Abstract
Nanoviruses are a family of plant viruses that posses a genome of multiple circular single-stranded DNA (ssDNA) components and are strikingly similar in their replication mode to the plant geminiviruses and to the circoviruses that infect birds or mammals. These viruses multiply by rolling circle replication using virus-encoded multifunctional replication initiator proteins (Rep proteins) that catalyze the initiation of replication on a double-stranded DNA (dsDNA) intermediate and the resolution of the ssDNA into circles. Here we report the solution NMR three-dimensional structure of the endonuclease domain from the Master Rep (M-Rep) protein of faba bean necrotic yellows virus (FBNYV), a representative of the nanoviruses. The domain comprises amino acids 2-95 (M-Rep2-95) and its global fold is similar to those previously described for the gemini- and circovirus Rep endonuclease domain, consisting of a central 5-stranded antiparallel β-sheet covered on one side by an α-helix and irregular loops and on the other, more open side of the domain, by an α-helix containing the catalytic tyrosine residue (the catalytic helix). Longer domain constructs extending to amino acids 117 and 124, were also characterized. They contain an additional α-helix, are monomeric and exhibit catalytic activity indistinguishable from that of M-Rep2-95. The binding site for the catalytic metal was identified by paramagnetic broadening and maps to residues on the exposed face of the central β-sheet. A comparison with the previously determined Rep endonuclease domain structures of tomato yellow leaf curl Sardinia virus (TYLCSV), a geminivirus, and that of porcine circovirus type 2 (PCV2) Rep allows the identification of a positively charged surface that is most likely involved dsDNA binding, and reveals common features shared by all endonuclease domains of nanovirus, geminivirus, and circovirus Rep proteins.
Nanoviruses possess the smallest known virions and the smallest individual genome components of all plant viruses (1) and are causative disease agents for a variety of important food and fodder legumes (2). The nanovirus genome is multipartite and consists of 6 to 8 molecules of circular ssDNA, ranging in size from 977 to 1111 nucleotides, individually encapsidated in small icosahedral virions of 17–20 nm in diameter. Each component encodes, in virion (+) sense polarity, a single protein and contains a common region that is part of the viral origin of replication (3). This region is partially conserved among all genomic components of a given nanovirus and contains sequence elements (including a common nonanucleotide sequence flanked by inverted repeats that may form a hairpin-loop structure) functionally equivalent to those found in the intergenic regions of plant geminiviruses and animal circoviruses (4, 5), that possess only one or two genomic components. The replication of the multiple nanovirus genomic DNAs is initiated by a single replication initiator protein, the so-called master Rep protein (M-Rep) (6), that cleaves a specific phosphodiester bond in the conserved nonanucleotide sequence at the viral replication origin. Despite the occurrence of multiple additional Rep proteins encoded by other DNA molecules associated with the nanoviruses, solely the M-Rep is required and sufficient to catalyze replication initiation of the nanovirus genomic DNAs (3, 7, 8). M-Rep is a multifunctional protein of ~33 kDa involved in both initiation and termination of rolling circle replication. It is related in sequence and biological function to the rolling circle replication initiator proteins of gemini- and circoviruses.
The distinct biochemical activities of Rep proteins from gemini-, circo- and nanoviruses are associated with different parts of the protein: (1) Sequence specific recognition of the replication origin and endonuclease activity, i.e. DNA cleavage and joining at the replication origin, reside in the N-terminal region (9–13). (2) Oligomerization of geminivirus Rep has been assigned to the central region, comprising amino acids 119-180 (12), although a corresponding domain has not yet been identified for nanovirus or circovirus Rep proteins. (3) The ATPase activity, essential for virus replication (14), resides in the C-terminal part of Rep from around amino acid 180 onwards (15), and very recently, helicase activity of Rep or its ATPase domain oligomers have been demonstrated for two different geminivirus Rep proteins (16, 17).
Amino acids involved in the different activities of the replication initiator proteins constitute characteristic motifs conserved throughout large families of genetic entities that multiply by rolling circle replication, including gemini-, circo- and nanoviruses (18, 19). In particular, three conserved sequence motifs of rolling circle replication initiator proteins (19) are localized in the N-terminal endonuclease domain of Rep. The recently determined structures of the endonuclease domains of the Rep proteins from the geminivirus tomato yellow leaf curl Sardinia virus (TYLCSV) (13) and from porcine circovirus type 2 (PCV2) (20) revealed structural and biochemical features corroborating the functional roles of these sequence motives (see below) and further rationalized their conservation in the RCR initiator proteins. Since nanovirus replication initiator proteins, including the M-Rep, are smaller than those of gemini- and circoviruses, and since their amino acid motifs are slightly different, it is instructive to determine and compare the structure of the endonuclease domain of a nanovirus M-Rep protein with those of the corresponding gemini- and circoviruses. Interestingly, infectivity of cloned viral DNAs has only been achieved for a single nanovirus to date, namely faba bean necrotic yellows virus (FBNYV), an expanding pathogen of legume crops, rendering it especially important and amenable to reverse genetics (21). Origin-specific DNA cleavage and nucleotidyl transfer activities have been demonstrated in vitro for the complete FBNYV M-Rep protein, with tyrosine 79 identified as the catalytic residue (3). In addition, divalent Mg2+ or Mn2+ cations were found to be essential for DNA cleavage by two different nanovirus M-Rep proteins (3, 22).
Here we report the three-dimensional solution NMR structure of the catalytic domain of the M-Rep protein of FBNYV and further characterize its in vitro endonuclease activity. In addition, we investigated the metal dependence of the DNA cleavage reaction, and identified residues involved in metal binding. Finally, a comparative analysis of the present structure with those of previously determined representative Rep endonuclease domains of a geminivirus (13) and a circovirus (20) is presented.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
A putative FBNYV Master Rep (GenBank accession Y11405) endonuclease domain comprising amino acids 1-95 was cloned by PCR introducing Ndel (5′) and BamHI (3′) restriction sites into twoplasmids, pET3a and pET15b. The protein overexpressed from pET15b contains a 20 amino acid polyhistidine-containing tag (Tag20) at its N-terminus (i.e. Tag20-M-Rep1-95). Two further constructs were designed, containing extensions of the putative domain past the catalytic tyrosine residue, comprising one or two potential α-helices. These consisted of residues 1-117 and 1-124 (M-Rep1-117 and M-Rep1-124), respectively, and were cloned into a pET28a vector (Novagen) which contains a prescission protease cleavage site after the polyhistidine sequence, resulting in a polypeptide with an extra N-terminal tag of 22 residues (Tag22). Another variant of the pET3a M-Rep1-95 construct that included an extra methionine preceding Met1 was also constructed by site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis kit (Stratagene).
All proteins were expressed in E.coli strain Rosetta BL21 (DE3) plysS (Novagen). Uniformly 15N and/or 13C-labeled proteins were prepared by growth in minimal media containing 15NH4Cl and/or 13C-glucose as the sole nitrogen and carbon sources, respectively. Cell mass was produced in rich medium, followed by exchange of cells into a small volume (typically 1/3) of labeled medium at high cell density (23). Cells were induced with 0.5 mM IPTG after one hour, allowing for growth recovery and unlabeled metabolite clearance, and grown overnight at 18 °C. For the M-Rep1-95 constructs the protein from the soluble fraction was purified in three steps: two ion exchange chromatography steps (GE Healthcare) using a Q-sepharose column (which excludes the protein) and a SP-sepharose column (which retains the protein), followed by size exclusion chromatography on Superdex 75 (GE Healthcare). The load and wash buffer for the Q column was 20 mM sodium phosphate (pH 6.6), 0.3 M NaCl and 1 mM DTT, and NaCl was reduced to 0.1 M NaCl for the SP column. For elution, the same buffer but with 1 M NaCl was used in both ion exchange columns. For the polypeptides Tag19-M-Rep1-95, Tag21-M-Rep1-117 and Tag21-M-Rep1-124 (see results) purification was achieved in a two step protocol: (1) affinity chromatography on Talon resin (Invitrogen) using immobilized cobalt, and (2) gel filtration on Superdex 75. Samples for NMR analysis contained 0.6–0.8 mM M-Rep2-95 protein (see results) in 20 mM sodium phosphate buffer (pH 6.6), 0.6 M NaCl, 0.01 % NaN3 and 1 mM DTT. In the case of Tag21-M-Rep1-117 and Tag21-M-Rep1-124 the highest concentration obtainable was 0.4 mM. Proteolytic cleavage with prescission protease (1 h at 4 °C) was carried out on Tag21-containing 15N-labelled proteins, and cleavage products were examined by 2D 1H-15N NMR, without separation, in order to identify resonances belonging to the tags. This enabled us to directly analyze the spectra of the Rep extension constructs, which were recorded using the non-cleaved, Tag21-containing proteins, because cleavage of the affinity tags resulted in a significantly lower solubility, insufficient for the 3D NMR experiments necessary to assign their NMR resonances.
