Abstract
The rice (Oryza sativa) gene Xa27 confers resistance to Xanthomonas oryzae pv oryzae, the causal agent of bacterial blight disease in rice. Sequence analysis of the deduced XA27 protein provides little or no clue to its mode of action, except that a signal-anchor-like sequence is predicted at the amino (N)-terminal region of XA27. As part of an effort to characterize the biochemical function of XA27, we decided to determine its subcellular localization. Initial studies showed that a functional XA27-green fluorescent protein fusion protein accumulated in vascular elements, the host sites where the bacterial blight pathogens multiply. The localization of XA27-green fluorescent protein to the apoplast was verified by detection of the protein on cell walls of leaf sheath and root cells after plasmolysis. Similarly, XA27-FLAG localizes to xylem vessels and cell walls of xylem parenchyma cells, revealed by immunogold electron microscopy. XA27-FLAG could be secreted from electron-dense vesicles in cytoplasm to the apoplast via exocytosis. The signal-anchor-like sequence has an N-terminal positively charged region including a triple arginine motif followed by a hydrophobic region. Deletion of the hydrophobic region or substitution of the triple arginine motif with glycine or lysine residues abolished the localization of the mutated proteins to the cell wall and impaired the plant's resistance to X. oryzae pv oryzae. These results indicate that XA27 depends on the N-terminal signal-anchor-like sequence to localize to the apoplast and that this localization is important for resistance to X. oryzae pv oryzae.
Plant disease resistance (R) genes confer race-specific resistance to pathogens that have cognate avirulence (Avr) genes (Flor, 1971). The R protein presumably functions as part of a receptor complex that recognizes the Avr protein and subsequently initiates defense responses (Hammond-Kosack and Jones, 1997; Martin et al., 2003). The majority of R proteins fall into five classes based primarily upon the combination of a limited number of structural motifs that they contain, while a few other R proteins have novel structures (Dangl and Jones, 2001; Martin et al., 2003). One of the interesting aspects of R protein function is their subcellular localization. Several examples of direct physical interactions between R and Avr proteins have been demonstrated (Scofield et al., 1996; Tang et al., 1996; Jia et al., 2000; Leister and Katagiri, 2000; Kim et al., 2002). Viral effectors are present inside the plant cell, and the predicted structures of all known R proteins against viruses indicate that they are also intracellular (Burch-Smith et al., 2007). The tomato (Solanum lycopersicum) Cf proteins, which recognize extracellular Cladosporium fulvum Avr proteins (Lauge and De Wit, 1998), are localized to the plasma membrane (Rivas and Thomas, 2005). Fungal pathogen-directed R proteins can also be intracellular, as fungal Avr proteins are delivered to and function inside plant cells (Jia et al., 2000). Similarly, all bacteria-directed R proteins are predicted to be intracellular, with the exception of XA21 (Song et al., 1995), because most of the bacterial Avr effector proteins are secreted into the host cells through the bacterial type III secretion system (He et al., 2004).
However, many R proteins do not carry recognizable subcellular targeting signatures, and their actual subcellular localization needs to be determined experimentally. For instance, Arabidopsis (Arabidopsis thaliana) RPM1 and RPS2 are associated with cellular membranes, although they do not possess any canonical membrane-targeting domains (Boyes et al., 1998; Axtell and Staskawicz, 2003). This subcellular localization is consistent with the membrane localization of their corresponding Avr elicitors, AvrRpm1 and AvrRpt2, respectively (Nimchuk et al., 2000; Axtell and Staskawicz, 2003). Apart from the plasma membrane, Arabidopsis RRS1-R and its cognate Avr protein PopP2 colocalize to the nucleus, and the nuclear localization of RRS1-R is dependent on the presence of PopP2 (Deslandes et al., 2003). Recently, both tobacco (Nicotiana tabacum) N and barley (Hordeum vulgare) MLA10 were found to localize to the cytoplasm as well as the nucleus, and the nuclear retention of either R protein is indispensable for downstream signaling and defense (Burch-Smith et al., 2007; Shen et al., 2007). In these three cases, translocation of the R proteins during signaling might take place upon activation of the R proteins by the cognate Avr proteins (Deslandes et al., 2003; Burch-Smith et al., 2007; Shen et al., 2007). Rice (Oryza sativa) XA21 is a transmembrane receptor kinase that presumably recognizes an elicitor localized to the apoplast of rice cells, with its extracellular Leu-rich repeat domain (Song et al., 1995). The AvrXa21 molecule(s) corresponding to XA21 has yet to be identified, although it appears that it might be a sulfated protein secreted to the apoplast through a type I secretion system and involved in quorum sensing (Lee et al., 2006).
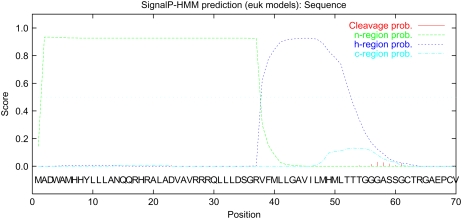
Bacterial blight, caused by Xanthomonas oryzae pv oryzae, is one of the most destructive bacterial diseases of rice (Mew, 1987). We previously reported the isolation of the resistance gene, Xa27, from rice (Gu et al., 2005). Xa27-dependent resistance is associated with the specific induction of the Xa27 gene by incompatible pathogens harboring avrXa27. Unlike other cloned R genes, the resistance specificity of Xa27 to various X. oryzae pv oryzae strains is determined by its promoter rather than by its gene product. Indeed, ectopic expression of the Xa27 coding region under the control of the rice PR1 promoter resulted in nonspecific resistance to both incompatible and compatible strains. The Xa27 gene encodes a predicted protein of 113 amino acid residues that lacks known functional domains that might provide a clue to its function. Interestingly, a signal-anchor-like sequence was predicted at the N-terminal region of XA27 by SignalP-HMM (http://www.cbs.dtu.dk/services/SignalP/; P = 0.790; Fig. 1). The signal-anchor-like sequence contains 37 positively charged amino acids, including a triple Arg motif from residues 27 to 29 (n-region), followed by a 20-amino acid hydrophobic region (h-region; Emanuelsson et al., 2007). Signal anchor sequences initiate translocation in the same manner as signal peptides, but they are not cleaved by signal peptidase after protein translocation, resulting in membrane association of the protein (von Heijne, 1988). As part of our effort to characterize the biochemical function of XA27, we carried out studies on its subcellular localization. We examined the function of its signal-anchor-like sequence and the relationship between XA27 localization and resistance to X. oryzae pv oryzae.
Figure 1.
Output of SignalP-HMM prediction of XA27 from the SignalP 3.0 server. A signal-anchor-like sequence was predicted at the N-terminal region of XA27, which contains an N-terminal positively charged region (n-region; residues 1–37) followed by a hydrophobic region (h-region; residues 38–57; Emanuelsson et al., 2007). The full-length amino acid sequence of XA27 was submitted to the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/) for prediction of the signal peptide or signal anchor of eukaryotes. Methods of both neural networks and hidden Markov models were used for prediction. Standard output format was selected.
