SUMMARY
Deubiquitinating enzymes (DUBs) are proteases that can antagonize ubiquitin-mediated signalling by disassembling ubiquitin-protein conjugates. How DUBs are regulated in vivo and how their substrate specificities are achieved are largely unknown. The conserved DUB Uch37 is found on proteasomes in organisms ranging from fission yeast to humans. Deubiquitination by Uch37 is activated by proteasomal binding, which enables Uch37 to process polyubiquitin chains. Here we show that in the nucleus Uch37 is also associated with the human Ino80 chromatin-remodeling complex (hINO80). In hINO80, Uch37 is held in an inactive state; however, it can be activated by transient interaction of the Ino80 complex with the proteasome. Thus, DUB activities can be modulated both positively and negatively via dynamic interactions with partner proteins. In addition, our findings suggest that the proteasome and the hINO80 chromatin-remodeling complex may cooperate to regulate transcription or DNA repair, processes in which both complexes have been implicated.
INTRODUCTION
Dynamic attachment and removal of the highly conserved 76-amino acid protein ubiquitin play critical roles in the regulation of many cellular processes including protein degradation, signal transduction, endocytosis, transcription, and DNA repair (Pickart, 2001). Protein ubiquitination is regulated by the opposing actions of two classes of enzyme systems: ubiquitin conjugating enzymes and ligases support ubiquitination of target proteins, and deubiquitinating enzymes (DUBs) catalyze removal of ubiquitin from target proteins. The balance between these competing processes determines the fate of the substrate.
Although ~100 different DUBs have been identified in the human genome, little is known about how the specificities and activities of these enzymes are regulated (Nijman et al., 2005). In particular, the mechanisms by which DUBs keep their enzymatic activites in check until they encounter their target proteins have been defined in only a few cases (e.g. van der Knapp et al., 2005; Lee et al. 2005b; Yao et al., 2006).
We have been investigating the mechanisms that underlie regulation of the human deubiquitinating enzyme Uch37 (also known as Uch-L5). Previous studies identified Uch37 as a subunit of the 19S regulatory particle (RP) of the proteasome, where it exhibits a robust deubiquitinating activity (Holzl et al., 2000; Lam et al., 1997; Li et al., 2000). Subsequently, we and others showed that Uch37 is linked to the 19S particle through a previously unknown proteasome subunit, hRpn13 (also known as Adrm1) (Hamazaki et al., 2006; Qiu et al., 2006; Yao et al., 2006). We further demonstrated that the carboxyl-terminal tail of Uch37 is auto-inhibitory and that hRpn13 activates the Uch37 deubiquitinating activity through interactions with this tail domain.
We recently found that Uch37 co-fractionated with an ATP-dependent chromatin remodeling complex known as the human Ino80 complex (hINO80) (Cai et al., 2007; Jin et al., 2005). Like the related yeast Ino80 complex (Shen et al., 2000), hINO80 catalyzes ATP-dependent nucleosome sliding in vitro and functions in transcriptional regulation and DNA repair in cells (Cai et al., 2007; Wu et al., 2007). hINO80 shares with the yeast Ino80 complex a set of eight core subunits, including the SNF2-like helicase Ino80, but contains additional metazoan-specific subunits, among which are nuclear factor related to κB (NFRKB) and the transcription factor YY1.
In this report, we demonstrate that Uch37 is a bona fide subunit of hINO80. In addition, we show that, although it is constitutively activated in the context of the proteasome, Uch37 deubiquitinating activity is subject to both positive and negative regulations in the context of hINO80. Our findings bring to light a previously unrecognized role for Uch37 and provide a model for the regulation of members of the large family of deubiquitinating enzymes.
RESULTS
The proteasome-associated Uch37 deubiquitinating enzyme is also a subunit of the human Ino80 chromatin remodeling complex
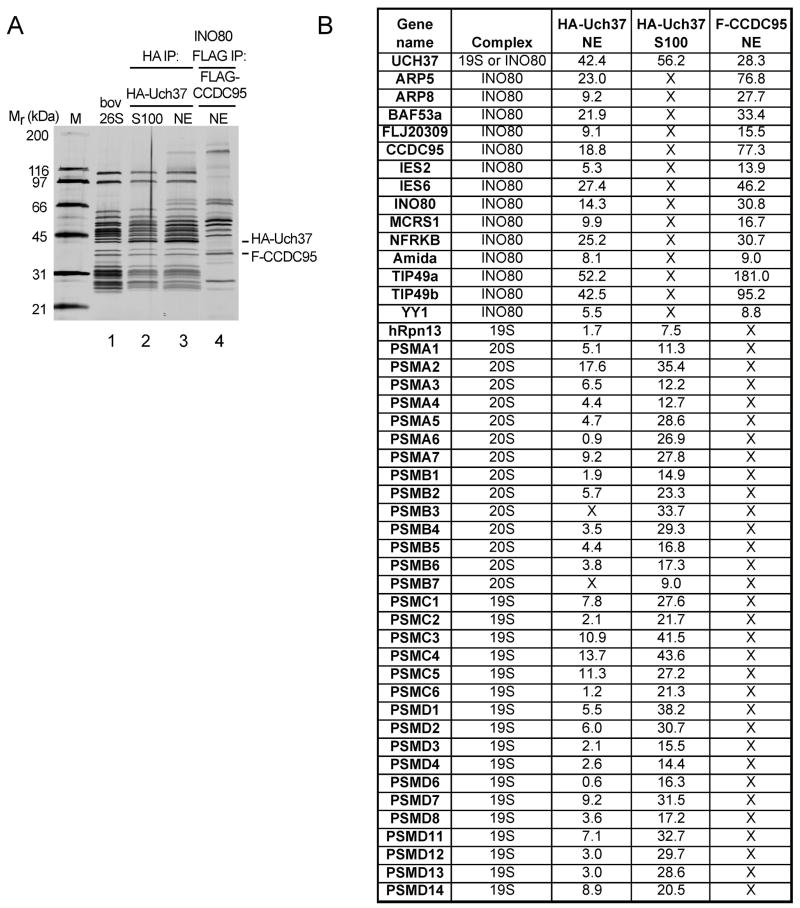
Uch37, an evolutionarily conserved DUB, is a subunit of the 19S regulatory particle (RP) of the proteasome. We recently found that Uch37 was also present in preparations of hINO80 immunopurified from any of 11 different HEK293 and HeLa cell lines stably expressing different Flag-tagged hINO80 subunits ((Cai et al., 2007) and data not shown), suggesting that Uch37 is associated with this complex. To explore this possibility further, we compared Uch37-associated proteins purified from cells that stably express HA-tagged Uch37 to those that copurified with Flag-tagged CCDC95, a subunit of hINO80. Immunoprecipitation (IP) with anti-HA-agarose revealed that Uch37 in the cytoplasmic fraction (S100) of cell lysates is associated exclusively with the proteasome (Figure 1A, lanes 1 and 2). In the nuclear extract fraction (NE), however, a set of proteins in addition to proteasome subunits co-precipitated with Uch37 under stringent high-salt wash conditions. These additional proteins comigrated with subunits of hINO80 (Figure 1A, lanes 3 and 4). All fourteen known hINO80 subunits were detected by mass spectrometry in the HA-Uch37 immunopurified fraction (Figure 1B).
