of nonspecific endoglucanases. However, parallel assays run without XGOs indicated that the measured values had negligible hydrolytic activity at the termination of the assay. The values measured, therefore, are indicative of transglycosylating activity and are expressed as percentages of xyloglucan-degrading activity per microgram of cell wall protein. Protein estimation was performed using the Bradford (1976) assay using a commercially available Bradford Reagent (Bio-Rad).
Cloning of S. longipes Expansin cDNA Fragments
The top internodes of plants from both ecotypes grown in different light conditions (low R/FR and green shade; see “Light Treatments” above) were harvested at different time points after the start of the light treatments. Total RNA was isolated from all of these tissue samples using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions. Potential genomic DNA contamination was removed using on-column DNase digestion (Qiagen). The concentration of recovered total RNA was measured using the Nanodrop spectrophotometer (Isogen LifeSciences). cDNA was synthesized from 2 μg of total RNA using oligo(dT) primers. cDNA synthesis was achieved using SuperScript III reverse transcriptase (Invitrogen). The reverse transcriptase reaction was carried out according to the manufacturer's instructions at 50°C, and the 20-μL reaction mixture contained 200 units of reverse transcriptase III, 4 μL of first-strand buffer, 40 units of RNase OUT recombinant RNase inhibiter, and 1 μL of 0.1 m dithiothreitol. Degenerate primers were designed from the conserved regions of known α-expansin gene sequences obtained from GenBank. The degenerate primer sequences were as follows: forward, 5′-GDGCHTGYTTTGARMTHARTG-3′, 5′-AAYGGYGGYTGGTGYAAYCC-3′, and 5′-ACNGCBYTVTTYAACAAHGG-3′; reverse, 5′-GCCAATTYTGHCCCCARTARC-3′. These degenerate primers were used to clone potential expansin cDNA fragments from the different cDNA pools obtained from the RNA of plants grown in different light treatments and at different time points during the treatments. This ensured to a certain extent the possibility of picking up as many expansin cDNA fragments as possible that were expressed in the internodes. The resulting cDNA fragments were cloned into the pGEMT Easy vector (Promega) and were then sent for sequencing (Macrogen). Among the more than 100 clones that were sent for sequencing, a number of sequences were identical. A total of 12 sequences were designated as fragments from different expansins, after sequencing these at least twice from both ends to ensure the accuracy of the sequences. Of these 12 expansin fragments, seven were different enough to allow the design of gene-specific primers (Table I).
Table I.
Sequences (5′ to 3′) of primer combinations and annealing temperatures for the seven α-expansin genes studied using real-time RT-PCR
These seven α-expansin gene fragments were cloned from the internodes of the alpine and prairie ecotypes of S. longipes. For each gene, sequences were identical in both ecotypes. The primers listed against a particular gene were used at the annealing temperatures indicated for the measurement of the transcript abundance of that gene using real-time RT-PCR. GenBank accession numbers for these genes are EU840703 to EU840714, EU840720, and EU840721.
| Gene | Forward Primer | Reverse Primer | Annealing Temperature |
|---|---|---|---|
| °C | |||
| SlEXPA1 | GGCCATGCCTATGTTCCTAA | CCCTTTGATGCTTACGCTCT | 62 |
| SlEXPA2 | GCATTGTCCCTGTTGCTTTT | CCTTGTCCCTTTAACCCACA | 62 |
| SlEXPA3 | GTACCATGCCGAAAACAAGG | ACCCCAGTAGCGACTCATTG | 62 |
| SlEXPA4 | GCTGGAATTGTCCCAGTCTC | ACCCCAGTAGCTTGACATGG | 65 |
| SlEXPA5 | CCCGTCCTCATTTCGACTTA | ACCCCAGTAACGACTCATGG | 65 |
| SlEXPA6 | TGTGCGAGAAAAGGAGGAGT | GCCAATTTTGTCCCCAGTAA | 62 |
| SlEXPA7 | TTTGAGCTTAAGTGCGCAGA | ATGGCCAAGTCAAAGTGAGG | 65 |
| Sl18S | CCGTTGCTCTGATGATTCATGA | GTTGATAGGGCAGAAATTTGAATGAT | 62 |
In Silico Analysis for Predicted Amino Acid Sequences of Expansin Gene Fragments
To determine the phylogenetic relationship between S. longipes α-expansins and α-expansin sequences with high identity from GenBank, deduced amino acid sequences were aligned using ClustalX software. A phylogenetic tree was then generated using this alignment file with TreeView software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) using default parameters.
