Abstract
Loss-of-function mutations in the Arabidopsis (Arabidopsis thaliana) ENHANCED DISEASE RESISTANCE1 (EDR1) gene confer enhanced resistance to infection by powdery mildew (Golovinomyces cichoracearum). EDR1 encodes a protein kinase, but its substrates and the pathways regulated by EDR1 are unknown. To identify components of the EDR1 signal transduction pathway(s), we conducted a forward genetic screen for mutations that suppressed edr1-mediated disease resistance. Genetic mapping and cloning of one of these suppressor mutations revealed a recessive missense mutation in the KEEP ON GOING gene (KEG; At5g13530), which we designated keg-4. KEG encodes a multidomain protein that includes a RING E3 ligase domain, a kinase domain, ankyrin repeats, and HERC2-like repeats. The KEG protein has previously been shown to have ubiquitin ligase activity and to negatively regulate protein levels of the transcription factor ABCISIC ACID INSENSITIVE5. KEG mRNA levels were found to be 3-fold higher in edr1 mutant plants compared to wild type. Loss-of-function mutations in KEG are seedling lethal and are hypersensitive to glucose and abscisic acid (ABA). The keg-4 mutation, in contrast, conferred resistance to 6% glucose and suppressed edr1-mediated hypersensitivity to ABA, suggesting that the keg-4 mutation suppresses ABA signaling by altering KEG function. Several ABA-responsive genes were found to be further up-regulated in the edr1 mutant following ABA treatment, and this up-regulation was suppressed by the keg-4 mutation. We conclude that edr1-mediated resistance to powdery mildew is mediated, in part, by enhanced ABA signaling.
Powdery mildew fungi are obligate biotrophic pathogens that can grow only on living plant tissues. These pathogens must evade or suppress host defenses until their life cycle is complete. A number of Arabidopsis (Arabidopsis thaliana) mutants displaying enhanced disease resistance to powdery mildew (in this case, Golovinomyces cichoracearum) have been characterized (Frye and Innes, 1998; Vogel and Somerville, 2000; Vogel et al., 2002; Tang et al., 2005a, 2006). These mutants can be grouped into two broad classes based on the presence or absence of mildew-induced lesions. The enhanced disease resistance1 (edr1), edr2, and edr3 mutants typify the former class (Frye and Innes, 1998; Tang et al., 2005a, 2006). In these mutants, fungal growth is inhibited at a very late stage of the infection process and resistance correlates with a more rapid activation of host defenses relative to wild-type plants, including programmed cell death (PCD). The most striking phenotypes caused by the edr1 mutation, besides powdery mildew-induced lesions, are enhanced drought-induced growth inhibition and enhanced ethylene-induced senescence (Frye et al., 2001; Tang et al., 2005b). The former two phenotypes require an intact salicylic acid (SA) signaling pathway, while the latter does not (Tang et al., 2005b). The general processes of PCD, drought responses, and senescence have all been linked to enhanced sensitivity to abscisic acid (ABA; Beaudoin et al., 2000; Ghassemian et al., 2000; Anderson et al., 2004; Mohr and Cahill, 2007; Xie et al., 2007), suggesting that EDR1 may be also be involved in ABA signaling (Frye et al., 2001).
ABA regulates many important events during both vegetative and reproductive growth of plants. These range from relatively slow effects, such as promotion of seed storage reserve synthesis, acquisition of desiccation tolerance and dormancy, and tolerance to drought, salt, and cold stresses (Leung and Giraudat, 1998), to rapid effects, such as stomatal closure (Leung and Giraudat, 1998; Finkelstein et al., 2002). Cumulative evidence suggests that the cross talk between ABA and SA is important for adaptation of plants to combinations of abiotic and biotic stresses (Kunkel and Brooks, 2002; Mauch-Mani and Mauch, 2005). SA inhibits ABA-induced stomatal closure (Rai et al., 1986), leaf abscission (Apte and Laloraya, 1982), and inhibition of seedling growth (Ray, 1986), while ABA increases susceptibility to biotrophic pathogens by counteracting SA-dependent defenses (Mohr and Cahill, 2003; de Torres-Zabala et al., 2007; Mohr and Cahill, 2007). Conversely, ABA-dependent priming of callose biosynthesis promotes enhanced resistance to some necrotrophic pathogens (Ton and Mauch-Mani, 2004).
The complex connections between SA signaling and ABA signaling are also observed during leaf senescence, which shares many physiological events with pathogen-induced defense responses, such as increases in ethylene and SA levels (Ryals et al., 1996; Morris et al., 2000), accumulation of hydrogen peroxide (Levine et al., 1994; Pastori and Del Rio, 1997), and accumulation of transcripts from pathogenesis-related (PR) genes (Hanfrey et al., 1996; Butt et al., 1998; Pontier et al., 1999; Quirino et al., 1999, 2000; Yoshida et al., 2001). ABA is considered a senescence promoter, although evidence for an in vivo role is rather poor compared with ethylene (Nooden and Leopold, 1988; Madhu et al., 1999; Panavas et al., 1999). Several mutations that inhibit defense responses in Arabidopsis also inhibit senescence (Morris et al., 2000). For example, the pad4 mutation, which enhances disease susceptibility and reduces SA accumulation (Jirage et al., 1999), displays a dramatic delay in PCD during senescence (Morris et al., 2000). Consistent with these observations, SA levels increase approximately 4-fold in senescing Arabidopsis leaves (Morris et al., 2000). Determining cause and effect in these processes is difficult, however, as SA-signaling pathways include positive feedback loops. For example, cell death promotes SA production, but SA production also promotes cell death (Glazebrook, 2005). Accordingly, it has been proposed that high concentrations of SA, such as those generated at the sites of pathogen entry, are required for cell death induction, whereas SA at low levels, detected beyond the margins of the initial infection sites, might lead to cell survival and lesion containment (Alvarez, 2000).
Because loss of EDR1 function leads to enhanced PCD and senescence, it is considered to be a negative regulator of these processes. The EDR1 protein belongs to a small family of protein kinases in Arabidopsis that includes the CTR1 protein (Frye et al., 2001), a negative regulator of ethylene responses (Kieber et al., 1993; Cao et al., 1997). Unlike loss of CTR1 function, however, loss of EDR1 does not activate ethylene-signaling pathways (Frye et al., 2001). The specific function of EDR1 thus remains unknown. To uncover additional genes in the EDR1 kinase pathway or identify other pathways that interact with the EDR1 pathway, we performed a suppressor screen to identify mutations that suppress the edr1 mutant phenotype. Here we describe one such suppressor mutation, which was found to be a missense mutation in the KEEP ON GOING (KEG) gene. KEG encodes a ubiquitin ligase thought to be involved in ABA signaling (Stone et al., 2006). Interestingly, we found that transgenic overexpression of KEG induces massive cell death in Arabidopsis.
RESULTS
The supp69 Mutation Blocks EDR1-Dependent Resistance to G. cichoracearum
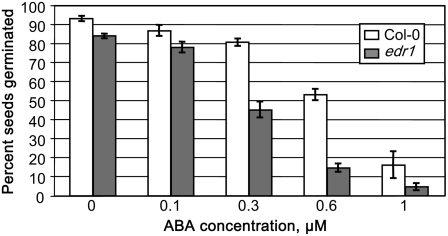
Because edr1 mutant plants show enhanced drought-induced growth inhibition (Tang et al., 2005b), we suspected that the edr1 mutation might be enhancing sensitivity to ABA. To test this hypothesis, we performed a seed germination assay on varying levels of ABA, which is known to inhibit germination (Finkelstein, 1994). Figure 1 shows that the edr1 mutant is indeed hypersensitive to exogenous ABA as the percent germination at 3 d of incubation on 0.6 μm ABA was only approximately 15% for edr1 seeds compared to greater than 50% for wild type. This enhanced ABA sensitivity suggested that we could enrich for suppressor mutants by germinating mutagenized seed on ABA-containing plates.