Endonuclease Activity Assays
Cleavage assays were carried out with each purified domain (M-Rep1-95, M-Rep2-95, Tag21-M-Rep1-117, and Tag21-M-Rep1-124, see results) using a 12mer ssDNA oligonucleotide containing the conserved nonamer sequence on the viral strand of FBNYV (F12=5′-TAGTATT^ACCCC-3′, where ^ denotes the cleavage site and the nonamer sequence is underlined). Reactions included 20 μM protein in 20 mM Tris pH 7.4, 0.1 M NaCl, in the absence or presence of 20 μM of the oligonucleotide F12, in a final volume of 10 μl. Samples were incubated at 37 °C for 5 minutes, followed by the addition of 2x SDS-PAGE sample buffer and heating for 5 minutes at 95–100 °C before loading onto the gel. Covalent Rep-DNA adducts were separated by SDS-PAGE (12% polyacrylamide) and stained with Coomassie for visualization. The effects of divalent metal ions on the cleavage by the different constructs were followed by including 2.5 mM of MgCl2, MnCl2, ZnCl2 or CaCl2 in the respective reaction buffer prepared without any metals. Each series contained one sample in which the divalent metal was prevented from interacting with protein and/or DNA by adding 30 mM EDTA before addition of the ssDNA substrate.
NMR Spectroscopy
All NMR experiments were carried out at 293 K (20 °C) on Bruker Avance 600 (with a cryogenic probe) and 700 MHz spectrometers. 1H, 15H and 13C backbone and side chain resonances of M-Rep2-95 and Tag21-M-Rep1-117 were assigned by three-dimensional (3D) double- and triple-resonance NMR experiments, as reported (24). Interproton distance constraints for M-Rep2-95 were derived from 3D and four-dimensional (4D) 15N- and 13C-separated nuclear Overhauser enhancement (NOE) experiments. Torsion angle restraints were derived from backbone chemical shifts using the program TALOS (25). Heteronuclear 3J couplings were measured by quantitative J-correlation Spectroscopy (26). Heteronuclear 15N{1H} NOE experiments were acquired at 700 MHz in an interleaved fashion using an interscan delay of 6 s, and echo-antiecho gradient selection for pure phase signals in the indirect dimension and water suppression. The 15N{1H} NOE value for each amide was determined as the intensity ratio of the correlation in the 1H-saturated experiment and that in the reference, non-saturated experiment. Errors bars for the 15N{1H} NOEs were determined from the baseline noise of the two spectra.
Structure Calculation
Structures were calculated from the experimental constraints in torsion angle space using CYANA 2.1. (27). Upper-limit distance constraints of 2.7, 3.3, 5.0 and 5.5 Å (with 0.5 Å added for NOEs involving methyl protons) were used, corresponding to strong-, medium-, weak- and very weak-intensity NOE cross-peaks, respectively. Additional NOEs were automatically assigned (in the 3D 15N-separated NOESY and 2D NOESY recorded in D2O) using the algorithm NOAH (28). All methyl resonances of Val and Leu residues were stereospecifically assigned using a sample produced with 10% 13C-glucose (29). Structural statistics were calculated with CYANA (30), MOLMOL (31), and PROCHECK (32), and figures were generated with MOLMOL and Swiss-PdbViewer 3.7 (33). Structural superpositions were carried out with Superpose (34).
Mapping of Divalent Metal Binding Site by NMR
The analysis of divalent metal binding was performed by comparing the perturbed (with added divalent metal) and the non-perturbed (reference) 2D 1H-15N HSQC spectra acquired under identical conditions. Titrations were performed at 700 MHz in the following manner: the protein was dialyzed into 50 mM Tris, 0.6 M NaCl, 1 mM DTT, pH 7.0 at a final concentration of 100 μM and a sample volume of 350 μl, and variable amounts of MnCl2 (1:0, 1:0.4, 1:0.8 and 1:1.6 protein:metal molar ratio) were added. Unpaired electrons are a particularly efficient source of relaxation, even when present at low concentration, resulting in a general broadening of resonances. Therefore, the number of scans was increased for successive additions (216, 216, 240, and 304) of the paramagnetic ion to compensate for signal loss due to broadening. Only well resolved and isolated peaks were used for metal binding analysis. Peak intensities were measured with NMR View (35) and normalized with respect to the number of scans and to the intensities of the corresponding signals in the reference spectrum. Amide correlations considered affected by divalent metal binding were those that exhibited a significant decrease in intensity for the three additions (1:0.4, 1:0.8 and 1:1.6) compared to the average decrease noted for all amides. In this manner, residues whose resonances broadened beyond the cut-off value defined by the average signal decrease minus one standard deviation were considered as strongly affected in a specific fashion: for the 1:0.4 titration point, this meant a reduction in signal size below 73%, for 1:0.8 below 55% and for 1:1.6 below 25%. Weakly but specifically affected residues were those whose intensities were below the cut-off only in the last two additions, 1:0.8 and 1:1.6. The rest of the residues are considered as non-affected by metal binding to the protein domain.
RESULTS AND DISCUSSION
Protein Production and Characterisation
The FBNYV M-Rep full length protein was expressed as a histidine-tagged polypeptide and, upon purification, exhibited significant aggregation resulting in high molecular weight complexes (data not shown). Therefore, we focused on accurately delineating the boundaries of the stably folded N-terminal endonuclease domain. After inconclusive results were obtained from limited proteolysis experiments performed using the full length protein, we designed three different protein truncations: the putative catalytic domain of M-Rep, comprising residues 1-95 (M-Rep1-95, with or without an N-terminal tag), a larger construct including one additional stretch, predicted to be α-helical, as well as an even larger protein that contained two of the α-helices predicted to form the potentialoligomeriz ation domain of Rep. Purity, labeling and identity of the expressed Rep domains were assessed by mass spectrometry (M-Rep1-95, Tag20-M-Rep1-95, Tag22-M-Rep1-117, and Tag22-M-Rep1-124 expected mass are 10956 Da., 13119 Da., 15997 Da. and 16871 Da., respectively). The observed masses were 10823 Da., 12987 Da., 15868 Da. and 16741 Da., respectively (Δ=−133, −132, −129, −130). Therefore, the non-tagged polypeptide comprises residues 2-95 (M-Rep2-95), after in vivo processing of the N-terminal methionine 1 (36). In the case of the tagged proteins, Met1 at the N-terminus of the tag was also not present, resulting in the polypeptides Tag19-M-Rep1-95, Tag21-M-Rep1-117, andTag21-M-Rep1-124.
In order to evaluate the possible role of methionine in position 1 on the stability of the domain and its DNA interaction, we also produced a plasmid coding for a M-Rep1-95 mutant containing an extra methionine residue before the ATG start codon of the native sequence, since the analogous residue in the biochemically and structurally related relaxases TrwC and Tral from conjugative plasmids (see discussion below) was shown to be critical for ssDNA binding (37, 38). The resulting protein was expected to contain an extra Met residue preceding the native Met1 residue in M-Rep. Mass spectroscopic analysis of the purified protein revealed a molecular mass of 10956 Da., corresponding precisely to the polypeptide M-Rep1-95. Therefore, in vivo processing led to the desired N-terminal sequence containing methionine at position 1.
Protein yields for all constructs were very similar (5–10 mg/l culture), decreasing when produced isotopically labeled for NMR (3–6 mg/l culture). The longer polypeptides, M-Rep1-117 and M-Rep1-124, with or without the Tag21 tag, exhibited solubility problems at concentrations above 0.4 mM, while the protein M-Rep2-95 could be concentrated up to ~0.8 mM and therefore was selected for structure determination. M-Rep1-95 became only available after the structure of M-Rep2-95 had been determined. All overexpressed proteins were monomeric in solution, as judged by gel filtration chromatography and analytical ultracentrifugation (data not shown). Therefore, the addition of 22 or 29 amino acids at the C-termini of the longer proteins, Tag21-M-Rep1-117 and Tag21-M-Rep1-124, are not sufficient for ordered oligomerization, although it can not be excluded that these regions, in the context of the complete Rep protein, are part of the oligomerization domain. The reduced solubility exhibited by these longer constructs as compared to their shorter counterpart may indeed indicate a tendency for multimerization by these C-terminal extensions.
Endonuclease Activity Assays
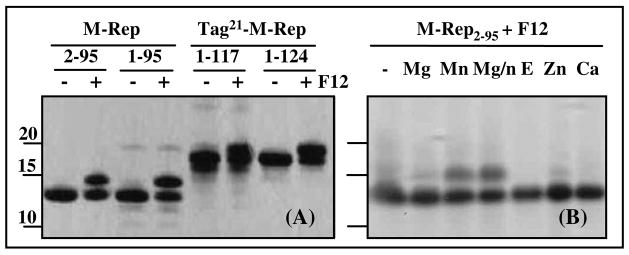
The intergenic region of all the DNA components of FBNYV contains the conserved nonanucleotide sequence that is an essential part of the origin for initiation of RCR. For the full length protein, the catalytic endonuclease activity has been described (3). In order to evaluate whether the FBNYV M-Rep domains investigated here were bona fide endonuclease domains possesig origin DNA cleavage activity, we used a 12 nucleotide fragment in the enzymatic assay (F12, see Experimental Procedures), that included the nonamer with the cleavage site. As shown in Figure 1A, domains M-Rep2-95, M-Rep1-95, Tag21-M-Rep1-117, and Tag21-M-Rep1-124 were catalytically active, cutting the F12 oligonucleotide at the T^A phosphodiester bond, and remaining bound to the 5′ end of the 5 nucleotides (ACCCC, Figure 1A) of the substrate oligonucleotide. This results in a slower migration on SDS-PAGE of the protein-DNA adduct than that of the free protein (Figure 1A). Cleavage efficiency was ca. 50 % for all three polypeptides, thus 50% of the protein was converted to the DNA-bound species. Since the shorter M-Rep2-95 construct was sufficient for catalytic activity, all subsequent structural work was carried out with this protein.
