Abstract
PspGI is a representative of a group of restriction endonucleases that recognize a pentameric sequence related to CCNGG. Unlike the previously investigated Ecl18kI, which does not have any specificity for the central base pair, PspGI prefers A/T over G/C in its target site. Here, we present a structure of PspGI with target DNA at 1.7 Å resolution. In this structure, the bases at the center of the recognition sequence are extruded from the DNA and flipped into pockets of PspGI. The flipped thymine is in the usual anti conformation, but the flipped adenine takes the normally unfavorable syn conformation. The results of this and the accompanying manuscript attribute the preference for A/T pairs over G/C pairs in the flipping position to the intrinsically lower penalty for flipping A/T pairs and to selection of the PspGI pockets against guanine and cytosine. Our data show that flipping can contribute to the discrimination between normal bases. This adds a new role to base flipping in addition to its well-known function in base modification and DNA damage repair.
INTRODUCTION
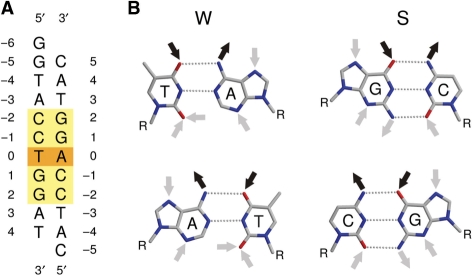
Many DNA binding proteins and enzymes recognize sequences in DNA that are 2-fold symmetric (palindromic). DNA duplexes that consist of an odd number of base pairs can be at best approximately symmetric, because exact 2-fold symmetry would require ‘like-with-like’ rather than Watson–Crick base pairing for the central nucleotide pair. Throughout this article, we refer to DNA sequences with an odd number of base pairs, which are symmetric except at the center of the duplex, as ‘pseudopalindromes’. Enzymes that modify both strands of such duplexes must be insensitive to the violation of 2-fold symmetry at the centers of their target sequences. In other words, they cannot distinguish A/T and T/A pairs, but may distinguish them from G/C and C/G pairs. In the usual shorthand notation for DNA bases (W stands for A or T; S stands for G or C; N stands for A, T, G or C), their specificity for this position must therefore be W, S or N (Figure 1).
Figure 1.
PspGI target sequence. (A) Oligonucleotide duplex used for co-crystallization with the D138A variant of PspGI. The recognition sequence is marked by the yellow box, and the symmetry breaking base pair is highlighted in orange. (B) Comparison of the hydrogen bonding requirements of a T/A (upper left) and A/T pair (lower left) with those of a G/C (upper right) and C/G pair (lower right).
In order to better understand this form of sequence recognition, we have focused on restriction endonuclease PspGI. This enzyme recognizes the pseudopalindromic sequence CCWGG and cleaves the two strands so that five nucleotide 5′-overhangs result (1). Sequence comparisons indicate that PspGI is similar to the catalytic domain of EcoRII (2), which has the same specificity (3,4) and to Ecl18kI, which recognizes the related sequence CCNGG, but displays no specificity for the central base pair (5).
Detailed sequence comparisons and biochemical studies indicate that Ecl18kI, PspGI and the catalytic domain of EcoRII have a shared dimeric architecture and conserved active sites (6–9). So far, only a structure of EcoRII without DNA (10) and the crystal structure of Ecl18kI with DNA have been published (11). The Ecl18kI structure indicates that this enzyme flips the central bases of its target sequence into hydrophobic pockets. Significant sequence similarity between Ecl18kI and PspGI/EcoRII suggests that these enzymes should also flip the nucleotide at the center of the CCXGG sequence, when X is adenine or thymine, and might or might not flip it when X is guanine or cytosine. Direct biochemical evidence for adenine or thymine flipping by PspGI/EcoRII is difficult to provide, but fluorescence experiments with 2-aminopurine (2AP) demonstrate that this adenine analog (which shares some hydrogen bonding requirements with guanine) can be flipped by PspGI/EcoRII (12,13). Evidence for flipping of a natural base in the CCNGG context by PspGI has been obtained for cytosine: this base in the central position of the CCNGG sequence was found to deaminate faster in the presence of the enzyme (14). Moreover, the accessibility of this base to chloroacetaldehyde modification in the PspGI-DNA complex provides additional evidence that cytosine can be unstacked by PspGI (15). At present, it is still unclear how PspGI and EcoRII distinguish between W and S at the center of their recognition sequence, and whether the lack of activity against CCSGG containing DNA goes hand-in-hand with a lack of flipping of at least one base of the central G/C pair. Here, we report the crystal structure of PspGI in complex with target DNA (with the CCWGG sequence). The structure provides a crystallographic demonstration that PspGI flips both adenine and thymine in its target sequence. In combination with biochemical results (16), it also sheds light on the specificity of the enzyme for the nucleotides at the center of its recognition sequence.
