Abstract
The aim of this work was to investigate the effect of decreased cytosolic pyruvate kinase (PKc) on potato (Solanum tuberosum) tuber metabolism. Transgenic potato plants with strongly reduced levels of PKc were generated by RNA interference gene silencing under the control of a tuber-specific promoter. Metabolite profiling showed that decreased PKc activity led to a decrease in the levels of pyruvate and some other organic acids involved in the tricarboxylic acid cycle. Flux analysis showed that this was accompanied by changes in carbon partitioning, with carbon flux being diverted from glycolysis toward starch synthesis. However, this metabolic shift was relatively small and hence did not result in enhanced starch levels in the tubers. Although total respiration rates and the ATP to ADP ratio were largely unchanged, transgenic tubers showed a strong decrease in the levels of alternative oxidase (AOX) protein and a corresponding decrease in the capacity of the alternative pathway of respiration. External feeding of pyruvate to tuber tissue or isolated mitochondria resulted in activation of the AOX pathway, both in the wild type and the PKc transgenic lines, providing direct evidence for the regulation of AOX by changes in pyruvate levels. Overall, these results provide evidence for a crucial role of PKc in the regulation of pyruvate levels as well as the level of the AOX in heterotrophic plant tissue, and furthermore reveal that these parameters are interlinked in vivo.
Pyruvate kinase (PK; EC 2.7.1.40) catalyzes the final reaction of the glycolytic pathway, converting ADP and phosphoenolpyruvate (PEP) to ATP and pyruvate, which is subsequently imported into mitochondria and used as a substrate for respiration. PK catalyzes an irreversible reaction in vivo in castor bean (Ricinus communis; Geigenberger et al., 1993) and is probably a primary regulation site of glycolysis and respiration, as indicated by various biochemical studies in mammalian and bacterial systems (Pilkis and Granner, 1992; Teusink et al., 2000) and plants (Thomas et al., 1997; Geigenberger et al., 1998; Schwender et al., 2004; Plaxton and Podesta, 2006). In plants, PK is believed to provide bottom-up control of plant glycolytic flux from hexose-phosphates to pyruvate owing to PEP's pronounced feedback allosteric inhibition of ATP- and inorganic pyrophosphate-dependent phosphofructokinases (Dennis and Greyson, 1987). PK in plants has been found to be subject to a number of regulatory mechanisms, including allosteric regulation by Glu and Asp (Turner et al., 2005), activation by changes in cytosolic pH (Podesta and Plaxton 1991, 1992, 1994), inhibition by ATP (Podesta and Plaxton, 1991, 1992, 1993), and phosphorylation and ubiquitination (Tang et al., 2003). While the role of PK in the regulation of glycolysis in response to hormone signals has been intensively studied in mammalian and bacterial systems (Pilkis and Granner, 1992; Teusink et al., 2000), much less is known about the signals regulating PK in plants.
Previous studies in vascular plants revealed that PK exists as tissue-specific isozymes that show pronounced differences in their respective physical and kinetic properties (Plaxton and Podesta, 2006). In plants, the situation is further complicated due to the presence of PK isozymes in both the cytosolic and plastidial compartments (denoted PKc and PKp, respectively; Plaxton, 1996; Givan, 1999). While the role of PKp has been studied recently and was shown to be critical for fatty acid biosynthesis in Arabidopsis (Arabidopsis thaliana) seeds (Andre and Benning, 2007; Andre et al., 2007; Baud et al., 2007), the impact of modified PKc levels on plant metabolism has been little studied. A set of tobacco (Nicotiana tabacum) plants lacking PKc in leaves was previously analyzed with respect to growth, photosynthesis, and respiration, and a number of conditional growth defects were identified, mainly as a consequence of altered regulation of sink-source metabolism due to a loss of PKc, specifically in leaves (Gottlob et al., 1992; Knowles et al., 1998; Grodzinski et al., 1999). However, no information on the effect of altered PKc on metabolism in heterotrophic tissues was provided. Moreover, targeted manipulation of PKc levels in heterotrophic tissues such as roots or tubers has not been reported yet (to our knowledge). For this reason, the aim of our study was to analyze the effect of altered PKc levels on potato (Solanum tuberosum) tuber growth and metabolism.
Given the likely importance of PKc for generating pyruvate for entry into the mitochondrial tricarboxylic acid (TCA) cycle, we focused our studies on understanding the effect of altered PKc activity on carbon metabolism, particularly on respiratory processes and the coordinated regulation of glycolysis, the TCA cycle, and the mitochondrial electron transport chain. It is becoming apparent that a coordinated regulation of these respiratory processes exists (Fernie et al., 2004). A recent example of this is the observation that enzymes of the cytosolic glycolytic pathway (including PK) are physically associated with the outer mitochondrial membrane (Giege et al., 2003) and that their degree of association is dynamically responsive to the respiratory burden imposed on the mitochondria (Graham et al., 2007). Furthermore, in vitro assays have shown that the addition of pyruvate or other α-keto acids to isolated mitochondrial preparations under conditions where the cytochrome c pathway is inhibited (e.g. in the presence of antimycin a or KCN) stimulates the activity of the mitochondrial alternative oxidase (AOX; Millar et al., 1993; Day et al., 1994). This effect has been demonstrated to be due to an interaction of pyruvate with the reduced form of the N-terminal Cys residue of the AOX, rather than a result of pyruvate metabolism per se (Millar et al., 1993, Millenaar and Lambers, 2003; Umbach et al., 2006). While direct evidence for endogenous changes in organic acid levels in regulating the AOX is currently lacking, the pyruvate addition experiments are suggestive of a regulatory role of glycolytic flux on the activity and dynamics of the mitochondrial electron transport chain.
In this study, we have generated and analyzed transgenic potato tubers with a decrease in PKc levels using an RNA interference (RNAi) approach. Decreased PKc levels led to decreased levels of pyruvate and several TCA cycle intermediates, which were accompanied by changes in carbon partitioning between glycolysis and starch and by a decrease in AOX protein level and respiratory capacity. The decrease in AOX-dependent respiratory capacity could be reverted by external feeding of pyruvate to tuber tissue. These results thus provide evidence for an important role of PKc in regulating pyruvate levels and the alternative pathway of respiration in heterotrophic potato tubers.
RESULTS
Generation of Potato Tubers with Decreased PKc Expression and Activity
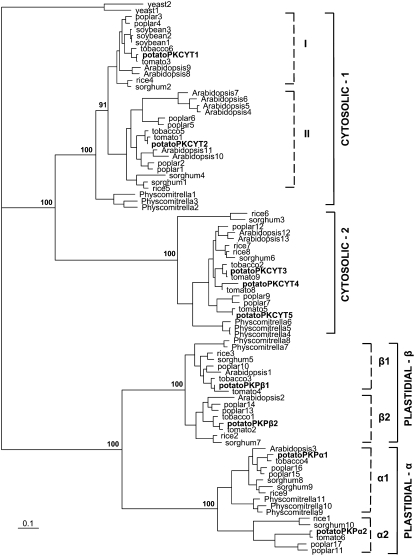
Plants contain multiple isozymes of PK (e.g. Arabidopsis contains 14 isozymes; Arabidopsis Genome Initiative 2000; Andre et al., 2007), but in potato only one gene (hereafter called PKCYT1) encoding a cytosolic isozyme has been cloned and sequenced to date (Blakeley et al., 1990; Cole et al., 1992). To identify additional potato genes encoding PK isozymes, we searched available cDNA and EST sequences from potato and other Solanaceous species, and compared these with the complete sets of PK proteins identified from sequenced plant genomes. A phylogenetic tree was constructed from the deduced protein sequences, including partial sequences and sequence gaps (Fig. 1). The same clustering and topology was obtained when more stringent analyses were performed on the sequences without gaps and excluding the partial sequences (Supplemental Fig. S1), and on the N- and C-terminal regions separately (to include the partial Solanaceae sequences), excluding the variable transit peptides and any C-terminal extensions (data not shown). In the phylogenetic tree, the cytosolic and plastidial isozymes of PK were clearly separated. The PKc isozymes were consistently split into two main classes (cytosolic-1 and -2; Fig. 1), with strong support for two subgroups within the angiosperm cytosolic-1 class (I and II; Fig. 1). As reported previously (Blakeley et al., 1995; Andre et al., 2007), there are two main classes of plastidial PK isozymes (α and β). Our phylogenetic analysis revealed two subgroups of the α and β classes in angiosperms (designated α1, α2, β1, and β2 according to Andre et al., 2007; Fig. 1). Nine sequences encoding PK isozymes were identified in potato. Based on the phylogenetic analysis, five are cytosolic (PKCYT1 [S53332; Blakeley et al., 1990], PKCYT2 [TC134535], PKCYT3 [TC141201], PKCYT4 [TC118783], and PKCYT5 [TC135950]) and four are plastidial (PKPα1 [TC135043], PKPα2 [TC159621], PKPβ1 [TC149988], and PKPβ2 [TC134739]). One of the Arabidopsis proteins (Arabidopsis 14, At3g49160) that has previously been annotated as a PK (Arabidopsis Genome Initiative, 2000) belongs to a rather divergent cluster of sequences that show only 25% to 31% identity with the other PK isozymes (PK-like; Supplemental Fig. S1). This cluster contains sequences from Physcomitrella patens, poplar (Populus trichocarpa), and tomato (Solanum lycopersicum) but not rice (Oryza sativa) or sorghum (Sorghum bicolor), and no potato ortholog was found in the available EST sequences. The role of these PK-related proteins in plants is not yet clear, and there is so far no experimental evidence for their function. In this context, it is interesting to note that de Bari et al. (2007) also suggest mitochondrial location of some of these proteins.
Figure 1.
Phylogenetic alignment of PK proteins. Protein sequences were deduced from cDNA or EST sequences obtained from the Joint Genome Institute and the National Center for Biotechnology Information. The tree shows a phylogram of PK protein sequences (including partial sequences and gaps, excluding the variable transit peptides and C-terminal extensions) using the JTT evolutionary model of amino acid substitution and rooted with the yeast sequences. Bootstrap values from 100 replicates are indicated. Potato PK proteins are shown in bold. The accession codes and protein sequences are given as supplemental material.