FIGURE 1.
(A) Endonuclease activity of FBNYV M-Rep protein domains. Oligonucleotide F12 (TAGTATT^ACCCC), containing the conserved nonamer sequence of the nanovirus replication origin (in italics) with five nucleotides after the scissile bond (^) was used as substrate. The reaction was monitored by SDS-PAGE (12%), followed by Coomassie staining. The change in electrophoretic mobility compared to the free proteins (−) after covalent adduct formation between the protein and the penta-nucleotide is easily seen (+). (B) Effect of divalent metals on the cleavage of oligonucleotide F12 by M-Rep2-95. Lanes are labelled by the added metal. Mg/n indicates addition of both Mg2+ and Mn2+; E indicates excess addition of EDTA to the Mn2+ containing reaction before addition of oligonucleotide F12.
Divalent Metal Effect on Endonuclease Activity
A requirement for Mg2+ or Mn2+ for efficient DNA cleavage had been reported for full length Rep of the geminivirus TYLCSV (39) and for M-Rep of the nanovirus FBNYV (3). We therefore tested several different divalent metals using M-Rep2-95 (Figure 1B). No cleavage occurred with CaCl2, while MgCl2 and ZnCl2 allowed the reaction to occur, but only about 5% of the protein was converted to the adduct. Most effective was MnCl2, either alone or simultaneously with MgCl2, resulting in conversion of about 50% of the free domain into the covalent adduct. EDTA addition, as expected, completely abrogated cleavage. This established that for M-Rep2-95 divalent metals are needed and that the order of efficiency was: Mn2+ > Mg2+ = Zn2+, while no cleavage was detected with Ca2+.
NMR Spectroscopy and Structure Determination
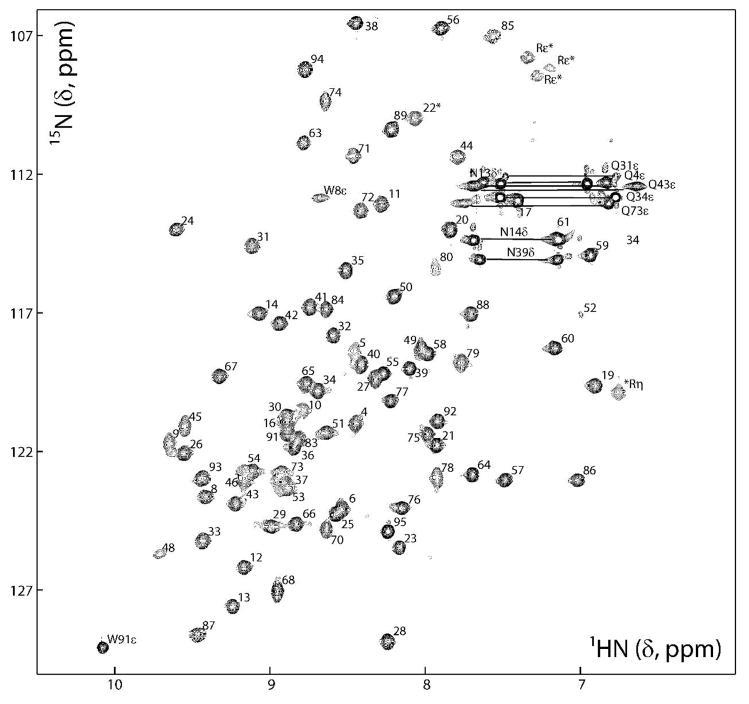
The structural integrity of the three proteins was assessed by 2D 1H- 15N HSQC Spectroscopy. The spectrum for M-Rep2-95 is shown in Figure 2, and a comparison with those of the longer Tag21-M-Rep1-117, and Tag21-M-Rep1-124 proteins is provided in Supplementary Figure S1 (Supporting Information). All three spectra display excellent dispersion and the number of observed signals corresponds approximately to the expected number in each case. Virtually complete assignments of 1H, 13C, and 15N resonances using standard 3D and 4D triple resonance experiments on U-15N, and U-13C,15N labeled samples have been reported for M-Rep2-95 and Tag21-M-Rep1-117 (24) and deposited (BMRB accession numbers 7112 and 7218, respectively). For M-Rep2-95 all expected backbone amide proton resonances are observed in the spectrum, with the exception of those of R3, C7, E47, R69, M81, and K82. The NHε1 resonance of the W8 indol group (Figure 2) appears broad and exhibits an unusual shift (HN,N:8.7,113 ppm), most likely caused by an interaction with the side chain of K68, producing a cation-π interaction, as observed in the structure of the folded domain (see below). Several Arg side chain HNε resonances were also observed (Figure 2). For the Tag21-M-Rep1-117, and Tag21-M-Rep1-124 proteins (Figure S1), the signals were less dispersed and significantly overlapped in the central region of the spectrum, caused by the presence of the unstructured affinity tags, as well as disordered parts of the extensions. This is reflected in the backbone assignments for these proteins (24). Given the overall superior spectral quality and solubility of the shortest construct M-Rep2-95, this protein was selected for a complete structure determination.
FIGURE 2.
2D 1H-15N HSQC spectrum of FBNYV M-Rep2-95 (0.6 mM in 20 mM sodium phosphate, pH 6.6, 0.6 M NaCl, 1 mM DTT, 8% D2O). Backbone amide resonances are labeled according to residue position in the protein sequence and the side chain NH2 signals are connected and labeled by residue type and number. Likewise, the side chain NH resonances of Trp and Arg residues are labeled by residue type and number, and position in the side chain. Folded signals in the 15N dimension are marked with asterisks.
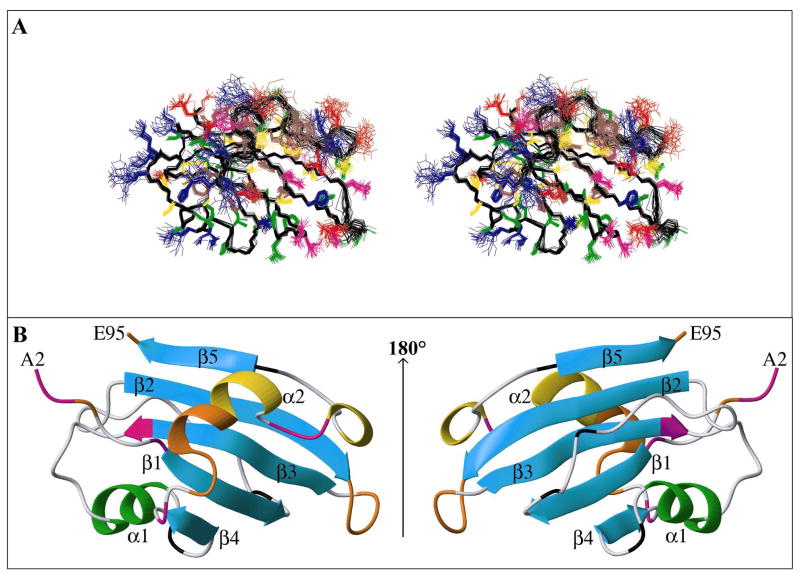
Structure calculations were carried out using a large number of interproton distance constrains obtained from analysis of 2D NOESY, 3D 15N-edited NOESY, 4D 13C-edited, 15N-edited NOESY, and 4D 13C-edited, 13C-edited NOESY spectra, supplemented by (φ,ϕ) dihedral angle constraints obtained from TALOS analysis of chemical shifts (and consistent 3JHNHα values from 3D HNHA), and χ1 dihedral angle constraints (Table 1). All X-Pro peptide bonds were found to be trans based on the observation of α-δ/δ′ sequential NOEs, with the exception of Pro90 that adopts a cis conformation, as revealed by the presence of α-α sequential NOEs. The two cysteine residues, Cys7 and Cys9, are reduced, as revealed by their characteristic 13Cβ chemical shifts of 30.3 and 29.7 ppm, respectively. β-strands as well as their arrangement within the β-sheet were identified by analysis of the inter-strand backbone NOEs and confirmed by H-D exchange analysis of amide backbone resonances after dissolving the sample in D2O (Figure S2). The final ensemble of 30 conformers is well defined (Figure 3A) and agrees well with all the NMR-derived geometrical constraints. No interproton distance or torsion angle violations >0.39 Å and >8.2°, respectively, were observed. The percentage of residues in the most favourable region of the Ramachandran map is 75.5% and only 1.3% of residues are found in disallowed regions. All pertinent structural statistics are provided in Table 1. A significant improvement of the structure quality is obtained (82% in most favorable regions and none in disallowed regions) when the disordered regions, 35–38 and 69–84, are omitted. This part of the structure contains the only residue (S80) found in a disallowed region of the (φ,ϕ) map and one of the two (A37 but not K50) in the generously allowed region. The 30-conformer ensemble, as well as the average minimized structure, have been deposited in the PDB under access code 2HWT.
Table 1.