MATERIALS AND METHODS
PspGI purification
PspGI-D138A was purified from ER2744 (fhuA2 glnV44 e14− rfbD1 relA1 spoT1 endA1 thi-1 Δ(mcrC-mrr)114::IS10 lacZ::T7gene1) cells containing pET21a-PspGI-D138A. Cultures were allowed to grow to an OD600 of 0.4 then induced with 0.3 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 12 h at 16°C after which they were spun down and frozen at −70°C. The cell pellets were thawed on ice and re-suspended in lysis buffer (20 mM Tris pH 8.5, 50 mM NaCl) containing protease inhibitors (Complete EDTA-free Protease Inhibitors, Roche Diagnostics GmbH, Mannheim, Germany). Cells were broken by sonication followed by centrifugation at 20 000g for 30 min to remove cell debris. The supernatant was heated to 70°C for 30 min followed by centrifugation at 20 000g for 30 min to remove precipitated proteins. The supernatant was loaded onto a 14 ml P11 column (Whatman, Maidstone, UK) and washed with 3 column volumes of wash buffer (20 mM Tris pH 8.5, 50 mM NaCl). PspGI-D138A was eluted from the column with elution buffer (20 mM Tris pH 8.5, 300 mM NaCl) and spin concentrated using Millipore 10 000 MWCO spin columns (Billerica, MA, USA). The concentrated protein was dialyzed into dilution buffer A (10 mM Tris pH 7.9, 50 mM KCl, 50% glycerol) and stored at −20°C. Protein concentrations were determined by a modified Bradford assay (Bio-Rad, Hercules, CA, USA) and purity was judged by SDS–PAGE.
Purification of PspGI-D138A labeled with selenomethionine followed a similar strategy except that expression was in B834 (F – ompT hsdS gal dcm met λDE3). Cultures were grown in 500 ml M9 minimal media supplemented with 5% LB and 50 μg/ml carbenicillin at 37°C until an OD600 of 0.7 was reached. Cells were spun down at 1000g and re-suspended in M9 minimal media. The cells were then grown for 2 h at 37°C in M9 minimal media with 50 μg/ml carbenicillin to use up any remaining l-methionine. After 2 h, l-selenomethionine (Acros Organics, Geel, Belgium) was added to 50 μg/ml and IPTG was added to 0.3 mM and PspGI-D138A was expressed for 10 h. Purification followed the same strategy as for unlabeled protein except that 1, 4-dithiothreitol (DTT) was added to all buffers to 1 mM. Quantification and purity estimation were the same as above and the MALDI-TOF spectrum was consistent with incorporation of selenomethionine.
PspGI crystallization
HPLC grade oligodeoxynucleotides 5′-CATCCAGGTAC-3′ (oligo 1) and 5′-GGTACCTGGAT-3′ (oligo 2) obtained from Metabion (Martinsried, Germany) were dissolved in 10 mM Tris pH 8.0, mixed in 1:1 molar ratio, heated to 95°C and cooled slowly overnight to 4°C. The (inactive) D138A mutant of the PspGI protein and its selenomethionine variant were concentrated in 10 mM Tris pH 7.5 and 50 mM KCl to 14.4 mg/ml and 16.4 mg/ml, respectively. Annealed oligoduplex and concentrated protein were mixed in stoichiometric ratio (one oligoduplex per PspGI dimer), supplemented with MnCl2 (final concentration of about 6 mM) and incubated at room temperature for 1 h prior to crystallization. Crystals were grown by the vapor diffusion technique. Sitting drops were set up automatically using a Lissy robot (Zinsser Analytic, Frankfurt, Germany) for pipetting reservoir buffers and a MicroSys 4000 robot (Genomic Solutions, Ann Arbor, Michigan, USA) for pipetting crystallization drops (at the 200 + 200 nl scale). The preliminary crystallization condition [20% MPD (2-methyl-2,4-pentandiol), 0.1 M citric acid, final pH 4.0] was scaled up to the 2 µl (protein) + 2 µl (buffer) and optimized by the addition of 0.4 µl of 0.1 M cobalt (II) chloride. Small crystals appeared overnight. Crystal growth lasted for about a week and then reached saturation. Crystals could be flash-cryocooled in the crystallization buffer (10 µl) mixed with 2 µl of 70% glycerol.
First crystal form
All crystals of PspGI-D138A with natural methionines and most with selenomethionines belonged to the same crystal form. The best specimens diffracted to 1.65 Å resolution, with a seemingly ‘clean’ diffraction pattern. Nevertheless, experimental phasing and molecular replacement with search models based on either the Ecl18kI-DNA complex structure (11) or the EcoRII apo-structure (10) did not succeed. In retrospect, this failure can be attributed to perfect merohedral twinning. As the twinning problem greatly complicates the interpretation of the data and degrades the ‘effective’ resolution, this crystal form will not be discussed further.