Figure 1.
Uch37 associates with both proteasome and hINO80. (A) Affinity-purified 2XHA-Uch37 and its co-purified proteins from cytoplasm (S100) or the nuclei (NE) of HEK293 cells were analyzed by SDS-PAGE and silver staining. Bovine 26S proteasome (0.5 μg) (Hoffman et al., 1992) and hINO80 affinity-purified through Flag-CCDC95 were included for comparison. To facilitate recovery of 26S proteasomes, 3 mM ATPγS was included in buffers during isolation of Uch37-containing complexes in lanes 2 and 3. (B) The samples shown in (A) were analyzed by mass spectrometry. Numbers are NSAFs (Normalized Spectral Count Abundance Factors) (Zybailov et al., 2006) multiplied by 1000. Proteins listed were not detected in mock purifications from parental HEK293 cells.
Uch37 is bound to hINO80 via its C-terminal tail and the N-terminal domain of NFRKB
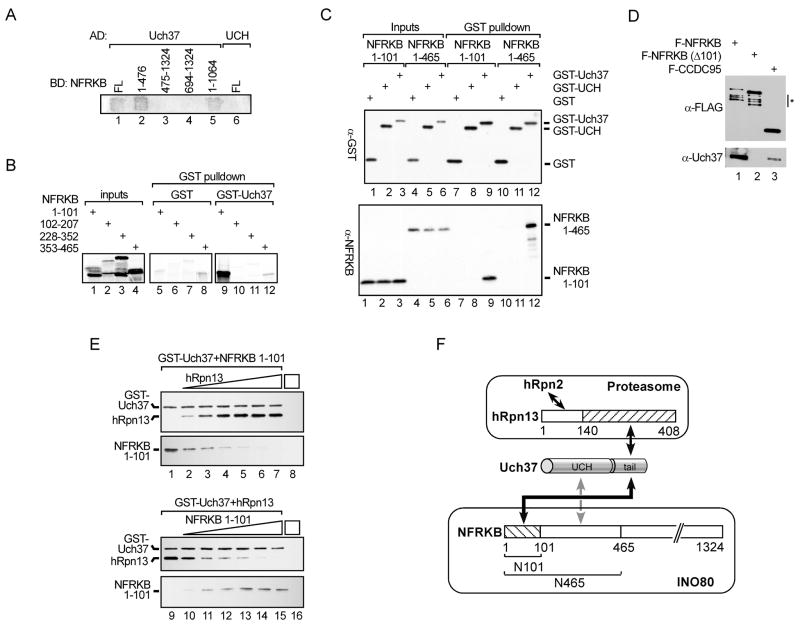
Using a yeast two-hybrid assay, we screened the fourteen hINO80 subunits for direct interactions with Uch37. The only interaction detected was with the 140 kDa protein NFRKB (Figure 2A and data not shown). NFRKB had been cloned previously as a DNA-binding protein that recognizes the interleukin-2 receptor alpha chain κB site (Adams et al., 1991); however, little is known about its function. Neither Uch37 nor NFRKB has recognizable homologs in Saccharomyces cerevisiae and, accordingly, both are absent from the yeast Ino80 complex (Shen et al., 2003).
Figure 2.
