Abstract
Pollen germination, along with pollen tube growth, is an essential process for the reproduction of flowering plants. The germinating pollen with tip-growth characteristics provides an ideal model system for the study of cell growth and morphogenesis. As an essential step toward a detailed understanding of this important process, the objective of this study was to comprehensively analyze the transcriptome changes during pollen germination and pollen tube growth. Using Affymetrix Arabidopsis (Arabidopsis thaliana) ATH1 Genome Arrays, this study is, to our knowledge, the first to show the changes in the transcriptome from desiccated mature pollen grains to hydrated pollen grains and then to pollen tubes of Arabidopsis. The number of expressed genes, either for total expressed genes or for specifically expressed genes, increased significantly from desiccated mature pollen to hydrated pollen and again to growing pollen tubes, which is consistent with the finding that pollen germination and tube growth were significantly inhibited in vitro by a transcriptional inhibitor. The results of Gene Ontology analyses showed that expression of genes related to cell rescue, transcription, signal transduction, and cellular transport was significantly changed, especially for up-regulation, during pollen germination and tube growth. In particular, genes of the calmodulin/calmodulin-like protein, cation/hydrogen exchanger, and heat shock protein families showed the most significant changes during pollen germination and tube growth. These results demonstrate that the overall transcription of genes, both in the number of expressed genes and in the levels of transcription, was increased. Furthermore, the appearance of many novel transcripts during pollen germination as well as tube growth indicates that these newly expressed genes may function in this complex process.
The primary function of pollen and the pollen tube is to deliver sperms to egg apparatus for double fertilization that is required for sexual reproduction in flowering plants. Pollen germination (PG) and pollen tube growth (PTG) are continuous and highly polarized processes with tip-growth characteristics; thus, they provide an ideal model system for the study of cell growth and morphogenesis in a broader sense (Feijó et al., 2001, 2004).
Using biochemical assays and mutant analysis, there were only a limited number of genes identified as being involving in pollen development, PG, and PTG, as well as in the interaction between pollen and female tissues (Scott et al., 1991; McCormick, 1993, 2004). In order to move toward a comprehensive understanding of molecular regulatory mechanisms for PG and PTG, further analysis has been needed of the potential functions for a number of genes that are expressed in pollen as well as in PG and PTG. Several previous studies have analyzed the transcriptional profiles of Arabidopsis (Arabidopsis thaliana) pollen on a more global scale by application of either Affymetrix GeneChips or serial analysis of gene expression (Becker et al., 2003; Honys and Twell, 2003, 2004; Lee and Lee, 2003; Pina et al., 2005). Affymetrix Arabidopsis 8 K GeneChips (representing about 8,000 genes) and ATH1 Genome Arrays (representing 22,591 genes) were used to compare the transcriptome differences between pollen grains and vegetative tissues (Becker et al., 2003; Honys and Twell, 2003) and to analyze the transcriptional changes during microgametogenesis and reproductive development in Arabidopsis (Hennig et al., 2004; Honys and Twell, 2004). Lee and Lee (2003) used the serial analysis of gene expression methods to profile the transcriptome in pollen under normal and chilling conditions. These studies revealed a high proportion of specifically and differentially expressed genes in pollen transcriptome (for review, see da Costa-Nunes and Grossniklaus, 2003).
Honys and Twell (2004) showed that there was a decline in the number of diverse transcripts accompanied by pollen maturity and an increase in the proportion of male gametophyte-specific transcripts. Pina et al. (2005) reported that the transcriptome of pollen grains was smaller and unique from any other vegetative tissue. Genes in functional categories of signaling, vesicle transport, and cytoskeleton were proportionally overrepresented in the pollen grain transcriptome, suggesting their commitment to PG and PTG (Pina et al., 2005). Although those previous studies drew the outline of the main characteristics of the pollen or male gametophyte transcriptome (Becker and Feijó, 2007; Grennan, 2007), the possible dynamic changes of the pollen transcriptome during PG and PTG remain unclear.
A number of very dynamic cellular events occur during PG and PTG, including calcium oscillation, vesicle transport, ion fluxes, cell wall biosynthesis, cytoskeleton dynamics, et cetera (for review, see Hepler et al., 2001). Thus, one may ask whether there are transcriptional changes during PG and PTG. Although it was previously proposed that the mature pollen grains might already contain most of the transcripts for PG and PTG in some plant species (Mascarenhas, 1989; Guyon et al., 2000), one may speculate that there may be transcriptional changes during the transition from mature pollen to germinating and germinated pollen. A more detailed and comprehensive transcriptome analysis during PG and PTG may reveal additional novel genes functionally involved in these processes. In this study, in vitro PG assays showed that the inhibition of transcription resulted in significant decreases in both PG and PTG rates. By application of Affymetrix Arabidopsis ATH1 Genome Arrays, the transcriptional changes during PG and PTG were further analyzed. Our results demonstrate that the overall transcription was increased along with PG and PTG, suggesting that some newly transcribed and/or transcriptionally changed genes may play roles in the regulation of PG and PTG.
RESULTS
Inhibition of PG and PTG by the Transcription Inhibitor Actinomycin D
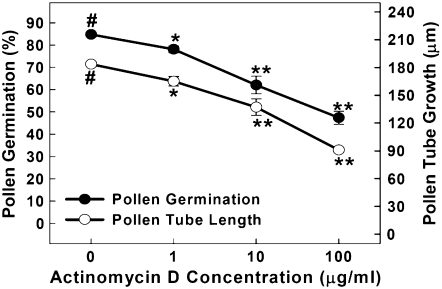
To test whether ongoing transcription plays any role in PG and PTG, the transcription inhibitor actinomycin D (Act D) was used during the in vitro PG assays. The results presented in Figure 1 clearly show that both PG and PTG were inhibited in a dose-dependent manner by the addition of Act D. As shown in Figure 1, treatment with 1 μg mL−1 Act D significantly inhibited both PG and PTG, and this inhibition increased to 44% and 50% for PG and PTG, respectively, when the concentration of Act D was increased to 100 μg mL−1. The inhibitory effects of Act D on PG and PTG shown in this study are similar to those in a previous report by Honys and Twell (2004), except for some differences in concentration dependence and the sensitivity of pollen to Act D (see details in “Discussion”).
Figure 1.
Effects of Act D on Arabidopsis PG and PTG. The experiments were repeated three times, and each treatment in one experiment had four replicates. Each data point is presented as mean ± se (n = 4). #, Data point for the control; *, significantly different from the control at P < 0.05 by Student's t test; **, significantly different from the control at P < 0.01 by Student's t test.