Figure 1.
ABA hypersensitivity of the edr1 mutant. Seeds of either wild-type Col-0 or the edr1 mutant were planted on one-half-strength Murashige and Skoog agar supplemented with the indicated concentrations of ABA. The percentage of seeds germinating after 3 d is shown.
To enrich for edr1 suppressor mutants, we screened an ethyl-methane sulfonate-mutagenized edr1 population on agar plates containing 0.7 μm ABA (60,000 M2 seeds derived from 3,500 M1 edr1 plants). Approximately 1,000 seedlings were identified that germinated within the first 3 d of incubation, a time period during which very few edr1 mutant seeds had germinated. Seedlings were transplanted to pots containing Metromix 360. Four to 5 weeks later these plants were inoculated with G. cichoracearum and scored for disease responses 8 d postinoculation. Seventy-four mutants displaying visible powder were selected and their response to G. cichoracearum retested in the next generation. Among these, 11 mutants were found to be fully susceptible to G. cichoracearum, lacking edr1-dependent necrotic lesions and allowing abundant development of G. cichoracearum conidiophores. Here we describe one mutant, which was designated supp69. Characterization of the other mutants is ongoing and will be described elsewhere.
The supp69 mutant displayed a wild-type Columbia-0 (Col-0)-like phenotype 8 d after infection with G. cichoracearum (Fig. 2). No other obvious developmental or morphological phenotypes of supp69 plants were observed when grown under normal conditions. Complementation tests revealed that supp69 is not allelic to pad4 or npr1 (data not shown), which have been previously shown to suppress the edr1 phenotype (Tang et al., 2005b). Segregation analysis of a backcross to the edr1 mutant revealed that susceptibility to G. cichoracearum was caused by a single recessive mutation. We therefore proceeded with genetic mapping of the supp69 mutation and a detailed characterization of the supp69 mutant phenotype.
Figure 2.
Suppression of the edr1 powdery mildew resistant phenotype. Col-0, edr1, and supp69 plants 8 d after powdery mildew infection. Abundant white powder visible on Col-0 and supp69 indicates asexual sporulation, thus a susceptible response; the lower leaves of the edr1 mutant display regions of chlorosis and necrosis and are free of visible powder.
The supp69 Mutation Maps to Chromosome 5
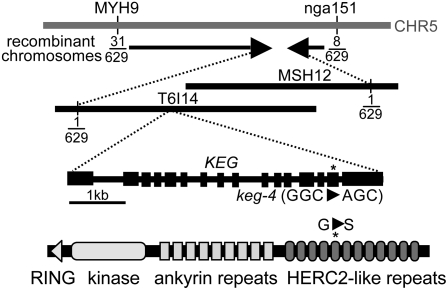
Genetic mapping of the supp69 mutation was complicated by a lack of edr1 mutant alleles in Arabidopsis accessions other than Col-0. We therefore crossed the supp69 mutant (edr1-supp69) to the Landsberg erecta (Ler) accession and identified F3 families that were homozygous for the edr1 mutation and segregating for the supp69 mutation (see “Materials and Methods”). Twenty-eight F3 families were selected and pooled for mapping purposes. F3 plants were scored for susceptibility to G. cichoracearum. Susceptibility segregated in an approximately 1:3 ratio, confirming that supp69 was caused by a single recessive mutation. DNA was isolated from 629 susceptible F3 plants and scored for microsatellite markers distributed across the Arabidopsis genome. Initially, the supp69 mutation was mapped to a region between microsatellite markers MYH9 and nga151 on chromosome 5 (Fig. 3). To further localize the mutated gene, we created PCR-based markers at intervals between MYH9 and nga151 using small insertions/deletions that are polymorphic between Ler and Col-0 (Jander et al., 2002). Fine-mapping localized the mutation to a 126-kb interval covering the 3′ end of bacterial artificial chromosome (BAC) clone T6I14 (GenBank accession AL391710) and the 5′ end of BAC MSH12 (GenBank accession AB006704) defined by one recombinant at each border (Fig. 3). This region harbors 31 loci (from At5g13470–At5g13750). Twenty-one of these were amplified by PCR and sequenced. A single G to A transition mutation was identified in At5g13540, which was recently found to be misannotated, with the full open reading frame encompassing both the At5g13530 and At5g13540 loci (Stone et al., 2006). The combined gene has been named KEG for “keep on going” (Stone et al., 2006). The supp69 mutation was located in the 15th exon of KEG, and causes a Gly-to-Ser substitution (G1144S) in the HERC2-like domain of the KEG protein (Fig. 3; Stone et al., 2006).
Figure 3.
Positional cloning of the supp69 mutation. The number of recombination events between the indicated markers and the supp69 mutation over the total number of chromosomes scored is shown. Horizontal lines indicate BAC clones spanning the region to which keg-4 was mapped. Sequencing of candidate genes in this interval revealed a G to A transition in the KEG gene. The genomic structure of the KEG gene with exons indicated by black boxes and the position of the keg-4 mutation are shown. The domain structure of the KEG protein and the location of the Gly to Ser substitution caused by keg-4 are also shown.
Loss-of-function keg mutants display a strong postgerminative growth arrest shortly after the emergence of the first true leaves (Stone et al., 2006), indicating that KEG is essential for plant development. Because supp69 plants show normal growth and development, we conclude that the mutation in KEG does not cause a complete a loss of function. As three keg mutants have been described previously (Stone et al., 2006), the keg mutation in supp69 was designated keg-4.
Complementation of the supp69 Mutation
To confirm that the mutation in At5g13530 was responsible for the suppression of the edr1 phenotype, we transformed supp69 plants with a genomic copy of the KEG gene under control of its native promoter and tested for restoration of the edr1 mutant phenotype. Thirteen independent T1 transgenic plants were inoculated with G. cichoracearum. All 13 were resistant to G. cichoracearum and showed necrotic lesions and almost no conidiation 8 d after infection, demonstrating that the KEG genomic construct complemented the keg-4 mutation (data not shown). The transgene did not cause any growth phenotypes, as all transgenic lines were indistinguishable from wild-type Col-0 plants prior to inoculation.
The keg-4 Mutation Suppresses the Enhanced Ethylene-Induced Senescence Phenotype of edr1
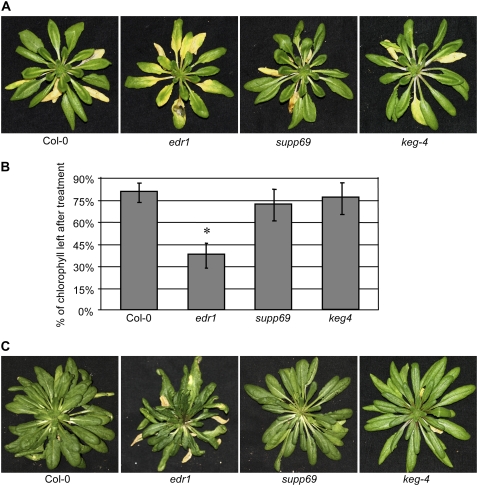
Besides showing enhanced resistance to powdery mildew, edr1 mutants display an enhanced ethylene-induced senescence phenotype (Frye et al., 2001). To test whether the keg-4 mutation also suppressed this trait, we exposed plants to ethylene (100 μL L−1) for 3 d. The supp69 plants showed the same rate of senescence as Col-0 plants in contrast to the enhanced senescence phenotype of edr1 (Fig. 4A). To determine whether the keg-4 mutation by itself affected senescence rates, we crossed out the edr1 mutation by backcrossing to wild-type Col-0 plants and selecting homozygous EDR1/EDR1 keg-4/keg-4 plants in the F2. Ethylene induced visible chlorosis (yellowing) on the oldest two to three leaves of wild-type Col-0, supp69, and keg-4 plants after 3 d exposure to ethylene. However, in edr1 mutant plants, chlorosis was observed on much younger leaves and appeared earlier (Fig. 4A). Quantification of chlorophyll levels revealed significant differences between ethylene-treated edr1 plants and the other three genotypes (Fig. 4B). Consistent with this finding, edr1 plants also showed a faster rate of senescence under standard short-day growth conditions, which became visibly obvious by 11 weeks of growth (Fig. 4C). We thus conclude that the keg-4 mutation by itself does not delay senescence in a wild-type background, but fully suppresses the enhanced senescence of the edr1 mutant.