NFRKB recruits Uch37 to hINO80. (A) NFRKB interacts with Uch37 in yeast two-hybrid assay. Full-length (FL) Uch37 or N-terminal UCH domain (UCH) was fused to Gal4 activation domain (AD); full-length or fragments of NFRKB were fused to Gal4 DNA binding domain (BD). (B) [35S]-methionine-labeled NFRKB fragments made by in vitro translation were incubated with purified GST or GST-Uch37 at 37 °C for 2 h. Inputs (15%) and proteins bound to glutathione-agarose were analyzed by SDS-PAGE and detected using a phosphorimager. Panels show similar exposures. (C) 2 μg purified recombinant N101 or N465 was incubated with 2 μg GST, GST-UCH, or GST-Uch37 at 30 °C for 1 h and incubated with glutathione-agarose. After four washes with PBS, bound proteins and inputs (20%) were analyzed by immunoblotting with anti-GST and anti-NFRKB antibodies. (D) Anti-Flag immunoprecipitates from extracts of HEK293 cells stably expressing N-terminally Flag-tagged full-length NFRKB, NFRKB lacking the N-terminal 101 amino acids (NFRKB Δ101), or CCDC95 analyzed by SDS-PAGE and blotted with anti-Flag or anti-Uch37 antibodies. Endogenous Uch37 runs as a doublet (Yao et al., 2006). Degradation products are marked by asterisk. (E) hRpn13 and NFRKB compete for binding to Uch37. GST-Uch37 (1 μM) was incubated with equimolar N101 and 0, 0.5, 1, 2, 3, 4, or 5 equivalents of hRpn13 (lanes 1–7) or equimolar hRpn13 and increasing N101 (lanes 9–15). GST-Uch37 and bound proteins were pulled-down with glutathione-agarose and detected by anti-His immunoblotting. GST-Uch37 contains a 6XHis-tag whereas N101 and hRpn13 contain 10XHis tags. (F) Model for Uch37 partitioning between proteasome and hINO80 complexes in cells. Solid black and dashed gray arrows indicate strong and weak interactions, respectively; hRpn2, the 19S RP subunit that binds hRpn13.
The Uch37-interacting region of NFRKB was localized by the two-hybrid assay to the N-terminal third (residues 1–1476) of NFRKB (Figure 2A). Importantly, two-hybrid assays detected an interaction between NFRKB and full-length Uch37, but not a truncated Uch37 that retains only the UCH domain. This is similar to what was previously observed for the interaction between Uch37 and hRpn13, the proteasome subunit that binds Uch37 via the C-terminal tail to recruit it to the 19S RP.
To define in more detail the interaction between Uch37 and NFRKB, we generated a series of fragments from the N-terminal third of NFRKB by in vitro translation using a bacterial cell-free system and tested their abilities to interact with GST-Uch37 in GST-pulldown assays. Only the fragment containing the N-terminal 101 residues of NFRKB (N101) bound selectively to GST-Uch37 (Figure 2B). In addition, purified recombinant N101 bound stably to full-length Uch37 but not the UCH domain alone (Figure 2C). Confirming the importance of the N-terminal 101 residues for stable association between Uch37 and NFRKB, Flag-NFRKB co-immunoprecipitated with endogenous Uch37 from HEK293 cells, whereas NFRKBΔN101, which lacked the first 101 amino acids, failed to do so (Figure 2D). Nevertheless, the N101 fragment appears to account only partially for NFRKB binding to Uch37. A larger fraction of the input NFRKB fragment was retained by GST-Uch37 when binding reactions contained an NFRKB fragment containing the first 465 residues (N465) than when they contained N101 (Figure 2C, compare lanes 3 and 6 to lanes 9 and 12). Furthermore, the interaction between N465 and Uch37 is more salt-resistant than that between N101 and Uch37 (data not shown). Thus, additional, weaker interactions between Uch37 and residues 102–465 of NFRKB are likely.
Suggesting that NFRKB contributes to recruitment of Uch37 to hINO80, when HA-Uch37-associated NFRKB was reduced by about 50% by treatment with NFRKB siRNA, HA-Uch37-associated hINO80 (detected via immunoblot of the subunit Ies2) was decreased by a similar amount (Figure S1). It is important to note, however, that although NFRKB was the only hINO80 subunit that interacted with Uch37 in our yeast two-hybrid assays, it is likely that Uch37 interacts with other hINO80 subunits in the context of the intact complex.
Arguing that Uch37 is not incorporated simultaneously into the proteasome and hINO80, we never detected co-purification of the proteasome and hINO80 under our purification conditions when proteasome or hINO80 subunits other than Uch37 were expressed as Flag-tagged bait proteins. Because hRpn13 and NFRKB interact with Uch37 in the proteasome and hINO80, respectively, we asked whether the isolated proteins bind to Uch37 in a mutually exclusive fashion. Indeed, bacterially expressed and purified hRpn13 and N101 compete for Uch37 binding in vitro (Figure 2F). Taken together, these results show that Uch37 is partitioned between two large multi-subunit complexes, the 19S RP of the proteasome and hINO80 (Figure 2G).