Arabidopsis Pollen Collection and in Vitro PG and PTG Assays in Liquid Medium
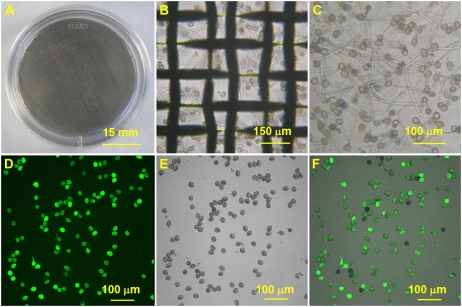
To investigate the changes of the Arabidopsis pollen transcriptome during PG and PTG, pollen samples from three different stages, desiccated mature pollen grains (MP), hydrated pollen grains (HP), and pollen tubes (PT), were collected for RNA extraction. The transitions from MP to HP and from HP to PT were regarded as PG and PTG, respectively. MP was collected from freshly anther-dehisced flowers using a vacuum, as described previously (Johnson-Brousseau and McCormick, 2004). In order to get enough HP and PT samples, a “thin liquid layer” germination method was developed (Fig. 2, A–C; see “Materials and Methods” for details). Using this method, the PG rate was approximately 70% (usually between 70% and 75%), and, more importantly, a large amount of hydrated pollen grains and pollen tubes can be processed at the same time so that sufficient HP and PT could be collected for RNA extraction. The viability of HP was assayed by fluorescein diacetate (FDA) staining. In our experiments, more than 90% of HP grains were positive for FDA staining, indicating the high viability of the cultured pollen grains (Fig. 2, D–F).
Figure 2.
Large-scale in vitro PG in liquid medium and pollen viability assay. A to C show the thin liquid layer method for PG and PTG. A, A 35-mm dish with a steel-wire net (80 μm) in it. The pollen grain suspension with 30 μL of basic medium was spread on the steel-wire net. (B) The pollen tubes under the steel-wire net after 4 h of incubation. (C) The collected pollen tubes after filtering through a 50-μm nylon mesh. D to F show FDA staining of the hydrated pollen grains after incubation in liquid medium for 45 min. D, FDA fluorescence image. E, Bright-field image. F, Merged image of D and E.
Genes Expressed in MP, HP, and PT
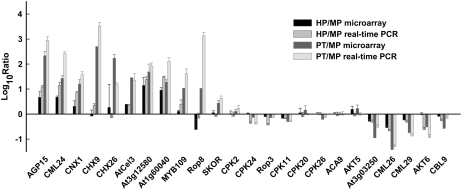
The Arabidopsis ATH1 Genome Array, containing 22,591 probe sets representing 73.6% genome coverage of Arabidopsis (Bock et al., 2006), was used to investigate the dynamics of transcription throughout PG and PTG. The microarrays were hybridized with the fragmented cRNA made from total RNA of MP, HP, and PT, respectively. Two biological replicates derived from independent plant populations were performed to ensure reproducibility and reliability. Raw data sets were normalized and analyzed using GCOS software (see “Materials and Methods” for details). By conducting real-time PCR analyses for 24 selected genes, most of which showed similar expression patterns compared with the ATH1 microarray data, the validity of the microarray data was further confirmed (Fig. 3). The raw data sets (CEL) and the normalized expression data sets have been deposited in the Gene Expression Omnibus (GSE6696) at the National Center for Biotechnology Information (Edgar et al., 2002; www.ncbi.nlm.nih.gov/geo/).
Figure 3.
Confirmation of transcriptional profiles of the selected genes by real-time PCR. HP-MP and PT-MP ratios in microarray data of 24 tested genes were compared with the real-time PCR results. The sequences of the gene-specific primers used for PCR are listed in Supplemental Table S8.
Our microarray data showed that 3,945 genes gave present calls in MP (representing 17.5% of the unigene targets on the ATH1 microarray), 4,637 genes were expressed in HP (20.5%), and 4,892 genes were expressed in PT (21.7%; Table I). In total, 5,640 genes (representing 25.0% of the unigene targets on the ATH1 microarray) were present in at least one of the three stages (MP, HP, and PT). The numbers of detected transcripts are similar to those in previous studies that used the same microarray chips (Honys and Twell, 2004; Pina et al., 2005). Among these genes, 3,440 (15.2%) genes were consistently expressed in all three stages (Table I; Supplemental Table S1) and another 2,200 genes were preferentially expressed in one or two stages. Further analysis of these 2,200 genes showed that 163 genes were specifically expressed in MP (4.1% of the expressed genes in MP), 352 genes in HP (7.6% of the expressed genes in HP), and 731 genes in PT (14.9% of the expressed genes in PT) (Table I).
Table I.
Numbers of expressed genes, specifically expressed genes, and transcriptionally changed genes during PG and PTG
The first column indicates the three stages (MP, HP, and PT) and two processes (PG and PTG). The “Number of Genes” and “Percentage” columns refer to the numbers of expressed genes, specifically expressed genes, and transcriptionally changed genes as well as the relative percentages.
| Gene Category | No. of Genes | Percentage |
|---|---|---|
| Genes expressed in | Percentage in ATH1 array (22,591 genes) | |
| MP | 3,945 | 17.5 |
| HP | 4,637 | 20.5 |
| PT | 4,892 | 21.7 |
| All three stages | 3,440 | 15.2 |
| At least one stage | 5,640 | 25.0 |
| Genes specifically expressed in | Percentage in respective stage | |
| MP | 163 | 4.1 |
| HP | 352 | 7.6 |
| PT | 731 | 14.9 |
| Transcriptionally changed genes | Percentage in total expressed genes (5,640 genes) | |
| Up-regulated in PG | 222 | 3.9 |
| Down-regulated in PG | 104 | 1.8 |
| Up-regulated in PTG | 804 | 14.3 |
| Down-regulated in PTG | 686 | 12.2 |
The genes that showed significant changes in expression levels during PG and PTG were analyzed. The genes that changed by more than 1.6-fold (P < 0.01) in both biological replicates during PG or PTG are listed in Supplemental Table S2 (see “Materials and Methods” for details). During PG, 222 genes were up-regulated, which was much more than were down-regulated (104 genes; Table I; Supplemental Table S2). Compared with PG, the overall change in transcription during PTG was more significant. There were 804 genes that were up-regulated and 686 genes that were down-regulated during PTG (Table I; Supplemental Table S2).
To identify the coregulated genes, all of the detected transcripts (5,640 genes) were further analyzed and distributed into nine clusters (I–IX) according to their expression profiles (Supplemental Fig. S1; Supplemental Table S3). Cluster I contained 704 genes (representing 12.5% of the 5,640 expressed genes) that were exclusively up-regulated during PTG (from HP to PT), while cluster V contained 119 genes (2.1%) that were up-regulated during PG (from MP to HP). Cluster III had 630 genes (11.2%) that were down-regulated during PTG. The expression of genes in cluster II (3,980 genes) was not significantly changed in PG and PTG processes.
Gene Ontology Analysis and Family Analysis of the Transcriptionally Changed Genes during PG and PTG
To evaluate the potential functions of genes with altered transcription levels during PG and PTG, a Gene Ontology (GO) analysis was conducted using the FunCat Scheme version 2.0 web service in the Munich Information Center for Protein Sequences (Ruepp et al., 2004; http://mips.gsf.de/projects/funcat). The genes whose expression levels changed more than 1.6-fold during PG or PTG were analyzed, and they were finally classified into 14 main GO categories, as shown in Figure 4. Functional categories such as cell rescue, transcription, subcellular localization, metabolism, proteins with binding function, and cellular transport were overrepresented during PG (Fig. 4A). During PTG, the up-regulated genes were mainly aggregated into the categories of metabolism, signal transduction, cellular transport, subcellular localization, cell rescue, and protein fate (Fig. 4B).