Real-Time Gene Expression Measurements
The top internodes of plants were harvested before the start of the treatments and at every day for 3 d after the start of treatments. Harvested material was immediately frozen and stored at −80°C. All of the data shown are means of four biological replicates each consisting of internodes harvested from at least three pots. Total RNA from these samples was extracted using the RNeasy Plant Mini Kit (Qiagen). Reverse transcription of total RNA using random hexamers was performed as described above. Real-time reverse transcription (RT)-PCR was performed using 18S rRNA as an internal standard in a 20-μL reaction that contained 11 μL of SYBR Green Supermix (Bio-Rad; no. 170-8882), 50 ng of cDNA (0.1 ng for 18S rRNA), and gene-specific primers (Table I). A Bio-Rad MyiQ single-color real-time PCR detection system was used. The following program was used for all of the genes tested: 3 min at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at gene-specific annealing temperature, and 60 s at 72°C. For each expansin, a number of primer pairs were designed using the Primer 3 software (http://frodo.wi.mit.edu/primer3/primer3_code.html). Of these, the primer pairs that resulted in no cross-amplification with other expansin sequences (as tested on plasmid sequences) and did not form primer dimers were chosen for the real-time RT-PCR measurements. The annealing temperature was also optimized for each primer pair to result in specific amplification of the transcript of interest. Primer sequences and annealing temperatures are given in Table I. In addition, for every primer combination used, efficiency and melting curves were obtained. PCR products were also resolved on 1% agarose gels in order to confirm single products of the expected size. The Ct value for each gene was normalized relative to the Ct value of 18S rRNA. Relative transcript levels were calculated using the comparative Ct method (Livak and Schmittgen, 2001) and expressed relative to the average value at day 0, which was set as 1.
Statistical Analysis
For growth rates and real-time RT-PCR measurements, treatments and their respective controls were analyzed using Student's t test. For xyloglucan-degrading activity and AIE values, two-way ANOVA followed by Tukey's b test were performed.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU840703 to EU840714, EU840720, and EU840721.
Acknowledgments
We thank Ankie Ammerlaan and Rob Welschen for technical assistance and Alex Boonman for help with the figures and helpful comments on the manuscript.
This work was supported by the National Science and Engineering Council of Canada (Discovery grant to C.C.C.) and the Netherlands Organisation for Scientific Research (VENI grant no. 86306001 to R.P.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Rashmi Sasidharan (r.sasidharan@uu.nl) and Ronald Pierik (r.pierik@uu.nl).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Albert M, Werner M, Proksch P, Fry SC, Kaldenhoff R (2004) The cell wall-modifying xyloglucan endotransglycosylase/hydrolase LeXTH1 is expressed during the defence reaction of tomato against the plant parasite Cuscuta reflexa. Plant Biol 6 402–407 [DOI] [PubMed] [Google Scholar]
- Alokam S, Chinnappa CC, Reid DM (2002) Red/far-red light mediated stem elongation and anthocyanin accumulation in Stellaria longipes: differential response of alpine and prairie ecotypes. Can J Bot 80 72–81 [Google Scholar]
- Antosiewicz DM, Purugganan MM, Polisensky DH, Braam J (1997) Cellular localization of Arabidopsis xyloglucan endotransglycosylase-related proteins during development and after wind stimulation. Plant Physiol 115 1319–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL, Casal JJ, Kendrick RE (1991) Responses of light-grown wild-type and long-hypocotyl mutant cucumber seedlings to natural and stimulated shade light. Photochem Photobiol 54 819–826 [Google Scholar]
- Ballaré CL, Scopel AL, Sanchéz RA (1990) Far-red radiation reflected from adjacent leaves: an early signal of competition in plant canopies. Science 247 329–332 [DOI] [PubMed] [Google Scholar]
- Belfield EJ, Ruperti B, Roberts JA, McQueen-Mason S (2005) Changes in expansin activity and gene expression during ethylene-promoted leaflet abscission in Sambucus nigra. J Exp Bot 56 817–823 [DOI] [PubMed] [Google Scholar]
- Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ (2002) Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 14 3073–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Caderas D, Muster M, Vogler H, Mandel T, Rose JK, McQueen-Mason S, Kuhlemeier C (2000) Limited correlation between expansin gene expression and elongation growth rate. Plant Physiol 123 1399–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P, Braam J (1999) Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci 4 361–366 [DOI] [PubMed] [Google Scholar]
- Chinnappa CC, Donald GM, Sasidharan R, Emery RJN (2005) The biology of Stellaria longipes (Caryophyllaceae). Can J Bot 83 1367–1383 [Google Scholar]