RESULTS
Functional XA27-GFP Localizes to the Apoplast
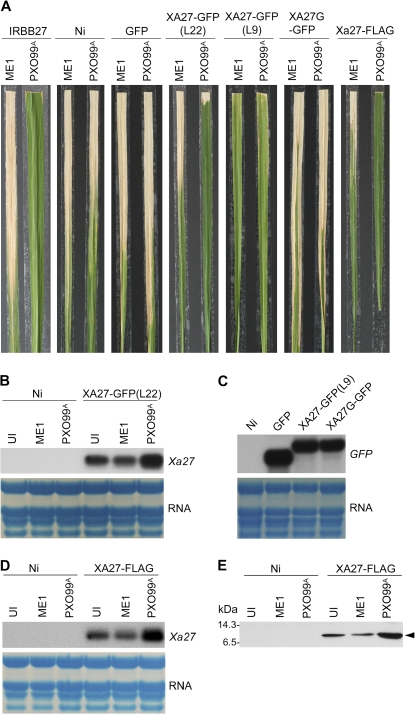
To investigate XA27 localization, we generated transgenic plants in a Nipponbare background that carried an XA27-GFP fusion gene under the control of either the native Xa27 promoter (PXa27:XA27-GFP:TXa27) or the maize (Zea mays) ubiquitin gene promoter (PUbi: XA27-GFP:TNos; Table I). Seven resistant transgenic PXa27:XA27-GFP:TXa27 lines were generated, and line 22 (L22) was selected for further analysis. L22 carried a single copy of the PXa27:XA27-GFP:TXa27 gene and retained race-specific resistance to PXO99A (Fig. 2A; Table II). The PXa27:XA27-GFP:TXa27 gene in L22 expressed at a low level constitutively, which might have resulted from leaky activity of the Xa27 promoter (Fig. 2B). The PXa27:XA27-GFP:TXa27 gene in L22 was induced after inoculation with PXO99A but not with compatible X. oryzae pv oryzae strain PXO99AME1, in which avrXa27 is disrupted (Gu et al., 2005; Fig. 2B). Thirty-three independent transgenic PUbi:XA27-GFP:TNos lines were produced, and their disease phenotype after inoculation with PXO99A varied from complete susceptibility to full resistance in the T0 generation (data not shown). Line 9 (L9) of PUbi:XA27-GFP:TNos was selected for further study. The PUbi:XA27-GFP:TNos gene in L9 was expressed constitutively at a high level (Fig. 2C). L9 conferred resistance to both PXO99A and PXO99AME1 in T1 and in subsequent generations (Fig. 2, A and C; Table II). Compared with L22 and line 8 (L8) of PUbi:GFP:TNos (Fig. 2A; Tables I and II), L9 displayed phenotypic changes, including inhibition of tillering, delay in flowering, stiff leaves, and early leaf senescence, which might have resulted from overexpression of the PUbi:XA27-GFP:TNos gene (data not shown).
Table I.
Constructs used in this study
| Construct | Gene of Interest in the Constructa | Reference |
|---|---|---|
| NA5.2 | Wild-type Xa27 (IRBB27 allele) | Gu et al. (2005) |
| pSSZ41 | PUbi:GFP:TNos | Kolesnik et al. (2004) |
| pCUXA27GFP | PUbi:XA27-GFP:TNos | This study |
| pC27XA27GFP | PXa27:XA27-GFP:TXa27 | This study |
| pC27XA27FLAG | PXa27:XA27-FLAG:TXa27 | This study |
| pCUN57GFP | PUbi:N57-GFP:TNos | This study |
| pCUN37GFP | PUbi:N37-GFP:TNos | This study |
| pCUN57GGFP | PUbi:N57G-GFP:TNos | This study |
| pCUN57KGFP | PUbi:N57K-GFP:TNos | This study |
| pCUXA27GGFP | PUbi:XA27G-GFP:TNos | This study |
| pC27XA27GGFP | PXa27:XA27G-GFP:TXa27 | This study |
| pC27XA27G | PXa27:XA27G:TXa27 | This study |
PUbi, Maize ubiquitin gene promoter; GFP, coding region of the GFP gene (accession no. AAB92477); TNos, terminator of the nopaline synthase gene (Nos); PXa27, Xa27 promoter; TXa27, Xa27 terminator; XA27-GFP, fusion gene encoding XA27-GFP; XA27-FLAG, fusion gene encoding XA27-FLAG; N57-GFP, fusion gene encoding N57-GFP; N37-GFP, fusion gene encoding N37-GFP; N57G-GFP, fusion gene encoding N57G-GFP; N57K-GFP, fusion gene encoding N57K-GFP; XA27G-GFP, fusion gene encoding XA27G-GFP; XA27G, Xa27 mutant encoding XA27G. For generation of fusion or mutated genes, see “Materials and Methods.”
Figure 2.
Generation of XA27-tagged lines. A, Disease phenotypes of XA27-tagged lines, control lines, and wild-type plants at 14 DAI with X. oryzae pv oryzae strains. Plants were inoculated with X. oryzae pv oryzae PXO99A harboring wild-type avrXa27 or avrXa27 mutant PXO99AME1 (ME1), and photographs were taken at 14 DAI. IRBB27, wild-type Xa27 plants; Ni, Nipponbare; GFP, L8 of PUbi:GFP:TNos; XA27-GFP(L22), L22 of PXa27:XA27-GFP:TXa27; XA27-GFP(L9), L9 of PUbi:XA27-GFP:TNos; XA27G-GFP, L18G of PUbi:XA27G-GFP:TNos; XA27-FLAG, L18F of PXa27:XA27-FLAG:TXa27. B, Expression of PXa27:XA27-GFP:TXa27 in uninoculated (UI) L22 plants or L22 plants at 3 DAI with X. oryzae pv oryzae strains PXO99AME1(ME1) and PXO99A. Nipponbare (Ni) was used as a control. C, Ubiquitin promoter-driven GFP, XA27-GFP, and XA27G-GFP gene expression in transgenic lines. GFP, L8 of PUbi:GFP:TNos; XA27-GFP(L9), L9 of PUbi:XA27-GFP:TNos; XA27G-GFP, L18G of PUbi:XA27G-GFP:TNos. Nipponbare (Ni) was used as a control. D, Expression of PXa27:XA27-FLAG:TXa27 in uninoculated (UI) L18F plants or L18F plants at 3 DAI with PXO99AME1(ME1) or PXO99A. Nipponbare (Ni) was used as a control. E, Expression of XA27-FLAG in uninoculated (UI) L18F plants or L18F plants at 3 DAI with PXO99AME1(ME1) or PXO99A. Nipponbare (Ni) was used as a control. XA27-FLAG was detected with anti-FLAG monoclonal antibodies. The molecular mass values of standard protein markers (Amersham Biosciences; RPN755) are shown in kilodaltons. The position of XA27-FLAG is indicated (arrowhead).