Figure 4.
Functional category distribution of transcriptionally changed genes in PG and PTG. The FunCat Scheme version 2.0 Web service at the Munich Information Center for Protein Sequences was used to analyze the GO categories of transcriptionally changed genes in PG (A) and PTG (B). The numbers of changed genes in 14 main biological categories are represented.
Pina et al. (2005) sorted the main gene families of Arabidopsis, and these gene families were further grouped according to their functional classes of signaling, transcription, transporter/channel, cell wall, cytoskeleton, vesicle trafficking, cell cycle, and small RNA pathways. To further analyze the potential functions of the genes with altered transcription during PG and PTG, the gene family tables organized by Pina et al. (2005) were used. In addition, in order to focus on transporter/channel-related genes, a comprehensive list of genes encoding membrane proteins from the Arabidopsis genome (Bock et al., 2006) was also applied. The contents of these two tables were integrated and used to identify the important functional categories and gene families that correlated with the processes of PG or PTG. Supplemental Table S4 showed that the categories of cell wall, transcription, and signaling were overrepresented during PG, whereas the overrepresented categories during PTG were transporters, signaling, and cell wall. To further reveal the differences between PG and PTG, the user-driven tool MapMan (Thimm et al., 2004) was applied to display the diagrams of various pathways related to these processes. The results showed that genes in some functional categories or gene families (such as abiotic stress, divalent cation transport, G-proteins, etc.) were significantly changed in their transcription levels during PG or PTG (Supplemental Fig. S2; Supplemental Table S5). The integrated results suggested remarkable functional differences between PG and PTG. The function terms of cell rescue and transcription were overrepresented in PG, and genes in the categories of transport and signaling were preferentially expressed in PTG. Furthermore, genes in the CaM/CML (for calmodulin/calmodulin-like protein), CHX (for cation/hydrogen exchanger), and Hsp (for heat shock protein) families were significantly changed, mainly up-regulated during PG and PTG (Tables II–IV; Supplemental Table S7), which suggested their roles in these processes.
Table II.
Numbers of CaM/CML, CHX, and Hsp genes expressed and transcriptionally changed during PG and PTG
The first column indicates the name of the gene family. The gene number in the column “Genome” represents the total number for each gene family in the Arabidopsis genome, and the number in the column “ATH1” shows the gene number included in the ATH1 chip for each family. The following three columns show the numbers of expressed genes in three different stages (MP, HP, and PT). The four right columns refer to the numbers of transcriptionally up-regulated (Up) and down-regulated (Down) genes during PG and PTG. For more details, see Supplemental Table S6 and S7.
| Gene Family | Genome | ATH1 | MP | HP | PT | Up PG | Down PG | Up PTG | Down PTG |
|---|---|---|---|---|---|---|---|---|---|
| CaM | 7 | 6 | 5 | 5 | 4 | 0 | 0 | 1 | 0 |
| CML | 50 | 48 | 14 | 17 | 19 | 4 | 2 | 11 | 3 |
| CHX | 28 | 25 | 11 | 11 | 17 | 0 | 2 | 9 | 3 |
| Hsp | 219 | 180 | 46 | 73 | 77 | 19 | 0 | 18 | 9 |
Table III.
Transcriptionally up-regulated genes of CML and Hsp families during PG
The first three columns show the gene family name, Affymetrix probe set, and The Arabidopsis Information Resource locus (Arabidopsis Genome Initiative identifier [AGI ID]) assigned to the corresponding probe. The gene annotation is given in the fourth column. For more details, see “Materials and Methods” and Supplemental Table S7.
| Gene Family | Probe Set | AGI ID | Annotation FC P Value |
|---|---|---|---|
| CML | 259866_at | At1g76640 | Calmodulin-related protein, putative (CML39) |
| 259064_at | At3g07490 | Calcium-binding protein, putative (CML3) | |
| 259137_at | At3g10300 | Calcium-binding EF hand family protein (CML49) | |
| 256755_at | At3g25600 | Calmodulin, putative (CML16) | |
| Hsp | 260248_at | At1g74310 | Heat shock protein 101 (HSP101) |
| 250351_at | At5g12030 | 17.7-kD class II heat shock protein 17.6A (HSP17.7-CII) | |
| 252515_at | At3g46230 | 17.4-kD class I heat shock protein (HSP17.4-CI) | |
| 248045_at | At5g56030 | Heat shock protein 81-2 (HSP81-2) | |
| 263150_at | At1g54050 | 17.4-kD class III heat shock protein (HSP17.4-CIII) | |
| 248332_at | At5g52640 | Heat shock protein 81-1 (HSP81-1)/heat shock protein 83 (HSP83) | |
| 266294_at | At2g29500 | 17.6-kD class I small heat shock protein (HSP17.6B-CI) | |
| 260978_at | At1g53540 | 17.6-kD class I small heat shock protein (HSP17.6C-CI; amino acids 1–156) | |
| 247691_at | At5g59720 | 18.1-kD class I heat shock protein (HSP18.1-CI) | |
| 261838_at | At1g16030 | Heat shock protein 70, putative/HSP70, putative | |
| 262911_s_at | At1g59860 | 17.8-kD class I heat shock protein (HSP17.8-CI) | |
| 250296_at | At5g12020 | 17.6-kD class II heat shock protein (HSP17.6-CII) | |
| 263374_at | At2g20560 | DNAJ heat shock family protein | |
| 256999_at | At3g14200 | DNAJ heat shock N-terminal domain-containing protein | |
| 256245_at | At3g12580 | Heat shock protein 70, putative/HSP70, putative | |
| 255811_at | At4g10250 | 22.0-kD ER small heat shock protein (HSP22.0-ER) | |
| 248043_s_at | At5g56000 | Heat shock protein 81-4 (HSP81-4) | |
| 252670_at | At3g44110 | DNAJ heat shock protein, putative (J3) | |
| 258984_at | At3g08970 | DNAJ heat shock N-terminal domain-containing protein |
Table IV.