Second crystal form
A single crystal of the PspGI-D138A selenomethionine variant appeared morphologically distinct from all other crystals. Crystallographic analysis showed that this crystal represented indeed a different crystal form that did not suffer from any twinning problems. Therefore, the complete crystallographic analysis reported in this work was based on this one crystal, which belonged to space group P212121 with cell constants 46.9 Å, 96.1 Å and 127.1 Å. It contained a PspGI dimer in complex with the 11-mer oligonucleotide duplex in the asymmetric unit and diffracted to 1.7 Å resolution at the beamline BW6 of the DORIS ring at the Deutsches Elektronensynchrotron (DESY) in Hamburg. In order to solve the structure by the multiple anomalous diffraction method (MAD), we collected datasets at a high energy remote wavelength (0.976 Å) and at the inflection point (0.9793 Å). The SHELXC program (17) indicated that the anomalous signals in the two datasets were over 60% correlated in the lowest resolution shell and over 30% correlated in the shell around 3.0 Å, suggesting that the anomalous differences should be readily interpretable. Based on the known amino acid sequence of PspGI-DNA, we expected either four or six selenium sites, depending on whether or not the initiator methionines were present and ordered in the crystals. Consistent with this expectation, the SHELXD program (17) readily identified four sites with significant occupancies (1.00, 0.87, 0.39 and 0.38), which stood out against the highest noise signal with an occupancy of 0.08. The correct handedness of the selenium substructure was determined by comparing quality parameters after 20 cycles of density modification with the SHELXE program (18). The contrast, a SHELXE measure of map quality, was 0.49 for the correct hand and only 0.31 for the incorrect hand. Although the strongly solvent flattened phases for the correct hand were essentially correct, they were of insufficient quality for automatic density improvement and model building by ARP/wARP (19). Inspection of the heavy atom sites suggested that the high- and low-occupancy sites were related by 2-fold non-crystallographic symmetry (NCS). Moreover, the direction of the putative 2-fold axis was supported by a peak of the self-rotation function. As pairs of atom coordinates are insufficient to derive a general NCS symmetry, we supplemented the two pairs of atom coordinates with the midpoint of the line connecting the two strongest sites, and assumed that this point should be a fixed point of the NCS operation. We then subjected a weakly modified SHELXE density (only three cycles of density modification by this program) to combined solvent flattening and averaging using the program DM. Averaging masks were obtained by automasking, and refinement of NCS operators was allowed to maximize density correlation. The resulting phases to 1.7 Å resolution were of sufficient quality for ARP/wARP to automatically build a model with altogether 509 residues, or 98.5% of the residues in the final structure submitted to the PDB. A model of B-DNA with the correct sequence was generated with the program 3DNA (20) and manually modified to fit the ARP/wARP generated density. Manual model building and modification was done with the programs O (21) and XtalView (22), and refinement was performed with REFMAC (23) and CNS (24). The resulting quality factors of the model appear satisfactory for a structure of 1.7 Å resolution (Table 1). Note that the diffraction data were collected for the selenomethionine variant of PspGI and that therefore all methionine residues are replaced by selenomethionines in the model. The final coordinates and corresponding structure factors were submitted to PDB with the accession code 3BM3.
Table 1.
Data collection and refinement statistics
| Data collection statistics | |
|---|---|
| Space group | P212121 |
| a (Å) | 46.9 |
| b (Å) | 96.1 |
| c (Å) | 127.1 |
| Wavelength (Å) | 0.976 |
| Total reflections | 207295 |
| Unique reflections | 63138 |
| Resolution range (Å) | 20.0–1.7 |
| Completeness (%) (last shell) | 98.7 (98.0) |
| I/σ (last shell) | 21.3 (3.2) |
| R(sym) (%) (last shell) | 6.5 (25.4) |
| B(iso) from Wilson (Å2) | 20.7 |
| Refinement statistics | |
| Resolution range (Å) | 20.0–1.7 |
| Reflections work/test | 59905/3230 |
| Protein atoms (excluding H) | 4552 |
| DNA atoms (excluding H) | 445 |
| Solvent atoms (excluding H) | 306 |
| R-factor (%) | 17.0 |
| R-free (%) | 19.2 |
| Rmsd bond lengths (Å) | 0.013 |
| Rmsd angles (°) | 1.4 |
| Ramachandran core region (%) | 94.7 |
| Ramachandran allowed region (%) | 4.4 |
| Ramachandran additionally allowed region (%) | 0.8 |
| Ramachandran disallowed region (%) | 0.0 |
RESULTS
Crystallization of the PspGI-DNA complex
Wild-type PspGI and an inactive variant of the enzyme, in which the predicted active site residue Asp138 was replaced with alanine, were co-crystallized with several different DNA duplexes. However, crystals with good diffraction patterns were obtained only for the inactive variant and an 11-mer duplex with the PspGI recognition sequence at its center and complimentary single nucleotide overhangs at its ends (Figure 1). In the same crystallization conditions, two different crystal forms appeared. One crystal form was unsuitable for crystallographic analysis due to perfect merohedral twinning. The other form was rare and could only be grown with the selenomethionine variant of the protein, but proved much easier to handle. In the following, we focus on this crystal form.
The PspGI dimer-DNA oligoduplex in the asymmetric unit
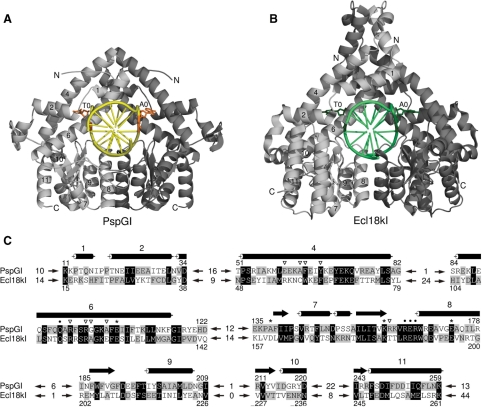
The asymmetric unit of the non-twinned crystal form contains two PspGI protomers and one undecamer duplex. A single (local, non-crystallographic) 2-fold axis relates the two protein subunits and the inner and outer C/G pairs of the PspGI recognition sequence. We therefore conclude that the crystallographic dimer is also the physiologically relevant dimer. As expected, the PspGI subunits assemble similarly as those Ecl18kI subunits that form the functional dimer [Figure 2, (11)]. In solution, the two subunits of the PspGI dimer are equivalent, and therefore there is only one way for the duplex to bind, even though the two DNA strands of the duplex differ at the center of the recognition sequence and in the non-recognized flanks of the duplex. In the crystal, different environments make the two PspGI subunits non-equivalent, creating two possible binding modes for the DNA duplex. The electron density for DNA bases that do not follow the 2-fold symmetry depends on whether or not binding is influenced by the crystal environment. This influence was minimal in the previous Ecl18kI–DNA co-crystal structure and bases appeared averaged (11). In contrast, a single binding mode predominates in the PspGI-DNA co-crystal structure. It stacks the cytosine at the 5′-end of the shorter flank against the guanidino group of Arg118 of a neighboring molecule. Due to the asymmetry of the flanks of the DNA duplex (Figure 1), the alternative binding mode would lead to a clash instead of this favorable interaction.