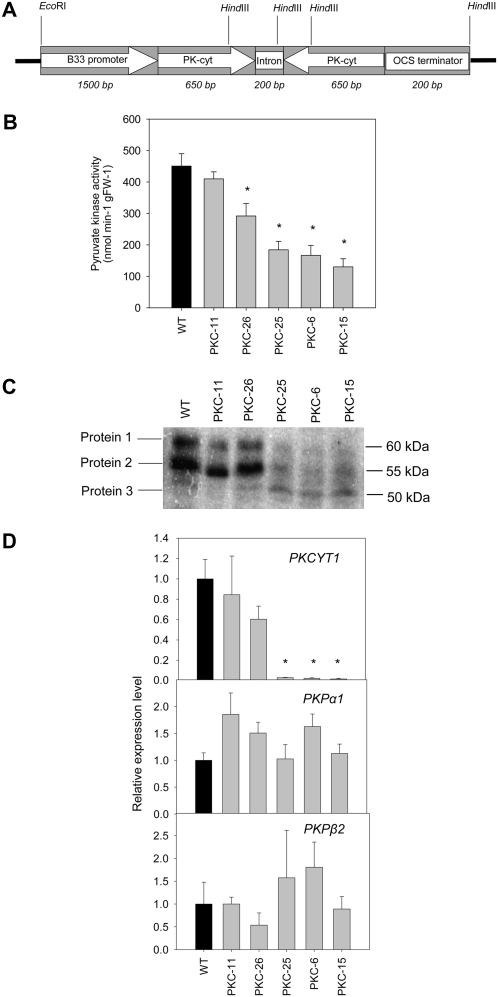
To generate potato tubers with decreased PKc expression, we targeted the PKCYT1 gene in an RNAi gene silencing approach. The PKCYT1 gene was chosen because our database searches indicated that transcripts of PKCYT1 are highly abundant in tubers and it is therefore likely to encode a major tuber isozyme. An RNAi construct was created, containing a PCR-amplified, 650-bp cDNA fragment homologous to the PKCYT1 gene in a hairpin orientation between the tuber-specific B33 promoter and the OCS terminator (Fig. 2A), and transformed into potato. The resultant transgenic lines were screened by measuring total PK activity in 10-week-old tubers using an enzyme assay at pH 6.9. Five lines were selected for further analysis: PKC-25, -6, and -15 had significantly reduced PK activities down to 40%, 37%, and 29% of wild-type level, respectively, PKC-26 had significantly reduced activity to 64% of wild-type activity, while activity in PKC-11 was not significantly different from wild type (Fig. 2B).
Figure 2.
Generation of plants with reduced PKc in tubers. A, RNAi construct used to transform potato plants, containing a PCR-amplified, 650-bp fragment homologous to PKCYT1 (PK-cyt) in a hairpin orientation under the control of the B33 patatin promoter. B, PK activity in tuber extracts from five resultant transgenic lines plus wild type assayed at pH 6.9 (*, significant difference from wild-type levels, n = six tubers from six different plants). C, Western blot of tuber extracts probed with an anti-PKc antibody. D, Quantitative real-time RT-PCR expression analysis of PK transcripts for PKCYT1, PKPα1, and PKPβ2 genes in tubers from the PKc transgenic lines and wild type (normalized to wild-type expression levels). *, Significant difference from wild-type levels (P < 0.05). Data represent the mean and se (n = four independent tuber RNA extractions from different plants).
PKc protein levels in the transgenic tubers were determined from western blots using a PKc-specific antibody raised against PKc from Brassica napus (Smith et al., 2000; Fig. 2C). This antibody has previously been shown to recognize only PKc proteins and not PKp proteins in different plant species (Smith et al., 2000; Plaxton et al., 2002; Rivoal et al., 2002). Analysis of wild-type potato showed that tubers contain two strongly immunoreactive proteins of approximately 60 and 55 kD (proteins 1 and 2; Fig. 2C) and one less immunoreactive protein (approximately 50 kD, protein 3; Fig. 2C). In the PKc transgenic lines, levels of the two major PKc immunoreactive polypeptides (proteins 1 and 2) are strongly decreased in the strongly silenced lines (PKC-25, -6, and -15) but not in the weakly silenced lines (PKC-26 and -11; Fig. 2C). Interestingly, the minor immunoreactive polypeptide (protein 3) appears to be induced in PKC-25, -6, and -15. It is difficult to assign the different immunoreactive bands to particular genes because the predicted molecular masses of the PKc polypeptides are similar (54–57 kD); nevertheless, it is likely that protein 2 corresponds to the PKCYT1 protein, based on the predicted molecular mass (55 kD) and the reduced levels of this protein in the transgenic tubers. Protein 3 appears to be the only PKc polypeptide abundant in potato leaves (data not shown) and could be a posttranslationally cleaved product of one of the PKc proteins, similar to that described in soybean (Glycine max; Tang et al., 2003).
To assess the transcript levels of PK genes in the transgenic lines, quantitative real-time reverse transcription (RT)-PCR was performed with cDNA derived from tuber RNA using primers specific for the potato PK transcripts. The results showed that the targeted PKCYT1 transcript was strongly reduced to 3%, 2%, and 1% of wild-type levels in the lines with low PK activity (PKC-25, -6, and -15, respectively), reduced to 56% in PKC-26, and not reduced in PKC-11 (Fig. 2D). The expression of two other PKCYT genes, PKCYT2 and PKCYT3, was also measured, with both showing minor but nonsignificant decreases in transcript level (data not shown). Two PKP genes, PKPα1 and PKPβ2, are expressed in tubers, and there were no significant differences in their transcript levels in the transgenic tubers compared to wild type (Fig. 2D). Transcript levels of PKPβ1 were undetectable in tubers, and PKPα2 is not represented in tuber EST collections, indicating that they are not major tuber isozymes. Taken together, these data show that the PKCYT1 gene has been strongly silenced in three of the transgenic lines, resulting in a large decrease in the levels of the two major PKc proteins and PK activity in tubers. Given that the transcript levels of PKP genes were unaffected, these lines were deemed to be suitable for further analysis to determine the effect of decreased PKc on tuber metabolism.
Effect of Decreased PKc Expression on Enzyme Activities
Measurement of maximal catalytic activities of the glycolytic enzymes hexokinase, fructokinase, phosphoglucomutase, and phosphofructokinase showed no significant difference in the PKc transgenics compared to wild type (Table I). The maximal activities of enzymes that bypass PKc to produce pyruvate via malate (PEP carboxylase [PEPC], malate dehydrogenase, and NAD malic enzyme) were also not significantly different from wild type, nor was the activity of PEP phosphatase, which is a vacuolar acid phosphatase that can convert PEP to pyruvate without requiring ATP (Plaxton, 1996; Table I). Furthermore, the activation state of PEPC was assessed by measuring the extent of inhibition of the enzyme by 15 mm malate at suboptimal assay conditions (pH 7.0, 0.25 mm PEP; see Schuller et al., 1990). The values (in percentage of control without malate) were 33.9 ± 6.9, 27.0 ± 3.9, 33.5 ± 5.4, 27.7 ± 2.9, 32.6 ± 5.1, and 33.7 ± 5.2 for wild type, PKC-6, PKC-11, PKC-15, PKC-25, and PKC-26, respectively (mean ± se, n = 4–8), suggesting that the level of PEPC activation is not different from wild type in the transgenic lines. For the analysis of PEPC activation state, frozen potato tuber tissue was extracted immediately using an extraction buffer that contained a cocktail of different protease inhibitors. The same extraction buffer has been used in previous studies to analyze the in vivo activation state of Suc-P synthase in growing potato tubers (Geigenberger et al., 1997). Overall, this indicates that there were no pleiotropic or compensatory changes of other enzymes in response to decreased PKc expression.
Table I.
Enzyme activities in tubers of transgenic plants with decreased PKc and wild type
Maximal catalytic activities were measured in 10-week-old tubers. Data show the mean and se (n = six tubers from six different plants per line).
| Enzyme | Enzyme Activity
|
|||||
|---|---|---|---|---|---|---|
| Wild Type | PKC-11 | PKC-26 | PKC-25 | PKC-6 | PKC-15 | |
| nmol min−1 mg protein−1 | ||||||
| Hexokinase | 23.6 ± 1.4 | 27.5 ± 0.7 | 25.9 ± 1.6 | 23.8 ± 2.1 | 21.3 ± 2.1 | 23.9 ± 1.3 |
| Fructokinase | 30.7 ± 2.3 | 35.4 ± 3.2 | 26.6 ± 2.9 | 22.6 ± 3.9 | 25.3 ± 3.1 | 26.7 ± 2.0 |
| Phosphoglucomutase | 1,255 ± 75 | 1,245 ± 48 | 1,364 ± 90 | 1,066 ± 113 | 1,037 ± 67 | 1,147 ± 62 |
| ATP-dependent phosphofructokinase | 15.2 ± 1.8 | 15.7 ± 0.3 | 13.2 ± 1.3 | 11.2 ± 1.9 | 12.3 ± 2.3 | 13.3 ± 1.4 |
| PEPC | 19.6 ± 1.0 | 21.4 ± 1.1 | 22.9 ± 1.5 | 22.2 ± 1.3 | 18.6 ± 1.1 | 20.4 ± 0.7 |
| NAD-malate dehydrogenase | 1,828 ± 202 | 1,982 ± 333 | 1,760 ± 342 | 1,418 ± 318 | 1,436 ± 292 | 2,072 ± 359 |
| NAD malic enzyme | 8.4 ± 0.5 | 8.2 ± 1.0 | 9.0 ± 0.7 | 9.2 ± 1.2 | 9.0 ± 0.2 | 8.3 ± 1.1 |
| PEP phosphatase | 107 ± 5 | 122 ± 13 | 127 ± 8 | 111 ± 10 | 114 ± 8 | 122 ± 12 |
Effect of Decreased PKc Expression on Growth and Metabolite Levels
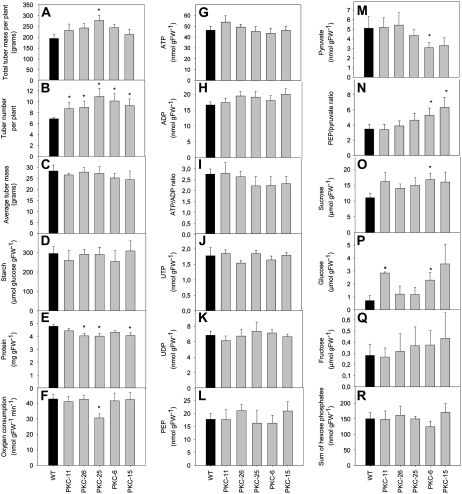
Wild-type and PKc transgenic plants were grown under greenhouse conditions for metabolic analysis of tubers from 10-week-old plants. Total tuber mass per plant was slightly elevated in all transgenic lines (significant for PKC-25; Fig. 3A), while tuber number per plant was significantly higher in all transgenic lines compared to wild type (Fig. 3B). The reason for the increase in tuber number upon reduction of PKc is not revealed by our data. However, the average tuber mass was not significantly different from wild type (Fig. 3C). Starch content in the transgenics was not different to wild type but protein content was reduced (significant for PKC-26, -25, and -15; Fig. 3, D and E). Respiration was measured as the rate of oxygen consumption by tuber discs cut from freshly harvested tubers. While PKC-25 showed a significant reduction in respiration, all other lines were not significantly different from wild type (Fig. 3F).