Uch37 activity can be inhibited by NFRKB alone or by incorporation into hINO80
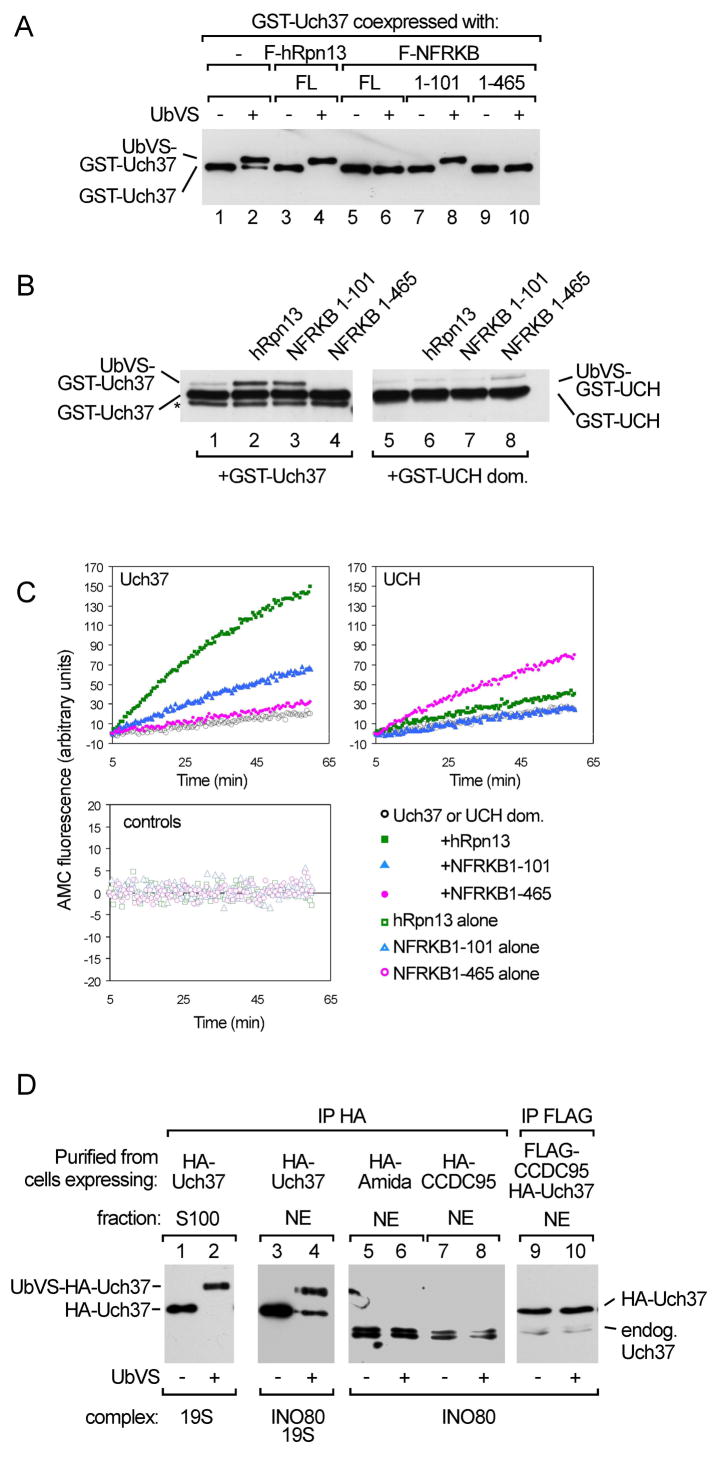
Uch37 is one of the four UCH-family DUBs in humans (Johnston et al., 1999). Unlike Uch-L1 and Uch-L3, it contains a unique C-terminal extension that inhibits its catalytic activity (Yao et al., 2006). Through interactions with the Uch37 C-terminal tail, hRpn13 both recruits Uch37 to the proteasome and activates deubiquitinating activity by relieving auto-inhibition. To determine whether NFRKB also modulates Uch37 activity, we affinity-purified from insect cells recombinant Uch37 in complexes with NFRKB, various NFRKB derivatives, or hRpn13 (Figure S2) and assayed Uch37 for its ability to react with ubiquitin vinylsulfone (UbVS). UbVS is a mechanism-based DUB inhibitor that irreversibly modifies the active-site cysteine and is used as a probe for deubiquitinating activity (Borodovsky et al., 2001). Consistent with our previous report that hRpn13 activates Uch37, the Uch37-hRpn13 complex reacts with UbVS more rapidly than Uch37 alone (Figure 3A). Similarly, the N101 fragment, which also binds to the Uch37 tail domain, enhanced the reaction of Uch37 with UbVS. Unexpectedly, however, both the longer N465 fragment and full-length NFRKB blocked formation of the Uch37-UbVS adduct.
Figure 3.
Regulation of Uch37 activity by interaction with NFRKB or incorporation into hINO80. (A) GST-Uch37 alone or in complex with Flag-tagged hRpn13, NFRKB, or NFRKB fragments expressed in insect cells, affinity purified using glutathione-agarose (lanes 1 and 2) or anti-Flag agarose (lanes 3–10), incubated with or without 2 μM UbVS at 37 °C for 1 h, and analyzed by anti-GST immunoblotting. For a coomassie-stained gel of the proteins, see Figure S2. (B) 0.1 μM GST-Uch37 or GST-UCH were pre-incubated for 1 h with or without 0.5 μM hRpn13, N101, or N465 at 21°C; 0.25 μM UbVS was then added. After 30 min, reaction products were analyzed by anti-GST immunoblotting. The asterisk denotes a degradation product. (C) UbAMC hydrolysis monitored by AMC fluorescence. As indicated, reactions contained 2.5 nM full-length GST-Uch37, 2.5 nM GST-UCH, 12.5 nM hRpn13, 12.5 nM N101, or 6.25 nM N465. A coomassie-stained gel of recombinant proteins is shown in Figure S3. (D) hINO80-associated Uch37 did not react with UbVS. Affinity-purified HA-Uch37 from cytoplasmic (lanes 1 and 2) or nuclear (lanes 3 and 4) fractions of HEK293 cells was incubated with or without 2 μM UbVS at 37 °C for 1 h. Lanes 5–10, hINO80 affinity-purified from stable cell lines expressing HA-tagged hINO80 subunit Amida, HA-tagged CCDC95, or Flag-tagged CCDC95 was incubated with or without UbVS. Reaction products were detected by anti-Uch37 immunoblotting.