Figure 4.
The keg-4 mutation suppresses enhanced ethylene-induced senescence in the edr1 mutant. A, Plants were photographed after 3 d of exposure to 100 μL L−1 ethylene. B, The chlorophyll content in leaves five to eight (leaf one being the first true leaf) of the plants shown in section A. Bars represent the relative mean and sd of values obtained from four plants in comparison to untreated controls (*, significantly different [P < 0.001] by one-way ANOVA with Tukey's multiple-comparison post hoc test). C, The edr1 mutant shows signs of accelerated senescence at 11 weeks. All plants were grown under standard short-day growth conditions. Similar results were obtained in two additional independent experiments.
The keg-4 Mutation Suppresses edr1-Mediated Drought-Induced Growth Inhibition
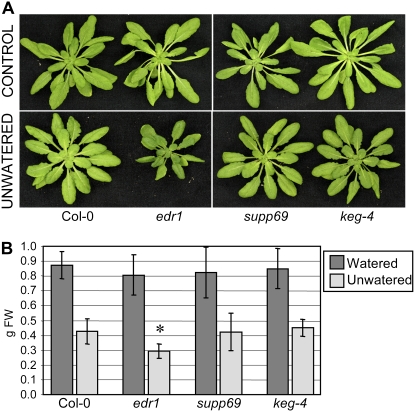
We previously reported that edr1 plants appeared more sensitive than wild-type Col-0 plants to under-watering, often growing slower than wild-type Col-0 plants (Tang et al., 2005b). To further characterize supp69, we grew plants under standard growth conditions for 3 weeks and then stopped watering them for 2 weeks. The edr1 plants were significantly smaller than wild-type Col-0, supp69, or keg-4 plants at the end of the 2-week drought period, although they were the same size at the start (Fig. 5A). Control edr1 plants grown with the standard watering regime did not significantly differ in size from Col-0, supp69, or keg-4 at 5 weeks (Fig. 5A). To quantify the edr1-mediated drought-induced growth phenotype, we weighed the individual plants (fresh weight) grown under standard or drought conditions. Figure 5B shows that edr1 mutant plants weighed almost the same as the other tested plants when grown under standard conditions, but weighed significantly less when grown under drought conditions. These data indicate that keg-4 also suppresses the edr1-mediated drought sensitivity.
Figure 5.
The keg-4 mutation suppresses edr1-mediated drought-induced growth inhibition. A, Plants were grown under standard growth conditions for 3 weeks and then watering was withheld for 2 weeks. B, Quantification of drought-induced growth inhibition. Aerial portions were weighed immediately after removal from soil. Bars represent the mean and sd of values from 10 plants (*, significantly different [P < 0.001] from others by one-way ANOVA with Tukey's multiple-comparison post hoc test). The experiment was repeated two additional times with similar results.
The keg-4 Mutation Suppresses the ABA Hypersensitivity of edr1
The suppression of edr1-mediated drought sensitivity suggested that the keg-4 mutation should also suppress the ABA hypersensitivity of edr1 mutants. We therefore tested the supp69 mutant for sensitivity to ABA using the seed germination assay described above. We plated seeds on Murashige and Skoog agar containing 0.7 μm ABA. As described above, this level of ABA inhibited germination of edr1 seeds more than wild-type seeds, which resulted in noticeably smaller seedlings at 5 d of incubation (Fig. 6). The supp69 and keg-4 mutant seedlings were indistinguishable from wild-type plants in this assay, confirming that keg-4 also suppresses edr1-mediated ABA hypersensitivity.
Figure 6.
The keg-4 mutation suppresses edr1-mediated ABA hypersensitivity. Plants were germinated on plates containing 0.7 μm ABA and the photo was taken at day 5. The experiment was repeated three additional times with similar results.
The supp69 and keg-4 Mutants Show Lowered Sensitivity to Glc
Null alleles of KEG4 have been shown previously to confer hypersensitivity to exogenous Glc (Stone et al., 2006), which causes growth arrest of wild-type Arabidopsis seedlings when added to agar at 6% (Zhou et al., 1998; To et al., 2002). Glc influences postgerminative growth of Arabidopsis via its ability to activate ABA biosynthesis genes and consequent activation of ABA-inducible genes (Cheng et al., 2002; Finkelstein and Gibson, 2002), and many Glc-insensitive mutants are also ABA insensitive. We therefore tested whether the keg-4 mutation affected Glc sensitivity. Significantly, the keg-4 mutation conferred Glc insensitivity (Fig. 7). After 15 d on Glc-containing media, the supp69 and keg-4 seedlings showed only a slight growth inhibition relative to plants grown in the absence of Glc, while wild-type, Col-0, and edr1 seeds showed almost no growth (Fig. 7). All mutants and wild-type plants grew similarly on plates containing 6% mannitol, a nonmetabolizable sugar, demonstrating that the response was not simply the result of osmotic stress (Fig. 7). There were also no significant differences between plants grown on Murashige and Skoog with 6% Suc (data not shown). These data indicate that the keg-4 allele confers phenotypes opposite to that of a keg loss-of-function mutation (Stone et al., 2006), and further support our conclusion that the keg-4 mutation inhibits ABA signaling.
Figure 7.
The keg-4 mutation confers Glc insensitivity. Col-0, edr1, supp69, and keg-4 seeds were germinated on solid Murashige and Skoog media with the addition of either 6% Glc or 6% mannitol. The photograph was taken 15 d after seeds germinated on the mannitol plates. The experiment was repeated four additional times with similar results.
The keg-4 Mutation Suppresses edr1-Mediated Changes in Gene Expression
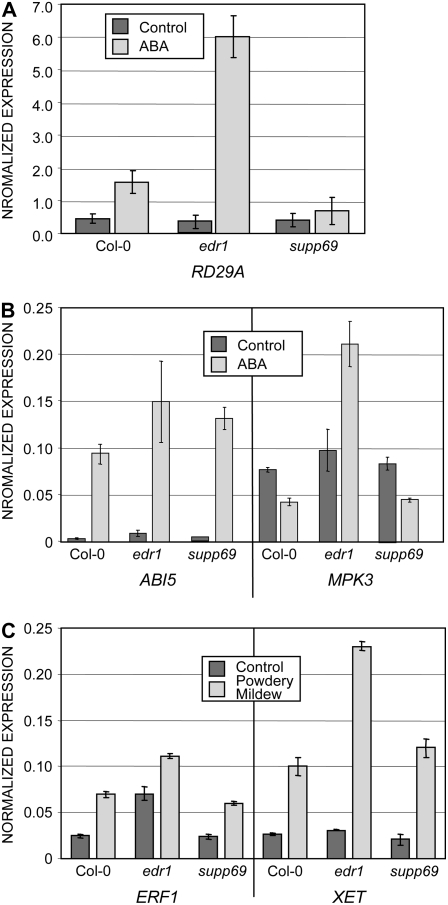
ABA induces the expression of many genes that are important for adaptation to stress. Based on the edr1 mutant phenotypes observed above and the ability of the keg-4 mutation to suppress them, we hypothesized that EDR1 might play a role in ABA-induced changes in gene expression. To test this hypothesis, we examined the expression of RESPONSIVE TO DESSICATION29A (RD29A; At5g52310), a well-characterized ABA-inducible gene (Yamaguchi-Shinozaki and Shinozaki, 2006). In wild-type Col-0, treatment with 100 μm ABA for 3 h induced RD29A (Fig. 8A). Induction of this gene was greater in the edr1 mutant, consistent with edr1 being hypersensitive to ABA. The keg-4 mutation fully suppressed the enhanced expression in edr1, indicating that keg-4 suppresses edr1-mediated hypersensitivity at the level of gene induction (Fig. 8A).