Figure 3.
Growth parameters and metabolite levels in wild-type and transgenic tubers with decreased PKc. Plants were grown under greenhouse conditions, and 10-week-old tubers were harvested for analysis. Data show the mean and se (n = six tubers from six different plants per line). *, Significant difference from wild-type levels (P < 0.05).
The levels of hexose-phosphates (sum of Glc-1-P, Fru-6-P, and Glc-6-P) and PEP were similar in all lines (Fig. 3, R and L), while pyruvate was significantly decreased in PKC-6 and -15 (Fig. 3M), resulting in a significant increase in the PEP to pyruvate ratio in these lines (Fig. 3N), which is consistent with the decrease in PK activity. Measurement of sugars revealed that PKC-6 had significantly higher Glc and Suc, while PKC-11 had significantly higher Glc (Fig. 3, O–Q). Measurement of adenine nucleotides showed that ATP levels were similar in all lines, while ADP levels showed a trend of being slightly higher than wild type, and the ATP to ADP ratio a trend of being slightly decreased in PKC-25, -6, and -15 (Fig. 3, G–I). These changes were, however, not significant. UTP and UDP levels were similar in all lines (Fig. 3, J and K).
Metabolite profiling using an established gas chromatography-mass spectrometry (GC-MS) method (Roessner et al., 2001) was performed to determine further changes in metabolite levels in tubers from the three most strongly silenced transgenic lines (PKC-25, -6, and -15) compared to wild type. The results are shown in Table II. The most striking changes observed were a significant reduction in the levels of several organic acids that are intermediates of the TCA cycle (aconitate, fumarate, and isocitrate) in the transgenics compared to wild type. Succinate, citrate, and malate were not significantly different in the transgenics. The levels of Asp (a direct product of oxaloacetate from the TCA cycle) were significantly reduced in all transgenic lines. Lys was increased in the transgenic lines. Glycerate, which is probably representative of 3-phosphoglycerate, a precursor of PEP, was significantly reduced in the transgenic lines. The other significant changes detected in these transgenic tubers were significant in only a single line (e.g. the significant increase in Ala in PKC-6) and therefore not deemed representative of all PKc transgenic lines.
Table II.
Metabolite levels in tubers of transgenic plants with decreased PKc and wild type
Metabolites were measured by GC-MS profiling in samples from the same 10-week-old tubers used for the growth and metabolite measurements shown in Figure 3. The data show the full set of metabolites that could be identified in the chromatograms. Results are the mean ± se normalized to wild type (n = six tubers from six different plants per line). Significant differences from wild type are shown in bold (P < 0.05).
| Metabolite | Wild Type | PKC-25 | PKC-6 | PKC-15 |
|---|---|---|---|---|
| Sugars | ||||
| Ara | 1.00 ± 0.39 | 1.05 ± 0.29 | 1.03 ± 0.25 | 2.14 ± 0.38 |
| Gal | 1.00 ± 0.14 | 0.78 ± 0.12 | 0.96 ± 0.11 | 1.65 ± 0.47 |
| Maltose | 1.00 ± 0.13 | 0.87 ± 0.18 | 1.13 ± 0.11 | 1.36 ± 0.26 |
| Man | 1.00 ± 0.14 | 0.89 ± 0.14 | 1.05 ± 0.15 | 1.59 ± 0.22 |
| Myoinositol | 1.00 ± 0.15 | 0.77 ± 0.16 | 0.73 ± 0.05 | 1.02 ± 0.17 |
| Xyl | 1.00 ± 0.18 | 1.19 ± 0.06 | 1.20 ± 0.16 | 1.06 ± 0.23 |
| Phosphorylated intermediates | ||||
| Glycerol-1-P | 1.00 ± 0.04 | 1.02 ± 0.03 | 1.03 ± 0.03 | 1.17 ± 0.06 |
| Myonositol-1-P | 1.00 ± 0.08 | 1.21 ± 0.05 | 1.20 ± 0.07 | 1.11 ± 0.11 |
| Organic acids | ||||
| Aconitate | 1.00 ± 0.33 | 0.40 ± 0.18 | 0.35 ± 0.05 | 0.51 ± 0.20 |
| Chlorogenic acid | 1.00 ± 0.25 | 0.35 ± 0.10 | 0.45 ± 0.13 | 0.72 ± 0.16 |
| Citrate | 1.00 ± 0.33 | 2.93 ± 1.97 | 2.49 ± 1.90 | 2.86 ± 1.68 |
| Dehydroascorbate | 1.00 ± 0.12 | 0.96 ± 0.05 | 1.09 ± 0.12 | 1.05 ± 0.14 |
| Fumarate | 1.00 ± 0.25 | 0.42 ± 0.04 | 0.31 ± 0.04 | 0.44 ± 0.09 |
| γ-Amino-butyric acid | 1.00 ± 0.03 | 0.98 ± 0.06 | 1.00 ± 0.04 | 0.93 ± 0.07 |
| Gluconic acid | 1.00 ± 0.15 | 0.87 ± 0.06 | 0.85 ± 0.04 | 0.91 ± 0.15 |
| Glycerate | 1.00 ± 0.13 | 0.55 ± 0.02 | 0.57 ± 0.01 | 0.61 ± 0.05 |
| Isocitrate | 1.00 ± 0.06 | 0.47 ± 0.17 | 0.36 ± 0.22 | 0.44 ± 0.15 |
| Malate | 1.00 ± 0.16 | 1.00 ± 0.12 | 1.10 ± 0.09 | 1.25 ± 0.21 |
| Putrescine | 1.00 ± 0.15 | 1.26 ± 0.06 | 1.44 ± 0.09 | 1.02 ± 0.15 |
| Quinate | 1.00 ± 0.03 | 1.18 ± 0.01 | 0.98 ± 0.17 | 0.96 ± 0.08 |
| Saccharic acid | 1.00 ± 0.21 | 0.85 ± 0.11 | 0.94 ± 0.03 | 0.99 ± 0.09 |
| Shikimate | 1.00 ± 0.10 | 1.26 ± 0.09 | 1.11 ± 0.03 | 1.07 ± 0.20 |
| Succinate | 1.00 ± 0.28 | 1.00 ± 0.22 | 1.11 ± 0.26 | 0.79 ± 0.32 |
| Amino acids | ||||
| Ala | 1.00 ± 0.18 | 1.28 ± 0.15 | 1.58 ± 0.07 | 1.22 ± 0.03 |
| Arg | 1.00 ± 0.25 | 1.20 ± 0.13 | 1.20 ± 0.11 | 1.01 ± 0.10 |
| Asn | 1.00 ± 0.31 | 0.90 ± 0.25 | 0.71 ± 0.25 | 0.84 ± 0.12 |
| Asp | 1.00 ± 0.03 | 0.73 ± 0.01 | 0.73 ± 0.01 | 0.71 ± 0.08 |
| β-Ala | 1.00 ± 0.05 | 0.95 ± 0.07 | 1.13 ± 0.06 | 0.94 ± 0.11 |
| Cys | 1.00 ± 0.41 | 1.61 ± 0.52 | 2.11 ± 0.46 | 1.66 ± 0.60 |
| Glu | 1.00 ± 0.05 | 1.05 ± 0.10 | 1.02 ± 0.07 | 1.05 ± 0.08 |
| Gln | 1.00 ± 0.29 | 1.51 ± 0.45 | 1.16 ± 0.39 | 1.38 ± 0.40 |
| Gly | 1.00 ± 0.07 | 1.08 ± 0.11 | 1.15 ± 0.04 | 1.08 ± 0.09 |
| Homo-Ser | 1.00 ± 0.07 | 1.19 ± 0.06 | 0.94 ± 0.07 | 1.31 ± 0.11 |
| Ile | 1.00 ± 0.20 | 1.23 ± 0.21 | 1.46 ± 0.18 | 1.17 ± 0.20 |
| Leu | Not detected | Not detected | Not detected | Not detected |
| Lys | 1.00 ± 0.27 | 3.45 ± 0.70 | 4.57 ± 0.61 | 2.43 ± 0.39 |
| Met | 1.00 ± 0.14 | 1.11 ± 0.13 | 1.16 ± 0.11 | 0.99 ± 0.09 |
| Phe | 1.00 ± 0.11 | 1.06 ± 0.16 | 1.20 ± 0.11 | 0.96 ± 0.09 |
| Pro | 1.00 ± 0.31 | 0.89 ± 0.23 | 0.74 ± 0.18 | 1.08 ± 0.25 |
| Ser | 1.00 ± 0.12 | 1.09 ± 0.10 | 1.08 ± 0.08 | 1.01 ± 0.09 |
| Thr | 1.00 ± 0.11 | 0.98 ± 0.07 | 0.90 ± 0.07 | 0.94 ± 0.10 |
| Trp | 1.00 ± 0.35 | 0.71 ± 0.20 | 0.92 ± 0.26 | 0.64 ± 0.07 |
| Tyr | 1.00 ± 0.23 | 1.34 ± 0.31 | 1.67 ± 0.31 | 1.00 ± 0.10 |
| Val | 1.00 ± 0.06 | 1.02 ± 0.05 | 1.03 ± 0.05 | 1.00 ± 0.03 |
Effect of Decreased PKc Expression on Metabolic Fluxes
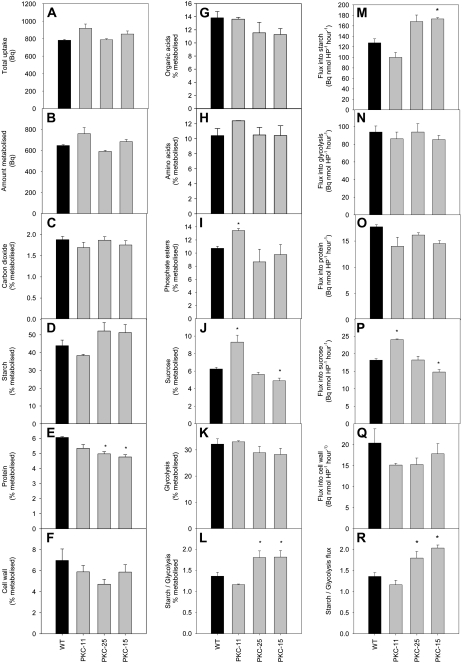
The effect of reduced PKc on carbon fluxes was determined by incubating tuber discs from wild type and lines PKC-11, -25, and -15 with [U-14C]Glc followed by fractionation of the labeled material to determine the label distribution into different pathways (Fig. 4). The transgenic tubers showed no difference from wild type in total label uptake (Fig. 4A), amount metabolized (Fig. 4B), or in the percentage of metabolized label incorporated into carbon dioxide (Fig. 4C), cell wall (Fig. 4F), or amino acids (Fig. 4H). However, there was a tendency of increased label incorporation into starch (Fig. 4D), significant reduction of label incorporated into protein (Fig. 4E, significant for lines PKC-25 and -15), and a tendency of reduction of label into organic acids (Fig. 4G). PKC-11 showed a significant increase of label into phosphate esters and Suc, while PKC-15 had reduced label incorporation into Suc (Fig. 4, I and J). The total sum of label incorporation into glycolysis was calculated as the sum of label in carbon dioxide, protein, organic acids, and amino acids (Fig. 4K). The ratio of label incorporation into starch versus glycolysis was significantly increased in PKC-25 and -15 (Fig. 4L).