Experimental Data and Structural Statistics of the 30 conformers representing the solution structure of M-Rep2-95.
| A. NMR-derived constraints | ||||
|---|---|---|---|---|
| Total interproton | 1248 | |||
| Intraresidue | 177 | |||
| Sequential (i-j=l) | 340 | |||
| Short range (l<i-j<5) | 168 | |||
| Long range (i-j>4) | 563 | |||
| Total dihedral angles | 110 | |||
| φ | 52 | |||
| ϕ | 52 | |||
| χ1 | 6 | |||
| Total number of constraints | 1358 | |||
| Total number of constraints per residue | 14.8 | |||
| B. Structural Quality
| ||||
| 1. Residual violationsa
| ||||
| Upper limits | Lower limits | Van der Waals | Torsions | |
|
| ||||
| Number of violationsb | 19±6 (8;33) | 0±0 (0;0) | 11±2(7;15) | 0±0 (0;1) |
| Maximum violation | 0.27 ±0.06 (0.17;0.39) | 0.02 ±0.01 (0.01;0.04) | 0.21 ±0.03 (0.17;0.31) | 3.76±1.08 (2.62;8.12) |
|
| ||||
| CYANA target function (Å2) | 1.77±0.23 (1.32;2.17) | |||
|
| ||||
| 2. Ramachandran statisticsc
| ||||
| 30-conformer ensemble | Regularized mean structure | |||
|
| ||||
| Residues in most favoured regions | 75.5/81.7 | 76.3/82.1 | ||
| Residues in additional allowed regions | 19.9/16.5 | 19.7/16.1 | ||
| Residues in generously allowed regions | 3.3/1.8 | 2.6/1.8 | ||
| Residues in disallowed regions | 1.3/0.0 | 1.3/0.0 | ||
|
| ||||
| C. Coordinate precision (Å, mean/pairwise)d
| ||||
| Folded domaine | Well-defined residuese | |||
|
| ||||
| N, Cα, C′ | 0.50±0.13/0.75±0.17 (0.32;0.83)/(0.31;1.24) | 0.23±0.05/0.40±0.08 (0.16;0.33)/(0.19;0.67) | ||
| All heavy atoms | 0.95±0.13/1.37±0.17 (0.78;1.28)/(0.88;1.79) | 0.70±0.12/1.03±0.15 (0.52;0.97)/(0.66;1.41) | ||
Average values, standard deviation, and maximum and minimum values (in brackets) for the 30 conformer ensemble. Upper limit, lower limit, and van der Waals violations are given in Å, and torsion violations in degrees.
Number of distance constraint violations larger than 0.1 Å (upper limits, lower limits, van der Waals), and torsional constraint violations larger than 5 degrees.
The 76/56 non-Gly, non-Pro residues in the folded/well-defined region 7–118/7–34,39–68,85–118 of the 30 conformers and those of the regularized mean structure are considered separately. Values are in percent.
Average r.m.s difference between the 30 conformer ensemble and the regularized mean structure/pairwise r.m.s difference between members of the ensemble.
The folded domain consists of 90 residues (5–94), excluding flexible N- and C-tails. The well-defined 70 residues include the folded domain except 20 residues (35–38 & 69–84) located in flexible loops.
FIGURE 3.
Three-dimensional structure of FBNYV M-Rep2-95. (A) Stereoview representation displaying best-fit superpositions of 30 conformers in the final ensemble (residues 5 to 95). Backbone heavy atoms are shown in black while side chain heavy atoms are displayed in blue (H,K,R), red (D,E), brown (F,Y,W), green (A,I,L,P,V), yellow (C,M,S,T), and magenta (N,Q). (B) Ribbon representations of M-Rep2-95. The central 5-stranded β-sheet is shown in cyan, the helix α1 in green, and the helix α2 carrying the catalytic tyrosine is colored in yellow. The strands and helices are numbered and the N- and C-termini labeled. Loop residues exhibiting substantial flexibility (15N{1H} NOE < 0.7) or non-detected NH resonances are colored in orange and magenta, respectively. Proline residues are colored in black.
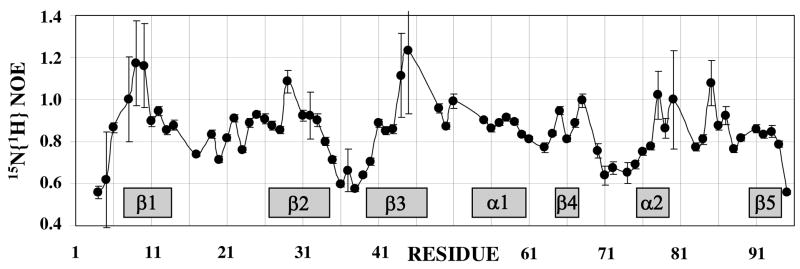
The boundaries of the folded domain of M-Rep2-95 were derived using the following criteria: flexible regions associated with low heteronuclear 15N{1H} NOEs (Figure 4) and devoid of long-range 1H-1H NOEs were considered to be outside the domain. In addition, relatively narrow resonance lines were present for the flexible N- and C-termini. However, the experimental 15N{1H} NOE values were significantly affected by saturation transfer (40), as several amides exceeded the maximum theoretical value of 0.84 at 70 MHz 15N frequency (Figure 4). Nevertheless, qualitative conclusions with respect to flexibility are not affected by this experimental artifact. Backbone amides with significant flexibility are represented in orange in the ribbon representation in Figure 3B. At the N-terminus Q4 and V5 (15N{1H} NOE < 0.6) are flexible, but I6 is clearly ordered (15N{1H} NOE > 0.7). Within the protein, two other relatively flexible regions (15N{1H} NOE values of 0.5–0.6) are noted. The first comprises residues 35-40 and connects strands β2 and β3. The second region followshelix α2, including residues 71-75. At the C-terminus, the E95 reveals increased flexibility (15N{1H} NOE of 0.55), although some tertiary interproton NOEs have been observed between E95 and L21, K26, and Y27. Taken these data together allows us to conclude that for M-Rep2-95 the well folded domain contains residues 6-94.
FIGURE 4.
Heteronuclear 15N{1H} NOE values of the backbone amides of FBNYV M-Rep2-95. Errors are indicated as vertical lines for each point. Secondary structure elements are indicated by grey boxes.
Three Dimensional Structure Description
The overall fold of M-Rep2-95 is shown as a ribbon diagram in Figure 3B. It consist of five β-strands and two α-helices, arranged in a central 5-stranded anti-parallel β-sheet (β1 (residues 8-13), β2 (27-34), β3 (40-47), β4 (65-67) and β5 (91-94); Figure S2), decorated on its periphery by two α-helices, each one on a different side of the β-sheet. Helix α1 (54-58), the loops connecting α1 to the strands in the central β-sheet (loop β3-α1 and loop α1-β4), and a long loop connecting strands β1 and β2 cover one face of the sheet. The other side of the β-sheet is much more exposed, only partially covered by helix α2 (76-79), preceded and followed by two 3 10 helical turns (73-75 and 83-85), and its flanking loops. Although the overall structure is quite well defined, the precision is lower in regions with a lower number of geometric restrains and these regions coincide with those for which increased motion was deduced from relaxation data. For example, the variability in structure observed for the region connecting the β4 strand to the α2 helix and the latter to the β5 strand (Figure 3A) correlates with the low 15N{1H} NOE values for amide resonances of residues 71-75 (in orange) and with the absence of backbone amide resonances for residues 68, 81, and 82 (in magenta), most likely resulting from exchange with H2O and/or conformational exchange. Indeed, one could envisage a “breathing” motion between the α2 helix and the β-sheet that may cause such conformational exchange. Removal of this region, residues 35-38 (loop β2-β3) and 69-84 (loop β4-β5, including α2), as well as the flexible terminal tails from the calculation of coordinate precision, results in a significant improvement (Table 1). In fact, within the 30 conformer ensemble, the helix α2 and the two flanking 310 helical turns are not populated consistently to 100%, but only 57%, 50%, and 43% of the conformers adopt the 76–79 helix, and the 73–75 and 83–85 turns, respectively.
Analysis of the chemical shifts for the extension constructs Tag21-M-Rep1-117, and Tag21-M-Rep1-124, in conjunction with angular information derived from 3JHNHα values, allowed to qualitatively assess whether additional secondary structure elements or structured regions are present in these larger proteins when compared to M-Rep2-95, for which the 3D structure was determined. TALOS (25) analysis of both longer proteins revealed that strand β5 is extended from residue 95 to residue 98 and the presence of an additional helical turn, α3, involving residues 103-106. No significant differences in chemical shifts were noted between Tag21-M-Rep1-124 and Tag21-M-Rep1-117. Apart from the C-terminal residues of M-Rep2-95 that are followed by the extensions 96-117 and 96-124 in the longer constructs, the largest variation in chemical shifts (illustrated in the 1H-15N HSQC spectra in Figure S1) between M-Rep2-95 and Tag21-M-Rep1-117 (or Tag21-M-Rep1-124) are consistently observed in three areas of the protein (illustrated in Supplementary Figure S3): (1) the N-terminal tail, including the first residue of strand β1 (residues 2-8); (2) the last half of strand β3 and the loop β3-α1 connecting it to helix α1 (residues 43-46 and 48-53, respectively); and (3) the loop β4-α2 and the catalytic helix α2(residues 70-80). These areas are located in the lower left region in the structure as displayed in the left hand side of Figure 3B, suggesting that elongation of strand β5 directs the polypeptide chain to run over the central β-sheet. In addition, helical turn α3 (103-106) likely packs against the left part of the exposed face (left hand side of Figure 3B) through its conserved leucine residue, possibly contacting the catalytic helix α2and the loops β3-α1 and β4-α2 and thereby displacing the N-terminal flexible tail. This potential packing of the helical turn α3 against the folded domain is transmitted to relatively remote sites, that also experience a significant variation in chemical shifts between the short and the longer constructs (see residues 63, 82, and 85 in Figure S3). This additional element not present in the structure determined here could influence the flexibility in the loop β4-α2 right before the catalytic helix α2 However, any such altered flexibility had little effect on the cleavage efficiency (Figure 1A). On the other hand, the α3 helical turn contains two basic amino acids conserved in the M-Rep of all members of the genus Nanovirus but not in those of BBTV. Together with other basic residues in that surface area of the protein these residues could potentially participate in dsDNA binding, thereby influencing activity (see below).