Table II.
Disease evaluation of selected transgenic lines and wild-type plants to X. oryzae pv oryzae strains PXO99A and PXO99AME1
| Line | Genea | Lesion Length (cm) and Disease Scoreb
|
|
|---|---|---|---|
| PXO99a | PXO99aME1 | ||
| IRBB27 | Xa27 | 0.3 ± 0.2 (R) | 25.4 ± 4.7 (S) |
| Nipponbare | xa27(Ni) | 14.5 ± 4.0 (S) | 15.0 ± 2.8 (S) |
| L8 | PUbi:GFP:TNos | 15.6 ± 2.7 (S) | 15.6 ± 2.6 (S) |
| L22 | PXa27:XA27-GFP:TXa27 | 0.7 ± 0.9 (R) | 13.3 ± 3.0 (S) |
| L18F | PXa27:XA27-FLAG:TXa27 | 0.2 ± 0.1 (R) | 6.3 ± 1.7 (MS) |
| L9 | PUbi:XA27-GFP:TNos | 0.9 ± 1.1 (R) | 3.0 ± 0.9 (R) |
| L12 | PUbi:N57-GFP:TNos | 16.0 ± 1.7 (S) | 16.6 ± 4.4 (S) |
| L2 | PUbi:N37-GFP:TNos | 14.3 ± 0.4 (S) | 15.9 ± 3.1 (S) |
| L5 | PUbi:N57G-GFP:TNos | 13.3 ± 2.8 (S) | 14.7 ± 1.6 (S) |
| L3 | PUbi:N57K-GFP:TNos | 15.0 ± 1.7 (S) | 15.2 ± 0.8 (S) |
| L18G | PUbi:XA27G-GFP:TNos | 10.4 ± 1.8 (S) | 13.9 ± 4.2 (S) |
| L46 | PXa27:XA27G-GFP:TXa27 | 11.6 ± 3.2 (S) | 14.9 ± 4.1 (S) |
For genes, refer to the genes of interest in the constructs in Table I. xa27(Ni), Susceptible allele of the Xa27 gene in Nipponbare.
The lesion length is the average of 16 infected leaves. The sd of the mean is indicated. MS, Moderately susceptible; R, resistant; S, susceptible.
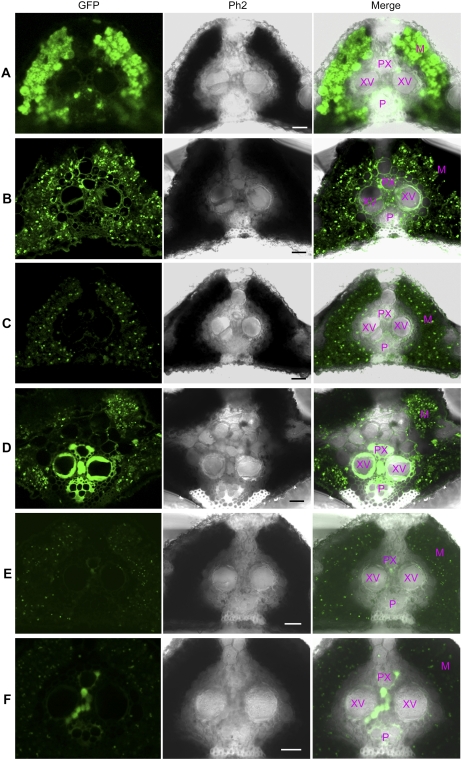
Leaf cross sections from the immediate vicinity of the infected sites in L8 of PUbi:GFP:TNos, L9 of PUbi:XA27-GFP:TNos, and L22 of PXa27:XA27-GFP:TXa27 were subjected to confocal microscopy. The GFP fluorescence in L8 was strong, especially in mesophyll parenchyma cells (Fig. 3A). Although the fluorescence intensity in L9 was weaker than that in L8, the XA27-GFP protein was evenly distributed among vascular bundles and mesophyll tissues (Fig. 3B). The expression of GFP in L8 or XA27-GFP in L9 did not change after bacterial inoculation with PXO99A (data not shown). A low level of XA27-GFP was detected in uninoculated L22 plants, due to leaky expression of the PXa27:XA27-GFP:TXa27 gene (Fig. 3C). XA27-GFP was strongly induced in vascular bundles of L22 plants at 3 d after inoculation (DAI) with PXO99A. It was mainly accumulated in the vascular elements, including xylem vessels, protoxylem, and phloem (Fig. 3D). However, the expression of PXa27:XA27-GFP:TXa27 in mesophyll parenchyma cells of L22 did not change significantly after bacterial inoculation (Fig. 3, C and D).
Figure 3.
Expression of GFP, XA27-GFP, and XA27G-GFP in leaf cross sections of transgenic plants. A, L8 of PUbi:GFP:TNos at 3 DAI with X. oryzae pv oryzae PXO99A. B, L9 of PUbi:XA27-GFP:TNos at 3 DAI with PXO99A. C, L22 of PXa27:XA27-GFP:TXa27. D, L22 at 3 DAI with PXO99A. E, L46 of PXa27:XA27G-GFP:TXa27. F, L46 at 3 DAI with PXO99A. M, Mesophyll parenchyma cells; P, phloem; Ph2, Phase Contrast 2; PX, protoxylem; XV, xylem vessel. Bars = 20 μm.