Figure 2.
A comparison of PspGI-DNA and Ecl18kI-DNA complex structures. (A) PspGI–DNA complex. The two PspGI subunits are shown in ribbon representation in dark and light gray. The DNA is represented by a smoothed backbone and sticks for the bases in yellow, except for the flipped bases, which are shown in orange in all atom representation. (B) Ecl18kI-DNA complex (11). Only a functional dimer is shown. The color coding is analogous to (A), except that the DNA backbone and non-flipped bases are presented in light green color and the flipped bases are shown in dark green color. The helices that are conserved between the two enzymes are numbered (the numbering published previously for Ecl18kI is used). (C) Structure-based alignment of PspGI and Ecl18kI. Conserved helices are numbered consistently in all panels. Stars indicate the active site residues: Glu105, Asp138 (mutated to Ala), Lys160 and Glu173, dots mark the residues involved in sequence recognition (Gln94, Arg164, Glu165 and Arg166). Inverted triangles point to the residues forming the flipped base binding pockets. The length of the unaligned fragments is stated between the arrows.
Gross PspGI structure
PspGI has the typical restriction endonuclease fold that is built around a mixed β-sheet with extensive helical decorations on either side. As in Ecl18kI, there is a long helical region at the N-terminus. The overall structural similarity of PspGI to Ecl18kI is readily apparent, but there are also notable structural differences, especially at the ends of the sequence and in loop regions (Figure 2). Instead of the long N-terminal α-helix 1 in Ecl18kI, PspGI has a shorter, strand-like irregular structure, a sharp kink and a 310 helix. At the C-terminus, full-length PspGI is 31 residues shorter than Ecl18kI. Mass spectrometry indicates that actual PspGI preparations are further truncated by several residues (data not shown). As a result, PspGI lacks (ordered) equivalents for the last two helices of Ecl18kI. PspGI occurs naturally in the hyperthermophile Pyrococcus GI-H, which grows at 85°C, while Ecl18kI is produced by the mesophilic Enterobacter cloaceae 18k, which lives at 37°C. Therefore, one might expect to find generally tighter loops in PspGI than in Ecl18kI, but this is only true in two cases. In between helices 4 and 6, PspGI lacks a connecting 310 helix and the α-helix 5, which makes PspGI more compact than Ecl18kI in the region most distant to the catalytic core. Moreover, the region between β-strands 1 and 2 is two residues shorter and better ordered in PspGI. Contrary to expectations, many PspGI loops are longer than their Ecl18kI counterparts. PspGI has additional residues in the region around Ser46 (seven additional residues), an extra winding of helix 8, a longer loop immediately following it (five additional residues) and a longer helix 10 plus an additional helix (14 additional residues; Figure 2).
Active site architecture
As we could not obtain diffracting crystals of wild-type PspGI with DNA, a variant of PspGI with alanine replacing the predicted active site residue Asp138 was used for the structural studies. In the crystals of this PspGI mutant with DNA, there are no metal ions, and the conformation of the other putative metal binding residues is distorted. Nevertheless, protein backbone superpositions of PspGI, Ecl18kI (11) and the related restriction endonuclease NgoMIV (25) can be used to confirm that the PspGI active site residues are Glu105, Asp138, Lys160 and Glu173 as predicted (7). The comparison further indicates that in a hypothetical complex of wild-type PspGI with two Mg2+ ions, Asp138 (the residue mutated to alanine in our structure) must serve as a bridging ligand; whereas, Glu105 and Glu173 coordinate one metal ion each. In the actual PspGI-D138A co-crystal structure with DNA, Asp138 is mutated and the rotamer conformations of Glu105 and Glu173 are not correct for metal binding. PspGI Lys160 is spatially equivalent to Ecl18kI Lys182 and NgoMIV Lys187 and must therefore be the active site lysine (Supplementary Figure S1). Note that in the Ecl18kI/PspGI/EcoRII group of restriction endonucleases, the active sites are fairly distant from the central nucleotides of the target pseudopalindrome, so that the active site mutation and the lack of metal ions are unlikely to have influence on the interaction of PspGI with the central bases.
Overall architecture of the bound DNA
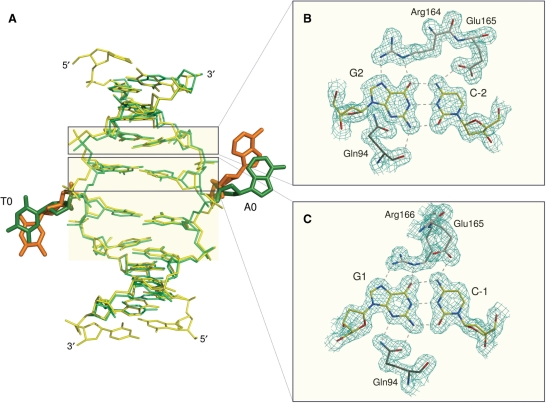
The conformation of the DNA duplex in the complexes with PspGI and Ecl18kI is generally similar. In both cases, the bases at the center of the recognition sequence are extruded from the DNA base stack and flipped into extrahelical positions. Unlike many other nucleotide flipping enzymes, which are not restriction endonucleases (26–29), PspGI and Ecl18kI flip nucleotides of both DNA strands and do not insert any amino acid residue into the DNA stack to replace the missing bases, at least not permanently. As there is no intercalation to keep bases adjacent to the flipped bases apart, these come closer towards each other than 6.8 Å, which would be the normal spacing between them in regular B-DNA. Moreover, PspGI and Ecl18kI kink the DNA so that the major groove gets compressed and the minor groove expanded. Despite these similarities, fine differences (mostly in dihedral angles) result in slightly different positions of the flipped bases: in the PspGI co-crystal structure, the flipped bases are displaced slightly more toward the 5′-ends of their DNA strands than in the Ecl18kI co-crystal structure (Figure 3). It is not clear whether the two complexes map out different positions along the trajectory of the base during flipping, or whether they represent genuinely different endpoints of flipping in the different enzymes, and, if the latter is true, whether this difference might account for their different specificities.