To extend these observations, we purified recombinant hRpn13 and NFRKB fragments (Figure S3) and compared their effects on the deubiquitinating activity of either full-length Uch37 or the UCH domain alone. Consistent with the results obtained with coexpressed proteins, the reactivity of Uch37 with UbVS is markedly increased by addition of either N101 or hRpn13, but is inhibited by addition of N465 (Figures 3B and S4). Consistent with the observation that neither hRpn13 nor N101 binds to the UCH domain, neither protein significantly affected the deubiquitinating activity of the isolated UCH domain; however, incubation of the UCH domain with N465 led to a small but reproducible increase in UbVS reactivity. To assess further the effect of the NFRKB fragments on Uch37 activity we measured Uch37-catalyzed hydrolysis of the fluorogenic substrate Ub-7-amino-4-methylcoumarin (UbAMC), a more quantitative assay for ubiquitin C-terminal hydrolase activity (Case and Stein, 2006). Using this assay, we confirmed the ability of N101 to activate full-length Uch37 but not the isolated UCH domain (Figure 3C). Also consistent with the UbVS assay results, the N465 fragment activated the UCH domain for UbAMC hydrolysis, providing further support for the idea that there are low affinity interactions between the UCH domain and residues 102–465 of NFRKB. N465 did not, however, inhibit UbAMC hydrolysis by full-length Uch37. Similarly, with both the purified Uch37-N465 complex (Figure S5) and the hINO80 complex (see below), we also observed inhibition of the UbVS reaction but no effect on UbAMC hydrolysis. We discuss this finding in more detail later.
We then compared the deubiquitinating activity of Uch37 in hINO80 and proteasomes from cells. As expected, proteasome-associated Uch37 immunopurified from cytosol reacted completely with UbVS (Figure 3D). In contrast, only a fraction of nuclear Uch37 was reactive. We suspected that the unreactive fraction was the nuclear Uch37 associated with hINO80. Indeed, there was very little UbVS reactivity of Uch37 associated with hINO80 affinity-purified through either of two other hINO80 subunits, CCDC95 or Amida. We estimate that less than 5% of the Uch37 in hINO80 reacted with 1 μM UbVS after a 2 h incubation, a result consistent with our observation that binding to NFRKB alone inhibits the activity of Uch37.
hRpn13 or proteasomes can transiently activate hINO80-associated Uch37
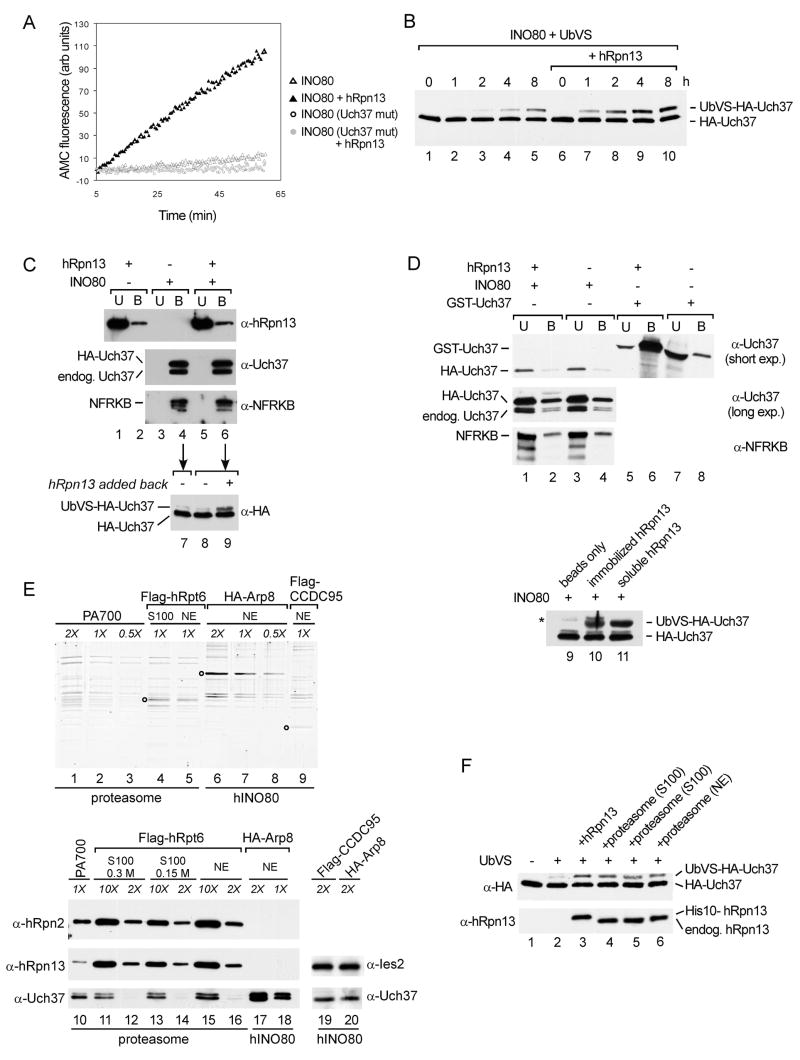
By sequential IP from HEK293 cells that express both Flag-tagged CCDC95 and HA-tagged Uch37, we isolated hINO80 containing either wild-type Uch37 or the catalytically-inactive C88A mutant. The wild-type Uch37-containing complex had only low activity against UbAMC. Interestingly, addition of exogenous recombinant hRpn13 to hINO80 caused a marked activation of Uch37, both in UbAMC hydrolysis and in UbVS reactivity (Figure 4A, B). Confirming that the observed activities were due to hINO80-associated Uch37 and not to other, contaminating DUB(s), there was no detectable activity in the mutant Uch37-containing complex in either the absence or presence of hRpn13.