Figure 8.
The keg-4 mutation suppresses edr1-mediated changes in gene expression. A, Quantitative RT-PCR analysis of RD29A expression in 8-d-old seedlings in response to 100 μm ABA treatment. Total RNA was isolated from whole seedlings 30 h after treatment. B, ABI5 and MPK3 expression in 3-d-old seedlings germinated on Murashige and Skoog agar containing 0.7 μm ABA. C, ERF and XET expression in 5-week-old plants inoculated with powdery mildew. Total RNA was isolated from leaves 30 hpi. All data represent the average and sd of three biological replicates from independent experiments.
We also analyzed expression of ABSCISIC ACID INSENSITIVE5 (ABI5; At2g36270) and MAP KINASE3 (MPK3; At3g45640). ABI5 is a key transcription factor required for the induction of many ABA-responsive genes, and is itself inducible by ABA (Lopez-Molina et al., 2001; Brocard et al., 2002). It is most highly expressed in germinating seeds (Lopez-Molina et al., 2001; Brocard et al., 2002). MPK3 has been linked to ABA signaling in seedlings because overexpression of MPK3 increases ABA sensitivity in ABA-induced postgermination growth arrest (Lu et al., 2002). In addition, Arabidopsis plants with guard cell-specific silencing of MPK3 display a partial insensitivity to ABA-mediated inhibition of stomatal opening (Gudesblat et al., 2007). Although ABA treatment does not activate MPK3 in leaf mesophyll protoplasts (Kovtun et al., 2000), ABA does appear to activate MPK3 in Arabidopsis seedlings (Lu et al., 2002). ABI5 was highly induced by ABA treatment in both wild-type and edr1 seedlings (Fig. 8B), indicating that there is a positive feedback loop regulating this gene. Induction appeared to be slightly enhanced in the edr1 mutant, but this difference was not statistically significant. In contrast, MPK3 transcript levels were suppressed approximately 2-fold in wild-type plants in response to ABA treatment, while in edr1 seedlings MPK3 was induced 2-fold, resulting in a 4-fold difference in MPK3 transcript levels between wild-type and edr1 mutant seedlings. This difference was fully suppressed by the keg-4 mutation.
We also examined the expression of two powdery mildew-inducible genes, ETHYLENE-RESPONSIVE BINDING FACTOR1 (ERF1; At4g17500) and XYLOGLUCAN ENDOTRANSGLYCOSYLASE (XET; At5g57560). At 30 h postinoculation (hpi), both genes were induced about 3-fold in wild-type Col-0 plants (Fig. 8C). The basal and induced levels of ERF1 were higher in the edr1 mutant, while only the induced levels of XET1 were higher. The keg-4 mutation restored the expression of both genes to wild-type levels. Thus the keg-4 mutation is able to suppress the effect of edr1-induced changes in gene expression for both ABA- and pathogen-responsive genes.
Overexpression of KEG Leads to Massive Cell Death
To further analyze the impact of KEG expression in plant development, we tested the effect of overexpressing the KEG gene in transgenic Arabidopsis plants. We constructed transgenic plants expressing the full-length KEG cDNA under the control of the constitutive cauliflower mosaic virus 35S promoter (35S∷KEG). We were unable to obtain any transformants containing the 35S∷KEG construct, which suggested that constitutive overexpression of KEG in Arabidopsis may be lethal. We therefore constructed transgenic Col-0 plants expressing the KEG cDNA under the control of a steroid-inducible promoter (Aoyama and Chua, 1997). We generated 24 transgenic lines containing this construct. All developed large necrotic lesions within 40 h of treatment with 50 μm dexamethasone (DEX; Fig. 9). In contrast, none of the DEX-treated leaves of control plants or ethanol-treated plants containing the DEX∷KEG transgene showed any visible cell death (data not shown). These data suggest that ectopic overexpression of KEG results in cell death and that the level of KEG is tightly controlled.
Figure 9.
Overexpression of the KEG gene is toxic to plants. Wild-type Col-0 and transgenic T1 supp69 plants carrying a DEX-inducible wild-type copy of KEG were sprayed with 50 μm DEX. The photos show the same plant at time 0 (before spraying) and 7 d after induction for three independent transgenic lines. Note that large necroses develop on all leaves irrespective of their ages.
Analysis of KEG Expression
To gain insight into the spatial and temporal pattern of KEG expression, we searched the Arabidopsis microarray data available through the Genevestigator Web interface (https://www.genevestigator.ethz.ch/). Microarray analyses showed that KEG is expressed in various tissues and organs at all developmental stages and is not specifically induced by any factor. To investigate KEG expression more directly, we constructed transgenic plants expressing a KEG promoter∷GUS fusion. A 1,044-bp fragment 5′ to the KEG start codon was fused to the GUS reporter gene and the construct was transformed into wild-type Col-0 plants. We obtained a number of pKEG∷GUS transformants and analyzed a total of 24 transgenic lines. Supplemental Figure S1 shows representative GUS staining patterns. GUS staining was observed in all tissues of 8-d-old seedlings (most prominent in the meristem parts), consistent with the microarray data. However, in 7-week-old flowering plants, GUS staining was only observed in the youngest parts of the stem, anthers, and the receptacle of immature siliques. No staining was observed in mature leaves, older parts of the stem, flower parts other than anthers, or mature siliques. These results suggest that the expression of KEG may be under developmental regulation and that KEG is expressed mainly in the actively growing and dividing cells.
To further investigate the expression KEG and EDR1, we performed quantitative reverse transcription (RT)-PCR analyses. In 1-week-old seedlings, treatment with 100 μm ABA for 3 h induced the expression of both EDR1 and KEG (Supplemental Fig. S2A). The edr1 mutation did not affect KEG transcript levels. We also examined expression of these genes in 5-week-old plants 30 h after powdery mildew treatment (Supplemental Fig. S2B). Both EDR1 and KEG were slightly induced by powdery mildew. The transcript level of EDR1 was reduced about 2-fold in the edr1 mutant compared to the wild-type Col-0. Because the edr1 mutation creates an early stop codon (Frye et al., 2001), this reduction in edr1 transcript level is likely due to nonsense-mediated decay, a cellular mechanism of mRNA surveillance that detects nonsense mutations and prevents the expression of truncated or erroneous proteins (Chang et al., 2007). Interestingly, the level of KEG transcript in 5-week-old plants was about 3-fold greater in edr1 than wild-type Col-0 plants (Supplemental Fig. S2B), suggesting that that EDR1 negatively regulates KEG transcription in rosette leaves.