Transcriptionally up-regulated genes of CML/CaM, CHX, and Hsp families during PTG
See Table III for the definition of each column. For more details, see Supplemental Table S7.
| Gene Family | Probe Set | AGI ID | Annotation FC P Value |
|---|---|---|---|
| CaM and CML | 252713_at | At3g43810 | Calmodulin-7 (CAM7) |
| 249583_at | At5g37770 | Calmodulin-related protein 2, touch-induced (TCH2; CML24) | |
| 259137_at | At3g10300 | Calcium-binding EF hand family protein (CML49) | |
| 254487_at | At4g20780 | Calcium-binding protein, putative (CML42) | |
| 259064_at | At3g07490 | Calcium-binding protein, putative (CML3) | |
| 256755_at | At3g25600 | Calmodulin, putative (CML16) | |
| 252206_at | At3g50360 | Caltractin/centrin (CML20) | |
| 254782_at | At4g12860 | Calcium-binding protein, putative (CML2) | |
| 259866_at | At1g76640 | Calmodulin-related protein, putative (CML39) | |
| 253963_at | At4g26470 | Calcium-binding EF hand family protein (CML21) | |
| 262639_at | At1g62820 | Calmodulin, putative (CML14) | |
| 255772_at | At1g18530 | Calmodulin, putative (CML15) | |
| CHX | 249884_at | At5g22910 | Cation/hydrogen exchanger, putative (CHX9) |
| 246337_at | At3g44910 | Cation/hydrogen exchanger, putative (CHX12) | |
| 266161_at | At2g28170 | Cation/hydrogen exchanger, putative (CHX7) | |
| 249883_at | At5g22900 | Cation/hydrogen exchanger, putative (CHX3) | |
| 246336_at | At3g44900 | Cation/hydrogen exchanger, putative (CHX4) | |
| 263469_at | At2g31910 | Cation/hydrogen exchanger, putative (CHX21) | |
| 251101_at | At5g01680 | Cation/hydrogen exchanger, putative (CHX26) | |
| 258408_at | At3g17630 | Cation/hydrogen exchanger, putative (CHX19) | |
| 263720_at | At2g13620 | Cation/hydrogen exchanger, putative (CHX15) | |
| Hsp | 264865_at | At1g24120 | DNAJ heat shock protein, putative |
| 263754_at | At2g21510 | DNAJ heat shock N-terminal domain-containing protein | |
| 258979_at | At3g09440 | Heat shock cognate 70-kD protein 3 (HSC70-3; HSP70-3) | |
| 256245_at | At3g12580 | Heat shock protein 70, putative/HSP70, putative | |
| 262911_s_at | At1g59860 | 17.8-kD class I heat shock protein (HSP17.8-CI) | |
| 248043_s_at | At5g56000 | Heat shock protein 81-4 (HSP81-4) | |
| 250351_at | At5g12030 | 17.7-kD class II heat shock protein 17.6A (HSP17.7-CII) | |
| 263150_at | At1g54050 | 17.4-kD class III heat shock protein (HSP17.4-CIII) | |
| 261838_at | At1g16030 | Heat shock protein 70, putative/HSP70, putative | |
| 248616_at | At5g49580 | DNAJ heat shock N-terminal domain-containing protein | |
| 250296_at | At5g12020 | 17.6-kD class II heat shock protein (HSP17.6-CII) | |
| 266294_at | At2g29500 | 17.6-kD class I small heat shock protein (HSP17.6B-CI) | |
| 260978_at | At1g53540 | 17.6-kD class I small heat shock protein (HSP17.6C-CI; amino acids 1–156) | |
| 261458_at | At1g21080 | DNAJ heat shock N-terminal domain-containing protein | |
| 249148_at | At5g43260 | Chaperone protein DNAJ-related | |
| 250934_at | At5g03030 | DNAJ heat shock N-terminal domain-containing protein | |
| 266858_at | At2g26890 | DNAJ heat shock N-terminal domain-containing protein | |
| 255811_at | At4g10250 | 22.0-kD ER small heat shock protein (HSP22.0-ER) |
Members of the CaM/CML, CHX, and Hsp Gene Families Functioned during PG and PTG
The Arabidopsis genome contains seven CaM genes and 50 additional CML genes (McCormack and Braam, 2003; McCormack et al., 2005). There are probe sets for six CaM genes and 48 CML genes included in the Arabidopsis ATH1 Genome Array (Table II), and five CaM genes and 19 CML genes were detected during PG and PTG in our experiments (Supplemental Table S6). As shown in Tables III and IV and Supplemental Table S7, some CaM and CML genes significantly increased in transcription levels during PG or PTG. Among these genes, four CML genes (CML39, CML49, CML3, and CML16) were up-regulated during PG and 12 genes (CaM7, CML49, CML3, CML16, etc.) increased their transcription during PTG (Tables II–IV; Supplemental Table S7). It should be noted that some other CaM/CML genes (CaM2, CaM3, CML6, CML13, CML25, CML28, etc.) were highly expressed during PG and PTG (Supplemental Table S6).
Along with the related KEA subfamily genes, the CHX family was predicted to comprise the CPA2 (for cation/proton antiporter) family (Maser et al., 2001). The Arabidopsis genome contains 28 CHX genes (AtCHX1–AtCHX28; Sze et al., 2004), and 25 CHX genes are included in the Affymetrix ATH1 array (Table II). Our data showed that 18 AtCHX genes were detected during PG and PTG, and the numbers of AtCHX genes expressed at the three stages (MP, HP, and PT) were 11, 11, and 17, respectively (Table II; Supplemental Table S6). Seven members of this family were specifically expressed in PT (Supplemental Table S6). It should be mentioned that the transcriptional levels of nine CHX genes were specifically or preferentially up-regulated during PTG (Tables II and IV; Supplemental Table S7), but no CHX genes were up-regulated during PG.
The Affymetrix Arabidopsis ATH1 Genome Array contains probe sets for 180 of 219 genes of Hsps and Hsp-related proteins in the Arabidopsis genome (Thimm et al., 2004; Table II). Our microarray data showed that the transcription of 85 genes of this family was detectable during PG and PTG (Supplemental Table S6). The number of expressed Hsp genes increased from MP (46 genes) to HP (73 genes) and PT (77 genes; Table II). There were 19 and 18 genes up-regulated in PG and PTG, respectively, and 10 of them overlapped (Tables II–IV; Supplemental Table S7). It should be emphasized that many genes dramatically increased their transcription during PG, and the transcriptional changes of some genes were even over 50-fold (Table III; Supplemental Table S7). On the other hand, there was no Hsp gene down-regulated in this process.
DISCUSSION
Transcriptional Effects on PG and PTG in Arabidopsis
Honys and Twell (2004) previously reported that Act D had an inhibitory effect on both PG and PTG in Arabidopsis (ecotype Landsberg erecta) and that the inhibition could reach 20% to 30% at concentrations between 0.1 and 1 μg mL−1 Act D. Using a different ecotype (Columbia) of Arabidopsis, we found similar inhibitory effects of Act D on PG and PTG. We observed a significant dose-dependent inhibition of PG and PTG by Act D when its concentration was increased from 1 to 100 μg mL−1 (Fig. 1). The differences in concentration dependence and sensitivity of PG and PTG to Act D in these two independent experiments may have resulted from different ecotypes of Arabidopsis, different media, or different methods used in PG assays. Honys and Twell (2004) used liquid medium (Hodgkin, 1983; Derksen et al., 2002), in which two ecotypes of Arabidopsis pollen showed different and relatively low germination rates, ranging from 64% (Landsberg erecta) to 76% (Columbia; Lalanne et al., 2004). Using the modified solid medium and methods (see “Materials and Methods” for details), we usually observed an approximately 85% germination rate for Arabidopsis (Columbia) pollen in our experiments (Fig. 1). The different germination rates in these two independent experiments may result in different sensitivities of PG and PTG to Act D.