Figure 3.
Specific DNA recognition. (A) Superposition of the DNA duplexes in the PspGI-DNA (yellow/orange) and Ecl18kI-DNA (light/dark green) complexes. DNA colors are consistent with Figure 2. Please note the different conformation of the central base pairs in the two structures. Hydrogen bonding interactions between PspGI and the (B) outer and (C) inner C/G pairs of the recognition sequence. The electron density is taken from the original ARP/wARP map and contoured at 1 σ.
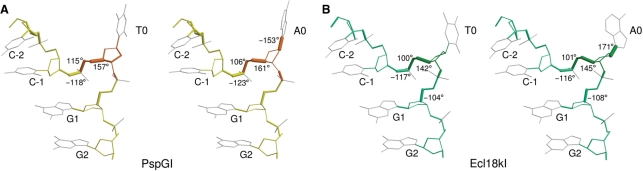
DNA distortions that lead to nucleotide flipping
In the crystal structure of Ecl18kI with DNA, some mechanistic details of nucleotide flipping were obscured by the coexistence of two DNA binding modes, which blurred the electron density in the key regions. In contrast, the PspGI-DNA crystal structure is largely free from this complication, allowing us to discuss nucleotide flipping in more detail (Figure 4). Independent refinement of the two DNA strands of the PspGI-DNA co-crystal structure shows that the dihedral angles χ, which describe the rotation around the glycosidic bond, are radically different between the two flipped nucleotides. The flipped T-nucleotide is present in the usual anti conformation, but the A-nucleotide assumes the normally disfavored syn conformation. All other backbone torsion angles are roughly similar for the two strands. Qualitatively, only the flipped nucleotides and their immediate neighbors appear to differ significantly in shape and position from their counterparts in regular B-DNA. Quantitatively, a comparison of actual dihedral angles in the crystal structures with values for idealized B-DNA shows that major differences are limited to nucleotides −2 to +1. For the flipped nucleotides, the most conspicuous feature is the syn conformation of the flipped adenine, but the dihedral angles α and γ differ radically and the ζ angles to a lesser extent from the values that would be expected for regular B-DNA. For the −2 and −1 nucleotides, the biggest differences are found for the dihedral ζ angles, and for the +1 nucleotides, the most unusual angles are the γ angles. The comparison of DNA distortions in the PspGI-DNA and Ecl18kI-DNA co-crystal structures is complicated by the need for double-conformation refinement in the case of Ecl18kI (11). On the basis of a slightly better fit to the density, we had previously modeled the flipped A- and T-nucleotides in this complex as syn and anti, respectively. This is consistent with the new and more reliable assignment for the PspGI-DNA co-crystals. Disregarding the dihedral angles of the flipped central nucleotides, DNA backbones within one complex are more similar than between different complexes (Figure 4).
Figure 4.
DNA distortion in the (A) PspGI and (B) Ecl18kI structures. The bond thickness corresponds to the angular difference with respect to the regular B-DNA. Discrepancies >100° are marked (ν3 angle difference is not shown since it overlaps with δ). Color coding is consistent with Figure 2. Idealized B-DNA values used: α −30°, β 136°, γ 31°, δ 143°, ε −141°, ζ −161°, χ −98°, ν0 −33°, ν1 45°, ν2 −40° and ν4 6° (20).
PspGI interactions with the specifically recognized bases
PspGI and Ecl18kI recognize the outer and inner G/C base pairs of their target sequences in very similar ways, via interactions that follow the 2-fold symmetry as predicted (7,8). Following a DNA strand in 5′–3′ direction, the scissile phosphoester bond is located upstream of two cytosine residues. The PspGI subunit that would catalyze the hydrolysis of this phosphoester bond approaches the two cytosines (C-2 and C-1) and their guanine hydrogen bonding partners (G2 and G1) from the minor groove side, and makes direct hydrogen bonds with both guanines via Gln94 (Figure 3). The other PspGI subunit mediates the major groove interactions via the guanidino groups of Arg164 and Arg166, which form two hydrogen bonds each with the outer (G2) and inner (G1) guanines, respectively. In addition, Glu165 accepts hydrogen bonds to its terminal carboxylate oxygen atoms from the exocyclic amino groups of the outer (C-2) and inner (C-1) cytosines. If either C-2 or C-1 was N4-methylated, the favorable hydrogen bonding interaction with one of these bases would be replaced by the unfavorable interaction with a non-polar group. As M.PspGI has been classified as an N4-methyltransferase (2), its protective action against the PspGI restriction endonuclease is accounted for, irrespective of which cytosine is the actual target of methylation.