Delineation of the Mn2+ Binding Site
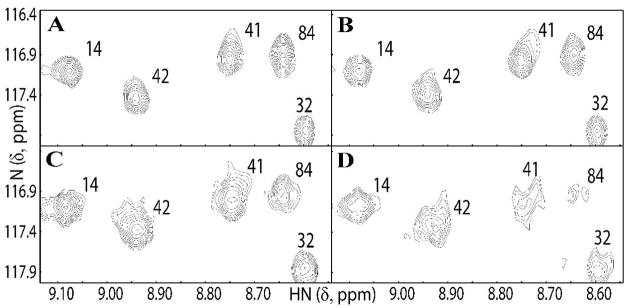
Since it was known that Mg2+ or Mn2+ ions were necessary for DNA cleavage by the intact TYLCSV geminivirus Rep (39) and for M-Rep of the nanovirus FBNYV (3) we delineated the binding site for Mn2+in the structure of the endonuclease domain of FBNYV M-Rep2-95. Titration of the protein with increasing amounts of MnCl2 (1:0, 1:0.4, 1:0.8, and 1:1.6 protein:Mn, molar ratio) by 2D 1H-15N HSQC spectroscopy readily revealed those resonances that specifically broadened (Figure 5) prior to an overall resonance broadening that made detection difficult. The latter occurs since paramagnetic ions also cause general broadening (due to intermolecular electron-spin relaxation) of all resonances, in addition to the specific line broadening experienced by amide resonances close to the metal binding site. Examples of strongly and weakly specifically affected amides as defined in Experimental Procedures are resonances 84 and 41, respectively, while resonances 14, 32, and 42 are not affected (Figure 5). Intensity changes for all backbone and side chain amide resonances upon manganese addition are provided in Supplementary Figure S4. Care was taken to normalize the intensity changes with respect to the number of scans and to the signal intensity in the absence of metal to unambiguously identify specific broadening effects beyond the general drop in intensity of 1:0.82:0.59:0.32 observed along the titration series for all the amide resonances (see Experimental Procedures). No chemical shift changes were noted, indicating Mn2+ binding is weak. Titration with Mg2+ ions was also carried out and again, no chemical shift changes were identified.
FIGURE 5.
Region of the 2D 1 H-15N HSQC spectra of FBNYV M-Rep2-95 illustrating the decrease in signal intensity in the free protein (A), upon addition of 0.4 (B), 0.8 (C), and 1.6 (D) molar equivalents of MnCl2. Intensities (contour levels) were normalized and resonances are labeled by residue number in the polypeptide sequence.
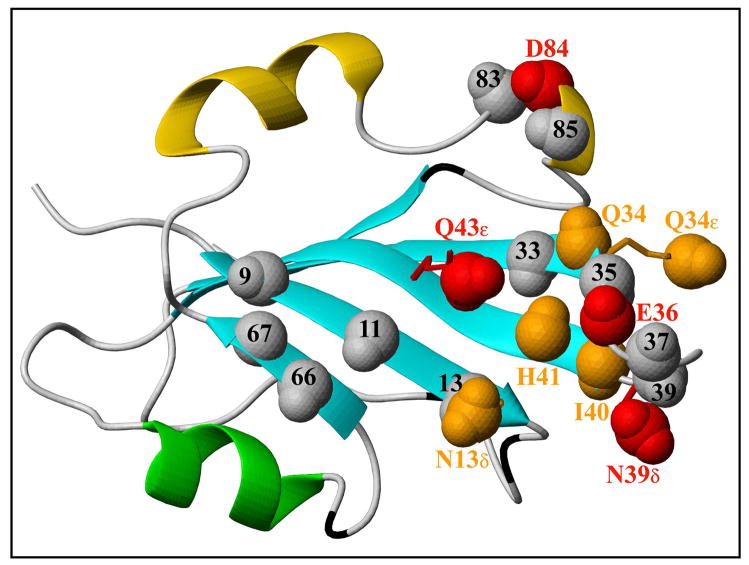
In the structure, the strongly and weakly specifically affected NH and NH2 atoms are shown as red (E36, N39δ, Q43ε and D84) and orange (N13δ, Q34, Q34ε, I40 and H41) spheres, respectively, with greek letters indicating the position of side chain NH2 groups (Figure 6). Amide sites that do not comply with the stringent cut-off criteria but show indications of some specific broadening are 9, 11, 13, 33, 35, 37, 39, 66, 67, 83, and 85 (Supplementary Figure S4) and are shown in grey in Figure 6. The parts of the structure affected by metal binding reside on the exposed side of the β-sheet close to the β2-β3- loop and the 310 helical turn following the catalytic helix α2. Given the fact that manganese is generally coordinated by six ligands in a regular octahedral arrangement, preferring oxygen atoms in ligands rather than nitrogen and sulphur (41), it is reasonable to propose that the side chains of residues E33, H41, and D84 coordinate the Mn2+ ion, given their spatial proximity (as well as the conservation in nanovirus Rep proteins, see below and Figure 7). Additional ligands could be H2O molecules (41), or the affected residues E36 and Q43, although the former amino acid would cause a significant conformational change, and the latter is, due to its chemical nature (Mn2+coordination is usually mediated by Asp, Glu, and His sidechains), a rare ligand for manganese. Nevertheless, given the established role of the two conserved histidines in divalent metal coordination for endonuclease domains belonging to the HUH family (37, 42, 43) and the dramatic broadening caused by Mn2+ in the sidechain resonances of Q43 from the FBNYV domain (Figure S4), the role of the glutamine residue conserved in nanovirus (and circovirus) Rep domains in metal binding (possibly in an alternative mode with respect the HUH-containing domains) can not be excluded. Studies with point mutants of the Rep domains may aid in determining the precise role of the conserved glutamine residue in divalent metal ion coordination. This, however, requires a quantitative metal binding assay which currently is lacking.
FIGURE 6.
Localization of amide sites affected by manganese binding in the structure of FBNYV M-Rep2-95. The strongly affected sites are represented as red spheres, less affected ones in orange, and borderline ones in grey (see text). Perturbed NH2 side chain sites are shown as sticks in red or orange with the NH2 atoms displayed as spheres.
FIGURE 7.
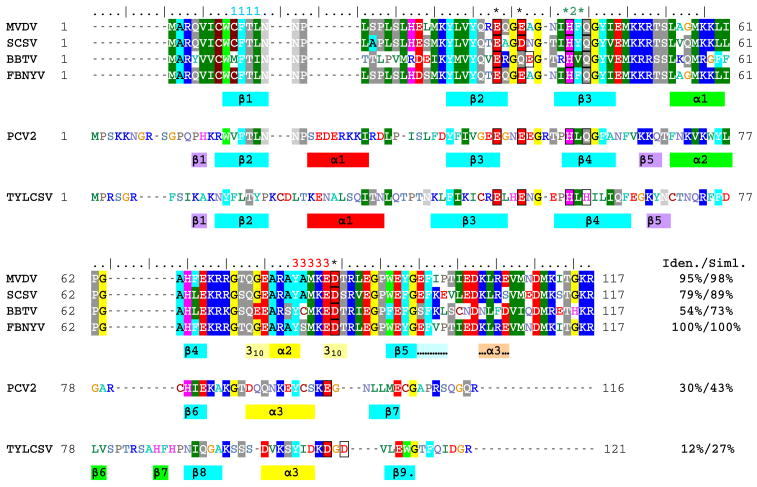
Structure-based alignment of the Rep endonuclease domain sequences from the nanoviruses faba bean necrotic yellows virus (FBNYV, accession number O39828), milk vetch dwarf virus (MVDV, NP_619769), subterranean clover stunt virus (SCSV, NP_620694), and banana bunchy top virus (BBTV, NP_604483)), the circovirus porcine circovirus type 2 (PCV2, AAQ94098)), and the geminivirus tomato yellow leaf curl Sardinia virus (TYLCSV, CAA43466). The secondary structure elements present in FBNYV M-Rep2-95 (identical color coding as in Figure 3B), as well as those in the extension Tag21-M-Rep1-.117 (indicated with “…” and highlighted in pale blue and orange), together with those of the previously determined structures of the Rep endonuclease domains of PCV2 (20) and TYLCSV (13) are displayed below the corresponding sequences. The conserved RCR initiator protein sequence motifs 1, 2, and 3 are labeled above the alignment by 1111, *2*, and 33333, respectively. Putative divalent metal binding residues are boxed and labeled above the alignment by an asterisk. The pale yellow bars flanking helix α2 in FBNYV indicate regions adopting a 310 helix conformation (see text). Sequence identities (Iden.) and similarities (Siml.) of the different sequences compared to the FBNYV Rep sequence are provided at the end of the alignment.