DISCUSSION
The edr1 mutant of Arabidopsis displays enhanced resistance to powdery mildew and undergoes more rapid senescence than wild-type plants when exposed to ethylene (Frye and Innes, 1998). In addition, edr1 mutants display enhanced growth inhibition and spontaneous cell death in response to drought (Tang et al., 2005b), and enhanced cell death mediated by the RPW8 powdery mildew resistance gene (Xiao et al., 2005). All of these phenotypes, except for ethylene-induced senescence, can be suppressed by mutations in the SA signaling pathway (sid2, npr1, pad4, and eds1; Frye et al., 2001; Tang et al., 2005b; Xiao et al., 2005). However, edr1 mutant plants do not show constitutive expression of SA-inducible genes when grown under optimal conditions (Frye and Innes, 1998), thus it has been unclear why loss of EDR1 function leads to these various phenotypes. The data presented above suggest that there may be a mechanistic link between EDR1 function, ABA signaling, and SA enhancement of PCD.
Several lines of evidence point to ABA as a central player in edr1-mediated phenotypes. The most direct is the hypersensitivity of edr1 mutant seeds to ABA-mediated inhibition of germination (Fig. 1). In addition, the enhanced drought-induced growth inhibition of edr1 plants is consistent with enhanced ABA sensitivity (Fig. 5), as is the enhanced induction of RD29A by exogenous ABA (Fig. 8A). Most compelling, however, is the identification of the keg-4 missense mutation in KEG, which suppresses all known edr1-mediated phenotypes. Because loss-of-function mutations in KEG cause accumulation of the ABI5 transcription factor and ABA hypersensitivity, and because KEG physically associates with ABI5, KEG is believed to be a central regulator of ABA signaling (Stone et al., 2006). Furthermore, loss-of-function mutations in ABI5 substantially suppress the phenotypes caused by loss-of-function mutations in KEG, indicating that a primary role of KEG is regulating ABI5 levels (Stone et al., 2006). The observation that keg-4 suppresses all known edr1-mediated phenotypes, including ethylene-induced senescence, appears to place KEG function upstream of SA signaling because mutations in SID2, NPR1, PAD4, and EDS1 do not suppress the ethylene induced senescence phenotype of edr1 (Tang et al., 2005b). Understanding the function of KEG and the nature of the keg-4 mutation thus appears key to understanding how the edr1 mutation confers its various phenotypes.
The KEG protein is quite large (178 kD) and contains multiple functional domains (Fig. 3). Starting at the N-terminal end, these are the RING (for really interesting new gene) E3 ligase domain, a kinase domain, nine tandem ankyrin repeats, and 12 HERC2-like (for HECT and RCC1 like) repeats. The RING domain of KEG has been shown to have E3 ubiquitin-ligase activity in vitro, and the kinase domain autophosphorylates in vitro (Stone et al., 2006). The ankyrin repeats are required for interaction between KEG and the ABI5 transcription factor, at least in in vitro pull down assays (Stone et al., 2006). The function of the HERC2-like repeats is unknown. The HERC2-like repeats is where the keg-4 mutation is located, however, suggesting that this domain plays a critical role in KEG function.
The keg-4 mutation causes a Gly-to-Ser substitution in the fifth HERC2-like repeat (Fig. 3). This Gly residue is highly conserved among the 12 HERC2-like repeats of KEG (Stone et al., 2006). The HERC2-like repeats were first defined in the Arabidopsis KEG protein as a 61 amino acid motif with similarity to the mammalian HERC2 protein, which is a HECT-type ubiquitin E3 ligase (Garcia-Gonzalo and Rosa, 2005; Stone et al., 2006). However, HERC2 contains only a single copy of this motif. The combination of multiple HERC2-like repeats and a RING E3-ligase domain appears to be unique to plants. Single KEG homologs have been identified in rice (Oryza sativa), Medicago truncatula, and Populus tricocarpa (Stone et al., 2006), thus KEG appears to be highly conserved among angiosperms.
The phenotypes conferred by the keg-4 mutation are generally opposite to the phenotypes conferred by loss-of-function mutations in KEG. In particular, the keg-4 mutant is resistant to high levels of exogenous Glc, while keg loss-of-function mutants are hypersensitive to Glc (Fig. 7; Stone et al., 2006), suggesting that keg-4 has reduced sensitivity to ABA rather than hypersensitivity. Consistent with this, keg-4 suppressed the ABA hypersensitivity of edr1 (Fig. 6). Also, keg loss-of-function mutants undergo a growth arrest shortly after germination, consistent with hypersensitivity to endogenous ABA, while keg-4 mutants germinate and grow similar to wild-type plants on half-strength Murashige and Skoog agar. One plausible explanation for these observations is that the HERC2-like repeat domain functions to regulate the E3 ligase activity of KEG in response to ABA. In the absence of ABA, KEG presumably keeps ABI5 protein levels low via ubiquitylating ABI5 and thus targeting it for degradation. In the presence of ABA, protein levels of ABI5 increase dramatically, thus ABA must somehow prevent the ubiquitylation of ABI5, presumably by modifying KEG activity (Stone et al., 2006). We speculate that the keg-4 mutation renders KEG insensitive to ABA by modifying the structure of the HERC2-like repeats, locking KEG in an on position relative to ABI5 ubiquitylation.
One observation that seems inconsistent with the above model is the recessive nature of the keg-4 mutation. If the keg-4 mutation causes KEG to constitutively ubiquitylate ABI5 even in the presence of ABA, then one would expect reduced ABI5 levels, and thus reduced responsiveness to ABA even in a heterozygous state. We attempted to assess ABI5 protein levels directly in edr1 and keg-4 mutant seedlings, both in the presence and absence of exogenous ABA, using a previously described antibody (Lopez-Molina et al., 2001), but were unable to detect any protein even in wild-type ABA-treated seedlings. It was thus not possible to compare ABI5 protein levels between the various mutant and wild-type plants. Given the recessive nature of keg-4, we speculate that the level of mutant KEG protein present in heterozygous plants is insufficient to ubiquitylate all of the ABI5 protein and that there may be a minimum threshold level of ABI5 that the plant must go below to suppress edr1-mediated signaling.
The edr1 mutant displays approximately 3-fold elevated levels of KEG mRNA in rosette leaves, both before and after powdery mildew inoculation (Supplemental Fig. S2). One would expect this to lead to increases in KEG protein levels and thus decreases in ABA sensitivity. It is possible, however, that this increase simply reflects an elevated steady-state level of ABA signaling in the edr1 mutant as exogenous application of ABA induces KEG transcript levels about 3-fold in wild-type seedlings (Supplemental Fig. S2). If EDR1 normally functions as a negative regulator of ABA signaling, then plants may compensate for loss of EDR1 function by increasing KEG levels. This compensation may allow for normal growth under nonstressed conditions, but under times of abiotic or biotic stress, is insufficient, leading to overactivation of ABA pathways.
KEG overexpression in mature rosettes leads to rapid cell death (Fig. 9). This phenotype cannot be explained by reduction in ABI5 protein levels alone, as ABI5 null mutants are viable (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000). This observation suggests that KEG may have other substrates in addition to ABI5 and/or that KEG overexpression leads to ubiquitylation and degradation of inappropriate substrates. The former hypothesis is supported by the finding that abi5 mutations do not fully suppress KEG loss-of-function mutant phenotypes (Stone et al., 2006). Furthermore, KEG loss-of-function mutations partially suppress the ABA insensitivity of abi5-1 mutants (Stone et al., 2006), which suggests that loss of KEG causes accumulation of an ABA-responsive transcription factor that can compensate for loss of ABI5 function. If KEG regulates the levels of multiple transcription factors, then keg-4 suppression of edr1 phenotypes may be a consequence of reducing the levels of these specific transcription factors in addition to ABI5.