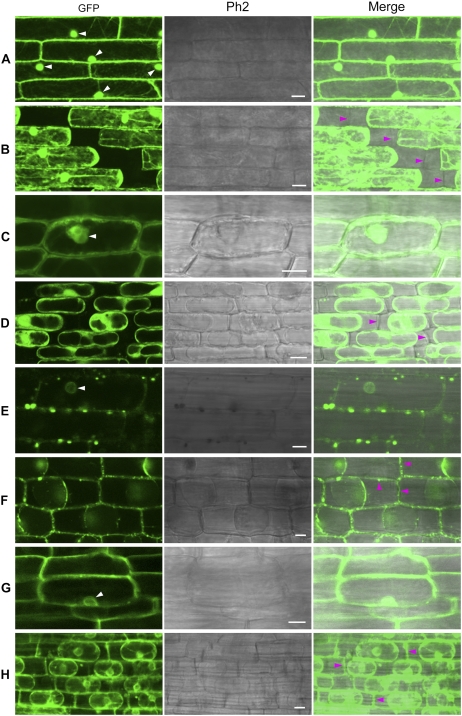
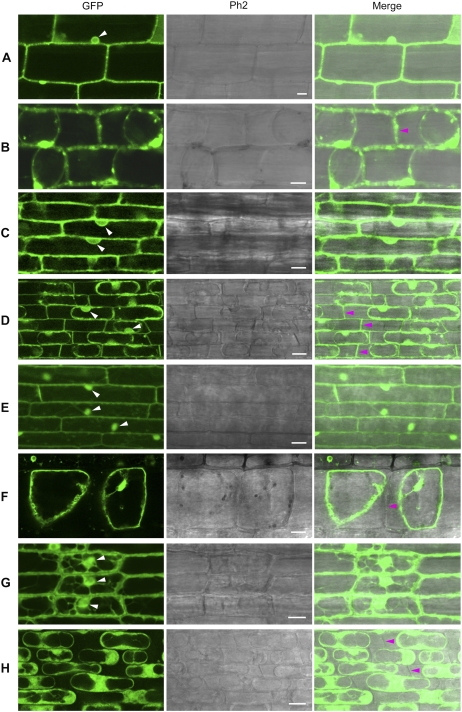
How does XA27-GFP come to be localized in xylem vessels and protoxylem, which consist of apoplast structures? One possibility is that it might be secreted from the neighboring xylem parenchyma cells. To test this possibility, we studied the localization of XA27-GFP in leaf sheath and root cells of L9 and L22. To make it easy to identify the cell wall, we induced plasmolysis in the leaf sheath or root tissues by treatment with a high-osmolarity solution. The control GFP protein in L8 was found in both the cytosol and the nucleus (Fig. 4, A and C; Supplemental Fig. S1A). After plasmolysis, no GFP protein was detected on the cell walls of leaf sheath or root cells of L8 (Fig. 4, B and D). In contrast, fluorescent spots appeared along the borders of leaf sheath cells of L9, which could be XA27-GFP-containing vesicles (see below; Fig. 4E). After plasmolysis, XA27-GFP was detected on the cell walls of both leaf sheath and root cells (Fig. 4, F and H). Compared with leaf sheath cells, XA27-GFP was more evenly distributed on root cell walls (Fig. 4G). In addition, XA27-GFP proteins were detected in the perinuclear region, producing a fluorescent nuclear halo (Fig. 4, E and G). 4′,6-Diamidino-2-phenylindole staining also indicated that XA27-GFP did not localize to the nucleus (Supplemental Fig. S1B). This perinuclear protein might be associated with the endoplasmic reticulum. Although a smaller amount of XA27-GFP was detected in L22, the localization patterns of XA27-GFP in the leaf sheath and the root cells of L22 were similar to that in L9 (Supplemental Fig. S2).
Figure 4.
Localization of GFP and XA27-GFP in leaf sheath and root cells of transgenic plants. A, Leaf sheath cells of L8 of PUbi:GFP:TNos. B, Leaf sheath cells of L8 after plasmolysis. C, Root cells of L8. D, Root cells of L8 after plasmolysis. E, Leaf sheath cells of L9 of PUbi:XA27-GFP:TNos. F, Leaf sheath cells of L9 after plasmolysis. G, Root cells of L9. H, Root cells of L9 after plasmolysis. The nuclei are indicated with white arrowheads. Cell walls are indicated with magenta arrowheads. Ph2, Phase Contrast 2. Bars = 10 μm.
XA27-FLAG Translocates to the Apoplast via Electron-Dense Vesicles
To further determine the subcellular localization of XA27, we fused XA27 with the FLAG epitope tag and performed immunogold electron microscopy to visualize XA27-FLAG in transgenic PXa27:XA27-FLAG:TXa27 plants (XA27-GFP could not be recognized by commercial anti-GFP antibodies). Forty-three independent transgenic PXa27:XA27-FLAG:TXa27 lines were generated. Line 18 (L18F) was selected for further analysis because it carried a single copy of the PXa27:XA27-FLAG:TXa27 gene. This line conferred complete resistance to PXO99A and displayed moderate susceptibility to PXO99AME1 (Fig. 2A; Table II). The PXa27:XA27-FLAG:TXa27 gene in L18F was expressed constitutively at a low level but was inducible upon inoculation with PXO99A (Fig. 2D). The XA27-FLAG protein in L18F was detected as a band with a molecular size of about 13 kD in immunoblot analysis with anti-FLAG monoclonal antibody (Fig. 2E).
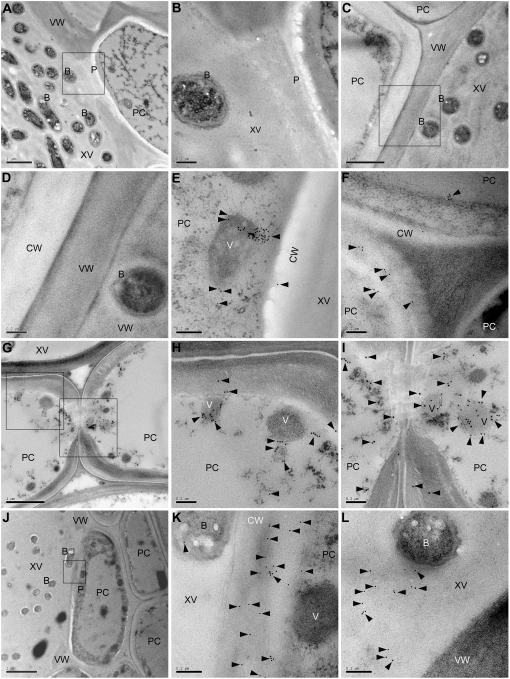
Leaf cross sections of L18F plants were subjected to immunogold electron microscopy. As depicted by the gold particles, XA27-FLAG was detected in the cytoplasm and the apoplast of the xylem parenchyma cells (Fig. 5, E–L). In uninoculated L18F plants, XA27-FLAG localized mainly to electron-dense vesicles in the cytoplasm and to the cell walls of xylem parenchyma cells (Fig. 5, E and F). After inoculation with PXO99A, XA27-FLAG protein was induced to higher levels in the xylem parenchyma cells of L18F plants. Electron-dense vesicles containing XA27-FLAG were abundant at the pits between xylem parenchyma cells (Fig. 5, G–I), between xylem vessels and parenchyma cells (Fig. 5, J–K), and between xylem parenchyma cells and mesophyll parenchyma cells (data not shown). The XA27-FLAG protein was also found in the xylem vessels of inoculated L18F plants, where it localized to lumina of the xylem vessels rather than being associated with bacteria or vessel walls (Fig. 5L). XA27-FLAG appears to be secreted from electron-dense vesicles in the cytoplasm to the apoplast, perhaps via exocytosis (Fig. 5H). No XA27-FLAG protein was found to localize to the nucleus or other organelles of xylem parenchyma cells in either uninoculated or inoculated L18F plants (data not shown). In control experiments, in which preimmune serum was used to replace anti-FLAG antibody, we did not observe concentrated labeling of vesicular structures in specimens of L18F (Fig. 5, A and B). Similar results were obtained with samples of Nipponbare lacking the transgene when anti-FLAG antibody was used (Fig. 5, C and D).
Figure 5.