Figure 4.
hRpn13 or proteasomes activate hINO80-associated Uch37. (A) hINO80 (~50 nM) containing either wild-type or C88A mutant Uch37 was isolated by sequential immunoprecipitation of Flag-CCDC95 and HA-Uch37. UbAMC hydrolysis was assayed with or without 50 nM hRpn13. (B) Affinity purified hINO80 (~0.25 μM) was incubated with 1 μM UbVS with or without 1 μM recombinant hRpn13 at 37 °C; an additional 1 μM UbVS was added every 2 h. Reactions were stopped at the indicated times and analyzed by immunoblotting with anti-HA antibody. (C) Exogenous hRpn13 does not displace Uch37 from hINO80. hINO80 (~35 pmol) was immobilized on anti-Flag agarose (lanes 3–6), and 100 pmol of hRpn13 was added (lanes 1, 2, 5, 6). After 2 h rotation at 4 °C, unbound proteins (U) were recovered, the agarose beads were washed, and bound proteins (B) were eluted with Flag peptide. These fractions were immunoblotted for hRpn13, Uch37, and NFRKB. Equivalent amounts of anti-Flag agarose without hINO80 (lanes 1 and 2) served as a control for non-specific binding. Aliquots of bound fractions shown in lanes 4 and 6 were further incubated for 1 h with 1 μM UbVS, and Uch37 was detected by immunoblotting with anti-HA antibody (lanes 7 and 8). hRpn13 was added back to the bound fraction shown in lane 6 and incubated with UbVS for 1 h (lane 9). (D) Purified recombinant His-Myc-hRpn13 was immobilized on anti-Myc agarose (lanes 1, 2, 5, 6); equivalent amounts of beads without hRpn13 served as controls (lanes 3, 4, 7, 8). Purified hINO80 (lanes 1–4) or GST-Uch37 (lanes 5–8) were added to beads and rotated at 4 °C for 3 h. Unbound (U) and bound (B) proteins were recovered as described in C except that elution was with Myc peptide. As a positive control, beads, bead-bound His-Myc-hRpn13, or soluble hRpn13 were incubated with hINO80 and UbVS at 37 °C for 30 min with rotation (lanes 9–11). Asterisk, immunoglobulin heavy chain. (E) Relative abundance of Uch37 in PA700 from bovine erythrocytes, HeLa cell proteasomes and hINO80. Proteasomes were affinity-purified from HeLa cells through a Flag-tagged hRpt6 subunit, and hINO80 was affinity-purified from HEK293 cells through HA-tagged Arp8 or Flag-tagged CCDC95 subunits. PA700, the 19S RP from bovine erythrocytes, contains 0.5–1 Uch37 polypeptides per complex (Lam et al., 1997; Yao et al., 2006) and was included as a standard. Purified complexes were normalized by SYPRO-Ruby staining after SDS-PAGE and measuring fluorescence intensities of similar sized bands using a Typhoon 8600 Imager (lanes 1–9). Circles mark the tagged subunits in each complex. To estimate relative molar concentrations of Uch37 in each complex, various amounts of each complex were analyzed by immunoblotting using antibodies against Uch37, hRpn13, hRpn2, and Ies2 (lanes 10–20). Uch37 appears as a single band in lanes 19 and 20 because the doublet was not resolved on the 4–15% gradient gel used in this experiment. (F) Proteasomes can activate hINO80-associated Uch37. hINO80 (~5 pmol) that had been affinity purified from a cell line stably expressing both Flag-tagged CCDC95 and HA-tagged Uch37 was incubated with 1 μM UbVS at 37 °C for 2 h. Reactions were supplemented with 5 pmol recombinant hRpn13 (lane 3), proteasomes immunoaffinity purified from S100 fraction in 150 mM NaCl (lane 4) or 300 mM NaCl (lane 5), or proteasomes from nuclear extract purified in 300 mM NaCl (lane 6).
To determine whether activation by hRpn13 was due to displacement of Uch37 from hINO80, we immobilized hINO80 on anti-Flag-agarose, and then added hRpn13 to activate hINO80-associated Uch37 (Figure S6). We then analyzed bound and unbound fractions by western blotting. Despite the presence of a 3-fold molar excess of hRpn13, we detected no dissociation of Uch37 from immobilized hINO80 (Figure 4C, lanes 1–6). Arguing that activation of hINO80-associated Uch37 requires the continuous presence of hRpn13, Uch37 from the bound fractions failed to react with UbVS after hRpn13 was removed (lanes 7 and 8); however, it could be re-activated in a control reaction in which hRpn13 was added back (lane 9). In a reciprocal experiment, we incubated either purified hINO80 or free GST-Uch37 with a large excess of His-Myc-tagged hRpn13 immobilized on anti-Myc-agarose. Again, we detected neither stable binding of hINO80 to hRpn13 nor dissociation of HA-Uch37 from hINO80. Neither HA-Uch37 nor NFRKB were specifically retained by immobilized hRpn13 (Figure 4D, lanes 1–4). Confirming that the activities of soluble and bead-bound hRpn13 are similar, immobilized hRpn13 retained a large amount of free GST-Uch37 (lanes 5–8), and both soluble and bead-bound hRpn13 activated hINO80-associated Uch37 (lanes 9–11). Thus, hRpn13 activates hINO80-associated Uch37 without displacing it from the hINO80 complex, and activation is a consequence of transient interactions between Uch37 and hRpn13. These experiments additionally show that the Uch37 subunit in hINO80 was not inactive due to covalent modification or damage during isolation; rather, the enzyme is held in check in the hINO80 complex and poised for activation.
In previous studies, we and others have used a combination of semi-quantitative western blotting and active site titration to show that about 80% of 19S regulatory particles from bovine blood (PA700) contain Uch37 (Koulich et al., 2008; Lam et al., 1997; Yao et al., 2006). To our surprise, Uch37 is much less abundant in affinity-purified HeLa cell proteasomes (Figure 4E). We estimated that HeLa cell proteasomes contain a 5 to 10-fold molar excess of hRpn13 over Uch37; therefore, most of these proteasomes have the potential to bind Uch37. In contrast, Uch37 is highly abundant in hINO80 complexes purified through either the CCDC95 or Arp8 subunits. This raised the possibility that proteasome-associated hRpn13, like free hRpn13, might stimulate hINO80-associated Uch37. We used Flag immunopurification to prepare proteasomes from cells stably expressing Flag-tagged hRpt6, and hINO80 was prepared from cells expressing Flag-CCDC95 and HA-Uch37. As shown in Figure 4F, HA-tagged Uch37, whose source was exclusively hINO80, could indeed be activated by proteasomes.