PspGI interactions with the flipped bases
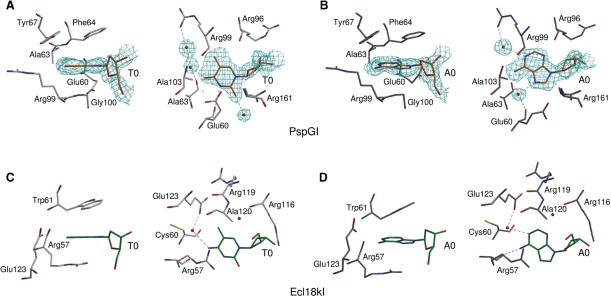
Prior modeling studies predicted the interaction of the flipped bases with Phe64 and Arg161 (14). The crystal structure confirms these contacts and identifies additional ones: according to our model, the flipped bases stack against the aromatic rings of Phe64 and the more distal Tyr67 ‘on top’, Gly100 ‘at the bottom’ and the hydrophobic part of the side chain of Glu60 ‘on the side’ (in the orientation of the left panels of Figure 5A,B). The side walls of the pocket are formed by Ala63 and Ala103, Glu60 and the arginines Arg96 and Arg99, which have their guanidino side chain groups pointing away from the base. In the plane of the bases, the N3 atom of thymine donates a hydrogen bond to the main chain carbonyl oxygen atom of Glu60. The O2 atom of thymine lies within hydrogen bonding distance of the guanidino group of Arg161, but because of poor geometry, it is not clear whether a hydrogen bond is formed. In contrast to the flipped thymine, the flipped adenine interacts with PspGI indirectly through solvent-mediated hydrogen bonds. One solvent molecule is bound to the N7 atom (and possibly the N6 atom) of adenine and also to the carbonyl oxygen atom of Glu60. A second solvent molecule is linked to the N1 atom (and possibly the N6 atom) of adenine and also to the guanidino group of Arg99.
Figure 5.
Flipped base binding pockets of (A and B) PspGI and (C and D) Ecl18kI. In case of PspGI (A and B), only residues closer than 4 Å to the flipped nucleosides are shown. In the case of Ecl18kI (C and D), spatial analogs of some PspGI residues (Ecl18kI residues Cys60 and Arg119) are included for comparison, even though they do not approach the base so closely. In each panel, the structure on the right is ∼90° horizontally rotated relatively to the one on the left. Only the hydrogen bonds in the planes of the bases are shown. The electron density in (A) and (B) is the 2Fo–Fc ARP/wARP generated map for the preliminary model (contoured at 1 σ cutoff). Color coding of DNA and protein molecules is consistent with Figure 2.
DISCUSSION
A and T flipping by PspGI
We have shown here that PspGI flips both bases at the center of its recognition sequence out of the double helix. This makes it the second structure of a restriction enzyme after Ecl18kI to cause this dramatic distortion in DNA. It is thus likely that other restriction endonucleases, which are similar to Ecl18kI and PspGI by primary sequence and cleave related targets analogously, will also be found to flip DNA bases. The DNA distortions caused by PspGI and Ecl18kI share considerable similarity and presumably serve the same purpose—they bring the scissile phosphates closer to a distance that would be found for an endonuclease with a four base-pair recognition sequence (11).
Pocket specificity for pyrimidines
PspGI cleaves oligoduplexes with a like-with-like T/T faster than otherwise identical duplexes with a C/C pair (16), suggesting that thymine fits the PspGI pocket better than cytosine. On the ‘sugar edge’, thymine and cytosine both have a 2-oxo group and are therefore difficult to distinguish. On the Watson–Crick edge of the flipped thymine the O4 atom interacts with a water molecule, and on its ‘C–H’ edge the C5 methyl group is surrounded by a fairly hydrophobic environment. Replacement of thymine with cytosine would convert the O4 hydrogen bond acceptor to an N4 hydrogen bond donor, but a hydrogen bond to the water in the same position could still be made. The replacement would also abolish some hydrophobic interactions between the C5 methyl group of thymine and PspGI. Finally, it would convert the N3 atom of the pyrimidine from a hydrogen bond donor to an acceptor and therefore change the favorable interaction (despite non-ideal geometry) with the carbonyl oxygen atom of Glu60 into an unfavorable one (Figure 5).
Pocket specificity for purines
The biochemical data on PspGI suggest that adenine fits the PspGI pocket better than guanine (16). On the ‘sugar edge’, guanine and adenine are distinguishable by the presence or absence of a 2-amino group on the purine ring. In order to test the effect of the 2-amino group, we superimposed a guanine base on the flipped adenine base in the PspGI-DNA co-crystal structure (Supplementary Figure S2). A few short contacts between the guanine base and amino acid residues in the protein pocket result, but these could be easily relieved by a minor repositioning of the base or by a slight relaxation of the PspGI structure. On the Hoogsteen edge of the flipped purine (which would be located on the major groove side in regular DNA), we found a water molecule bound to the carbonyl oxygen atom of Glu60 and the 6-amino group and N7 nitrogen atom of the flipped adenine. This interaction could favor adenine over guanine, because a water molecule, unless it is protonated, would not allow the same network of hydrogen bonds if the adenine base was replaced with a guanine base containing 6-oxo group (Figure 5).