Ylstra and McCormick (1999) showed that Act D inhibited transcription in developing tobacco (Nicotiana tabacum) pollen in a concentration-dependent manner, with a concentration range from 0 to 200 μg mL−1, and 98.6% of de novo RNA synthesis was inhibited by 100 μg mL−1 Act D with no decrease in pollen viability. In our in vitro PG assays, we observed that 100 μg mL−1 Act D inhibited PG and PTG of Arabidopsis by 44% and 50%, respectively (Fig. 1). Meanwhile, the viability of pollen tubes was not remarkably decreased. Pollen tubes treated with 100 μg mL−1 Act D still kept growing slowly, although their growth rates decreased dramatically. The length of pollen tubes reached 85 ± 2 μm at 4.5 h and 113 ± 1 μm at 6 h when the concentration of Act D was 100 μg mL−1. All of these findings indicated that the effects of Act D on PG and PTG resulted from the inhibition of transcription but not from the decrease in pollen viability. It is suggested that transcription, especially of newly transcribed genes and/or the transcriptionally increased genes, is potentially important for the regulation of PG and PTG in Arabidopsis.
A New Method for the Collection of Germinating Pollen and Pollen Tubes for RNA Extraction
Although Pereira et al. (2005) reported a method to obtain RNA samples from Arabidopsis pollen tubes for reverse transcription PCR analysis, the amount of RNA was not sufficient for a microarray experiment. We developed a thin liquid layer in vitro PG method in this study, by which Arabidopsis pollen grains can germinate well in vitro (germination rate can reach 70% or higher) and a large amount of germinated pollen can be collected for microarray experiments. Recently, Boavida and McCormick (2007) also optimized a liquid medium and developed a method to isolate large amounts of Arabidopsis pollen tubes. Both methods may further facilitate future pollen transcriptomic and proteomic analyses.
A pollen grain with the emergence of a pollen tube may be considered to be entering the germination process. Given that transcriptional change is an extremely rapid process, it should appear before the observation of any morphological changes. In our experiment, we collected pollen grains at 45 min after they were incubated in liquid medium as the HP sample. At this time, about 15% to 20% of pollen grains showed the emergence of pollen tubes (Fig. 2, D–F). Although most of the pollen grains did not exhibit significant morphological changes (the emergence of a pollen tube) at this stage, it is reasonable to speculate that those hydrated pollen grains may have initiated germination-related gene transcription. Taking this into account, the RNA extracted at this stage should contain and represent the transcriptome information for PG.
Increased Transcription during PG and PTG
Our data showed that the total number of transcribed genes gradually increased from MP to HP to PT (Table I), suggesting that newly initiated transcription occurs during PG and PTG. This result is consistent with the observation that PG and PTG were inhibited by the transcription inhibitor Act D (Fig. 1). Honys and Twell (2004) reported a dramatic decrease in the number of transcripts expressed during microgametogenesis in Arabidopsis, which may be associated with reduced pollen activity during pollen maturation. Subsequently, the desiccated mature pollen becomes “silent” or “resting” at the end of microgametogenesis. However, some unique or male gametophyte-specific transcripts were selectively activated during pollen maturation (Honys and Twell, 2004). In particular, a set of de novo synthesized transcription factors emerged at the late stage of microgametogenesis, suggesting ongoing transcription after pollen maturation (Honys and Twell, 2004). In this study, microarray data clearly showed that a large number of genes initiated transcription during PG and/or PTG (Table I). It is assumed that potential functions of these genes may be to prepare essential materials and energy for PTG and to increase the physiological and biochemical activities of germinating pollen and pollen tubes. In addition, a specific analysis showed that the proportion of specifically expressed genes was remarkably increased from MP to HP to PT (Table I), which suggested that PTG may require many cellular components or regulatory factors for its dynamic activity.
Besides newly transcribed genes, many other genes already present in MP significantly increased their transcriptional levels during PG or PTG. Analysis of these transcriptionally changed genes (changed by more than 1.6-fold) demonstrated that up-regulated genes were dominant during both PG and PTG compared with down-regulated genes (Table I). These results suggest that the newly transcribed genes as well as the up-regulated genes might play important roles in PG or PTG.
Genes That May Be Functionally Involved in PG and PTG
Because overall transcription was significantly increased in this study, further analyses were concentrated on the functional distribution of up-regulated genes. The results derived from both FunCat 2.0 and MapMan analyses showed that the cell rescue-related (or stress/abiotic/heat-related) genes, mainly Hsps, were overrepresented among the up-regulated genes during PG (Fig. 4A; Supplemental Fig. S2; Supplemental Table S5). Meanwhile, many genes in the transcription category (transcription factors or transcription-related regulators) were also markedly up-regulated during PG (Fig. 4A; Supplemental Tables S4 and S5). The genes in these two categories (cell rescue and transcription) were assumed to sense or raise germination signals, activate pollen from the resting (silent) stage, and mobilize materials and energy for PG.
In contrast to PG, PTG had a distinct gene function distribution. Signaling-related genes and transporter genes were dominantly transcribed and/or up-regulated during PTG (Fig. 4B; Supplemental Tables S4 and S5). These findings are consistent with previous reports that genes in these two categories are involved in the regulation of polarized tip growth of pollen tubes (for review, see Hepler et al., 2001). It has been demonstrated that growing pollen tubes are regulated under a complex signal cross talk for polarized growth, and plenty of transporters supply sufficient materials for the rapid growth of pollen tubes (Hepler et al., 2001; Holdaway-Clarke and Hepler, 2003).
Members of several gene families (CaM/CML, CHX, and Hsp) from different functional categories (signaling, transport, and cell rescue) seem to be involved in the regulation of PG and/or PTG (Tables II–IV; Supplemental Tables S6 and S7). It is well accepted that cytosolic free calcium is a key messenger in the regulation of PG and PTG (for review, see Steer and Steer, 1989; Taylor and Hepler, 1997; Franklin-Tong, 1999). Ca2+-dependent modulation of cellular activities occurs via intracellular Ca2+-binding proteins or known Ca2+ sensors, such as CaMs and CMLs. Previous studies showed that some CaMs or CaM-related proteins had similar localization with cytosol-free calcium in growing pollen tubes (Tirlapur et al., 1994; Rato et al., 2004) and had essential roles in the regulation of PG (Golovkin and Reddy, 2003). Our results showed that some CaMs or CMLs were up-regulated or highly transcribed during PG or PTG, suggesting their potential roles in Ca2+ signaling pathways for the regulation of PG and/or PTG in Arabidopsis. Further functional characterization of these CaM/CML genes may increase our understanding of the Ca2+ signaling mechanism during PG and PTG.
Previous studies have shown that H+ (Feijó et al., 1999, 2001) and K+ (Feijó et al., 1995; Fan and Wu, 2000; Fan et al., 2001) fluxes across the pollen or pollen tube plasma membrane are critical for PTG and/or PG, but the molecular identification of transporters mediating the H+ and K+ fluxes in pollen and/or pollen tubes remains unclear. Sze et al. (2004) demonstrated that there were 18 genes of the CHX family that were specifically or preferentially expressed in the male gametophyte. In addition, by analyzing promoter∷GUS activities of AtCHXs, they suggested that some AtCHXs may be functionally involved in osmotic adjustment and K+ homeostasis during mature pollen desiccation and rehydration (Sze et al., 2004; Bock et al., 2006). Our data show that seven AtCHX genes were specifically expressed in PT during PG and PTG, and nine genes were preferentially up-regulated during PTG (Table IV; Supplemental Tables S6 and S7). Further attention should be given to analysis of these AtCHXs for their possible involvement in regulating the local pH gradient immediately adjacent to the PM of pollen tubes, in adjusting ion homeostasis, as well as in maintaining turgor and membrane potential of pollen tubes (for review, see Holdaway-Clarke and Hepler, 2003).