- Cho HT, Cosgrove DJ (2000) Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 97 9783–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14 3237–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H (1997) Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell 9 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Kim JE, Park JA, Eom TJ, Kim WT (2006) Constitutive expression of abiotic stress inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett 580 3136–3144 [DOI] [PubMed] [Google Scholar]
- Choi D, Lee Y, Cho HT, Kende H (2003) Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell 15 1386–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmer TD, Peeters AJ, Wagemaker CA, Vriezen WH, Ammerlaan A, Voesenek LACJ (2004) Expression of alpha-expansin genes during root acclimations to O2 deficiency in Rumex palustris. Plant Mol Biol 56 423–437 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (1989) Characterization of long-term extension of isolated cell walls from growing cucumber hypocotyls. Planta 177 121–130 [PubMed] [Google Scholar]
- Cosgrove DJ (1996) Plant cell enlargement and the action of expansins. Bioessays 18 533–540 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (2000) Loosening of plant cell walls by expansins. Nature 407 321–326 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6 850–861 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA (2003) A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol 133 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S (2008) Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta KM, Pontin MA, Karlin-Neumann G, Bottini R, Spalding EP (2003) Genomic and physiological studies of early cryptochrome 1 action demonstrates roles for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J 36 203–214 [DOI] [PubMed] [Google Scholar]
- Franklin KA (2008) Shade avoidance. New Phytol 170 930–944 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC (2005) Phytochromes and shade-avoidance responses in plants. Ann Bot (Lond) 96 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ (1992) Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J 282 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudali S, Janakowski S, Sobczak M, Griesser M, Grundler FMW, Golinowski W (2008) Two tomato alpha-expansins show distinct spatial and temporal expression patterns during development of nematode-induced syncytia. Physiol Plant 132 370–383 [DOI] [PubMed] [Google Scholar]
- Hyodo H, Yamakawa S, Takeda Y, Tsuduki M, Yokota A, Nishitani K, Kohchi T (2003) Active gene expression of a xyloglucan endotransglucosylase/hydrolase gene, XTH9, in inflorescence apices is related to cell elongation in Arabidopsis thaliana. Plant Mol Biol 52 473–482 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Ma L, Strickland E, Deng XW (2005) Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17 3239–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, McQueen-Mason S (2004) A role for expansins in dehydration and rehydration of the resurrection plant Craterostigma plantagineum. FEBS Lett 559 61–65 [DOI] [PubMed] [Google Scholar]
- Kaku T, Tabuchi A, Wakabayashi K, Hoson T (2004) Xyloglucan oligosaccharides cause cell wall loosening by enhancing xyloglucan endotransglucosylase/hydrolase activity in azuki bean epicotyls. Plant Cell Physiol 45 77–82 [DOI] [PubMed] [Google Scholar]
- Kaku T, Tabuchi A, Wakabayashi K, Kamisaka S, Hoson T (2002) Action of xyloglucan hydrolase within the native cell wall architecture and its effect on cell wall extensibility in azuki bean epicotyls. Plant Cell Physiol 43 21–26 [DOI] [PubMed] [Google Scholar]
- Kende H, Bradford K, Brummell D, Cho HT, Cosgrove D, Fleming A, Gehring C, Lee Y, McQueen-Mason S, Rose J, et al (2004) Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol Biol 55 311–314 [DOI] [PubMed] [Google Scholar]
- Kurepin LV, Pharis RP, Reid DM, Chinnappa CC (2006. a) Involvement of gibberellins in the stem elongation of sun and shade ecotypes of Stellaria longipes that is induced by low light irradiance. Plant Cell Environ 29 1319–1328 [DOI] [PubMed] [Google Scholar]
- Kurepin LV, Walton LJ, Reid DM, Pharis RP, Chinnappa CC (2006. b) Growth and ethylene evolution by shade and sun ecotypes of Stellaria longipes in response to varied light quality and irradiance. Plant Cell Environ 29 647–652 [DOI] [PubMed] [Google Scholar]
- Lee DK, Ahn JH, Song SK, Choi YD, Lee JS (2003) Expression of an expansin gene is correlated with root elongation in soybean. Plant Physiol 131 985–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald SE, Chinnappa CC, Reid DM (1988) Phenotypic plasticity in the Stellaria longipes complex (Caryophyllaceae). Acta Oecol-Oec Plant 9 103–104 [DOI] [PubMed] [Google Scholar]
- Matsui A, Yokoyama R, Seki M, Ito T, Shinozaki K, Takahashi T, Komeda Y, Nishitani K (2005) AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant J 42 525–534 [DOI] [PubMed] [Google Scholar]
- McQueen-Mason S, Cosgrove DJ (1994) Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci USA 91 6574–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osato Y, Yokoyama R, Nishitani K (2006) A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. J Plant Res 119 153–162 [DOI] [PubMed] [Google Scholar]