DISCUSSION
We have shown that Uch37 is partitioned in vivo between two structurally distinct multi-protein complexes and that these complexes have different effects on deubiquitination activity. Recent studies have reported several examples of DUBs that associate with other proteins; these include Ubp8p in the SAGA complex (Lee et al., 2005b), Ubp6p (Usp14 in mammals) in the 19S RP (Leggett et al., 2002), and Usp7/HAUSP with GMP synthase (van der Knaap et al., 2005). In many cases, binding partners have the dual function of targeting DUBs to specific cellular locations and enhancing their catalytic activities. These regulatory mechanisms ensure substrate specificity, which is especially important with DUBs given that a vast number of different proteins throughout the cell are mono- and polyubiquitinated.
It was unexpected, however, to discover a mode of regulation in which a DUB is held in a latent state by its association with a complex. As we have shown previously, proteasome-associated Uch37 is activated via interactions with the hRpn13 subunit. In contrast, INO80-associated Uch37 or Uch37 in complex with the NFRKB 1–465 fragment is either inhibited (as assessed by UbVS reactivity) or held in a relatively inactive state similar to that of free Uch37 (as assessed by its ability to hydrolyze UbAMC). The difference between these two assays was observed under all assay conditions tested, independent of whether reactions contained intact hINO80 complex, co-expressed and co-purified Uch37 and NFRKB 1–465 fragment, or Uch37 and NFRKB 1–465 fragment purified separately and mixed. We do not understand the mechanistic basis for this difference; however, we note that Uch37 is not the only DUB where association with a regulatory protein results in different behavior measured by reactivity with UbVS and the ability to hydrolyze UbAMC (L.S. and R.E.C., unpublished results). Until the physiological substrate(s) for hINO80-associated Uch37 is identified, we cannot know whether the assays using UbVS or UbAMC better reflect the physiological reaction.
Nevertheless, based on the results of both assays we have used, Uch37 in hINO80 has poor deubiquitinating activity. This low activity is most likely due to an inability to bind ubiquitin efficiently. Several atomic-resolution structures of UCH-type DUBs closely related to Uch37 have been solved, either for the isolated enzyme or for complexes with ubiquitin derivatives (Johnston et al., 1997; Johnston et al., 1999; Misaghi et al., 2005; Rajesh et al., 1999). All suggest that the activities of these enzymes are regulated, at least in part, by the movement of an active-site crossover loop that can block substrate entry. Among the UCH enzymes, only Uch37’s DUB activity is known to be regulated. We have shown previously that the C-terminal tail of Uch37 is auto-inhibitory and that hRpn13 relieves this auto-inhibition by binding to the tail (Yao et al., 2006). Activation most likely is achieved by facilitating substrate access, as we showed that association of Uch37 with hRpn13 lowers the Km for UbAMC without affecting kcat. A likely mechanism is that, until displaced by hRpn13, the Uch37 C-terminal tail stabilizes blockade of the active-site by the crossover loop. That N465 activates the isolated UCH domain (Figure 3B, C) suggests that inhibition of the full-length enzyme depends on the Uch37 tail and does not involve rearrangements of active-site residues. We speculate that N465 inhibits Uch37 reactivity with UbVS by enhancing the auto-inhibitory effect of the C-terminal tail, most likely via interactions that stabilize the active-site crossover loop in a conformation that prevents substrate binding. Blocking substrate entry would prevent Uch37 from indiscriminate binding to ubiquitinated proteins; this model provides an attractive rationale for why Uch37 is kept in a latent state in the hINO80 complex. We cannot, however, exclude the possibility that assembly of Uch37 into hINO80 affects DUB activity by a different mechanism, since our attempts to measure the binding of ubiquitin to Uch37 within hINO80 have been unsuccessful.
Our dissection of interactions between hINO80 and Uch37 shows that one polypeptide, NFRKB, has elements that can inhibit and activate deubiquitinating activity. We do not yet understand the signal that controls the switch between these two modes. Activation of bound Uch37 could involve remodeling of hINO80 or association with its cognate substrate(s). A variety of transcription factors are mono- or polyubiquitinated, and in some cases ubiquitination is required for transcription activation (Muratani and Tansey, 2003). Moreover, multiple histones in chromatin, most notably H2A and H2B, are reversibly ubiquitinated (Weake and Workman, 2008). These all are potential substrates for deubiquitination by Uch37.
Our observation that hRpn13 (Figure 4A, B) or proteasomes (Figure 4E) can activate hINO80-associated Uch37 is particularly intriguing. Although the best known functions of the proteasome involve proteolysis, many recent studies have suggested a direct role for the proteasome in transcriptional regulation (Gonzalez et al., 2002; Lassot et al., 2007; Lee et al., 2005a). Uch37 activation via transient association of 19S RP- or proteasome-bound hRpn13 with hINO80 is one possible mechanism. Whereas PA700, the 19S RP from enucleated bovine erythrocytes, contains near stoichiometric Uch37, we find that proteasomes purified from HEK293 or HeLa cells contain abundant hRpn13 but only small amonts of Uch37 (Figure 4E). Although a recent report suggested the presence of greater amounts of Uch37 in a proteasome fraction from HeLa cells (Koulich et al., 2008), much of the Uch37 in the fraction could have been due to hINO80 that comigrated with proteasomes during glycerol gradient sedimentation of the extract. Thus, it is possible that hINO80-associated Uch37 is activated in vivo by Uch37-free proteasomes that contain hRpn13. This model predicts that deubiquitination requires co-localization of both the hINO80 chromatin remodeling complex and the proteasome (or 19S RP) to chromatin. Such a process could dramatically enhance the targeting of DUB activity to specific sites on chromatin and, consequently, aid in the regulation of gene expression or DNA repair.