Figure 4.
Metabolism of [14C]Glc in wild-type and transgenic tubers with decreased PKc. Tuber discs were cut from freshly harvested 10-week-old tubers and incubated with 2 mm [U-14C]Glc (specific activity = 11.54 kBq μmol−1) for 2 h, followed by fractionation of the material and determination of label distribution. Data show the mean of three biological replicates per line and error bars represent ses. *, Significant difference from wild-type levels (P < 0.05).
Metabolic fluxes were calculated according to Geigenberger et al. (1997) by determining the specific activity of the hexose phosphate pool, which was similar in all lines (wild type = 1.11 ± 0.06, PKC-11 = 1.46 ± 0.14, PKC-25 = 0.91 ± 0.21, PKC-15 = 1.17 ± 0.25 Bq nmol−1). There was an increased flux of carbon into starch in transgenic lines PKC-25 and -15 compared to wild type (significant for line PKC-15; Fig. 4M). Fluxes into protein and glycolysis were not significantly different from wild type, but the ratio of flux into starch versus glycolysis was significantly higher than wild type in PKC-25 and -15 (Fig. 4, N, O, and R). Flux into Suc was significantly higher than wild type in PKC-11 but lower in PKC-15, while flux into cell wall was not significantly different from wild type (Fig. 4, P and Q). These data show that reduced PKc causes an altered redistribution of carbon fluxes, with slightly less flux through glycolysis and greater flux into starch synthesis.
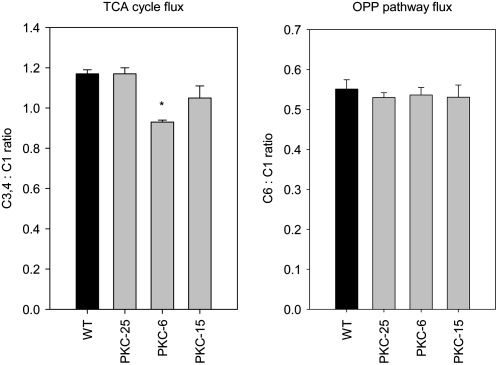
The relative flux of carbon through the TCA cycle was measured by incubating tuber discs in positionally labeled Glc isotopomers ([1-14C]Glc, [3,4-14C]Glc, and [6-14C]Glc) and measuring the label released as CO2. Carbon from the C3,4 positions can only be released as CO2 via the mitochondrial TCA cycle, whereas carbon from the C6 position can be released as CO2 from both mitochondrial respiration and the oxidative pentose phosphate (OPP) pathway. The relative TCA cycle flux can be compared among the plant lines from the ratio of label evolution from the C3,4 to C1 incubations, while the C6 to C1 ratio is a measurement of flux through the OPP pathway relative to the other pathways of carbon oxidation (ap Rees and Beevers, 1960; Nunes-Nesi et al., 2005). The results revealed that tubers from lines PKC-6 and -15 have 20% and 10% lower rates of flux through the TCA cycle flux compared to wild type (significant for PKC-6), while TCA cycle flux in PKC-25 exhibited equivalent flux rates to those observed for wild type (Fig. 5A). Flux through the OPP pathway in the transgenics was not significantly different from wild type (Fig. 5B). These data imply that there were no major alterations in the relative flux through the TCA cycle and the OPP pathway in the transgenics (Fig. 5).
Figure 5.
Measurement of flux through the TCA cycle and OPP pathway in wild-type and transgenic tubers with decreased PKc by incubation of tuber discs in positionally labeled isotopomers of [14C]Glc. Tuber discs were cut from freshly harvested 10-week-old tubers and incubated in [14C]Glc labeled in either the C1, C3,4, or C6 position for 6 h. The evolved CO2 gas was continuously captured in 10% KOH and the amount of label released as CO2 was measured. TCA cycle flux was determined as the ratio of label evolved as CO2 from the C3,4 incubation compared to the C1 incubation, while OPP pathway flux was determined as the ratio of label evolved as CO2 from the C6 incubation compared to the C1 incubation. Data shows the mean of three replicate incubations per Glc isotopomer per line, and error bars represent ses. *, Significant difference from wild-type levels (P < 0.05); n = six tubers from six different plants.
Respiration was measured as the overall rate of oxygen consumption by tuber discs cut from freshly harvested tubers. While PKC-25 showed a significant reduction in respiration, all other lines were not significantly different from wild type (Fig. 3F).
We also measured internal oxygen concentrations in intact tubers. Because changes in respiration rates will lead to inverse changes in internal oxygen concentrations (see Bologa et al., 2003), internal oxygen concentrations can be used as an indirect measure of respiration rates in intact tubers. Internal oxygen concentrations were measured with a needle-type, 230-μm-diameter, minimally invasive oxygen micro-optode, which was directly inserted into growing tubers of similar size. Consistent with the lack of changes in respiration rates in discs, reduction of PKc did not lead to significant changes in internal oxygen levels of growing tubers. Oxygen concentrations as percentage of normal air were as follows: 47 ± 4, 39 ± 6, and 50 ± 5 for wild type, PKC-25, and PKC-15, respectively (mean ± se, n = 19 tubers for wild type, n = 13 tubers for PKC-25, and n = 17 tubers for PKC-15 from three different plants per line). This provides indirect evidence that overall respiration was not substantially changed in tubers of the transgenic lines.
Effect of Decreased PKc Expression on AOX Capacity
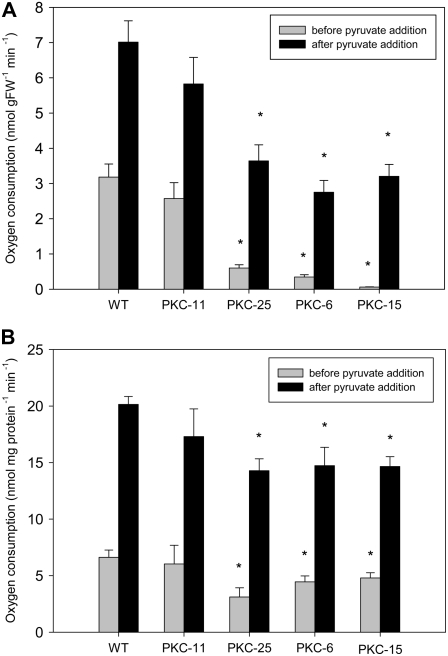
The AOX capacity was calculated as the rate of KCN-insensitive, salicylhydroxamic acid (SHAM)-sensitive oxygen consumption in tuber slices, and the level of stimulation by pyruvate was also determined. Without exogenous pyruvate addition, AOX capacity was significantly lower than wild type in the strongest transgenic lines (PKC-25, -6, and -15) but no different than wild type in the nonsilenced line PKC-11 (Fig. 6A). Upon addition of pyruvate, respiration was significantly increased in all genotypes but remained significantly lower than wild type in PKC-25, -6, and -15 (Fig. 6A). As reported previously (Millar et al., 1993), the stimulation of KCN-insensitive respiration by pyruvate occurred within seconds and resulted in a linear rate of oxygen consumption for the duration of the experiment (at least 10 min) until SHAM was added. The initial levels of oxygen consumption before KCN addition were similar in all lines (wild type = 27.1 ± 4.1, PKC-11 = 28.8 ± 5.7, PKC-25 = 23.6 ± 3.2, PKC-6 = 24.5 ± 2.0, PKC-15 = 27.1 ± 4.1 nmol oxygen g fresh weight [FW]−1 min−1), and the residual rates of respiration (measured as KCN-, SHAM-insensitive respiration) were very low, as has previously been reported for potato tubers (Hiser et al., 1996), and not significantly different between the lines (wild type = 2.5 ± 0.5, PKC-11 = 2.0 ± 0.3, PKC-25 = 1.2 ± 0.6, PKC-6 = 2.4 ± 0.4, PKC-15 = 2.0 ± 0.3 nmol oxygen g FW−1 min−1). The reverse experiment was also performed to measure the rate of SHAM-insensitive, KCN-sensitive respiration. In the absence of KCN, the addition of SHAM and pyruvate had no effect on the respiration rate (data not shown).
Figure 6.
Measurement of AOX capacity in intact tissue slices (A) and isolated mitochondria (B) from 10-week-old wild-type and transgenic tubers with decreased PKc. Data show the average KCN-insensitive, SHAM-sensitive respiration in the absence (light gray bars) and presence (black bars) of pyruvate. Respiration rates were calculated from oxygen consumption rates measured using a Clark electrode in the presence of KCN (to inhibit the COX), before and after the addition of pyruvate (to stimulate the AOX), followed by SHAM (to inhibit the AOX and calculate residual respiration rates, which were subtracted to obtain KCN-sensitive, SHAM-insensitive respiration). Error bars represent ses (n = 12 tubers from six different plants per line [A] or four mitochondrial preparations per line, representing four independent tuber harvests [B]). *, Significant difference from wild-type levels (P < 0.05).