- Palmer SJ, Davies WJ (1996) An analysis of relative elemental growth rate, epidermal cell size and xyloglucan endotransglycosylase activity through the growing zone of ageing maize leaves. J Exp Bot 47 339–347 [Google Scholar]
- Pierik R, Whitelam GC, Voesenek LACJ, de Kroon H, Visser EJ (2004) Canopy studies on ethylene-insensitive tobacco identify ethylene as a novel element in blue light and plant-plant signalling. Plant J 38 310–319 [DOI] [PubMed] [Google Scholar]
- Pritchard J, Hetherington PR, Fry SC, Tomos AD (1993) Xyloglucan endotransglycosylase activity, microfibril orientation and the profiles of cell wall properties along growing regions of maize roots. J Exp Bot 44 1281–1289 [Google Scholar]
- Redgwell RJ, Fry SC (1993) Xyloglucan endotransglycosylase activity increases during kiwifruit (Actinidia deliciosa) ripening: implications for fruit softening. Plant Physiol 103 1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy B, McQueen-Mason S, Nosberger J, Fleming A (2001) Differential expression of alpha- and beta-expansin genes in the elongating leaf of Festuca pratensis. Plant Mol Biol 46 491–504 [DOI] [PubMed] [Google Scholar]
- Rochange SF, Wenzel CL, McQueen-Mason SJ (2001) Impaired growth in transgenic plants over-expressing an expansin isoform. Plant Mol Biol 46 581–589 [DOI] [PubMed] [Google Scholar]
- Rose JK, Lee HH, Bennett AB (1997) Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci USA 94 5955–5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43 1421–1435 [DOI] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC (2003) Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 11 680–683 [DOI] [PubMed] [Google Scholar]
- Smith H (1990) Signal perception, differential expression within multigene families and the molecular basis of phenotypic plasticity. Plant Cell Environ 13 585–594 [Google Scholar]
- Soga K, Wakabayashi K, Hoson T, Kamisaka S (1999) Hypergravity increases the molecular mass of xyloglucans by decreasing xyloglucan-degrading activity in azuki bean epicotyls. Plant Cell Physiol 40 581–585 [DOI] [PubMed] [Google Scholar]
- Sulova Z, Lednicka M, Farkas V (1995) A colorimetric assay for xyloglucan-endotransglycosylase from germinating seeds. Anal Biochem 229 80–85 [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Kamisaka S, Hoson T (1997) Purification of xyloglucan hydrolase/endotransferase from cell walls of azuki bean epicotyls. Plant Cell Physiol 38 653–658 [Google Scholar]
- Tabuchi A, Mori H, Kamisaka S, Hoson T (2001) A new type of endo-xyloglucan transferase devoted to xyloglucan hydrolysis in the cell wall of azuki bean epicotyls. Plant Cell Physiol 42 154–161 [DOI] [PubMed] [Google Scholar]
- Takeda T, Furuta Y, Awano T, Mizuno K, Mitsuishi Y, Hayashi T (2002) Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. Proc Natl Acad Sci USA 99 9055–9060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Pierik R, Millenaar FF, Voesenek LACJ, Van Der Straeten D (2005) Reaching out of the shade. Curr Opin Plant Biol 8 462–468 [DOI] [PubMed] [Google Scholar]
- Van Sandt VST, Stieperaere H, Guisez Y, Verbelen JP, Vissenberg K (2007. a) XET activity is found near sites of growth and cell elongation in bryophytes and some green algae: new insights into the evolution of primary cell wall elongation. Ann Bot (Lond) 99 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sandt VST, Suslov D, Verbelen JP, Vissenberg K (2007. b) Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann Bot (Lond) 100 1467–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeburg RA, Benschop JJ, Peeters AJ, Colmer TD, Ammerlaan AH, Staal M, Elzenga TM, Staals RH, Darley CP, McQueen-Mason SJ, et al (2005) Ethylene regulates fast apoplastic acidification and expansin A transcription during submergence-induced petiole elongation in Rumex palustris. Plant J 43 597–610 [DOI] [PubMed] [Google Scholar]
- Wu Y, Jeong BR, Fry SC, Boyer JS (2005) Change in XET activities, cell wall extensibility and hypocotyl elongation of soybean seedlings at low water potential. Planta 220 593–601 [DOI] [PubMed] [Google Scholar]
- Zenko C, Yokoyama R, Nishitani K, Kamisaka S (2004) Effect of hypergravity stimulus on XTH gene expression in Arabidopsis thaliana. Biol Sci Space 18 162–163 [PubMed] [Google Scholar]
- Zenoni S, Reale L, Tornielli GB, Lanfaloni L, Porceddu A, Ferrarini A, Moretti C, Zamboni A, Speghini A, Ferranti F, et al (2004) Downregulation of the Petunia hybrida alpha-expansin gene PhEXP1 reduces the amount of crystalline cellulose in cell walls and leads to phenotypic changes in petal limbs. Plant Cell 16 295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Alvarez S, Marsh EL, LeNoble ME, Cho IJ, Sivaguru M, Chen S, Nguyen HT, Wu Y, Schachtman DP (2007) Cell wall proteome in the maize primary root elongation zone. II. Region-specific changes in water soluble and lightly ionically bound proteins under water deficit. Plant Physiol 145 1533–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]