Subcellular localization of XA27-FLAG in leaf vascular elements of L18F of PXa27:XA27-FLAG:TXa27 detected by immunogold electron microscopy. A, Xylem vessel and xylem parenchyma cells of L18F at 3 DAI with X. oryzae pv oryzae PXO99A. Bar = 1 μm. B, High magnification of the pit area indicated with a box in A. Bar = 0.2 μm. C, Xylem vessel and xylem parenchyma cells of Nipponbare at 3 DAI with PXO99A. Bar = 1 μm. D, High magnification of the cell wall between xylem vessel and xylem parenchyma cells indicated with a box in C. Bar = 0.2 μm. E, Electron-dense vesicles in xylem parenchyma cells near the pit between xylem vessel and xylem parenchyma cells of L18F. Bar = 0.2 μm. F, Cell walls of xylem parenchyma cells of L18F. Bar = 0.2 μm. G, Xylem vessel and xylem parenchyma cells of L18F at 3 DAI with PXO99A. Bar = 1 μm. H, High magnification of the area indicated with a box in G. Bar = 0.2 μm. I, High magnification of the pit area between two xylem parenchyma cells indicated with a box in G. Bar = 0.2 μm. J, Xylem vessel and xylem parenchyma cells of L18F at 3 DAI with PXO99A. Bar = 2 μm. K, High magnification of the pit area between xylem vessel and xylem parenchyma cells indicated with a box in J. Bar = 0.2 μm. L, Xylem vessel and cell wall of xylem parenchyma cells of L18F at 3 DAI with PXO99A. Bar = 0.2 μm. Samples in A and B were probed with preimmune serum. Samples in C to L were detected with anti-FLAG monoclonal antibody. The immune reaction was labeled with 10-nm (A–F, J–L) or 15-nm (G–I) gold-conjugated goat anti-mouse IgG antibody. Gold particles are indicated by arrowheads. B, Bacteria; CW, cell wall; P, pit; PC, xylem parenchyma cell; V, vesicle; VW, vessel wall; XV, xylem vessel.
The N-Terminal Signal-Anchor-Like Sequence Is Required for XA27 Localization
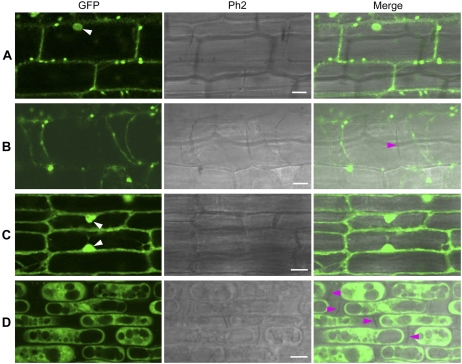
To determine whether the signal-anchor-like sequence is sufficient to explain the observed localization of XA27 to the apoplast and cell wall, we fused this sequence to GFP (N57-GFP) and generated 56 transgenic plants with the PUbi:N57-GFP:TNos gene (Table I). Line 12 (L12) of PUbi:N57-GFP:TNos was selected to study N57-GFP localization (Table II). The expression of the PUbi:N57-GFP:TNos gene in L12 was comparable to that of the PUbi:GFP:TNos gene in L8 (Supplemental Fig. S3). Like XA27-GFP, N57-GFP localized to the cell walls of leaf sheath and root cells (Fig. 6, A to D).
Figure 6.
Localization of N57-GFP and N37-GFP in leaf sheath and root cells of transgenic plants. A, Leaf sheath cells of L12 of PUbi:N57-GFP:TNos. B, Leaf sheath cells of L12 after plasmolysis. C, Root cells of L12. D, Root cells of L12 after plasmolysis. E, Leaf sheath cells of L2 of PUbi:N37-GFP:TNos. F, Leaf sheath cells of L2 after plasmolysis. G, Root cells of L2. H, Root cells of L2 after plasmolysis. The nuclei are indicated with white arrowheads. Cell walls are indicated with magenta arrowheads. Ph2, Phase Contrast 2. Bars = 10 μm.
To determine whether the h-region of the signal-anchor-like sequence is required for this localization, we deleted the h-region from the signal-anchor-like sequence and fused the remaining 37-amino acid n-region with GFP (N37-GFP). We generated 66 transgenic plants with the PUbi:N37-GFP:TNos gene (Table I). Line 2 (L2) of PUbi:N37-GFP:TNos was selected to study N37-GFP localization (Table II; Supplemental Fig. S3). After plasmolysis, no N37-GFP was detected on the cell walls of leaf sheath or of root cells (Fig. 6, E and F).
The triple Arg motif in the signal-anchor-like sequence of XA27 is conserved between XA27 and its paralogs in rice (Gu et al., 2005). To determine the function of this triple Arg motif in protein localization, we replaced it with Gly or Lys residues and generated transgenic plants with the mutated fusion genes PXa27:XA27G-GFP:TXa27, PUbi:XA27G-GFP:TNos, PUbi:N57G-GFP:TNos, and PUbi:N57K-GFP:TNos (Table I). Line 46 (L46) of PXa27:XA27G-GFP:TXa27, line 18 (L18G) of PUbi:XA27G-GFP:TNos, line 5 (L5) of PUbi:N57G-GFP:TNos, and line 3 (L3) of PUbi:N57K-GFP:TNos were selected to study the localization of these fusion proteins (Table II). Like the PXa27:XA27-GFP:TXa27 gene in L22, the PXa27:XA27G-GFP:TXa27 gene in L46 showed weak expression in uninoculated plants but was induced at xylem parenchyma cells after inoculation of L46 plants with PXO99A. However, the induced XA27G-GFP protein was trapped in xylem parenchyma cells and was not secreted into the apoplast or xylem vessel (Fig. 3, E and F). In addition, XA27G-GFP localized to the nucleus, detected by 4′,6-diamidino-2-phenylindole staining (Supplemental Fig. S1C). Although the expression of the PUbi:XA27G-GFP:TNos gene in L18G, the PUbi:N57G-GFP:TNos gene in L5, and the PUbi:N57K-GFP:TNos gene in L3 was comparable to that of the PUbi:XA27-GFP:TNos gene in L9 and the PUbi:N57-GFP:TNos gene in L12 (Fig. 2C; Supplemental Fig. S3), XA27G-GFP, N57G-GFP, and N57K-GFP failed to localize to the cell walls of either leaf sheath or root cells (Fig. 7; Supplemental Fig. S4).
Figure 7.
Localization of XA27G-GFP in leaf sheath and root cells of L18G of PUbi:XA27G-GFP:TNos. A, Leaf sheath cells. B, Leaf sheath cells after plasmolysis. C, Root cells. D, Root cells after plasmolysis. The nuclei are indicated with white arrowheads. Cell walls are indicated with magenta arrowheads. Ph2, Phase Contrast 2. Bars = 10 μm.