Our data indicate that edr1 plants have enhanced ABA signaling and that this is causally related to enhanced resistance to powdery mildew. This conclusion would seem to be at odds with recent work showing that the hemibiotrophic bacterial pathogen Pseudomonas syringae strain DC3000 specifically induces ABA biosynthesis in Arabidopsis to promote virulence and that ABA-insensitive Arabidopsis mutants have enhanced resistance to strain DC3000 (de Torres-Zabala et al., 2007). If our model is correct, one would predict that the edr1 mutant should have enhanced susceptibility to P. syringae, but in fact, edr1 was originally isolated in a screen for mutants with enhanced resistance to P. syringae (Frye and Innes, 1998). Further analysis of the edr1 mutant phenotype, however, revealed that the enhanced resistance to P. syringae was variable, and in nonstressed plants was not reproducible (Frye and Innes, 1998). We speculate that the enhanced responses of edr1 to abiotic stresses such as drought may indirectly promote resistance to P. syringae, and that the edr1 mutant was isolated in the original screen because it had undergone a stress response prior to inoculation.
The role of ABA in regulating defense responses is still poorly understood and, depending on the pathogen studied, ABA may enhance resistance or enhance susceptibility (Mauch-Mani and Mauch, 2005). For example, both salt stress and exogenous ABA enhance the resistance of barley (Hordeum vulgare) to the biotrophic powdery mildew fungus Blumeria graminis (Wiese et al., 2004). Likewise, ABA application enhances resistance of Arabidopsis to the oomycete Pythium irregulare, while ABA-deficient and ABA-insensitive mutants are more susceptible (Adie et al., 2007). Similarly, ABA application protects Arabidopsis against the necrotrophic fungi Alternaria brassicicola and Plectosphaerella cucumerina, while the ABA-deficient and ABA-insensitive mutants display enhanced susceptibility to these pathogens (Ton and Mauch-Mani, 2004; Adie et al., 2007). Finally, ABA-deficient and ABA-insensitive Arabidopsis mutants also display enhanced susceptibility to soilborne bacterium Ralstonia solanacearum (Hernandez-Blanco et al., 2007). We are currently testing whether the abi-5 mutation and other mutations that confer ABA insensitivity or ABA deficiency affect edr1-mediated resistance to powdery mildew. We predict that abi-5 should at least partially suppress edr1.
Although the above examples show that ABA can positively regulate resistance against some pathogens, including powdery mildews, the majority of studies have shown that ABA promotes susceptibility to pathogens. Early studies showed that exogenous application of ABA enhanced susceptibility of potato (Solanum tuberosum) tubers to Phytophthora infestans and Cladosporium cucumerinum (Henfling et al., 1980), the susceptibility of soybean (Glycine max) to Phytophthora megasperma f. sp. glycinea (Ward et al., 1989), the susceptibility of rice to rice blast (Magnaportha grisea; Matsumoto et al., 1980), and the susceptibility of tobacco (Nicotiana tabacum) to blue mold (Peronospora tabacina; Salt et al., 1986). More recently, ABA treatment has been shown to increase the susceptibility of Arabidopsis to Peronospora parasitica and the avirulent bacterium P. syringae pathovar tomato strain 1065 (Mohr and Cahill, 2003), but not to the virulent strain DC3000. As described above though, strain DC3000 up-regulates endogenous ABA levels in Arabidopsis (de Torres-Zabala et al., 2007); thus, exogenous ABA may have little effect on this strain. ABA application also enhances the susceptibility of Arabidopsis to Fusarium oxysporum (Anderson et al., 2004) and the susceptibility of tomato (Solanum lycopersicum) to Botrytis cineria (Audenaert et al., 2002). Consistent with this, ABA-deficient Arabidopsis mutants are more resistant to F. oxysporum (Anderson et al., 2004) and B. cinerea (Adie et al., 2007), and the ABA-deficient sitiens mutant of tomato is more resistant to B. cineria (Audenaert et al., 2002) and to P. syringae (Thaler and Bostock, 2004).
The molecular mechanisms underlying ABA regulation of defense responses are just beginning to be defined. In Arabidopsis, exogenous application of ABA suppresses transcription of defense genes induced by jasmonic acid (JA) and ethylene (Anderson et al., 2004), as well as defenses induced by SA (Yasuda et al., 2008); thus, the enhanced resistance of ABA-deficient mutants could be explained by up-regulation of JA/ethylene- and/or SA-mediated defense pathways. The enhanced resistance of the sitiens mutant of tomato also correlated with enhanced SA-dependent defense signaling (Thaler and Bostock, 2004), suggesting that ABA negatively regulates SA-dependent defenses in tomato as it does in Arabidopsis. In light of these data, it is difficult to reconcile how the edr1 mutant could have both enhanced ABA sensitivity (this study) and enhanced expression of PR-1 (Frye and Innes, 1998), and why resistance of edr1 plants to powdery mildew is dependent on SA signaling (Frye et al., 2001).
The regulatory interactions between ABA, JA, and ethylene are clearly complex, as SA signaling is usually considered antagonistic to JA/ethylene signaling (Kunkel and Brooks, 2002; Thaler et al., 2002; Li et al., 2004), yet both signaling pathways are up-regulated in ABA-deficient mutants. To add to this complexity, transcriptional profiling analyses have revealed a large number of genes that respond similarly to exogenous methyl jasmonate and SA application (Schenk et al., 2000), despite their generally antagonistic effects on well-characterized defense genes such as PR-1 and PDF1.2. It may thus be an oversimplification to conclude that ABA antagonizes all JA- and SA-induced defenses, when most studies to date have focused on just a few well-characterized genes (Anderson et al., 2004; Yasuda et al., 2008). It is plausible that ABA acts synergistically with some SA-inducible genes and antagonistically with others, with the former set being particularly important for resistance to powdery mildew.
The challenge in front of us is to determine which ABA-regulated responses contribute to resistance to some pathogens, and which responses contribute to susceptibility to others. This will require careful analyses, including transcriptional profiling of mutants blocked in more defined defense signaling steps than analyzed to date, and these mutants need to be tested against a diverse collection of pathogens.
MATERIALS AND METHODS
ABA Germination Assay
For testing ABA sensitivity (Fig. 1), seeds were sterilized then plated on one-half-strength Murashige and Skoog salts (Sigma-Aldrich) supplemented with varying concentrations of ABA and 0.8% agar. Plates were placed at 4°C for 72 h then transferred to a growth room set to 23°C and a 9 h light (150 mE m−2 s−1)/15 h dark cycle for 3 d, at which time seeds were scored for germination (a root emerging from the seed coat).
Plant Growth Conditions and Mutant Screening
Ethyl methanesulfonate-mutagenized edr1 plants (M2 generation) were planted on one-half-strength Murashige and Skoog plates supplemented with 0.7 μm ABA, 1% Suc, and 0.8% agar and grown in growth rooms as described in the previous paragraph. Seedlings germinating by day 3 were transplanted to MetroMix 360 and allowed to grow for 5 weeks in the same growth rooms, at which time they were inoculated with powdery mildew (Golovinomyces cichoracearum strain UCSC1). Disease phenotypes were scored 8 d after inoculation. Plants displaying powder and no necrotic lesions were selected and allowed to set seeds. Approximately 60,000 M2 plants derived from 3,500 M1 plants were screened. For liquid cultures, seeds were put into half-strength Murashige and Skoog without agar and shaken continuously at 200 rpm under continuous light.
Powdery Mildew Infections
G. cichoracearum strain UCSC1 was maintained on hypersusceptible Arabidopsis (Arabidopsis thaliana) pad4-2 mutant plants. Plants were inoculated between 4 and 6 weeks of age by gently brushing the leaves of diseased plants and healthy plants together to pass the conidia (asexual spores). The disease phenotype was scored 8 d after inoculation.
Ethylene-Induced Senescence Assay
Five-week-old plants were placed in a sealed chamber containing 100 μL L−1 of ethylene for 3 d. Leaves five to eight (leaf one being the oldest true leaf) were removed and chlorophyll was extracted and measured as previously described (Frye et al., 2001).