Comparison of the FBNYV Master Rep Endonuclease Domain with those from Gemini- and Circoviruses
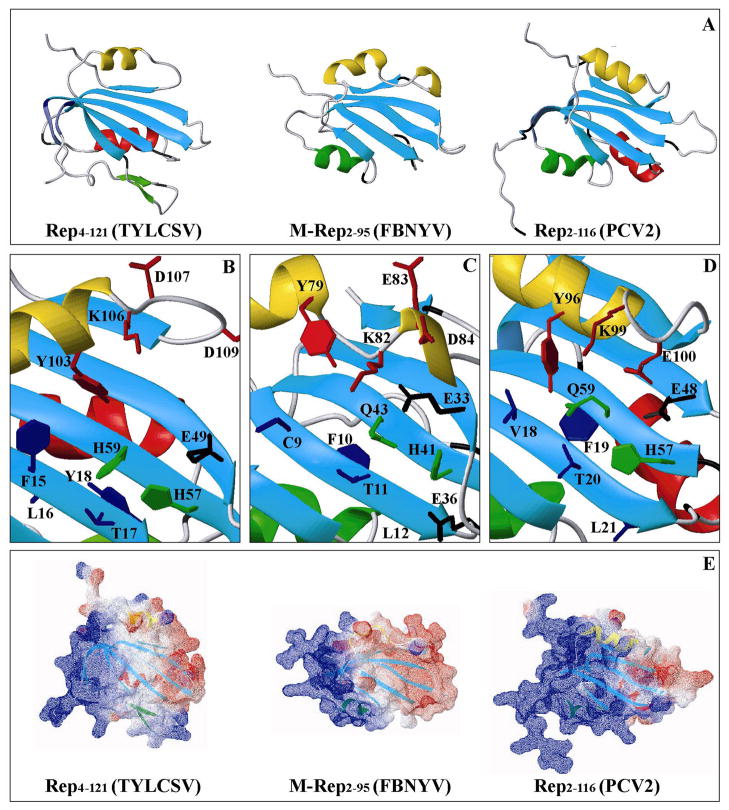
The present structure is the prototype for a nanovirus Master Rep protein, given the high level of sequence similarity between FBNYV and a large number of nanovirus Rep endonuclease domains, including that from BBTV, a virus from another genus in the Nanoviridae family (Figure 7).. In contrast, the M-Rep endonuclease domain of FBNYV exhibits low sequence identity to the Rep sequence of the geminivirus TYLCSV (12%), while that of the circovirus PCV2 occupies an intermediate position with 30% sequence identity. Structurally, however, all endonuclease domains of these Rep proteins are similar, with conservation of the secondary structure elements β1, β2, β3, β4, β5, and α2 (FBNYV nomenclature, Figure 7). A major difference is the absence of both the α1 helix (red) and the β1-β5 mini β-sheet extension (purple) in FBNYV Rep, compared to the TYLCSV and PCV2 Rep domains, while helices α1 in FBNYV and α.2 in PCV2 (green) are substituted by the β6-β7 hairpin in TYLCSV Rep (Figure 8A). The nanovirus Rep endonuclease domain is the least decorated structure and remains the core structure for all viral Rep endonuclease domains known to date, including that of the Rep from the dependovirus AAV5 (42), which possesses a linear ssDNA genome. This basic fold is also common to the HUH motif-containing relaxase proteins from conjugative plasmids, but they appear circularly permuted with respect to the viral Rep domain structures (43), resulting in much lower DALI Z-scores (44) between relaxases and Rep domains (Z=0.7–1.8), in relation to the comparison among relaxases (Z=28.0) or among the viral Reps (Z=3.8–7.9; Supplementary Table S1). A quantitative evaluation of the structural similarity in the central 5-stranded β-sheet between the four different viral Rep domains and the two relaxases is provided in Supplementary Table S2, revealing a highly similar arrangement of strands β1 to β5 (FBNYV nomenclature), with rmsd values of backbone heavy atom coordinates below ~2.0 Å for all pairwise comparisons. Inclusion of the catalytically essential tyrosine and lysine residues (except for the relaxase TrwC, in which this residue is not present) in the structural comparison consistently increases the rmsd values (see below). This indicates that the positioning of these two residues at i and i+3 positions (at i+4 for AAV5 Rep and Tral) with respect to the conserved β-sheet introduces a real and potentially important variation in the different structures. In fact, a striking structural difference between the circovirus Rep endonuclease domain on the one hand and those of the gemini- and nanovirus Rep counterparts on the other, is the orientation of the catalytic helix relative to the central β-sheet (Figure 8A): in the circovirus endonuclease domain this helix runs almost parallel to the β-strands, whereas in the gemini- and nanovirus domains the catalytic helix axis is oriented at angles of ~45 and ~70 degrees relative to the central β-strand, respectively. This may be related to the different positioning of dsDNA recognition elements (iteron sequences (45)) relative to the conserved nonamer sequence, the target of the endonucleolytic action. On the other hand, the currently available Rep endonuclease domains representing four virus families (and the relaxases) are structurally equally distant from each other with no transparent evolutionary relationship.
FIGURE 8.
Comparison of the three dimensional structures of the viral RCR initiator endonuclease domains from the geminivirus TYLCSV (PDB 1L2M, left), the nanovirus FBNYV (PDB 2HWT, center) and the circovirus PCV2 (PDB 2HWO, right). Ribbon representation (A): β-strands in the central β-sheet are depicted in cyan and α-helices in yellow, red and green. Active site regions of TYLCSV (B), FBNYV (C), and PCV2 (D) endonuclease domains, including residues in the conserved sequence motifs I (blue), II (green), and III (red), as well as putative metal binding residues not present in these motifs (black). Electrostatic surface representation (E) of TYLCSV (left), FBNYV (center) and PCV2 (right). Negative and positive potentials are colored red and blue, respectively, with neutral regions in white. Secondary structure elements are visible through the partially transparent surface and color coded as in (A) (generated with Swiss-PdbViewer 3.7 (33)).
As can be seen from the comparison of structures (Figures 7 and 8B-D), residues in the conserved motifs I, II and III (46) are generally located in structurally equivalent positions, although their side chain conformation differs between the three structures. Motif I residues are located on the β1 strand (CFTL in FBNYV) with two of the residues pointing towards the exposed surface of the β-sheet (C9 and T11) and the other two (F10 and L12) towards the hydrophobic core of the domain. As suggested previously for TYLCSV (13), the hydrophobic residues in motif I contribute to the domain architecture and stability while the two exposed residues (F/C/V and T for the gemini-, nano- and circovirus Rep, respectively) can participate in the recognition of the nucleic acid. The first, variable position, may allow for some degree of specificity towards parts of the viral DNA other than the conserved nonanucleotide sequence.
Motif II (HUQ for nano and circovirus, HUH for geminivirus, where U denotes a large, hydrophobic amino acid) is located on the central β3 strand, with the two conserved residues residing on the exposed face of the central β-sheet, and residue U buried in the hydrophobic core. The two conserved histidine residues were proposed to be involved in divalent metal coordination (19), a concept that was supported by the structure of the endonuclease domain of the dependovirus AAV5 Rep (42), that contained a Zn2+ ion coordinated by structurally homologous residues to the two histidines and a glutamic acid (H57, H59, and E49 in TYLCSV, Figure 8B). Alternatively, results presented here for the structure of the nanovirus domain as well as data recently reported for the circovirus domain (20), suggest that the first coordinating histidine could be replaced by an acidic side chain in the Rep proteins of these viruses, reaching down to the exposed surface of the β-sheet from the loop following the catalytic helix β2 (shown in yellow). In this way, metal coordination in nanovirus Rep would involve the side chains of residues H41, E33, and D84 in FBNYV (Figure 8C), and H57, E48, and E100 in the circovirus PCV2 (Figure 8D). In the model geminivirus structure shown in Figure 8B none of the acidic side chains following the catalytic helix (D107 or D109) points towards the presumed divalent metal site, therefore most likely will not be involved. This is partially supported by the properties of TYLCSV Rep mutant D107A that is unaffected in their endonuclease and nucleotidyl transferase activities (B. Gronenborn and J. Brevet, unpublished results). The general location of the divalent metal site is also consistent with the electrostatic surface potential of the proteins (Figure 8E). Motif III (YXXKE/D) contains the active site tyrosine (Y79 in the nanovirus), catalyzing DNA cleavage and nucleotidyl transfer reactions (3, 47), an essential lysine (48) and an acidic residue, the function of which remains undetermined. The catalytic tyrosine is located in all three cases in the so-called catalytic helix (yellow in Figure 8A–D). The Tyr and Lys side chains point in a similar direction towards the exposed side of the β-sheet, although in the nanovirus structure (Figure 8C) the tyrosine is slightly further away from the β-sheet. The higher mobility observed for this region in the FBNYV Rep domain structure, compared to the corresponding gemini- and circovirus ones, may allow for this residue to reach an equivalent position, even if it is not structurally locked in.