Localization of XA27 to the Apoplast Is Required for Disease Resistance
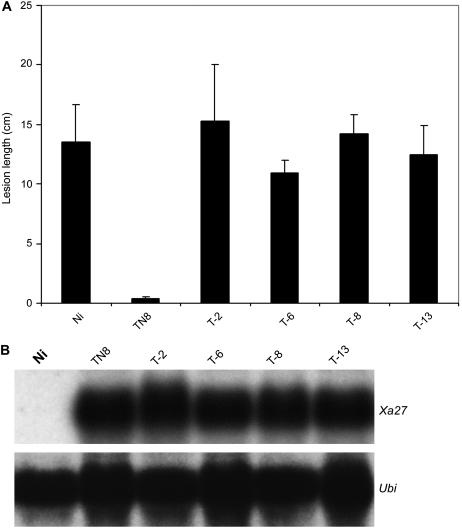
The failure of localization of XA27G-GFP to the apoplast in L18G of PUbi:XA27G-GFP:TNos and in L46 of PXa27:XA27G-GFP:TXa27 allowed us to ask whether the proper localization of the fusion protein is required for its function in conferring disease resistance. Plants expressing the XA27G-GFP proteins were not resistant to bacterial blight pathogens (Fig. 2A; Table II). These results suggest that the localization of XA27 to the apoplast is required for XA27-mediated disease resistance to X. oryzae pv oryzae. To further test this, we replaced the triple Arg motif in XA27 with Gly residues and generated transgenic plants with the PXa27:XA27G:TXa27 gene (Table I). Sixty-one independent transgenic lines were obtained, and they all carried the PXa27:XA27G:TXa27 gene (data not shown). Although the expression of PXa27:XA27G:TXa27 in these transgenic plants was comparable to that of the wild-type Xa27 transgene in transgenic line TN8 after inoculation with PXO99A (Fig. 8B), none of the XA27G transgenic plants conferred resistance to the incompatible strain (Fig. 8A).
Figure 8.
Disease evaluation of transgenic PXa27:XA27G:TXa27 plants to X. oryzae pv oryzae PXO99A and transgene expression after inoculation. A, Lesion length of bacterial blight on transgenic PXa27:XA27G:TXa27 plants at 14 DAI with PXO99A. Lesion lengths are average values from 16 inoculated leaves with sd. B, Expression of PXa27:XA27G:TXa27 in different transgenic plants (T-2, T-6, T-8, and T-13) at 3 DAI with PXO99A. Ni, Nipponbare; TN8, Xa27 transgenic line in the Nipponbare background (Gu et al., 2005).
DISCUSSION
Xa27 is specifically induced by AvrXa27 from incompatible pathogens (Gu et al., 2005). In vivo in IRBB27, the expression of Xa27 is normally tightly controlled and is not induced by any other biotic and abiotic stresses (Gu et al., 2005). Nonetheless, we found that constructs containing the Xa27 promoter showed constitutive low-level expression in transgenic plants. This was true whether the transgenic plants contained genomic clones of Xa27 under the control of its native promoter (L. Wu and Z. Yin, unpublished data) or fusion genes under the control of the Xa27 promoter. We speculate that the Xa27 promoter is sensitive to its position on the rice chromosome and that the constitutive expression might be due to a position effect on the transgenes in rice chromosomes.
Leaky expression of XA27 or its functional derivatives was found to provide enhanced resistance to compatible pathogen strains. This resistance was dosage dependent. In this study, we selected transgenic lines that retained resistance specificity to incompatible pathogens. The transgenes in these lines were inducible by AvrXa27. We believe that these lines are suitable for determining the localization of XA27 or its fusion proteins, because these fusion proteins confer resistance to the pathogen, indicating their functionality. In addition, the localization of XA27-GFP to the cell walls of leaf sheath and root cells in L22 of PXa27:XA27-GFP:TXa27 and in L9 of PUbi:XA27-GFP:TNos gave similar results, which indicates that subcellular localization of XA27-GFP is determined by the nature of the protein. In this case, the localization cannot be explained by any tissue specificity in transgene expression due to the promoter.
The apoplast is the extraprotoplastic matrix of plant cells, consisting of all compartments from the external face of the plasmalemma to the cell wall (Dietz, 1997). The apoplast provides a physical barrier against pathogen attack. It also plays an important role in signaling and defense during plant-pathogen interaction, through the presence of several extracellular enzymes and proteins located in the apoplast or associated with the cell wall (Dietz, 1997; Edreva, 2005; Huckelhoven, 2007). X. oryzae pv oryzae is a vascular pathogen that enters the rice leaf typically through hydathodes at the leaf tip and leaf margin. It multiplies in the intercellular spaces of the underlying epitheme and thereafter moves to the xylem vessels to cause systemic infection (Noda and Kaku, 1999). The bacteria rely on a type III secretion system to inject virulence effectors into xylem parenchyma cells, probably through the pits between xylem vessels and xylem parenchyma cells. Xylem vessels consist of giant intercellular spaces and form a special apoplast structure. Therefore, the local resistance at xylem parenchyma cells and xylem vessels is important for rice to counterattack the vascular pathogen. For example, the increase in peroxidase activity in extracellular spaces and the accumulation of cationic peroxidase in xylem vessels have been detected during interactions of rice and X. oryzae pv oryzae, especially during incompatible interactions (Reimers et al., 1992; Young et al., 1995; Hilaire et al., 2001).
Although the biochemical function of XA27 remains to be determined, its localization to xylem vessels may contribute to local resistance to the pathogen at the infection site. XA27 appears to be secreted to the lumina of xylem vessels through the pits between xylem vessels and xylem parenchyma cells. Since xylem parenchyma cells were not suitable for plasmolysis and analysis by confocal microscopy, the localization of XA27-GFP and its derivatives to the apoplast was examined in the leaf sheath cells and in roots of transgenic plants. As the extracellular space or apoplast of leaf sheath cells and root cells is limited, compared with that of xylem vessels, the secreted XA27-GFP protein mainly localizes to the cell wall in these cases. This suggests an extracellular or apoplastic localization of XA27.
Although we lack the means to detect the location of the endogenous XA27 protein, our studies indicate that XA27-FLAG and XA27-GFP proteins localized to the apoplast. XA27 might be secreted to the apoplast by exocytosis, as XA27-FLAG was detected in both cytoplasmic electron-dense vesicles and in the apoplast. The signal-anchor-like sequence of XA27 contains a triple Arg motif, which appears to be essential for both its localization and function. Mutation of this sequence abolished XA27 localization and impaired its ability to confer disease resistance. Similar Arg motifs are also found in the signal peptides of proteins translocated through the twin Arg translocation (Tat) pathway (Robinson and Bolhuis, 2004; Muller and Klosgen, 2005). The Tat pathway is responsible for the export of folded proteins across the cytoplasmic membrane of bacteria or thylakoid membranes of chloroplast in higher plants. Protein transported by the Tat pathway possesses a cleavable signal peptide harboring a twin Arg consensus motif (Robinson and Bolhuis, 2004). The only exception is the Rieske protein from Paracoccus denitrificans, which depends on an uncleavable N-terminal Tat signal sequence to anchor it to the cytoplasmic membrane (Bachmann et al., 2006). However, we failed to identify a Tat signal sequence at the N-terminal region of XA27 with the Tat signal prediction server TatP 1.0 (http://www.cbs.dtu.dk/services/TatP/; data not shown).