In isolated tuber mitochondria, similar results were obtained, with a significantly lower AOX capacity in PKC-25, -6, and -15 compared to wild type and PKC-11 (Fig. 6B). However, the difference in AOX capacity between the transgenics and wild type was not as marked as in intact tuber tissue. Mitochondria in intact cells are subject to other influences, such as redox poise, that are known to influence AOX activity (Umbach and Siedow, 1993; Day et al., 1995; Mackenzie and McIntosh, 1999; Vanlerberghe et al., 1999).
Effect of Decreased PKc Expression on AOX and Cytochrome c Oxidase Protein Levels
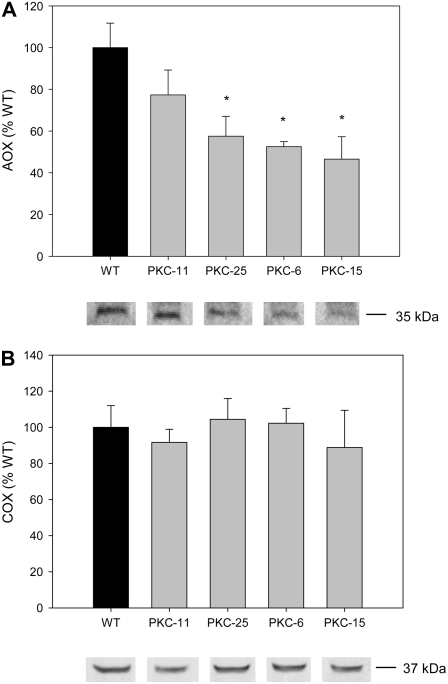
The results presented above indicate that decreased PKc led to a decrease in the capacity of the alternative pathway of respiration. To confirm these results, AOX protein levels were measured in isolated mitochondria by western blots of proteins separated under reducing conditions, using an AOX-specific antibody (Elthon and McIntosh, 1987). The results showed that levels of the AOX protein were significantly lower in mitochondria from the PKc transgenic tubers in lines PKC-25, -6, and -15 compared to wild type (Fig. 7A). In contrast, the levels of cytochrome c oxidase (COX) in the transgenics were not significantly different to wild type (Fig. 7B). The expression level of the AOX1 gene in tubers was assessed by quantitative real-time RT-PCR. The results showed that transcriptional abundance of this gene was reduced to 51% ± 11%, 42% ± 20%, and 66% ± 15% of wild-type levels in PKC-25, -6, and -15, respectively (wild type = 100% ± 13%) but not reduced in PKC-11 or PKC-26 (141% ± 32% and 108% ± 45%, respectively; n = 4).
Figure 7.
Quantification of AOX and COX protein levels in wild-type and PKc transgenic tubers. Protein from isolated tuber mitochondria was electrophoresed under reducing conditions and western blots were probed with antibodies against AOX (A) or COX (B). The graphs show the mean protein levels obtained from quantification of the western blots (n = three tuber mitochondrial isolations per line, representing three independent tuber harvests. Representative western blot results are shown beneath graph bars (bands taken from the same western blot are shown for each antibody). Error bars represent ses. *, Significant difference from wild-type levels (P < 0.05).
DISCUSSION
The role of PKc in plants has not yet been fully resolved, despite a number of physiological and biochemical approaches published in the past. In this article, we have studied the effect of decreased PKc levels in potato tubers on tuber metabolism using an RNAi silencing approach. Silencing the PKCYT1 gene in tubers resulted in a large decrease in total PK activity and decreased levels of the two major immunoreactive PKc polypeptides present in wild-type tubers (of sizes approximately 55 and 60 kD; see Fig. 1). The large reduction of PKCYT1 transcript levels to 1% to 3% of wild type levels in the PKc transgenic tubers probably accounts for the strongly decreased level of the 55-kD PKc immunoreactive band. However, the strong reduction of the 60-kD protein does not appear to be solely due to reduced transcription of the corresponding gene, because there was only weak cross-silencing of other PKC genes. It has previously been demonstrated that PKc in tobacco leaves and castor oil leaves is a heterotetramer consisting of two subunits of 57 kD and 56 kD (Plaxton, 1989; Knowles et al., 1998) and our results indicate PKc in potato tubers may also be composed of two different subunits. While this remains to be investigated, it is possible that the presence of the 55-kD protein is necessary to maintain levels of the 60-kD protein, as has been described for the heterotetrameric plant AGPase enzyme, in which the presence of the 49-kD subunit is required to maintain stability of the 52-kD subunit (Wang et al., 1998). Furthermore, analysis of tobacco plants lacking PKc activity and protein levels in leaves showed that the 56-kD and 57-kD PKc polypeptides were either both present or both absent (Gottlob et al., 1992). In the PKc transgenic plants analyzed here, the third PKc immunoreactive PKc polypeptide detected in tubers (approximately 50 kD) was slightly induced in the transgenics, but this also does not appear to be due to increased transcription of the PKC genes measured. This protein band may represent either another isozyme or a posttranslationally cleaved product of one of the larger isozymes, similar to that described by Tang et al. (2003).
Decreased PKc expression led to an increased PEP to pyruvate ratio in growing tubers, indicating that PEP to pyruvate conversion was indeed inhibited in the transgenic lines (Fig. 3N). This was mainly due to a decrease in pyruvate levels, while PEP levels remained rather unchanged. The fact that the levels of PEP and 3-phosphoglycerate were not substantially increased could be due to a coordinated inhibition of additional regulation sites upstream in glycolysis and Suc degradation. The decrease in pyruvate level was accompanied by a decrease in the levels of some other organic acids involved in the TCA cycle and decreases in the level of Asp (Table II). There was also a decrease in the level of total protein in the tubers. Labeling experiments using [14C]Glc showed a slight decrease in the partitioning of carbon toward protein and organic acid synthesis upon reduction of PKc levels. These results indicate that PKc plays an important role in the regulation of the levels of organic acids in tubers, as well as preferentially partitioning carbon toward the TCA cycle. This contention is strongly supported by the finding that PKc is part of a functional association of glycolytic enzymes with the mitochondrial membrane in potato as well as Arabidopsis and that cyanide treatment reduces the association of these enzymes while increasing the demand on the alternative pathway of respiration (Graham et al., 2007).
Surprisingly, the strong decrease in PKc levels did not lead to substantial changes in overall respiration rates and TCA cycle flux in tubers (Figs. 3F and 5A), confirming previous studies with transgenic tobacco plants lacking PKc in leaves (Grodzinski et al., 1999), This is in contrast to the proposed role of PKc as primary control site of respiration, which can be found in many textbooks (see Kruger, 1997). There are two possible explanations. First, the remaining activity of PKc might still be high enough to maintain overall respiration rates in the tissue. Secondly, plants have alternative enzymes to PKc that may be able to continue to supply the mitochondria with pyruvate or other carbon sources even when PKc is severely reduced, including PKp, PEPC, or PEP phosphatase. The contribution of these enzymes to the rate of pyruvate formation in vivo is difficult to determine. However, we can exclude that the expression of PKp, the activity and activation state of PEPC, and the activity of PEP phosphatase were increased in compensation to the decrease in PKc. Moreover, pyruvate levels were significantly decreased by almost 50% in the strongest PKc lines, compared to wild type, which indicates that if compensation occurred, it was only of a partial nature. Clearly, a moderate decrease in pyruvate and other organic acids did not affect overall respiration rates. This is supported by previous studies where activities of enzymes of the TCA cycle were severely decreased without displaying a massive effect on overall respiration rates (Carrari et al., 2003; Nunes-Nesi et al., 2005, 2007; Studart-Guimarães, 2007). When taken together, these observations indicate that levels of TCA cycle intermediates are not strictly limiting overall flux into respiration.
While a decrease in PKc did not affect the overall rate of oxygen consumption (Fig. 3F), there was a clear effect on the capacity of the AOX-mediated alternative pathway of mitochondrial electron transport (Fig. 6). AOX capacity was measured as the level of KCN-insensitive, SHAM-sensitive respiration, which has previously been used to assess AOX capacity in Arabidopsis and potato (Hiser et al., 1996; Umbach et al., 2005). In comparison to wild type, the PKc transgenics had substantially lower rates of KCN-insensitive, SHAM-sensitive respiration, both in whole tuber tissue and isolated mitochondria, even after external pyruvate addition. Direct feeding of pyruvate to tuber tissue led to activation of the AOX-dependent pathway in both the wild type and the transgenics, providing direct evidence for a regulation of AOX by pyruvate in vivo. The pyruvate-induced stimulation of the alternative pathway of respiration was evident in both wild-type and transgenic tubers, which suggests that the lower AOX capacity in the transgenic lines could be due to a reduction in the amount of AOX protein, but a modification of regulatory properties of the AOX cannot be discarded. Indeed, quantification of the levels of both the AOX transcript and protein demonstrated a large decrease of AOX protein in the transgenics compared to wild type (Fig. 7).
Our results are consistent with previous studies showing that AOX activity is increased after addition of pyruvate or other organic acids in vitro (Millar et al., 1993; Day et al., 1994). In short-term experiments, it has been shown that exogenous addition of pyruvate can act rapidly by stabilizing the active monomerization state of AOX (Vanlerberghe et al., 1999). Moreover, long-term additions of citrate for 1 d have been reported to lead to induction of AOX protein levels in Poa annua roots and tobacco cell cultures (Vanlerberghe et al., 1995; Millenaar et al., 2002). This study confirms and extends this work. To our knowledge, it is the first report demonstrating that pyruvate and AOX levels are correlated in planta. This study shows that alterations in glycolytic metabolism in the cytosol lead to changes in AOX protein levels in mitochondria that are probably due to changes in pyruvate levels. This may have consequences for redox-signaling, because previous studies have shown that loss of AOX protein in plant tissues or cell cultures results in an elevated level of mitochondrial reactive oxygen species and programmed cell death (Robson and Vanlerberghe, 2002; Umbach et al., 2005). However, measurement of thiobarbituric acid-reactive substances as an indicator of reactive oxygen species in the transgenic tubers revealed no major changes in cellular redox parameters (data not shown). This result together with the recent observation that the ucp1 knockout of Arabidopsis UNCOUPLING PROTEIN1, another element of the alternative respiratory pathway, was in no way compromised in its response to oxidative stress (Sweetlove et al., 2006), casts some doubts on previous suggestions of the importance of this pathway in protection of cellular components against oxidative damage. Suppression of the alternative pathway may even be beneficial in the context of the growing tuber, which is characterized by low internal oxygen concentrations and low adenylate energy states (Geigenberger et al., 2000; Geigenberger, 2003). Despite the fact that the exact physiological role of the AOX-mediated alternative pathway remains somewhat enigmatic, results presented here provide strong evidence of the ability of PKc to regulate its capacity via modulating pyruvate levels.