Models for discrimination between W and S
PspGI distinguishes A/T pairs from G/C pairs with almost million-fold specificity; whereas, the related enzyme Ecl18kI does not make this distinction at all (16). We speculated that difference in the pocket walls might account for this difference and therefore mutated Phe64 in PspGI to a tryptophan as in Ecl18kI or to alanine as a control, but both mutants retained specificity for CCWGG sites and did not cleave CCSGG sites (data not shown). Fortunately, some conclusions about PspGI specificity can be drawn without experimentally converting PspGI into an enzyme with Ecl18kI-like activity and vice versa.
The biochemical results suggest that PspGI senses differences between A/T and G/C pairs already in the binding step and greatly amplifies them in the catalytic step (16). Qualitatively, the disruption of an A/T pair with two Watson–Crick hydrogen bonds is ‘easier’ than the disruption of a G/C pair with three hydrogen bonds, even though stacking interactions contribute to base pair stability as well (30). Quantitative experiments show that enthalpic effects are partially offset by entropic effects (31), but the general conclusion that A/T base pairs are easier to flip than G/C pairs still holds. In the absence of an enzyme, this is reflected by differences in the rates and equilibrium constants for spontaneous flipping of A/T pairs and G/C pairs (32). We believe that these differences also explain (at least in part) the different binding constants for the interaction of PspGI with CCWGG and CCSGG duplexes.
The data for the cleavage rates of oligoduplexes with G/C, G/G and C/C instead of the A/T pair at the center of the recognition sequence indicate that the PspGI pocket selects against guanine, and to a lesser extent also against cytosine (16). According to the crystallographic data and 2AP cleavage experiments (12,16), no single feature of cytosine or guanine has ‘veto’ power over PspGI cleavage and can alone explain PspGI specificity. It rather seems that weak and dispersed interactions between the flipped base and PspGI pocket collectively ensure discrimination against guanine and cytosine.
In the absence of a crystal structure of PspGI in complex with a non-substrate oligoduplex with G/C instead of the A/T, the drastic differences in PspGI catalytic rates could have at least three different explanations. Guanine and cytosine in the central position could not be flipped at all, flipped only transiently, or stably flipped in a manner that indirectly inhibits DNA cleavage. The first possibility, a failure of PspGI to flip guanine or cytosine entirely, appears unlikely, because it would interfere with the compression of the DNA and its fit to PspGI, and should therefore decrease the binding constant for the duplex with central G/C more drastically than experimentally observed. Moreover, the fast deamination rate of cytosine in the CCSGG sequence (14) and its accessibility to chloroacetaldehyde modification (15) suggest that PspGI can unstack this base, even if it does not unstack guanine. For the second possibility, it remains to be explained why transient flipping suppresses catalytic rates far more drastically than the binding constants. The third possibility, binding of the flipped bases to the PspGI pockets in a manner that either allosterically inactivates the enzyme or distorts the phosphodiester backbone remains to be reconciled with the distances of the PspGI pockets to the active sites. In other words, the way in which transient or incorrect stable flipping of bases within the central base pair prevents catalysis at distant scissile phosphates remains unclear.
Our explanation for the PspGI specificity has much in common with interpretation of the specificity of uracil DNA glycosylase (UDG) and other DNA repair enzymes (33). Like PspGI, these enzymes also flip nucleotides and distinguish lesions with much higher specificity than would be expected on the basis of base pair strength alone. However, in contrast to these well-studied DNA repair enzymes, PspGI is specific for natural, undamaged base pairs in regular hydrogen bonding arrangements prior to the flipping event. Consequently, its sequence specificity is even more remarkable.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
EMBO/HHMI Young Investigator Award (to M.B.); Polish Ministry of Science and Higher Education (0295/B/P01/2008/34 to M.B.); National Institutes of Health of the United States (GM 57200 and CA 97899 to A.B.); Polish Ministry of Science and Higher Education (PBZ/MEiN/01/2006/24 to H.C.); Lithuania State Science and Studies Foundation (to V.S.). Funding for open access charge: Polish Ministry of Science and Higher Education (0295/B/PO1/2008/34)
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank New England Biolabs (Ipswitch, MA, USA) and Vera and Alfred Pingoud (Justus-Liebig-University, Giessen, Germany) for providing PspGI clones. They are grateful to Hans Bartunik for generous allocation of beamtime on BW6 (DESY, Hamburg) and to John SantaLucia (Wayne State University, Detroit, USA) for useful discussions regarding the article.