In conclusion, we have found that the inducible R protein XA27 localizes to the apoplast. This localization depends on a signal-anchor-like sequence at the N-terminal region of XA27 and is required for XA27-mediated resistance to X. oryzae pv oryzae. The identification of XA27 localization to the apoplast and its localization signal will facilitate further characterization of the biochemical function of the R protein.
MATERIALS AND METHODS
Constructs
The constructs used in this study (Table I) were made based on binary vectors pC1300 or pC1305.1 and verified by DNA sequencing. The XA27-GFP fusion gene was generated by PCR with the Xa27 coding region amplified from NA5.2 (Gu et al., 2005) and the GFP coding region amplified from pSSZ41 (Kolesnik et al., 2004). The fused PCR products were cloned into pC1305.1 to generate pCXA27GFP. The PstI fragment of the maize (Zea mays) ubiquitin promoter from pSSZ41 was inserted 5′ of the XA27-GFP fusion gene in pCXA27GFP to generate pCUXA27GFP. A fragment containing a partial XA27-GFP fusion gene was amplified from pCUXA27GFP and cloned into the SacI site of NA5.2 to generate pC27XA27GFP. The XA27-FLAG fusion gene was generated by PCR and cloned into the SpeI and SacI sites of NA5.2 to generate pC27XA27FLAG. The N57-GFP fusion gene was generated by fusing the 5′ coding region of Xa27, encoding the first 57 amino acid residues, with the GFP gene. The fusion gene was used to replace XA27-GFP in pCUXA27GFP to generate pCUN57GFP. Similar methods were used to produce pCUN37GFP, in which the N-terminal 37 amino acid residues of XA27 were fused with GFP. The triple Arg residues in XA27 (residues 27–29) were replaced with triple Gly residues by PCR. The mutated Xa27 gene was used to replace wild-type Xa27 in NA5.2 to generate pC27XA27G. The mutated Xa27 gene was tagged with GFP to give XA27G-GFP. This XA27G-GFP fusion gene was used to replace XA27-GFP in pCUXA27GFP and pC27XA27GFP to produce pCUXA27GGFP and pC27XA27GGFP, respectively. Similar methods were used to generate constructs pCUN57GGFP and pCUN57KGFP, in which the triple Arg residues in the N57-GFP fusion gene in pCUN57GFP were replaced with Gly and Lys residues, respectively.
Plant Material and Growth Conditions
Rice (Oryza sativa) IRBB27 is a near-isogenic line of Xa27 in the IR24 background (Gu et al., 2004; Table I). Nipponbare is a japonica variety carrying a susceptible Xa27 allele [xa27(Ni); Table I]. TN8 is an Xa27 transgenic line in the Nipponbare background carrying an Xa27 genomic subclone of the PstI fragment (Gu et al., 2005; Table I). All rice plants, including those inoculated with Xanthomonas oryzae pv oryzae strains, were grown in a greenhouse at temperatures ranging from 32°C during the day to 26°C at night, relative humidity averaged 85%, and the photoperiod was 12 to 13 h.
Rice Transformation
Agrobacterium tumefaciens-mediated transformation of Nipponbare rice was carried out as described previously (Yin and Wang, 2000).
Bacterial Blight Inoculation and Disease Scoring
The X. oryzae pv oryzae strains were cultured on PSA medium (10 g L−1 peptone, 10 g L−1 Suc, 1 g L−1 Glu, and 16 g L−1 bacto-agar, pH 7.0) for 2 to 3 d at 28°C. Bacteria were collected and suspended in sterile water at an optical density of 0.5 at 600 nm. Bacterial blight inoculation was carried out using the leaf-clipping method (Kauffman et al., 1973). The disease symptoms were scored according to the criteria described previously (Gu et al., 2004).
Confocal Laser Scanning Microscopy
GFP fluorescence in transgenic plants was examined on the cells of leaves, leaf sheaths, and roots. Leaves of inoculated or uninoculated plants were manually and transversely sectioned with a surgical blade (no. 10). Leaf sheath sections of about 3 × 3 mm2 and root tips of about 1 cm long were cut from 10-d-old plants. Samples were mounted in water for examination and photography. Plasmolysis was induced on leaf sheath or root cells by incubating the samples in 10% (w/v) mannitol solution for 1 h and mounting in the same solution for examination. Samples were examined for GFP fluorescence with a confocal microscope (Zeiss LSM 510 META) using the excitation/emission combination of 488/505 to 530 nm band pass. The bright-field image was taken simultaneously using Phase Contrast 2 optics.
RNA Gel-Blot Analysis
Total RNA was isolated from rice leaves using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions. About 20 μg of total RNA was loaded in each lane for RNA gel-blot analysis. The RNA loading was assessed by staining RNA blots with methylene blue or hybridizing RNA blots with the rice Ubiquitin2 gene (Ubi). DNA probes were labeled with [32P]dCTP using the Rediprime II random prime labeling system (Amersham Biosciences).
Western-Blot Analysis
About 20 μg of total proteins from transgenic plants was separated on 12% SDS polyacrylamide gels followed by blotting onto nitrocellulose membranes. XA27-FLAG proteins were detected with an anti-FLAG-M2 monoclonal antibody (Sigma) and horseradish peroxidase-coupled secondary antibody (Bio-Rad).
Immunogold Electron Microscopy
Immunogold electron microscopy was carried out according to the procedure described previously (Chye et al., 1999). Leaf cross sections of about 3 mm in length were fixed in a solution of 0.5% glutaraldehyde and 2% paraformaldehyde in 0.1 m phosphate buffer for 4 h under vacuum. Specimens were washed with 50 mm phosphate buffer for 45 min. After dehydration in a graded ethanol series, specimens were infiltrated in LR White resin (EMS) and embedded in gelatin capsules. Specimens were sectioned at 80 nm using a Leica Ultracut microtome and mounted on Formvar-coated slotted grids. XA27-FLAG proteins were detected with anti-FLAG monoclonal antibody (Sigma) followed by labeling with 10- or 15-nm gold-conjugated goat anti-mouse IgG antibody (EMS). Mouse preimmune serum was used to substitute anti-FLAG monoclonal antibody in control experiments. Samples on grids were further stained with 2% uranyl acetate and 1% lead citrate. Samples were examined with a transmission electron microscope (JEM-1230; JEOL) operating at 120 kV and photographed with a digital microphotography system (Gatan).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AAY54164.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Localization of GFP, XA27-GFP, and XA27G-GFP in leaf sheath cells of transgenic plants and 4′,6-diamidino-2-phenylindole staining of nuclei.
Supplemental Figure S2. Localization of XA27-GFP in leaf sheath and root cells of L22 of PXa27:XA27-GFP:TXa27.
Supplemental Figure S3. Transgene expression in GFP-tagged lines.
Supplemental Figure S4. Localization of N57G-GFP and N57K-GFP in leaf sheath and root cells of transgenic plants.