The results presented provide evidence for a role of PKc to regulate both the level and the subsequent use of pyruvate via the AOX-mediated alternative pathway of respiration. Moderate in vivo changes in pyruvate levels were shown to have a strong impact on the alternative pathway of respiration, while the COX-dependent pathway was hardly affected. Obviously, plants have to regulate cytosolic pyruvate levels very carefully to keep alternative respiration to a minimum. The role of PKc thus clearly contrasts with that of the plastidial PK isozyme, which is crucial for provision of pyruvate for fatty acid synthesis in the plastid (Andre and Benning, 2007; Andre et al., 2007; Baud et al., 2007), although it is interesting to note that both the potato PKc transgenics and the Arabidopsis PKp mutants had lower protein content. The main reason for the differences between these plants is likely to be the different subcellular location of the isozymes, because the primary effect of the genetic manipulation, an alteration of the overall PEP to pyruvate ratio, is similar in both instances (contrast Andre and Benning [2007] with this study). Nevertheless, we must be cautious when comparing potato and Arabidopsis because of the morphological and physiological differences between these two species. For example, potato tubers are nonphotosynthetic, starch-storing organs, whereas Arabidopsis seeds store oil and have some photosynthetic capacity during part of their development.
In conclusion, the study presented here provides new insights into the specific role of a major PKc isozyme in plants and complements previous detailed analyses of the plastidial isozymes in Arabidopsis. However, further studies will be required to fully understand the function of the other plant PK isozymes revealed by our phylogenetic analysis.
MATERIALS AND METHODS
Database Searching and Phylogenetic Analysis
PK cDNA or EST sequences were obtained from publicly available data from the Joint Genome Institute (http://www.phytozome.net) and the National Center for Biotechnology Information (http://www.ncbi.nml.nih.gov). A basic alignment of the deduced protein sequences was created using the ClustalW program (Chenna et al., 2003) and then manually edited using the GeneDoc program (Nicholas and Nicholas, 1997). Maximum likelihood trees were created using the PHYML program with the JTT or DCMut evolutionary model of amino acid substitution (Guindon et al., 2005) according to Lunn (2007) and displayed using TreeView (Page, 1996).
Plant Material
Potato (Solanum tuberosum) cv Desiree was obtained from Saatzucht Lange AG. Plants were maintained in tissue culture and then transferred to greenhouse conditions as described by Fernie et al. (2002). Developing tubers (<10 g FW) were harvested from healthy 10-week-old plants. For respiration analyses and mitochondrial isolations, tubers were used immediately. For enzyme, metabolite, and RNA analyses, tuber cores were immediately frozen in liquid nitrogen, ground into a fine powder using a ball-mill (Retsch), and stored at −80°C until further analysis.
Generation of PKc Transgenic Plants
A 650-bp fragment of the tomato (Solanum lycopersicum) cDNA clone (TC172280) showing 97% nucleotide identity to the potato PKCYT1 gene was amplified by PCR (5′ primer, CACCCGTGCTGAGGCTAC; 3′ primer, CAGGTGTGGGGGAGTTCA) and cloned via the GATEWAY method into a modified GATEWAY binary vector (based on pK7GWIWG2(I)0; Karimi et al., 2002) between the B33 patatin promoter and the OCS terminator using kanamycin selection. The construct was introduced into plants (cv Desiree) via Agrobacterium-mediated transformation, and plants were selected and maintained as described previously (Tauberger et al., 2000).
Enzyme Activity and Protein Measurements
Proteins were extracted and desalted according to Trethewey et al. (1998) using an extraction buffer containing 50 mm HEPES-KOH, pH 7.4, 5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 5 mm dithiothreitol, 2 mm benzamidine, 2 mm ɛ-amino-n-caproic acid, 0.5 mm phenylmethylsulfonyl fluoride, 0.1% (w/v) fatty-acid-free bovine serum albumin, 10% (v/v) glycerol, and 0.1% (v/v) Triton X-100, as in Geigenberger and Stitt (1993). Enzyme activities were measured according to the following: hexokinase (Renz et al., 1993), phosphoglucomutase, ATP-dependent phosphofructokinase and fructokinase (Sweetlove et al., 1996), PK (using an assay at pH 6.9) and PEP phosphatase (Plaxton, 1990), and NAD malic enzyme and malate dehydrogenase (Hatch et al., 1982). Total PEPC activity was measured as in Merlo et al. (1993) and activation state as in Schuller et al. (1990). Protein was quantified using the Bio-Rad Protein Assay Reagent (Bio-Rad Laboratories) according to the manufacturer's instructions using bovine serum albumin as a quantification standard.
Metabolite Measurements
For measurement of PEP, pyruvate, nucleotides, and hexose phosphates, 200 mg tuber material was extracted with trichloroacetic acid according to Trethewey et al. (1998). PEP and pyruvate were measured in fresh extracts according to Merlo et al. (1993). For nucleotide measurements, the TCA extracts were subjected to HPLC according to Geigenberger et al. (1997). Hexose phosphates were measured according to Merlo et al. (1993). Glc, Fru, Suc, and starch were measured after ethanol extraction according to Geigenberger et al. (1996). Extraction and measurement of all other metabolites (organic acids, amino acids, and sugars) was performed by GC-MS analysis of methanol extracts as described in Roessner et al. (2000). Recovery experiments were performed for the various metabolites and are documented in Merlo et al. (1993) for hexose-phosphates, pyruvate, and PEP, in Geigenberger et al. (1997) for nucleotides, and in Roessner et al. (2000) for metabolites analyzed by GC-MS.
[14C]Glc Feeding with [U-14C]Glc
Tuber discs were prepared from growing plants and [U-14C]Glc labeling experiments were performed as described by Geigenberger et al. (1997). Tuber discs were incubated with 2 mm Glc in 10 mm MES-KOH buffer, pH 6.5, containing [U-14C]Glc (Amersham-Buchler; 10 discs per 4 mL in 100-mL Erlenmeyer flasks; specific activity, 11.54 kBq μmol−1) for 2 h with shaking at 90 rpm, then briefly rinsed with 10 mm MES-KOH buffer, pH 6.5, and frozen in liquid nitrogen. Tissue extractions and fractionation of soluble components were carried out according to Geigenberger et al. (1997) and fractionation of insolubles into starch, protein, and cell wall was carried out according to Merlo et al. (1993).
[14C]Glc Feeding with Positionally Labeled Glc Isotopomers
TCA cycle flux analysis was performed based on Nunes-Nesi et al. (2005). Tuber discs were prepared from growing plants and incubated with 0.2 mm Glc in 10 mm MES-KOH buffer, pH 6.5, containing [1-14C]Glc, [3,4-14C]Glc, or [6-14C]Glc (Amersham-Buchler; 10 discs per 4 mL in 100-mL Erlenmeyer flasks; specific activity 1.85 kBq mL−1) with shaking at 90 rpm for 6 h. Released CO2 was captured in 10% KOH sampled every 2 h and quantified by liquid scintillation counting.
Tuber Disc Respiration Measurements
Tuber discs (8 mm × 2 mm) were prepared from freshly harvested, 10-week-old tubers and briefly rinsed in buffer (10 mm MES-KOH, pH 6.5) to remove broken cells, then two discs were immediately transferred to the temperature-controlled chamber of a Clark-type electrode (Hansatech) containing 1 mL buffer at 25°C. AOX capacity was calculated as the KCN-insensitive, SHAM-sensitive respiration rates according to Umbach et al. (2005) and Millar et al. (1993) by measuring oxygen consumption rates after the sequential addition of KCN (1 mm), pyruvate (4 mm), and SHAM (10 mm).
To investigate whether there is a possible wound-induced respiration during the time course of our tuber-disc experiments (<3 h; see above), we measured the rates of oxygen consumption at different time points after slicing the tubers (Supplemental Fig. S2). The data show that there is no increase in respiration rates within the first 3 h of incubation. Previous studies on respiration rates in tuber slices are consistent with this (Hajirezaei and Stitt, 1991).
Mitochondrial Isolation and Respiratory Analysis
Mitochondria were isolated from freshly-harvested, 10-week-old tubers according to Jenner et al. (2001) but with at least 100 g tuber material ground in 200 mL buffer in a Waring blender, and thereafter according the published protocol (Jenner et al., 2001). Protein was quantified as described above. Oxygen consumption was measured with a temperature-controlled, Clark-type electrode (Hansatech) using 300 mg protein in 1 mL reaction buffer (Sweetlove et al., 2002) with the sequential addition of NADH (1 mm), ADP (0.2 mm), KCN (1 mm), pyruvate (4 mm), and SHAM (10 mm) at 25°C to determine mitochondrial respiration rates according to Millar et al. (1993).
Analysis of Internal Oxygen Concentration
Pots were removed from plants and tubers were freed from soil and immediately fixed in a micromanipulator (Saur Laborbedarf) while remaining attached to the plant without previous washing with water. Without delay, a 230-μm diameter, minimally invasive oxygen micro-optode (Löhmannsröben et al., 2006) was phased into a depth of 4 mm under the tuber skin and the oxygen concentration recorded. The optical measurement of molecular oxygen was based on phosphorescent dyes with decay times dependent on the ambient oxygen content.