EXPERIMENTAL PROCEDURES
Cell culture, generation of stable cell lines, and RNA interference
Generation of stable cell lines using the Invitrogen Flp-in system has been described (Cai et al., 2007). Cell lines stably expressing 2XHA-Uch37, Flag-CCDC95, and Flag-NFRKB have been described in Yao et al., 2006, Cai et al., 2005, and Cai et al., 2007. DNA encoding the NFRKB ORF was obtained by PCR with cDNA prepared from human poly-A+ mRNA and shown by sequencing to match GenBank locus BC063280.
NFRKB knockdown was performed essentially as described (Cai et al., 2007). 2XHA-Uch37 expressing cells were transfected with 100 nM ON-TARGETplus SMARTpool siRNA (Dharmacon, Non-targeting pool or against NM_006165). After 48 h, cells were split and transfected again with the same amounts of siRNA; 48 h post-2nd transfection, cells were harvested and whole cell lysates prepared.
Recombinant proteins, antibodies and affinity purifications
Details of preparation of the recombinant proteins used in this study and antibody production are presented in the Supplementary Data. Anti-Flag, anti-HA, and GST- affinity purifications followed published procedures (Yao et al., 2006).
Deubiquitination assays
For reactions with ubiquitin-AMC (Boston Biochem), 0.1 μM GST-Uch37 or GST-UCH was preincubated at 21°C for 1 h with or without various binding partners in 50 mM Na HEPES, pH 7.5, 0.5 mM EDTA, 1 mM DTT, 0.1 M NaCl, and 0.1 mg/ml ovalbumin. Reactions were inititated by dilution to 2.5 nM in the same buffer with 0.5 μM ubiquitin-AMC at 30°C. Alternatively, purified hINO80 complex, with or without hRpn13, was added to assay buffer containing 0.5 μM ubiquitin-AMC. Reactions were monitored continuously for 1 h by fluorescence at 30 °C (SpectraMAX Gemini XPS; λex = 380 nm, λem = 460 nm).
Yeast two-hybrid assays and mass spectrometry
These methods are described in the Supplementary Data.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health grants R01 GM37666 (R.E.C.) and R37 GM41628 (R.C.C.). T. Y. is a fellow of the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams BS, Leung KY, Hanley EW, Nabel GJ. Cloning of R kappa B, a novel DNA-binding protein that recognizes the interleukin-2 receptor alpha chain kappa B site. New Biol. 1991;3:1063–1073. [PubMed] [Google Scholar]
- Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, et al. YY1 functions with INO80 to activate transcription. Nat Struct Mol Biol. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- Case A, Stein RL. Mechanistic studies of ubiquitin C-terminal hydrolase L1. Biochemistry. 2006;45:2443–2452. doi: 10.1021/bi052135t. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Delahodde A, Kodadek T, Johnston SA. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science. 2002;296:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman L, Pratt G, Rechsteiner M. Multiple forms of the 20 S multicatalytic and the 26 S ubiquitin/ATP-dependent proteases from rabbit reticulocyte lysate. J Biol Chem. 1992;267:22362–22368. [PubMed] [Google Scholar]
- Holzl H, Kapelari B, Kellermann J, Seemuller E, Sumegi M, Udvardy A, Medalia O, Sperling J, Muller SA, Engel A, et al. The regulatory complex of Drosophila melanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J Cell Biol. 2000;150:119–130. doi: 10.1083/jcb.150.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, Gutierrez JL, Coleman MK, Workman JL, Mushegian A, et al. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- Johnston SC, Larsen CN, Cook WJ, Wilkinson KD, Hill CP. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution. EMBO J. 1997;16:3787–3796. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SC, Riddle SM, Cohen RE, Hill CP. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulich E, Li X, Demartino GN. Relative Structural and Functional Roles of Multiple Deubiquitylating Proteins Associated with Mammalian 26S Proteasome. Mol Biol Cell. 2008;19:1072–1082. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- Lassot I, Latreille D, Rousset E, Sourisseau M, Linares LK, Chable-Bessia C, Coux O, Benkirane M, Kiernan RE. The proteasome regulates HIV-1 transcription by both proteolytic and nonproteolytic mechanisms. Mol Cell. 2007;25:369–383. doi: 10.1016/j.molcel.2006.12.020. [DOI] [PubMed] [Google Scholar]
- Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell. 2005a;123:423–436. doi: 10.1016/j.cell.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol. 2005b;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Li T, Naqvi NI, Yang H, Teo TS. Identification of a 26S proteasome-associated UCH in fission yeast. Biochem Biophys Res Commun. 2000;272:270–275. doi: 10.1006/bbrc.2000.2767. [DOI] [PubMed] [Google Scholar]
- Misaghi S, Galardy PJ, Meester WJ, Ovaa H, Ploegh HL, Gaudet R. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J Biol Chem. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajesh S, Sakamoto T, Iwamoto-Sugai M, Shibata T, Kohno T, Ito Y. Ubiquitin binding interface mapping on yeast ubiquitin hydrolase by NMR chemical shift perturbation. Biochemistry. 1999;38:9242–9253. doi: 10.1021/bi9903953. [DOI] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Shen X, Ranallo R, Choi E, Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, Verrijzer CP. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell. 2005;17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.