Supplemental Protocol S1. 4′,6-Diamidino-2-phenylindole staining and GFP fluorescence.
Supplementary Material
Acknowledgments
We thank Xuezhi Ouyang, Ao Yin, Qingwen Lin, Dongjiang Wang, and Tze Yee Teo for technical assistance, the Center for the Application of Molecular Biology to International Agriculture for binary vectors pC1300 and pC1305.1, and Stephen Cohen and Mithilesh Mishra for critical reading of the manuscript.
This work was supported by intramural research funds from the Temasek Life Sciences Laboratory (to Z.Y.) and by a grant from the Agri-Food and Veterinary Authority of Singapore (to Z.Y.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Zhongchao Yin (yinzc@tll.org.sg).
The online version of this article contains Web-only data.
References
- Axtell MJ, Staskawicz BJ (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112 369–377 [DOI] [PubMed] [Google Scholar]
- Bachmann J, Bauer B, Zwicker K, Ludwig B, Anderka O (2006) The Rieske protein from Paracoccus denitrificans is inserted into the cytoplasmic membrane by the twin-arginine translocase. FEBS J 273 4817–4830 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Nam J, Dangl JL (1998) The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA 95 15849–15854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP (2007) A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol 5 e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chye ML, Huang BQ, Zee SY (1999) Isolation of a gene encoding Arabidopsis membrane-associated acyl-CoA binding protein and immunolocalization of its gene product. Plant J 18 205–214 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG (2001) Plant pathogens and integrated defense responses to infection. Nature 411 826–833 [DOI] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ (1997) Function and responses of the leaf apoplast under stress. Prog Bot 58 221–254 [Google Scholar]
- Edreva A (2005) Pathogenesis-related proteins: research progress in the last 15 years. Gen Appl Plant Physiol 31 105–124 [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protocols 2 953–971 [DOI] [PubMed] [Google Scholar]
- Flor HH (1971) Current status of the gene-for-gene concept. Annu Rev Phytopathol 9 275–296 [Google Scholar]
- Gu K, Tian D, Yang F, Wu L, Sreekala C, Wang D, Wang GL, Yin Z (2004) High-resolution genetic mapping of Xa27(t), a new bacterial blight resistance gene in rice, Oryza sativa L. Theor Appl Genet 108 800–807 [DOI] [PubMed] [Google Scholar]
- Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, et al (2005) R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435 1122–1125 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JDG (1997) Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48 575–607 [DOI] [PubMed] [Google Scholar]
- He SY, Nomura K, Whittam TS (2004) Type III protein secretion mechanism in mammalian and plant pathogens. Biochim Biophys Acta 1694 181–206 [DOI] [PubMed] [Google Scholar]
- Hilaire E, Young SA, Willard LH, McGee JD, Sweat T, Chittoor JM, Guikema JA, Leach JE (2001) Vascular defense responses in rice: peroxidase accumulation in xylem parenchyma cells and xylem wall thickening. Mol Plant Microbe Interact 14 1411–1419 [DOI] [PubMed] [Google Scholar]
- Huckelhoven R (2007) Cell wall-associated mechanisms of disease resistance and susceptibility. Annu Rev Phytopathol 45 101–127 [DOI] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman HE, Reddy APK, Hsieh SPY, Merca SD (1973) An improved technique for evaluating resistance to rice varieties of Xanthomonas oryzae. Plant Dis Rep 57 537–541 [Google Scholar]
- Kim YJ, Lin NC, Martin GB (2002) Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109 589–598 [DOI] [PubMed] [Google Scholar]
- Kolesnik T, Szeverenyi I, Bachmann D, Kumar CS, Jiang S, Ramamoorthy R, Cai M, Ma ZG, Sundaresan V, Ramachandran S (2004) Establishing an efficient Ac/Ds tagging system in rice: large-scale analysis of Ds flanking sequences. Plant J 37 301–314 [DOI] [PubMed] [Google Scholar]
- Lauge R, De Wit PJ (1998) Fungal avirulence genes: structure and possible functions. Fungal Genet Biol 24 285–297 [DOI] [PubMed] [Google Scholar]
- Lee SW, Han SW, Bartley LE, Ronald PC (2006) Unique characteristics of Xanthomonas oryzae pv. oryzae AvrXa21 and implications for plant innate immunity. Proc Natl Acad Sci USA 103 18395–18400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister RT, Katagiri F (2000) A resistance gene product of the nucleotide binding site-leucine rich repeats class can form a complex with bacterial avirulence proteins in vivo. Plant J 22 345–354 [DOI] [PubMed] [Google Scholar]
- Martin GB, Bogdanove AJ, Sessa G (2003) Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol 54 23–61 [DOI] [PubMed] [Google Scholar]
- Mew TW (1987) Current status and future prospects of research on bacterial blight of rice. Annu Rev Phytopathol 25 359–382 [Google Scholar]
- Muller M, Klosgen RB (2005) The Tat pathway in bacteria and chloroplasts: review. Mol Membr Biol 22 113–121 [DOI] [PubMed] [Google Scholar]
- Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL (2000) Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101 353–363 [DOI] [PubMed] [Google Scholar]
- Noda T, Kaku H (1999) Growth of Xanthomonas oryzae pv. oryzae in planta and in guttation fluid of rice. Ann Phytopathological Soc Jpn 65 9–14 [Google Scholar]
- Reimers PJ, Guo A, Leach JE (1992) Increased activity of a cationic peroxidase associated with an incompatible interaction between Xanthomonas oryzae pv. oryzae and rice (Oryza sativa). Plant Physiol 99 1044–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas S, Thomas CM (2005) Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu Rev Phytopathol 43 395–436 [DOI] [PubMed] [Google Scholar]
- Robinson C, Bolhuis A (2004) Tat-dependent protein targeting in prokaryotes and chloroplasts. Biochim Biophys Acta 1694 135–147 [DOI] [PubMed] [Google Scholar]
- Scofield SR, Tobias CM, Rathjen JP, Chang JH, Lavelle DT, Michelmore RW, Staskawics BJ (1996) Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274 2063–2065 [DOI] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315 1098–1103 [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, et al (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270 1804–1806 [DOI] [PubMed] [Google Scholar]
- Tang X, Frederick RD, Zhou J, Halterman DA, Jia Y, Martin GB (1996) Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274 2060–2063 [DOI] [PubMed] [Google Scholar]
- von Heijne G (1988) Transcending the impenetrable: how proteins come to terms with membranes. Biochim Biophys Acta 947 307–333 [DOI] [PubMed] [Google Scholar]
- Yin Z, Wang GL (2000) Evidence of multiple complex patterns of T-DNA integration into the rice genome. Theor Appl Genet 100 461–470 [Google Scholar]
- Young SA, Guo A, Guikema JA, White FF, Leach JE (1995) Rice cationic peroxidase accumulates in xylem vessels during incompatible interactions with Xanthomonas oryzae pv. oryzae. Plant Physiol 107 1333–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.