RNA Isolation and Quantitative Real-Time RT-PCR Analysis
Total RNA was isolated from 100 mg ground tuber material using the Qiagen RNeasy Plant Minikit (Qiagen). RNA quality was assessed by agarose gel electrophoresis prior to DNaseI digestion (Promega) and cDNA synthesis using the Superscript III RT-PCR kit (Invitrogen). Quantitative real-time RT-PCR was performed as described by Czechowski et al. (2004). The quality of cDNA synthesis was assessed using primer pairs for the 5′ and 3′ ends of glyceraldehyde-3-P dehydrogenase (GAPDH) according to Czechowski et al. (2005). A set of control housekeeping genes was assessed using primers for GAPDH and ELONGATION FACTOR 1α as described in Czechowski et al. (2004) and for UBIQUITIN and β-TUBULIN as described by Diretto et al. (2006). From these, UBIQUITIN and GAPDH were selected as the best housekeeping genes used for normalization in this study, because their expression was identical (variation less than one cycle), and expression was also constant across the different cDNA samples (variation less than 1.5 cycles). Primers for the PK and AOX1 (GenBank accession no. AB176953) genes were designed across exon-intron boundaries; when genomic sequence was unavailable, a series of primers was designed based on the cDNA sequence and tested to identify those that would amplify across introns. All primer pairs used for quantification produced a single band of the expected size as assessed by agarose gel electrophoresis, and sequencing confirmed that the primers amplified the expected product. Furthermore, only one product for each primer pair was detected by melt curve analysis in all amplifications; this was distinguishable from the product of genomic DNA control amplifications. No genomic DNA contamination was detected in the cDNA samples used for quantification. The primers used for quantification analyses were: UBIQUITIN (Diretto et al., 2006), 5′-ACAATGTCAAAGCCAAGATCCA, 3′-CGGAGACGGAGCACGAGA; GAPDH, 5′-TTCAACATCATCCCTAGCAGCACT, 3′-TAAGGTCGACAACAGAAACATCAG; PKCYT1, 5′-CATTGATATGATAGCGCTTTCG, 3′-GAGATCACCTCGAGCAACC; PKCYT2, 5′-GGAGTTCCAAATCATATCGAC, 3′-CCATAAATGCATCAGAGTTGGC; PKCYT3, 5′-GGCAATCTCTAATTGTCCGAG, 3′-AACGACAACACGATCGTGTG; PKPα1, 5′-TGAATTGCCATCCATAGCATC, 3′-GTGACAACAGAGATGCCATGTG; PKPβ2, 5′-GGAGTCGGGTGACACACTAC, 3′-CCGTCTTGATTTCAGTTC; and AOX1, 5′-CCATGGGAGACTTATGAAGC, 3′-CATACCTCCCACCATTCC.
Western Blots
A total of 25 μg of protein from mitochondria isolated from tubers (for AOX and COX) or from tuber tissue (for PKc) in Laemmli buffer (Laemmli, 1970) were subjected to 10% SDS-PAGE under reducing conditions, then transferred to nitrocellulose membranes. Blots were briefly stained with Ponceau Red (0.1% Ponceau S [Sigma-Aldrich], 5% acetic acid) to confirm even loading and transfer, and then probed with primary antibodies raised against Brassica napus PKc (Smith et al., 2000), Sauromatum gutarum AOX (Elthon and McIntosh, 1987), or COX subunit II (Agrisera). The secondary antibody for PKc was alkaline phosphatase-conjugated anti-mouse IgG (Bio-Rad Laboratories), which was detected using the CDP-Star reagent (Roche) followed by exposure to BioMax XAR film (Kodak, Sigma-Aldrich). The secondary antibodies for AOX and COX were affinity-purified IRDye800-conjugated goat antibodies (anti-rabbit IgG for COX, anti-mouse IgG for AOX, Rockland Immunochemicals), and detection was performed using the Odyssey Infrared Imager system (LI-COR Biosciences). The AOX and COX western blots gave a specific band of the expected size in isolated mitochondria (35 kD for AOX, 37 kD for COX subunit II). Bands were quantified using the Scion Image Software (Scion Corporation), band sizes were estimated by comparison with size markers, and band intensity was proportional to the amount of extract loaded.
Statistical Analysis
Data were analyzed using the Student's t test and deemed significant if P < 0.05.
Sequence data from this article can be found in the GenBank/EMBL or TIGR transcript contig data libraries under accession numbers S53332 (potato PKCYT1), TC134535 (potato PKCYT2), TC141201 (potato PKCYT3), TC118783 (potato PKCYT4), TC135950 (potato PKCYT5), TC135043 (potato PKPα1), TC159621 (potato PKPα2), TC149988 (potato PKPβ1), and TC134739 (potato PKPβ2).
Supplemental Data
The following materials are available in the on-line version of this article.
Supplemental Figure S1. Phylogenetic alignment of PK and PK-like proteins.
Supplemental Figure S2. Respiration rates at different time points after tuber slicing.
Supplementary Material
Acknowledgments
We gratefully acknowledge Anett Doering for excellent technical assistance, Helga Kulka for plant cultivation, Romy Baran for plant transformation, and Prof. Bill Plaxton (Queen's University, Ontario, Canada) for kindly providing the anti-PKc antibody, and Dr. Thomas Elthon (University of Nebraska, Lincoln, NE) for kindly providing the anti-AOX antibody.
This work was supported by the Deutsche Forschungsgemeinschaft and by the Max Planck Society.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Peter Geigenberger (geigenberger@igzev.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Andre C, Benning C (2007) Arabidopsis seedlings deficient in a plastidic pyruvate kinase are unable to utilize seed storage compounds for germination and establishment. Plant Physiol 145 1670–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre C, Froehlich JE, Moll MR, Benning C (2007) A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19 2006–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ap Rees T, Beevers H (1960) Pathways of glucose dissimilation in carrot slices. Plant Physiol 35 830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Baud S, Wuilleme S, Dubreucq B, de Almeida A, Vuagnat C, Lepiniec L, Miquel M, Rochat C (2007) Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J 52 405–419 [DOI] [PubMed] [Google Scholar]
- Blakeley S, Gottlob-McHugh S, Wan J, Crews L, Miki B, Ko K, Dennis DT (1995) Molecular characterization of plastid pyruvate kinase from castor and tobacco. Plant Mol Biol 27 79–89 [DOI] [PubMed] [Google Scholar]
- Blakeley SD, Plaxton WC, Dennis DT (1990) Cloning and characterization of a cDNA for the cytosolic isozyme of plant pyruvate kinase: the relationship between the plant and non-plant enzyme. Plant Mol Biol 15 665–669 [DOI] [PubMed] [Google Scholar]
- Bologa KL, Fernie AR, Leisse A, Ehlers Loureiro M, Geigenberger P (2003) A bypass of sucrose synthase leads to low internal oxygen and impaired metabolic performance in growing potato tubers. Plant Physiol 132 2058–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Ehlers-Loureiro M, Fernie AR (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole KP, Blakeley SD, Dennis DT (1992) Structure of the gene encoding potato cytosolic pyruvate kinase. Gene 122 255–261 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible W, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38 366–379 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DA, Millar AH, Wiskich JT, Whelan J (1994) Regulation of alternative oxidase activity by pyruvate in soybean mitochondria. Plant Physiol 106 1421–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DA, Whelan J, Millar AH, Siedow JN, Wiskich JT (1995) Regulation of the alternative oxidase in plants and fungi. Aust J Plant Physiol 22 497–509 [Google Scholar]
- de Bari L, Valenti D, Pizzuto R, Altante A, Passarella S (2007) Phosphoenolpyruvate metabolism in Jerusalem artichoke mitochondria. Biochim Biophys Acta 1767 281–294 [DOI] [PubMed] [Google Scholar]
- Dennis DT, Greyson M (1987) Fructose-6-phosphate metabolism in plants. Physiol Plant 69 395–404 [Google Scholar]
- Diretto G, Tavazza R, Welsch R, Pizzichini D, Mourgues F, Papacchiloi V, Beyer P, Giuliano G (2006) Metabolic engineering of potato tuber carotenoids through tuber-specific silencing of lycopene epsilon cyclase. BMC Plant Biol 6 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elthon TE, McIntosh L (1987) Identification of the alternative terminal oxidase of higher plant mitochondria. Proc Natl Acad Sci USA 84 8399–8403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Carrari F, Sweetlove LJ (2004) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol 7 254–261 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Tiessen A, Stitt M, Willmitzer L, Geigenberger P (2002) Altered metabolic fluxes result from shifts in metabolite levels in sucrose phosphorylase-expressing potato tubers. Plant Cell Environ 25 1219–1232 [Google Scholar]
- Geigenberger P (2003) Response of plant metabolism to too little oxygen. Curr Opin Plant Biol 6 247–256 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Fernie AR, Gibon Y, Christ M, Stitt M (2000) Metabolic activity decreases as an adaptive response to low internal oxygen in growing potato tubers. Biol Chem 381 723–740 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Geiger M, Stitt M (1998) High-temperature perturbation of starch synthesis is attributable to inhibition of ADP-glucose pyrophosphorylase by decreased levels of glycerate-3-phosphate in growing potato tubers. Plant Physiol 117 1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Langenberger S, Wilke I, Heineke D, Heldt HW, Stitt M (1993) Sucrose is metabolized by sucrose synthase and glycolysis within the phloem complex of Ricinus communis L. seedlings. Planta 190 446–453 [Google Scholar]
- Geigenberger P, Lerchl J, Stitt M, Sonnewald U (1996) Phloem-specific expression of pyrophosphatase inhibits long-distance transport of carbohydrates and amino acids in tobacco plants. Plant Cell Environ 19 43–55 [Google Scholar]
- Geigenberger P, Reimholz R, Geiger M, Merlo L, Canale V, Stitt M (1997) Regulation of sucrose and starch metabolism in potato tubers in response to short-term water deficit. Planta 201 502–518 [Google Scholar]
- Geigenberger P, Stitt M (1993) Sucrose synthase catalyses a readily reversible reaction in vivo in developing potato tubers and other plant tissues. Planta 189 329–339 [DOI] [PubMed] [Google Scholar]
- Giege P, Heazlewood JL, Roessner-Tunali U, Millar AH, Fernie AR, Leaver CJ, Sweetlove LJ (2003) Enzymes of glycolysis are functionally associated with the mitochondrion of Arabidopsis cells. Plant Cell 15 2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givan CV (1999) Evolving concepts in plant glycolysis: two centuries of progress. Biol Rev Camb Philos Soc 74 277–309 [Google Scholar]