Hsps belong to a large family that has been classified into several groups, including Hsp60, Hsp70, Hsp90, Hsp100, and the small Hsps (for review, see Vierling, 1991; Winter and Sinibaldi, 1991; Miernyk, 1999). It is well known that Hsps are significantly induced when cells or organisms are exposed to supraoptimal temperatures or other stresses. Although some previous studies failed to detect heat shock responses in microspores or mature pollen in several species of plants, such as maize (Zea mays; Dupuis and Dumas, 1990; Young et al., 2001), rape (Brassica napus; Young et al., 2004), and tobacco (Volkov et al., 2005), other studies have shown that many Hsps are expressed in microspores and mature pollen even at normal temperature in maize (Magnard et al., 1996), Arabidopsis (Haralampidis et al., 2002), and tobacco (Volkov et al., 2005). The data presented in this study showed that the transcription of many Hsps and related genes was significantly changed (mainly up-regulated) during PG and/or PTG, although most of them had undetectable levels of transcription in desiccated mature pollen grains (Tables III and IV; Supplemental Tables S6 and S7). It is further proposed that these Hsps may function as molecular chaperones (for review, see Miernyk, 1999) involved in the regulation of protein modification (such as protein folding) during PG and PTG, which is associated with the rapid protein synthesis and high physiological activity in germinating pollen and pollen tubes.
In addition to the genes that showed significant transcriptional changes during PG and PTG, as discussed above, some other genes were constitutively transcribed at constant and high levels during PG and PTG, such as ROP1 (Li et al., 1999; Gu et al., 2003), ACA9 (Schiott et al., 2004), and VGD1 (Jiang et al., 2005). These genes have all been reported to play essential roles in the regulation of PG and PTG, suggesting that the constantly and highly transcribed genes are also important for the regulation of PG and PTG.
CONCLUSION
The data presented in this study provide, to our knowledge for the first time, a genome-wide view of the dynamic changes in the transcriptome during the transition from mature pollen grains to germinating pollen and to growing pollen tubes in Arabidopsis. Overall, transcription is increased during PG and PTG in terms of both the total number of transcribed genes and the transcriptional levels of some genes. A number of genes were significantly up-regulated, and, more importantly, some new groups of genes were preferentially transcribed during PG and/or PTG. The GO analysis of these up-regulated genes revealed a function skew toward cell rescue, transcription, signal transduction, and cellular transport during PG and PTG. Most of the functional categories correlate well with the known cellular dynamic activities in germinating pollen and/or growing pollen tubes. The members of the CaM/CML, CHX, and Hsp gene families, whose transcription was remarkably regulated during PG and PTG, were further analyzed and discussed in detail. This work provides a comprehensive transcriptional overview toward the further understanding of molecular networks for the regulation of PG and PTG.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana ecotype Columbia) plants were grown in mixed soil in a growth chamber. The light intensity was 120 to 150 μmol m−2 s−1 for a 16-h daily light period, and day and night temperatures were 22°C ± 2°C and 18°C ± 2°C, respectively. Plants were watered once every 5 d with tap water, and the relative humidity in the growth chamber was kept near 70%.
In Vitro PG Assay
In vitro Arabidopsis PG experiments were conducted as described previously (Fan et al., 2001), except that the basic medium was slightly modified. Briefly, the basic medium was composed of 1 mm KCl, 5 mm CaCl2, 0.8 mm MgSO4, 1.5 mm boric acid, 1% (w/v) agarose, 19.8% (w/v) Suc, 0.05% (w/v) lactalbumin hydrolysate, 10 μm myoinositol, and 5 mm MES, and the pH was adjusted to 5.8 with Tris. The heated solution (100°C, 2 min) was poured into small petri dishes (1.5 mL per dish; dish diameter was 35 mm) and cooled down to form a medium layer. For assays with the addition of Act D, the stock solution of Act D was added after the temperature of the medium dropped to below 60°C. The dehisced anthers were carefully dipped onto the surface of the medium to make pollen grains stick. The dishes were incubated for 4.5 h in a climate chamber (25°C ± 0.2°C, 100% relative humidity), frozen at −20°C for 3 min to quickly terminate the PG and PTG, and kept on ice for counting the PG rate and measuring pollen tube length. Pollen grains with emerging tubes equivalent to or longer than their diameters were considered germinated. An approximately 85% germination rate for Arabidopsis (Columbia) pollen was usually observed (Fig. 1). All of the experiments were repeated three times, and four replicates (dishes) were carried out for each treatment in every experiment. For each replicate or each dish, there were more than 400 pollen grains counted in order to calculate the PG rate, and approximately 80 pollen tubes were measured for length.
Preparation and Collection of Pollen and Pollen Tube Material for RNA Extraction
The desiccated mature pollen grains were collected from freshly anther-dehisced flowers using a vacuum as described previously (Johnson-Brousseau and McCormick, 2004) and then transferred directly into liquid nitrogen for triturating to prevent pollen hydration. After this, the triturated pollen samples were transferred to extraction buffer for total RNA extraction. In order to get enough hydrated pollen grains and especially pollen tubes, the thin liquid layer germination method was developed (Fig. 2, A–C). First, the freshly anther-dehisced flowers were collected into 1.5-mL microfuge tubes filled with 800 μL of basic medium (the contents of this medium are the same as the basic medium for in vitro PG assays described above, and the concentration of Suc is modified to 15% [w/v]). The microfuge tubes were strongly vortexed for 2 min to release the mature pollen grains into the medium. The mixture was then transferred into new microfuge tubes and centrifuged at 11,000 rpm for 1 min. The pollen pellet was resuspended in 30 μL of the basic medium and subsequently cultured in petri dishes. We noticed that making a well-spread thin layer of pollen grains is a key point in getting high rates (70%−75%) of PG. In order to well spread a tiny drop of pollen grains on the petri dish (35 mm diameter) surface, a steel-wire net (∅80 μm) was placed (Fig. 2B) in the petri dish. The covered petri dishes were transferred into a chamber and incubated at 25°C for 45 min, and then the hydrated pollen grains were collected for total RNA extraction. For the collection of pollen tube samples, the pollen grains were incubated for 4 h, which resulted in an average pollen tube length of about 150 μm. After removing the steel-wire net, the culture mixture was collected and washed using the basic medium (19.8% [w/v] Suc). The pollen tubes were filtered using a 50-μm nylon mesh to remove the ungerminated pollen grains. The pollen tube samples were collected from the nylon mesh for total RNA extraction.