Drought Stress Assay
Plants were grown in growth rooms as described above for 3 weeks and then watering was stopped for 2 weeks. The aerial portions of plants were then weighed and photographed. The mean weight from 10 plants of each line was used to represent the growth phenotype.
Genetic and Physical Mapping of supp69
Genetic mapping was accomplished using an F2 population derived from a cross between the supp69 mutant (carrying the edr1 mutation in the Columbia genotype, Col-0) and Ler. F2 seeds were planted and inoculated with G. cichoracearum as described above and plants displaying an edr1 phenotype were selected for collection of F3 seeds. Fifty-seven F3 families were planted (12 plants per family) and scored for disease susceptibility to G. cichoracearum. Twenty-eight of these families segregated susceptible plants, indicating that they contained the supp69 mutation. Genomic DNA was isolated from 84 susceptible F3 plants chosen from these 28 F3 families and scored with published microsatellite markers. This initial mapping localized the supp69 mutation between molecular markers MYH9 and nga151 on chromosome 5. New molecular markers at intervals between these two markers were next developed using the Monsanto Col-0 and Ler polymorphism database (http://www.arabidopsis.org/Cereon/index.jsp; primer sequences available upon request). We then selected 629 susceptible F3 plants representing 1,258 meioses and scored them for recombination between markers MYH9 and nga151. Ultimately, the supp69 mutation was localized to BAC clone T6I14. This analysis defined a 126-kb region that cosegregated with the supp69 mutation.
Sequencing of Candidate Genes
The genetic interval to which the supp69 mutation was mapped contained 31 loci within a 126-kb region. We amplified 21 of these genes from the supp69 mutant using the PCR and directly sequenced the PCR products. Once a mutation was identified, sequencing was stopped and the identity of supp69 confirmed by complementation. All sequencing reactions were performed using BigDye Terminator kits (Applied Biosystems) and separated on an ABI 3730 automated DNA sequencer (Applied Biosystems).
Complementation of supp69 by KEG and Overexpression of KEG
The genomic sequence of KEG together with its promoter (1,044 bp upstream of ATG) was PCR amplified from BAC T6I14 using primers to create attB end products and inserted into the pDONR207 vector using an Invitrogen BP Clonase kit (Invitrogen). The insert was next recloned into the pGWB19 vector (Nakagawa et al., 2007). All cloning products were checked for proper sequences.
The full-length KEG cDNA was amplified by PCR from a plasmid containing a KEG cDNA (a kind gift of Judy Callis, University of California, Davis) using primers with attB sites for recombination. The PCR product was introduced into the pDONR207 vector. The resulting clone was sequence verified and inserts recombined into the C-terminal hemagglutinin-tagged, DEX-inducible Gateway destination vector pBAV154 (Vinatzer et al., 2006) using the Invitrogen LR Clonase kit.
Plant Transformation
Plasmids were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation with selection on Luria-Bertani plates containing 50 μg mL−1 kanamycin sulfate (Sigma). Arabidopsis plants were transformed using the floral-dip method (Clough and Bent, 1998). Transgenic plants were selected either by growing on one-half-strength Murashige and Skoog salts plus 0.8% agar and 50 μg mL−1 kanamycin or by spraying 1-week-old seedlings grown in soil with 300 μm BASTA (Finale; Fornam Companies Inc.) five times in 2-d intervals. Transformants were transplanted to soil and allowed to set seeds.
Construction of the KEG Promoter∷GUS Reporter and GUS Activity Assays
A 1,044-bp promoter fragment of KEG was amplified by PCR from genomic DNA of wild-type Col-0 using primers to create attB end products. The resulting PCR products were then gel purified using the QIAquick Gel extraction kit (Qiagen) and the Invitrogen BP Clonase kit was then used to recombine the products into the Gateway donor vector pDONR207 (Invitrogen). The resulting clones were sequence verified and inserts recombined into the C-terminal GUS-tagged pGWB3 vector (constructed by Tsuyoshi Nakagawa, Shimane University, Izumo, Japan). The clone was also verified by sequencing and transformed into Agrobacterium strain GV3101 by electroporation. Plant transformation was conducted as described above. GUS activity analysis was performed as described (Jefferson et al., 1987) using 8-d-old seedlings and 7-week-old flowering plants.
Quantitative RT-PCR Analysis
For G. cichoracearum treatment, plants were grown and inoculated as described above. Leaves were removed from plants at 30 hpi. For ABA treatment seedlings were grown in liquid Murashige and Skoog media for 1 week in a room at 25°C under constant light. The seedlings were exposed to 100 μm ABA for 3 h before RNA was extracted. For checking ABI5 and MPK3 expression, seeds were germinated for 72 h on half-strength Murashige and Skoog plates with or without addition of 0.7 μm ABA.
Total RNA was isolated using the Qiagen RNeasy kit and treated with DNase (Invitrogen) to remove DNA contamination. The High Capacity reverse transcriptase kit (Applied Biosystems) was utilized to obtain cDNA, and the samples purified with Qiagen QIAquick PCR purification kit. Quantitative RT-PCR was performed using primers listed in Supplemental Table S1. A tubulin gene (At5g19770) was used as a control for normalizing the amount of cDNA. The Takara SYBR Premix Extaq was used for all quantitative RT-PCR runs and the Mx3000P (Stratagene) protocol was followed.
Statistical Analysis
Statistical significance of observed differences in datasets was determined using one-way ANOVA as implemented in the Analyze-it add in to Microsoft Excel (Analyze-It Software, Ltd.). The Tukey post hoc test was used to identify differences between single treatments when the ANOVA was significant.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. KEG is expressed in all tissues of seedlings but not in mature leaves.
Supplemental Figure S2. EDR1 and KEG are induced by ABA and powdery mildew.
Supplemental Table S1. Oligonucleotides used for quantitative RT-PCR experiments.
Supplementary Material
Acknowledgments
We would like to thank Jean Greenburg for the kind gift of the pBAV154 plasmid and Judy Callis for a KEG cDNA clone. We also thank the Arabidopsis Biological Resource Center at Ohio State University for providing the T6I14 BAC clone.