- Gottlob SG, Sangwan RS, Blakeley SD, Vanlerberghe GC, Ko K, Turpin DH, Plaxton WC, Miki BL, Dennis DT (1992) Normal growth of transgenic tobacco plants in the absence of cytosolic pyruvate kinase. Plant Physiol 100 820–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JWA, Williams TCR, Morgan M, Fernie AR, Ratcliffe RG, Sweetlove LJ (2007) Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling. Plant Cell 19 3723–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodzinski B, Jiao J, Knowles VL, Plaxton WC (1999) Photosynthesis and carbon partitioning in transgenic tobacco plants deficient in leaf cytosolic pyruvate kinase. Plant Physiol 120 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O (2005) PHYML Online: a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33 W557–W559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajirezaei MR, Stitt M (1991) Contrasting roles for pyrophosphate: fructose-6-phosphate phosphotransferase during aging of tissues from potato tubers and carrot storage tissues. Plant Sci 77 177–183 [Google Scholar]
- Hatch MD, Tsuzuki M, Edwards GE (1982) Determination of NAD malic enzyme in leaves of C4 plants. Plant Physiol 69 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiser C, Kapranov P, McIntosh L (1996) Genetic modification of respiratory capacity in potato. Plant Physiol 110 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner HL, Winning BM, Millar AH, Tomlinson KL, Leaver CJ, Hill SA (2001) NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol 126 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) Gateway vectors for Agrobacterium-mediated transformation. Trends Plant Sci 7 193–195 [DOI] [PubMed] [Google Scholar]
- Knowles VL, McHugh SG, Hu Z, Dennis DT, Miki BL, Plaxton WC (1998) Altered growth of transgenic tobacco lacking leaf cytosolic pyruvate kinase. Plant Physiol 116 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger NJ (1997) Carbohydrate synthesis and degradation. In DT Dennis, DH Turpin, DD Lefebvre, DB Layzell, eds, Plant Metabolism. Longman, Harlow, UK, pp 83–104
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Löhmannsröben H-G, Beck M, Hildebrandt N, Schmälzlin E, van Dongen JT (2006) New challenges in biophotonics: laser-based fluoroimmuno analysis and in-vivo optical oxygen monitoring. Proc Soc Photo Opt Instrum Eng 6157 61570E [Google Scholar]
- Lunn JE (2007) Gene families and evolution of trehalose metabolism in plants. Funct Plant Biol 34 550–563 [DOI] [PubMed] [Google Scholar]
- Mackenzie S, McIntosh L (1999) Higher plant mitochondria. Plant Cell 11 571–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo L, Geigenberger P, Hajirezaei M, Stitt M (1993) Changes of carbohydrates, metabolites and enzyme activities in potato tubers during development, and within a single tuber along a stolon-apex gradient. J Plant Physiol 142 392–402 [Google Scholar]
- Millar AH, Wiskich JT, Whelan J, Day DA (1993) Organic acid activation of the alternative oxidase of plant mitochondria. FEBS Lett 329 259–262 [DOI] [PubMed] [Google Scholar]
- Millenaar FF, Gonzalez-Meler MA, Siedow JN, Wagner AM, Lambers H (2002) Role of sugars and organic acids in regulating the concentration and activity of the alternative oxidase in Poa annua roots. J Exp Bot 53 1081–1088 [DOI] [PubMed] [Google Scholar]
- Millenaar FF, Lambers H (2003) The alternative oxidase: in vivo regulation and function. Plant Biol 5 2–15 [Google Scholar]
- Nicholas KB, Nicholas HB Jr (1997) GeneDoc. A tool for editing and annotating multiple sequence alignments: multiple sequence alignment editor and shading utility version 2.6.002. http://www.nrbsc.org/gfx/genedoc (October 15, 2008)
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50 1093–1106 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AMO, Loureiro M, Ratcliffe RG, Sweetlove LJ, Fernie AR (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137 611–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12 357–358 [DOI] [PubMed] [Google Scholar]
- Pilkis SJ, Granner DK (1992) Molecular physiology of the regulation of hepatic gluconeogenesis and glycolysis. Annu Rev Physiol 54 885–909 [DOI] [PubMed] [Google Scholar]
- Plaxton WC (1989) Molecular and immunological characterization of plastid and cytosolic pyruvate kinase isoenzymes from castor-oil plant endosperm and leaf. Eur J Biochem 181 443–451 [DOI] [PubMed] [Google Scholar]
- Plaxton WC (1990) Glycolysis. In PM Dey, JB Harborne, eds, Methods in Plant Biochemistry, Vol 3. Academic Press, London, pp 145–173
- Plaxton WC (1996) The organization and regulation of plant glycolysis. Annu Rev Plant Physiol Plant Mol Biol 47 185–214 [DOI] [PubMed] [Google Scholar]
- Plaxton WC, Podesta FE (2006) The functional organization and control of plant respiration. Crit Rev Plant Sci 25 159–198 [Google Scholar]
- Plaxton WC, Smith CR, Knowles VL (2002) Molecular and regulatory properties of leucoplast pyruvate kinase from Brassica napus (rapeseed) suspension cells. Arch Biochem Biophys 400 54–62 [DOI] [PubMed] [Google Scholar]
- Podesta FE, Plaxton WC (1991) Kinetic and regulatory properties of cytosolic pyruvate kinase from germinating castor oil seeds. Biochem J 279 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podesta FE, Plaxton WC (1992) Plant cytosolic pyruvate kinase: a kinetic study. Biochim Biophys Acta 1160 213–220 [DOI] [PubMed] [Google Scholar]
- Podesta FE, Plaxton WC (1993) Activation of cytosolic pyruvate kinase by polyethylene glycol. Plant Physiol 103 285–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podesta FE, Plaxton WC (1994) Regulation of cytosolic carbon metabolism in germinating Ricinus communis cotyledons. II. Properties of phosphoenolpyruvate carboxylase and cytosolic pyruvate kinase associated with the regulation of glycolysis and nitrogen assimilation. Planta 194 381–387 [Google Scholar]
- Renz A, Merlo L, Stitt M (1993) Partial purification from potato tubers of three fructokinases and three hexokinases which show differing organ and developmental specificity. Planta 190 156–165 [Google Scholar]
- Rivoal J, Smith CR, Moraes TF, Turpin DH, Plaxton WC (2002) A method for activity staining after native polyacrylamide gel electrophoresis using a coupled enzyme assay and fluorescence detection: application to the analysis of several glycolytic enzymes. Anal Biochem 300 94–99 [DOI] [PubMed] [Google Scholar]
- Robson CA, Vanlerberghe GC (2002) Transgenic plant cells lacking mitochondrial alternative oxidase have increased susceptibility to mitochondria-dependent and –independent pathways of programmed cell death. Plant Physiol 129 1908–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L (2000) Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23 131–142 [DOI] [PubMed] [Google Scholar]
- Schuller KA, Turpin DH, Plaxton WC (1990) Metabolite regulation of partially purified soybean nodule phosphoenolpyruvate carboxylase. Plant Physiol 94 1429–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB, Shachar-Hill Y (2004) Understanding flux in plant metabolic networks. Curr Opin Plant Biol 7 309–317 [DOI] [PubMed] [Google Scholar]
- Smith CR, Knowles VL, Plaxton WC (2000) Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell cultures: implications for the integration of glycolysis with nitrogen assimilation. Eur J Biochem 267 4477–4485 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Burrell MM, ap Rees T (1996) Characterization of transgenic potato (Solanum tuberosum) tubers with increased ADPglucose pyrophosphorylase. Biochem J 320 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32 891–904 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Lytovchenko A, Morgan M, Nunes-Nesi A, Taylor NL, Baxter CJ, Eickmeier I, Fernie AR (2006) Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc Natl Acad Sci USA 103 19587–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studart-Guimarães C, Fait A, Nunes-Nesi A, Carrari F, Usadel B, Fernie AR (2007) Reduced expression of succinyl coenzyme A ligase can be compensated for by up-regulation of the γ-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol 145 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Hardin SC, Dewey R, Huber SC (2003) A novel C-terminal proteolytic processing of cytosolic pyruvate kinase, its phosphorylation and degradation by the proteasome in developing soybean seeds. Plant J 34 77–93 [DOI] [PubMed] [Google Scholar]
- Tauberger E, Fernie AR, Emmermann M, Renz A, Kossmann J, Willmitzer L, Trethewey RN (2000) Antisense inhibition of plastidial phosphoglucomutase provides compelling evidence that potato tuber amyloplasts import carbon from the cytosol in the form of glucose-6-phosphate. Plant J 23 43–53 [DOI] [PubMed] [Google Scholar]
- Teusink B, Passarge J, Reijenga CA, Esgalhad E, van der Weijden CC, Schepper M, Walsh MC, Bakker BM, van Dam K, Westerhoff HV, et al (2000) Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur J Biochem 267 5313–5329 [DOI] [PubMed] [Google Scholar]
- Thomas S, Mooney PJF, Burrell MM, Fell DA (1997) Metabolic control analysis of glycolysis in tuber tissue of potato (Solanum tuberosum): explanation for the low control coefficient of phosphofructokinase over respiratory flux. Biochem J 322 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trethewey RN, Geigenberger P, Riedel K, Hajirezaei MR, Sonnewald U, Stitt M, Riesmeier JW, Willmitzer L (1998) Combined expression of glucokinase and invertase in potato tubers leads to a dramatic reduction in starch accumulation and a stimulation of glycolysis. Plant J 15 109–118 [DOI] [PubMed] [Google Scholar]
- Turner WL, Knowles VL, Plaxton WC (2005) Cytosolic pyruvate kinase: subunit composition, activity, and amount in developing castor and soybean seeds, and biochemical characterization of the purified castor seed enzyme. Planta 222 1051–1062 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Fiorani F, Siedow JN (2005) Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol 139 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach AL, Ng VS, Siedow JN (2006) Regulation of plant alternative oxidase activity: a tale of two cysteines. Biochim Biophys Acta 1757 135–142 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN (1993) Covalent and noncovalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiol 103 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, Day DA, Wiskich JT, Vanlerberghe AE, McIntosh L (1995) Alternative oxidase activity in tobacco leaf mitochondria. Plant Physiol 109 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, Yip JYH, Parsons HL (1999) In organelle and in vivo evidence of the importance of the regulatory sulfhydryl/disulfide system and pyruvate for alternative oxidase activity in tobacco. Plant Physiol 121 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Lue W, Uy T, Long J, Wang C, Eimert K, Chen J (1998) Characterization of ADG1, an Arabidopsis locus encoding for ADPG pyrophosphorylase small subunit, demonstrates that the presence of the small subunit is required for large subunit stability. Plant J 13 63–70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.