Catalytic Mechanism and Substrate Recognition by Viral RCR Rep Endonuclease Domains
Based on all the available structural results and inspired by the catalytic mechanism of the biochemically and structurally related bacterial relaxases (38, 49) we propose a model for the cleavage reaction performed by the viral Rep proteins. It consists of three steps: (1) binding and precise positioning of the phosphate group at the scissile bond, aided by the divalent metal ion and the positively charged side chain of the conserved lysine (K82 in Figure 8C). Interaction with the lysine will also induce the appropriate polarization of the phosphodiester bond. This is followed by (2) the nucleophilic attack of the hydroxyl group of the catalytic tyrosine residue (Y79 in Figure 8C) on the phosphate with formation of a penta-coordinated phosphate intermediate. In the last step (3) the tyrosine hydroxyl proton is abstracted by a general base, for example the metal coordinating histidine residue (H57, H41, and H57 in TYLCSV, FBNYV, and PCV2 Rep, respectively), and transferred to a nearby base (for example E36 in FBNYV, Figure 8C, or homologous residues in gemini and circovirus Rep, Figure 7). This results in formation of the phosphotyrosine bond and the covalent adduct between the protein and the 3′-fragment of the substrate ssDNA. The leaving 5′-fragment of the substrate ssDNA is protonated by a properly positioned residue acting as a general acid (for example, H59 in TYLCVS, Figure 8.B, although there is no corresponding residue that could play that role in the nanovirus and circovirus structures) creating the free 3′-OH. Mutagenesis studies based on the three available Rep domain structures may lead to further insights into the mechanistic details of the endonuclease reaction catalyzed by Rep and provide proof of this structure-based hypothesis.
Prior to the cleavage reaction, Rep proteins have to locate the conserved nonanucleotide in the context of the replication competent viral dsDNA intermediate. As proposed for TYLCSV (13), partially validated for PCV2 by the characterization of a complex with viral dsDNA (20), and clearly established in a AAV5 Rep-dsDNA complex structure (50), a cluster of positively charged residues and positive electrostatic surface potential is responsible for high affinity binding to specific dsDNA sequences, be it the iterons (45) or other sequence elements. This positively charged surface area is present in all three RCR viral Rep endonuclease domains (Figure 8E) and involves the N-terminal and C-terminal tails of the domains, the mini β-sheet extensions of the gemini- and circovirus Rep proteins, and the corresponding region of the nanovirus Rep (Figure 8A). Remarkably, the first β-strand of the mini β-sheet extension that is present in the TYLCSV geminivirus Rep protein and preceding the motif I sequence (β1, purple in Figure 7) was predicted to constitute an iteron related domain involved in specific recognition of the viral dsDNA (51). For the nanovirus domain the surface electrostatic potential could be even more positive in the context of the complete Rep protein as evidenced by the potential packing of the α3 helical turn containing two conserved basic amino acids that are present in the proteins extending beyond amino acid 95 (Figure 7; see above). Although direct experimental evidence by EMSA experiments on complex formation is currently only available for the circovirus PCV2, it is reasonable to assume that similar dsDNA recognition and binding exists for all viral RCR Rep proteins. Recognition of dsDNA sequences that confer specificity of a given Rep protein for its cognate genome may also position the conserved nonanucleotide (that is spatially distinct from the bound dsDNA site) such that it contacts the exposed side of the central p-sheet close to the catalytic residues (Figure 8B–D). The electrostatic surface potential in this region is slightly negative (Figure 8E); therefore the presence of a bound divalent cation would be essential to allow binding of the highly negatively charged nonanucleotide at the catalytic site. The combination of high affinity binding of nearby dsDNA sequences coupled with a transient and low affinity metal-mediated binding of the single stranded region would result in repulsion of the 5′ part of the nonanucleotide sequence following Repmediated DNA cleavage and aid to displace this sequence from the protein. In this manner, it would be available to serve as a primer for subsequent DNA synthesis.
Supplementary Material
Comparison of the 2D1H-15N HSQC spectra of M-Rep2-95, Tag21-M-Rep1-117 and Tag21-M-Rep1-124 (Supplementary Figure S1). Schematic representation of the five β-strands in M-Rep2-95, indicating inter-strand NOEs between backbone protons that allow to establish the topology and register of β-strands in the 5-stranded antiparallel β-sheet, as well as backbone amides resistant to D2O exchange (Supplementary Figure S2). Consensus chemical shift differences and ribbon representation of the M-Rep2-95 structure, indicating those residues that experience large differences in chemical shifts in the extension construct Tag21-M-Rep1-117 with respect to M-Rep2-95 (Supplementary Figure S3). Histograms depicting intensity changes observed for the backbone amide and side chain amide resonances the 1H-15N HSQC spectrum of M-Rep2-95 for increasing amounts of MnCl2. with final protein:MnCl2 molar ratios of 1:0.4, 1:0.8, and 1:1.6 (Supplementary Figure S4). DALI structural comparison among the endonuclease domains from viral RCR Rep proteins and with those from the relaxases of bacterial conjugative plasmids (Supplementary Table S1). Atomic coordinate differences of common structural elements in the endonuclease domains of viral RCR Rep proteins and from the relaxases of bacterial conjugative plasmids (Supplementary Table S2). This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
D. Padro and FJ. Blanco (CNIO) are acknowledged for technical support and continuous scientific support, respectively. SVR acknowledges a FPI predoctoral fellowship, and RCO is a Ramón y Cajal fellow, both from the Spanish Ministry of Science and Technology.
Abbreviations
- AAV5
adeno-associated virus 5
- BBTV
banana bunchy top virus
- dsDNA
double-stranded DNA
- EMSA
electrophoretic mobility shift assay
- FBNYV
faba bean necrotic yellows virus
- M-Rep
master Rep
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- PCV2
porcine circovirus type 2
- RC
rolling circle
- RCR
RC replication
- Rep
replication initiator protein
- rmsd
root-mean-squared deviation
- SDS-PAGE
polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulphate
- ssDNA
single-stranded DNA
- TYLCSV
tomato yellow leaf curl Sardinia virus
Footnotes
This work was supported in part by Plan Nacional de Biotecnologia grant BIO2001–2287 from the Spanish Ministry of Science and Technology (to R.C.O) and the Intramural AIDS Targeted Antiviral Program of the Office of the Director of the National Institute of Health (to A.M.G.).
BMRB and PDB accession codes: The assignment of NMR signals for the proteins M-Rep2-95 and Tag21-M-Rep1-117 have been deposited at the BMRB with access numbers 7112 and 7218, respectively. The 30-conformer ensemble, as well as the average minimized structure, have been deposited in the PDB with access code 2HWT.
References
- 1.Vetten HJ, Chu PWG, Dale JL, Harding R, Hu J, Katul L, Kojima M, Randles JW, Sano Y, Thomas JE. Nanoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy, VIII Report of the ICTV. Academic Press; Elsevier, London: 2005. pp. 343–52. [Google Scholar]
- 2.Katul L, Vetten HJ, Maiss E, Makkouk KM, Leseman DE, Casper R. Characterisation and serology of virus-like particles associated with faba bean necrotic yellows. Ann Appl Biol. 1993;123:629–647. [Google Scholar]
- 3.Timchenko T, de Kouchkovsky F, Katul L, David C, Vetten HJ, Gronenborn B. A single rep protein initiates replication of multiple genome components of faba bean necrotic yellows virus, a single-stranded DNA virus of plants. J Virol. 1999;73:10173–10182. doi: 10.1128/jvi.73.12.10173-10182.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazarowitz SG, Wu LC, Rogers SG, Elmer JS. Sequence-specific interaction with the viral AL1 protein identifies a geminivirus DNA replication origin. Plant Cell. 1992;4:799–809. doi: 10.1105/tpc.4.7.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mankertz A, Persson F, Mankertz J, Blaess G, Buhk HJ. Mapping and characterization of the origin of DNA replication of porcine circovirus. J Virol. 1997;71:2562–2566. doi: 10.1128/jvi.71.3.2562-2566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katul L, Timchenko T, Gronenborn B, Vetten HJ. Ten distinct circular ssDNA components, four of which encode putative replication-associated proteins, are associated with the faba bean necrotic yellows virus genome. J Gen Virol. 1998;79:3101–3109. doi: 10.1099/0022-1317-79-12-3101. [DOI] [PubMed] [Google Scholar]
- 7.Timchenko T, Katul L, Sano Y, de Kouchkovsky F, Vetten HJ, Gronenborn B. The master rep concept in nanovirus replication: identification of missing genome components and potential for natural genetic reassortment. Virology. 2000;274:189–195. doi: 10.1006/viro.2000.0439. [DOI] [PubMed] [Google Scholar]
- 8.Horser CL, Harding RM, Dale JL. Banana bunchy top nanovirus DNA-1 encodes the ‘master’ replication initiation protein. J Gen Virol. 2001;82:459–464. doi: 10.1099/0022-1317-82-2-459. [DOI] [PubMed] [Google Scholar]