For the FDA staining assay, pollen grains were incubated for 45 min in the basic medium with the addition of 2 μg mL−1 FDA. After washing out FDA with basic medium, the fluorescence of pollen grains was observed on a Zeiss LSM 510 META confocal microscope. The stock solution of FDA (5 mg mL−1) was made using acetone.
RNA Extraction, Target Preparation, and Hybridization to Affymetrix GeneChips
The collected pollen grains or pollen tubes were transferred to liquid nitrogen for trituration. The triturated samples were transferred to TRIzol reagent (Invitrogen) for extraction of total RNA. RNA concentration, purity, and integrity were determined and confirmed using an Eppendorf Biophotometer and electrophoresis. The total RNA was further purified using the RNeasy Mini Kit (Qiagen). The purified RNA (8 μg) was used to generate first-strand cDNA in a reverse transcription reaction (One-Cycle Target Labeling and Control Reagents; Affymetrix). After second-strand synthesis, the double-stranded cDNAs were used to generate cRNA via an in vitro transcriptional reaction. The cRNA was labeled with biotin, and 20 μg of the labeled cRNA was fragmented. The size distribution of the cRNAs and fragmented cRNAs was assessed using an Eppendorf Biophotometer and electrophoresis. Fragmented cRNA (15 μg) was added to 300 μL of hybridization solution, and 200 μL of this mixture was used for hybridization on Arabidopsis ATH1 Genome Arrays for 16 h at 45°C. The standard wash and double-stain protocols (EukGE-WS2v5-450) were applied using an Affymetrix GeneChip Fluidics Station 450. The arrays were scanned on an Affymetrix GeneChip scanner 3000.
GeneChip Data Analysis
To ensure reproducibility and reliability, two biological replicates were conducted and the RNAs that were used for microarray experiments were extracted from two independent plant populations. The scanned arrays were analyzed first with Affymetrix GCOS 1.0 (MAS 5.0) software to generate detection calls. All six data sets (three different samples from two biological replicates) were normalized using Affymetrix GCOS software, and the TGT value was set to 100. When analyzing the transcriptionally changed genes, the signal ratio between two stages was calculated to represent the fold change of this gene for its transcription in the corresponding process, and the fold change P value for each gene was obtained at the same time using GCOS software.
A gene was considered to be expressed when it had a detection call “P” in both replicates. A gene that had detection call P only in one stage (MP, HP, or PT) was defined as a specifically expressed gene in this stage. When determining the transcriptionally changed genes, the exclusive approach was used. A gene was considered to be transcriptionally changed when it was up-regulated or down-regulated more than 1.6-fold in both replicates (FC_rep1 > 1.6 and FC_rep2 > 1.6 or FC_rep1 < −1.6 and FC_rep2 < −1.6). A negative or positive value represents downward or upward regulation, respectively. Meanwhile, the fold change P values of this gene in two replicates must both be less than 0.01 (p-value_rep1 < 0.01 and p-value_rep2 < 0.01).
The FunCat Scheme version 2.0 Web service (http://mips.gsf.de/projects/funcat) and MapMan software were used to analyze the functional categories of genes with altered expression levels and the differences between the PG and PTG transcriptomes.
Microsoft Excel and Access were used to extract and manage the microarray data. The annotations of the 22,746 genes represented on the Arabidopsis ATH1 Genome Array were downloaded from NetAffx (http://www.affymetix.com/analysis/index.affx).
Real-Time PCR Analysis
Total RNA extraction and purification were performed as described above. The RNA extract was treated with DNase I (Takara Bio) using standard protocols to eliminate genomic DNA. RNAs were reverse transcribed with SuperScript II RNase H Reverse Transcriptase (Invitrogen) using Random Hexamer Primer (Promega). The cDNA samples were diluted to 8 ng μL−1. Triplicate quantitative assays were performed on 1 μL of each cDNA dilution using the Power SYBR Green PCR Master Mix (Applied Biosystems; P/N 4368577) with the Opticon II system according to the manufacturer's protocols (MJ Research). The gene-specific primers (Supplemental Table S5) were designed using Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi ). The amplification of 18S rRNA was used as an internal control to normalize the data.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Cluster analysis of the expressed genes during PG and PTG.
Supplemental Figure S2. MapMan analysis of the difference in transcription between PG and PTG.
Supplemental Table S1. Genes with detectable expression during PG and PTG.
Supplemental Table S2. Genes with altered transcription (up-regulated or down-regulated more than 1.6-fold) during PG and PTG.
Supplemental Table S3. Cluster analysis results of the expressed genes during PG and PTG.
Supplemental Table S4. Results of functional category and gene family analyses of genes with altered expression levels during PG and PTG.
Supplemental Table S5. MapMan analysis of genes with altered expression levels during PG and PTG.
Supplemental Table S6. Expressed genes in the CaM/CML, CHX, and Hsp gene families.
Supplemental Table S7. Transcriptionally up-regulated genes of the CaM/CML, CHX, and Hsp families during PG and PTG.
Supplemental Table S8. Sequences of the specific primers for the genes tested in real-time PCR experiments.
Supplementary Material
Acknowledgments
We thank Dr. Wenying Xu (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for helpful discussions and suggestions for the experimental protocols and data analysis as well as for technical assistance. We thank Dr. Mark Stitt (Max-Planck-Institut für Molekulare Pflanzenphysiologie) for providing us with the functional catalogue lists of the AHT1 chip. We also thank Dr. Yajun Wu (Department of Plants, Soils, and Biometeorology, Utah State University) and Dr. Andrey Pirozhenko (Invitrogen Bioinformatics) for critical reading of the manuscript.