REFERENCES
- 1.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 2005;33:D230–D232. doi: 10.1093/nar/gki029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan R, Xiao J, Xu S. Characterization of an extremely thermostable restriction enzyme, PspGI, from a Pyrococcus strain and cloning of the PspGI restriction-modification system in Escherichia coli. Appl. Environ. Microbiol. 1998;64:3669–3673. doi: 10.1128/aem.64.10.3669-3673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bigger CH, Murray K, Murray NE. Recognition sequence of a restriction enzyme. Nat. New Biol. 1973;244:7–10. doi: 10.1038/newbio244007a0. [DOI] [PubMed] [Google Scholar]
- 4.Boyer HW, Chow LT, Dugaiczyk A, Hedgpeth J, Goodman HM. DNA substrate site for the EcoRII restriction endonuclease and modification methylase. Nat. New Biol. 1973;244:40–43. doi: 10.1038/newbio244040a0. [DOI] [PubMed] [Google Scholar]
- 5.Denjmukhametov MM, Brevnov MG, Zakharova MV, Repyk AV, Solonin AS, Petrauskene OV, Gromova ES. The Ecl18kI restriction-modification system: cloning, expression, properties of the purified enzymes. FEBS Lett. 1998;433:233–236. doi: 10.1016/s0014-5793(98)00921-1. [DOI] [PubMed] [Google Scholar]
- 6.Pingoud V, Kubareva E, Stengel G, Friedhoff P, Bujnicki JM, Urbanke C, Sudina A, Pingoud A. Evolutionary relationship between different subgroups of restriction endonucleases. J. Biol. Chem. 2002;277:14306–14314. doi: 10.1074/jbc.M111625200. [DOI] [PubMed] [Google Scholar]
- 7.Pingoud V, Conzelmann C, Kinzebach S, Sudina A, Metelev V, Kubareva E, Bujnicki JM, Lurz R, Luder G, Xu SY, et al. PspGI, a type II restriction endonuclease from the extreme thermophile Pyrococcus sp.: structural and functional studies to investigate an evolutionary relationship with several mesophilic restriction enzymes. J. Mol. Biol. 2003;329:913–929. doi: 10.1016/s0022-2836(03)00523-0. [DOI] [PubMed] [Google Scholar]
- 8.Tamulaitis G, Solonin AS, Siksnys V. Alternative arrangements of catalytic residues at the active sites of restriction enzymes. FEBS Lett. 2002;518:17–22. doi: 10.1016/s0014-5793(02)02621-2. [DOI] [PubMed] [Google Scholar]
- 9.Tamulaitis G, Mucke M, Siksnys V. Biochemical and mutational analysis of EcoRII functional domains reveals evolutionary links between restriction enzymes. FEBS Lett. 2006;580:1665–1671. doi: 10.1016/j.febslet.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Zhou XE, Wang Y, Reuter M, Mucke M, Kruger DH, Meehan EJ, Chen L. Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold. J. Mol. Biol. 2004;335:307–319. doi: 10.1016/j.jmb.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 11.Bochtler M, Szczepanowski RH, Tamulaitis G, Grazulis S, Czapinska H, Manakova E, Siksnys V. Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease. Embo. J. 2006;25:2219–2229. doi: 10.1038/sj.emboj.7601096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamulaitis G, Zaremba M, Szczepanowski RH, Bochtler M, Siksnys V. Nucleotide flipping by restriction enzymes analyzed by 2-aminopurine steady-state fluorescence. Nucleic Acids Res. 2007;35:4792–4799. doi: 10.1093/nar/gkm513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carpenter MA, Bhagwat AS. DNA base flipping by both members of the PspGI restriction-modification system. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carpenter M, Divvela P, Pingoud V, Bujnicki J, Bhagwat AS. Sequence-dependent enhancement of hydrolytic deamination of cytosines in DNA by the restriction enzyme PspGI. Nucleic Acids Res. 2006;34:3762–3770. doi: 10.1093/nar/gkl545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daujotyte D, Liutkeviciute Z, Tamulaitis G, Klimasauskas S. Chemical mapping of cytosines enzymatically flipped out of the DNA helix. Nucleic Acids Res. 2008;36:e57. doi: 10.1093/nar/gkn200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamulaitis G, Zaremba M, Szczepanowski RH, Bochtler M, Siksnys V. How PspGI, EcoRII catalytic domain and Ecl18kI acquire specificities for different DNA targets. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uson I, Sheldrick GM. Advances in direct methods for protein crystallography. Curr. Opin. Struct. Biol. 1999;9:643–648. doi: 10.1016/s0959-440x(99)00020-2. [DOI] [PubMed] [Google Scholar]
- 18.Sheldrick GM. Macromolecular phasing with SHELXE. Z Kristallogr. 2002;217:644–650. [Google Scholar]
- 19.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 20.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 22.McRee DE. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Deibert M, Grazulis S, Sasnauskas G, Siksnys V, Huber R. Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nat. Struct. Biol. 2000;7:792–799. doi: 10.1038/79032. [DOI] [PubMed] [Google Scholar]
- 26.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 27.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 28.Lariviere L, Gueguen-Chaignon V, Morera S. Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with UDP-glucose: glucose binding and identification of the catalytic base for a direct displacement mechanism. J. Mol. Biol. 2003;330:1077–1086. doi: 10.1016/s0022-2836(03)00635-1. [DOI] [PubMed] [Google Scholar]
- 29.Mol CD, Parikh SS, Putnam CD, Lo TP, Tainer JA. DNA repair mechanisms for the recognition and removal of damaged DNA bases. Annu. Rev. Biophys. Biomol. Struct. 1999;28:101–128. doi: 10.1146/annurev.biophys.28.1.101. [DOI] [PubMed] [Google Scholar]
- 30.Hunter CA. Sequence-dependent DNA structure. The role of base stacking interactions. J. Mol. Biol. 1993;230:1025–1054. doi: 10.1006/jmbi.1993.1217. [DOI] [PubMed] [Google Scholar]
- 31.Petruska J, Goodman MF. Enthalpy-entropy compensation in DNA melting thermodynamics. J. Biol. Chem. 1995;270:746–750. doi: 10.1074/jbc.270.2.746. [DOI] [PubMed] [Google Scholar]
- 32.Giudice E, Varnai P, Lavery R. Base pair opening within B-DNA: free energy pathways for GC and AT pairs from umbrella sampling simulations. Nucleic Acids Res. 2003;31:1434–1443. doi: 10.1093/nar/gkg239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krosky DJ, Schwarz FP, Stivers JT. Linear free energy correlations for enzymatic base flipping: how do damaged base pairs facilitate specific recognition? Biochemistry. 2004;43:4188–4195. doi: 10.1021/bi036303y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.