- 9.Heyraud-Nitschke F, Schumacher S, Laufs J, Schaefer S, Schell J, Gronenborn B. Determination of the origin cleavage and joining domain of geminivirus Rep proteins. Nucl Acids Res. 1995;23:910–916. doi: 10.1093/nar/23.6.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jupin I, Hericourt F, Benz B, Gronenborn B. DNA replication specificity of TYLCV geminivirus is mediated by the amino-terminal 116 amino acids of the Rep protein. FEBS Lett. 1995;362:116–120. doi: 10.1016/0014-5793(95)00221-t. [DOI] [PubMed] [Google Scholar]
- 11.Choi IR, Stenger DC. Strain-specific determinants of beet curly top geminivirus DNA replication. Virology. 1995;206:904–912. doi: 10.1006/viro.1995.1013. [DOI] [PubMed] [Google Scholar]
- 12.Orozco BM, Kong LJ, Batts LA, Elledge S, Hanley-Bowdoin L. The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J Biol Chem. 2000;275:6114–6122. doi: 10.1074/jbc.275.9.6114. [DOI] [PubMed] [Google Scholar]
- 13.Campos-Olivas R, Louis JM, Clerot D, Gronenborn B, Gronenborn AM. The structure of a replication initiator unites diverse aspects of nucleic acid metabolism. Proc Natl Acad Sci USA. 2002;99:10310–10315. doi: 10.1073/pnas.152342699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desbiez C, David C, Mettouchi A, Laufs J, Gronenborn B. Rep protein of tomato yellow leaf curl geminivirus has an ATPase activity required for viral DNA replication. Proc Natl Acad Sci USA. 1995;92:5640–5644. doi: 10.1073/pnas.92.12.5640. [erratum appears in Proc Natl Acad Sci U S A 1995 Nov 21 ;92(24): 11322] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orozco BM, Miller AB, Settlage SB, Hanley-Bowdoin L. Functional domains of a geminivirus replication protein. J Biol Chem. 1997;272:9840–9846. doi: 10.1074/jbc.272.15.9840. [DOI] [PubMed] [Google Scholar]
- 16.Clérot D, Bernardi F. DNA helicase activity is associated with the replication initiator protein rep of tomato yellow leaf curl geminivirus. J Virol. 2006;80:11322–11330. doi: 10.1128/JVI.00924-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choudhury NR, Malik PS, Singh DK, Islam MN, Kaliappan K, Mukherjee SK. The oligomeric Rep protein of Mungbean yellow mosaic India virus (MYMIV) is a likely replicative helicase. Nucleic Acids Res. 2006;34:6362–6377. doi: 10.1093/nar/gkl903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbalenya AE, Koonin EV, Wolf YI. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- 19.Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucl Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vega-Rocha S, Byeon IL, Gronenborn B, Gronenborn AM, Campos-Olivas R. Solution Structure, Divalent Metal and DNA Binding of the Endonuclease Domain from the Replication Initiation Protein from Porcine Circovirus 2. J Mol Biol. 2007;367:473–487. doi: 10.1016/j.jmb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Timchenko T, Katul L, Aronson M, Vega-Arreguin JC, Ramirez BC, Vetten HJ, Gronenborn B. Infectivity of nanovirus DNAs: induction of disease by cloned genome components of Faba bean necrotic yellows virus. J Gen Virol. 2006;87:1735–1743. doi: 10.1099/vir.0.81753-0. [DOI] [PubMed] [Google Scholar]
- 22.Hafner GJ, Stafford MR, Wolter LC, Harding RM, Dale JL. Nicking and joining activity of banana bunchy top virus replication protein in vitro. J Gen Virol. 1997;78:1795–1799. doi: 10.1099/0022-1317-78-7-1795. [DOI] [PubMed] [Google Scholar]
- 23.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 24.Vega-Rocha SV, Gronenborn B, Gronenborn AM, Campos-Olivas R. 1H, 13C, and 15NNMR Assignment of the Master Rep Protein Nuclease Domain from the Nanovirus FBNYV. J Biomol NMR. 2006 doi: 10.1007/s 10858-006-9085-y. [DOI] [PubMed] [Google Scholar]
- 25.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 26.Bax A, Vuister GW, Grzesiek S, Delaglio F, Wang AC, Tschudin R, Zhu G. Measurement of homo- and heteronuclear J couplings from quantitative J correlation. Methods Enzymol. 1994;239:79–105. doi: 10.1016/s0076-6879(94)39004-5. [DOI] [PubMed] [Google Scholar]
- 27.Güntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 28.Mumenthaler C, Güntert P, Braun W, Wüthrich K. Automated combined assignment of NOESY spectra and three-dimensional protein structure determination. J Biomol NMR. 1997;10:351–362. doi: 10.1023/a:1018383106236. [DOI] [PubMed] [Google Scholar]
- 29.Neri D, Szyperski T, Otting G, Senn H, Wiithrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 30.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 31.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski RA, Rullmannn JA, Mac Arthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 33.Kaplan W, Littlejohn TG. Swiss-PDB Viewer (Deep View) Brief Bioinform. 2001;2:195–197. doi: 10.1093/bib/2.2.195. [DOI] [PubMed] [Google Scholar]
- 34.Maiti R, Van Domselaar GH, Zhang H, Wishart DS. SuperPose: a simple server for sophisticated structural superposition. Nucl Acids Res. 2004;32:W590–594. doi: 10.1093/nar/gkh477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 36.Hirel PH, Schmitter MJ, Dessen P, Fayat G, Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc Natl Acad Sci U S A. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guasch A, Lucas M, Moncalian G, Cabezas M, Perez-Luque R, Gomis-Ruth FX, de la Cruz F, Coll M. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat Struct Biol. 2003;10:1002–1010. doi: 10.1038/nsb1017. [DOI] [PubMed] [Google Scholar]
- 38.Larkin C, Datta S, Harley MJ, Anderson BJ, Ebie A, Hargreaves V, Schildbach JF. Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure. 2005;13:1533–1544. doi: 10.1016/j.str.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 39.Laufs J, Traut W, Heyraud F, Matzeit V, Rogers SG, Schell J, Gronenborn B. In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc Natl Acad Sci U S A. 1995;92:3879–3883. doi: 10.1073/pnas.92.9.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grzesiek S, Bax A. The importance of not saturating water in protein NMR. Application to sensitivity enhancement and NOE measurements. J Am Chem Soc. 1993;115:12593–12594. [Google Scholar]
- 41.Glusker JP. Structural aspects of metal liganding to functional groups in proteins. Adv Protein Chem. 1991;42:1–76. doi: 10.1016/s0065-3233(08)60534-3. [DOI] [PubMed] [Google Scholar]
- 42.Hickman AB, Ronning DR, Kotin RM, Dyda F. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol Cell. 2002;10:327–337. doi: 10.1016/s1097-2765(02)00592-0. [DOI] [PubMed] [Google Scholar]
- 43.Datta S, Larkin C, Schildbach JF. Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure. 2003;11:1369–1379. doi: 10.1016/j.str.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 45.Argüello-Astorga GR, Guevara-Gonzalez RG, Herrera-Estrella LR, Rivera-Bustamante RF. Gemini virus replication origins have a group-specific organization of iterative elements: a model for replication. Virology. 1994;203:90–100. doi: 10.1006/viro.1994.1458. [DOI] [PubMed] [Google Scholar]
- 46.Koonin EV, Ilyina TV. Computer-assisted dissection of rolling circle DNA replication. Biosystems. 1993;30:241–268. doi: 10.1016/0303-2647(93)90074-m. [DOI] [PubMed] [Google Scholar]
- 47.Laufs J, Schumacher S, Geisler N, Jupin I, Gronenborn B. Identification of the nicking tyrosine of geminivirus Rep protein. FEBS Letters. 1995;377:258–262. doi: 10.1016/0014-5793(95)01355-5. [DOI] [PubMed] [Google Scholar]
- 48.Hoogstraten RA, Hanson SF, Maxwell DP. Mutational analysis of the putative nicking motif in the replication- associated protein (AC1) of bean golden mosaic geminivirus. Mol Plant Microbe Interact. 1996;9:594–599. doi: 10.1094/mpmi-9-0594. [DOI] [PubMed] [Google Scholar]
- 49.Boer R, Russi S, Guasch A, Lucas M, Blanco AG, Perez-Luque R, Coll M, de la Cruz F. Unveiling the molecular mechanism of a conjugative relaxase: The structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site. J Mol Biol. 2006;358:857–869. doi: 10.1016/j.jmb.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 50.Hickman AB, Ronning DR, Perez ZN, Kotin RM, Dyda F. The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces. Mol Cell. 2004;13:403–414. doi: 10.1016/s1097-2765(04)00023-1. [DOI] [PubMed] [Google Scholar]
- 51.Arguello-Astorga GR, Ruiz-Medrano R. An iteron-related domain is associated to Motif 1 in the replication proteins of geminiviruses: identification of potential interacting amino acid-base pairs by a comparative approach. Archives of Virology. 2001;146:1465–1485. doi: 10.1007/s007050170072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of the 2D1H-15N HSQC spectra of M-Rep2-95, Tag21-M-Rep1-117 and Tag21-M-Rep1-124 (Supplementary Figure S1). Schematic representation of the five β-strands in M-Rep2-95, indicating inter-strand NOEs between backbone protons that allow to establish the topology and register of β-strands in the 5-stranded antiparallel β-sheet, as well as backbone amides resistant to D2O exchange (Supplementary Figure S2). Consensus chemical shift differences and ribbon representation of the M-Rep2-95 structure, indicating those residues that experience large differences in chemical shifts in the extension construct Tag21-M-Rep1-117 with respect to M-Rep2-95 (Supplementary Figure S3). Histograms depicting intensity changes observed for the backbone amide and side chain amide resonances the 1H-15N HSQC spectrum of M-Rep2-95 for increasing amounts of MnCl2. with final protein:MnCl2 molar ratios of 1:0.4, 1:0.8, and 1:1.6 (Supplementary Figure S4). DALI structural comparison among the endonuclease domains from viral RCR Rep proteins and with those from the relaxases of bacterial conjugative plasmids (Supplementary Table S1). Atomic coordinate differences of common structural elements in the endonuclease domains of viral RCR Rep proteins and from the relaxases of bacterial conjugative plasmids (Supplementary Table S2). This material is available free of charge via the Internet at http://pubs.acs.org.