This work was supported by a competitive research grant for creative research groups sponsored by the National Science Foundation of China (grant no. 30421002).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Wei-Hua Wu (wuwh@public3.bta.net.cn).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA (2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JD, Feijó JA (2007) How many genes are needed to make a pollen tube? Lessons from transcriptomics. Ann Bot (Lond) 100 1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boavida LC, McCormick S (2007) Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J 52 570–582 [DOI] [PubMed] [Google Scholar]
- Bock KW, Honys D, Ward JM, Padmanaban S, Nawrocki EP, Hirschi KD, Twell D, Sze H (2006) Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiol 140 1151–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa-Nunes JA, Grossniklaus U (2003) Unveiling the gene-expression profile of pollen. Genome Biol 5 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen J, Knuiman B, Hoedemaekers K, Guyon A, Bonhomme S, Pierson ES (2002) Growth and cellular organization of Arabidopsis pollen tubes in vitro. Sex Plant Reprod 15 133–139 [Google Scholar]
- Dupuis I, Dumas C (1990) Influence of temperature stress on in vitro fertilization and heat shock protein synthesis in maize (Zea mays L.) reproductive tissues. Plant Physiol 94 665–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan LM, Wang YF, Wang H, Wu WH (2001) In vitro Arabidopsis pollen germination and characterization of the inward potassium currents in Arabidopsis pollen grain protoplasts. J Exp Bot 52 1603–1614 [PubMed] [Google Scholar]
- Fan LM, Wu WH (2000) External pH regulates the inward K+ channels in the plasma membranes of Brassica pollen protoplasts. Prog Nat Sci 10 68–73 [Google Scholar]
- Feijó JA, Costa SS, Prado AM, Becker JD, Certal AC (2004) Signalling by tips. Curr Opin Plant Biol 7 589–598 [DOI] [PubMed] [Google Scholar]
- Feijó JA, Malhó R, Obermeyer G (1995) Ion dynamics and its possible role during in vitro pollen germination and tube growth. Protoplasma 187 155–167 [Google Scholar]
- Feijó JA, Sainhas J, Hackett GR, Kunkel JG, Heper PK (1999) Growing pollen tubes possess a constitutive alkaline band in the clear zone and a growth-dependent acidic tip. J Cell Biol 144 483–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijó JA, Sainhas J, Holdaway-Clarke TL, Cordeiro MS, Kunkel JG, Hepler PK (2001) Cellular oscillations and the regulation of growth: the pollen tube paradigm. Bioessays 23 86–94 [DOI] [PubMed] [Google Scholar]
- Franklin-Tong VE (1999) Signaling and the modulation of pollen tube growth. Plant Cell 11 727–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin M, Reddy AS (2003) A calmodulin-binding protein from Arabidopsis has an essential role in pollen germination. Proc Natl Acad Sci USA 100 10558–10563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grennan AK (2007) An analysis of the Arabidopsis pollen transcriptome. Plant Physiol 145 3–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Vernoud V, Fu Y, Yang Z (2003) ROP GTPase regulation of pollen tube growth through the dynamics of tip-localized F-actin. J Exp Bot 54 93–101 [DOI] [PubMed] [Google Scholar]
- Guyon VN, Astwood JD, Garner EC, Dunker AK, Taylor LP (2000) Isolation and characterization of cDNAs expressed in the early stages of flavonol-induced pollen germination in petunia. Plant Physiol 123 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralampidis K, Milioni D, Rigas S, Hatzopoulos P (2002) Combinatorial interaction of cis elements specifies the expression of the Arabidopsis AtHsp90-1 gene. Plant Physiol 129 1138–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Gruissem W, Grossniklaus U, Kohler C (2004) Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol 135 1765–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK, Vidali L, Cheung AY (2001) Polarized cell growth in higher plants. Annu Rev Cell Dev Biol 17 159–187 [DOI] [PubMed] [Google Scholar]
- Hodgkin T (1983) A medium for germinating Brassica pollen in vitro. Cruciferae Newsletter 8 62–63 [Google Scholar]
- Holdaway-Clarke TL, Hepler PK (2003) Control of pollen tube growth: role of ion gradients and fluxes. New Phytol 159 539–563 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5 R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Yang SL, Xie LF, Puah CS, Zhang XQ, Yang WC, Sundaresan V, Ye D (2005) VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Brousseau SA, McCormick S (2004) A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J 39 761–775 [DOI] [PubMed] [Google Scholar]
- Lalanne E, Honys D, Johnson A, Borner GHH, Lilley KS, Dupree P, Grossniklaus U, Twell D (2004) SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Lee DH (2003) Use of serial analysis of gene expression technology to reveal changes in gene expression in Arabidopsis pollen undergoing cold stress. Plant Physiol 132 517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lin Y, Heath RM, Zhu MX, Yang Z (1999) Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11 1731–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnard JL, Vergne P, Dumas C (1996) Complexity and genetic variability of heat-shock protein expression in isolated maize microspores. Plant Physiol 111 1085–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas JP (1989) The male gametophyte of flowering plants. Plant Cell 1 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJM, Sanders D, et al (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126 1646–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack E, Braam J (2003) Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol 159 585–598 [DOI] [PubMed] [Google Scholar]
- McCormack E, Tsai YC, Braam J (2005) Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci 10 383–389 [DOI] [PubMed] [Google Scholar]
- McCormick S (1993) Male gametophyte development. Plant Cell 5 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S (2004) Control of male gametophyte development. Plant Cell 16 S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk JA (1999) Protein folding in the plant cell. Plant Physiol 121 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira LG, Coimbra S, Oliveira H, Monteiro L, Sottomayor M (2005) Expression of arabinogalactan protein genes in pollen tubes of Arabidopsis thaliana. Planta 223 374–80 [DOI] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijó JA, Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rato C, Monteiro D, Hepler PK, Malho R (2004) Calmodulin activity and cAMP signalling modulate growth and apical secretion in pollen tubes. Plant J 38 887–897 [DOI] [PubMed] [Google Scholar]
- Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, Munsterkotter M, et al (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res 32 5539–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiott M, Romanowsky SM, Baekgaard L, Jakobsen MK, Palmgren MG, Harper JF (2004) A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc Natl Acad Sci USA 101 9502–9507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R, Hodge R, Paul W, Draper J (1991) The molecular biology of anther differentiation. Plant Sci 80 167–191 [Google Scholar]
- Steer MW, Steer JM (1989) Pollen tube tip growth. New Phytol 111 323–358 [DOI] [PubMed] [Google Scholar]
- Sze H, Padmanaban S, Cellier F, Honys D, Cheng NH, Bock KW, Conejero G, Li X, Twell D, Ward JM, et al (2004) Expression patterns of a novel AtCHX gene family highlight potential roles in osmotic adjustment and K+ homeostasis in pollen development. Plant Physiol 136 2532–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LP, Hepler PK (1997) Pollen germination and tube growth. Annu Rev Plant Physiol Plant Mol Biol 48 461–491 [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–939 [DOI] [PubMed] [Google Scholar]
- Tirlapur UK, Scali M, Moscatelli A, Del Casino C, Cai G, Tiezzi A, Cresti M (1994) Confocal image analysis of spatial variations in immunocytochemically identified calmodulin during pollen hydration, germination and pollen tube tip growth in Nicotiana tabacum L. Zygote 2 63–68 [DOI] [PubMed] [Google Scholar]
- Vierling E (1991) The heat shock response in plants. Annu Rev Plant Physiol Plant Mol Biol 42 579–620 [Google Scholar]
- Volkov RA, Panchuk II, Schoffl F (2005) Small heat shock proteins are differentially regulated during pollen development and following heat stress in tobacco. Plant Mol Biol 57 487–502 [DOI] [PubMed] [Google Scholar]
- Winter J, Sinibaldi R (1991) The expression of heat shock protein and cognate genes during plant development. In L Nover, ed, Results and Problems in Cell Differentiation: Heat Shock and Development, Vol 17. Hightower, Springer-Verlag, Berlin, pp 85–105 [DOI] [PubMed]
- Ylstra B, McCormick S (1999) Analysis of mRNA stabilities during pollen development and in BY2 cells. Plant J 20 101–108 [DOI] [PubMed] [Google Scholar]
- Young LW, Wilen RW, Bonham-Smith PC (2004) High temperature stress of Brassica napus during flowering reduces micro- and megagametophyte fertility, induces fruit abortion, and disrupts seed production. J Exp Bot 55 485–495 [DOI] [PubMed] [Google Scholar]
- Young TE, Ling J, Geisler-Lee CJ, Tanguay RL, Caldwell C, Gallie DR (2001) Developmental and thermal regulation of the maize heat shock protein, HSP101. Plant Physiol 127 777–791 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.