This work was supported by the National Institutes of Health (grant no. R01 GM063761 to R.W.I.) and a Howard Hughes Medical Institute undergraduate research award (to Y.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Roger Innes (rinnes@indiana.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adie BA, Perez-Perez J, Perez-Perez MM, Godoy M, Sanchez-Serrano JJ, Schmelz EA, Solano R (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez ME (2000) Salicylic acid in the machinery of hypersensitive cell death and disease resistance. Plant Mol Biol 44 429–442 [DOI] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua NH (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11 605–612 [DOI] [PubMed] [Google Scholar]
- Apte P, Laloraya M (1982) Inhibitory action of phenolic compounds on abscisic acid-induced abscission. J Exp Bot 33 826–830 [Google Scholar]
- Audenaert K, De Meyer GB, Hofte MM (2002) Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol 128 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard IM, Lynch TJ, Finkelstein RR (2002) Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol 129 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt A, Mousley C, Morris K, Beynon J, Can C, Holub E, Greenberg JT, Buchanan-Wollaston V (1998) Differential expression of a senescence-enhanced metallothionein gene in Arabidopsis in response to isolates of Peronospora parasitica and Pseudomonas syringae. Plant J 16 209–221 [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63 [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF (2007) The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76 51–74 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bogre L, Grant M (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J 26 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR (1994) Maternal effects govern variable dominance of two abscisic acid response mutations in Arabidopsis thaliana. Plant Physiol 105 1203–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14 S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gibson SI (2002) ABA and sugar interactions regulating development: cross-talk or voices in a crowd? Curr Opin Plant Biol 5 26–32 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Innes RW (1998) An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Tang D, Innes RW (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA 98 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Rosa JL (2005) The HERC proteins: functional and evolutionary insights. Cell Mol Life Sci 62 1826–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43 205–227 [DOI] [PubMed] [Google Scholar]
- Gudesblat GE, Iusem ND, Morris PC (2007) Guard cell-specific inhibition of Arabidopsis MPK3 expression causes abnormal stomatal responses to abscisic acid and hydrogen peroxide. New Phytol 173 713–721 [DOI] [PubMed] [Google Scholar]
- Hanfrey C, Fife M, Buchanan-Wollaston V (1996) Leaf senescence in Brassica napus: expression of genes encoding pathogenesis-related proteins. Plant Mol Biol 30 597–609 [DOI] [PubMed] [Google Scholar]
- Henfling JWDM, Bostock R, Kuc J (1980) Effect of abscisic acid on rishitin and lubimin accumulation and resistance to Phytophthora infestans and Cladosporium cucumerinum in potato tuber tissue slices. Phytopathology 70 1074–1078 [Google Scholar]
- Hernandez-Blanco C, Feng DX, Hu J, Sanchez-Vallet A, Deslandes L, Llorente F, Berrocal-Lobo M, Keller H, Barlet X, Sanchez-Rodriguez C, et al (2007) Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19 890–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Bevan M, Kavanagh T (1987) The use of the Escherichia coli beta-glucuronidase as a gene fusion marker for studies of gene expression in higher plants. Biochem Soc Trans 15 17–18 [DOI] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72 427–441 [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 1997 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5 325–331 [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49 199–222 [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79 583–593 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua NH (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol 41 541–547 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Han MH, Guevara-Garcia A, Fedoroff NV (2002) Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc Natl Acad Sci USA 99 15812–15817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhu A, Thomas G, Edward N (1999) The roles of abscisic acid and ethylene in the abscission and senescence of cocoa flowers. Plant Growth Regul 27 149–155 [Google Scholar]
- Matsumoto K, Suzuki Y, Mase S, Watanabe T, Sekizawa Y (1980) On the relationship between plant hormones and rice blast resistance. Ann Phytopathological Soc Jpn 46 307–314 [Google Scholar]
- Mauch-Mani B, Mauch F (2005) The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol 8 409–414 [DOI] [PubMed] [Google Scholar]
- Mohr PG, Cahill DM (2003) Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct Plant Biol 30 461–469 [DOI] [PubMed] [Google Scholar]
- Mohr PG, Cahill DM (2007) Suppression by ABA of salicylic acid and lignin accumulation and the expression of multiple genes, in Arabidopsis infected with Pseudomonas syringae pv. tomato. Funct Integr Genomics 7 181–191 [DOI] [PubMed] [Google Scholar]
- Morris K, MacKerness SA, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23 677–685 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104 34–41 [DOI] [PubMed] [Google Scholar]
- Nooden LD, Leopold AC (1988) Senescence and Aging in Plants. Academic Press, San Diego
- Panavas T, Pikula A, Reid PD, Rubinstein B, Walker EL (1999) Identification of senescence-associated genes from daylily petals. Plant Mol Biol 40 237–248 [DOI] [PubMed] [Google Scholar]
- Pastori GM, Del Rio LA (1997) Natural senescence of pea leaves (an activated oxygen-mediated function for peroxisomes). Plant Physiol 113 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Gan S, Amasino RM, Roby D, Lam E (1999) Markers for hypersensitive response and senescence show distinct patterns of expression. Plant Mol Biol 39 1243–1255 [DOI] [PubMed] [Google Scholar]
- Quirino BF, Noh YS, Himelblau E, Amasino RM (2000) Molecular aspects of leaf senescence. Trends Plant Sci 5 278–282 [DOI] [PubMed] [Google Scholar]
- Quirino BF, Normanly J, Amasino RM (1999) Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol Biol 40 267–278 [DOI] [PubMed] [Google Scholar]
- Rai V, Sharma S, Sharma S (1986) Reversal of ABA-induced stomatal closure by phenolic compounds. J Exp Bot 37 129–134 [Google Scholar]
- Ray S (1986) GA, ABA, phenol interaction and control of growth: phenolic compounds as effective modulators of GA-ABA interaction in radish seedlings. Biol Plant 28 361–369 [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt SD, Tuzun S, Kuc J (1986) Effect of β-inonone and abscisic acid on the growth of tobacco and resistance to blue mold: mimicry of effects of stem infection by Peronospora tabacina Asam. Physiol Mol Plant Pathol 28 287–297 [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Ade J, Frye CA, Innes RW (2005. a) Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J 44 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Ade J, Frye CA, Innes RW (2006) A mutation in the GTP hydrolysis site of Arabidopsis dynamin-related protein 1E confers enhanced cell death in response to powdery mildew infection. Plant J 47 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Christiansen KM, Innes RW (2005. b) Regulation of plant disease resistance, stress responses, cell death, and ethylene signaling in Arabidopsis by the EDR1 protein kinase. Plant Physiol 138 1018–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler J, Bostock R (2004) Interactions between abscisic-acid-mediated responses and plant resistance to pathogens and insects. Ecology 85 48–58 [Google Scholar]
- Thaler JS, Fidantsef AL, Bostock RM (2002) Antagonism between jasmonate- and salicylate-mediated induced plant resistance: effects of concentration and timing of elicitation on defense-related proteins, herbivore, and pathogen performance in tomato. J Chem Ecol 28 1131–1159 [DOI] [PubMed] [Google Scholar]
- To JP, Reiter WD, Gibson SI (2002) Mobilization of seed storage lipid by Arabidopsis seedlings is retarded in the presence of exogenous sugars. BMC Plant Biol 2 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Mauch-Mani B (2004) Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J 38 119–130 [DOI] [PubMed] [Google Scholar]
- Vinatzer BA, Teitzel GM, Lee MW, Jelenska J, Hotton S, Fairfax K, Jenrette J, Greenberg JT (2006) The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol Microbiol 62 26–44 [DOI] [PubMed] [Google Scholar]
- Vogel J, Somerville S (2000) Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc Natl Acad Sci USA 97 1897–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Raab TK, Schiff C, Somerville SC (2002) PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward EW, Cahill DM, Bhattacharyya MK (1989) Abscisic acid suppression of phenylalanine ammonia-lyase activity and mRNA, and resistance of soybeans to Phytophthora megasperma f.sp. glycinea. Plant Physiol 91 23–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese J, Kranz T, Schubert S (2004) Induction of pathogen resistance in barley by abiotic stress. Plant Biol (Stuttg) 6 529–536 [DOI] [PubMed] [Google Scholar]
- Xiao S, Calis O, Patrick E, Zhang G, Charoenwattana P, Muskett P, Parker JE, Turner JG (2005) The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J 42 95–110 [DOI] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Hanzlik S, Cook E, Shen QJ (2007) Salicylic acid inhibits gibberellin-induced alpha-amylase expression and seed germination via a pathway involving an abscisic-acid-inducible WRKY gene. Plant Mol Biol 64 293–303 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57 781–803 [DOI] [PubMed] [Google Scholar]
- Yasuda M, Ishikawa A, Jikumaru Y, Seki M, Umezawa T, Asami T, Maruyama-Nakashita A, Kudo T, Shinozaki K, Yoshida S, et al (2008) Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 20 1678–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Ito M, Nishida I, Watanabe A (2001) Isolation and RNA gel blot analysis of genes that could serve as potential molecular markers for leaf senescence in Arabidopsis thaliana. Plant Cell Physiol 42 170–178 [DOI] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J (1998) Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA 95 10294–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.