Abstract
The DNA damage induced SOS response in Escherichia coli is initiated by cleavage of the LexA repressor through activation of RecA. Here we demonstrate that overexpression of the SOS-inducible tisAB gene inhibits several SOS functions in vivo. Wild-type E. coli overexpressing tisAB showed the same UV sensitivity as a lexA mutant carrying a noncleavable version of the LexA protein unable to induce the SOS response. Immunoblotting confirmed that tisAB overexpression leads to higher levels of LexA repressor and northern experiments demonstrated delayed and reduced induction of recA mRNA. In addition, induction of prophage λ and UV-induced filamentation was inhibited by tisAB overexpression. The tisAB gene contains antisense sequences to the SOS-inducible dinD gene (16 nt) and the uxaA gene (20 nt), the latter encoding a dehydratase essential for galacturonate catabolism. Cleavage of uxaA mRNA at the antisense sequence was dependent on tisAB RNA expression. We showed that overexpression of tisAB is less able to confer UV sensitivity to the uxaA dinD double mutant as compared to wild-type, indicating that the dinD and uxaA transcripts modulate the anti-SOS response of tisAB. These data shed new light on the complexity of SOS regulation in which the uxaA gene could link sugar metabolism to the SOS response via antisense regulation of the tisAB gene.
INTRODUCTION
Exposure of Escherichia coli to agents that damage DNA or interfere with DNA replication induces expression of a set of genes termed the SOS response (1,2). Many of the SOS-inducible genes encode proteins that are involved in DNA repair, recombination, DNA replication and cell division. If E. coli contains a λ prophage, the lytic cycle of phage λ is also induced as part of the SOS response (3,4). RecA protein and the LexA repressor are the key regulators of SOS induction. Following DNA damage RecA and single-stranded DNA form a nucleoprotein filament, which catalyzes autocleavage of LexA repressor, leading to induction of more than 30 genes including recA and lexA (2). The kinetics of SOS induction in E. coli, measured by monitoring LexA cleavage after UV irradiation, is understood in great detail. The induction starts 1 min after UV irradiation, reaches a maximum after 4 min and stays fully induced for at least 20 min (1,5). Genes containing operators that bind LexA repressor weakly, such as the promoter of the uvrAB genes, are first to be turned on during SOS induction. As the level of LexA declines, genes with operators binding LexA more strongly, like recA and sulA, will be induced (1). This rapid induction of SOS functions reflects the need for increased capacity to tolerate and to repair DNA damage.
Less is known about how the SOS response is turned off. It is believed that repairing DNA damage removes or reduces the amount of RecA nucleoprotein filament leading to accumulation of uncleaved LexA repressor and repression of the SOS genes. Turning off the SOS response can also be an active process and several factors inhibiting SOS induction have been described. The plasmid encoded PsiB protein inhibits SOS induction during transfer of conjugative plasmids (6–8), and overexpression of the chromosomally encoded DinI and RecX proteins directly interferes with the formation of the RecA nucleoprotein filament inhibiting LexA cleavage (9–11).
In addition to the more than 30 genes under direct control of LexA repressor, the transcriptional level of more than 1000 genes in E. coli are affected by treatment with DNA damaging agents (12). The translational levels of several proteins are also known to be upregulated by DNA damage in a LexA-independent but RecA-dependent manner. One such example is the Eda protein, the last enzyme in the Entner–Doudoroff pathway, whose function is necessary for the recovery of respiration following the SOS response (13,14).
The SOS-inducible tisAB gene (synonym gene name ysdAB), was first described as an SOS-inducible transcript containing the two open reading frames YsdA and YsdB (2), and later by mutational analysis shown to encode the toxin TisB (15). Recently it has been shown that in vitro translation of TisB is mediated by a shorter endonucleolytically processed tisAB transcript, termed +42 tisAB. Translation of +42tisAB RNA is initiated by upstream ribosome binding to a ribosome standby site and may involve sliding of the ribosome to a downstream translational initiation region. Binding of the sRNA istR-1 to the ribosome standby site prevents loading of ribosomes and inhibits translation of +42 tisAB RNA (16).
In this article we describe how overexpression of plasmid encoded tisAB inhibits and slows down induction of the SOS response. In addition, we find that overexpression of tisAB is necessary for cleavage of the uxaA mRNA, encoding altronate dehydratase, an enzyme specific for feeding the sugar acid galacturonate into the Entner–Doudoroff pathway (14). The presence of antisense sequences to uxaA and also to the SOS-inducible gene, dinD, suggests additional regulatory complexity in the tisAB function.
MATERIALS AND METHODS
Strains, plasmids and media
The experiments were carried out in strains AB1157 (arg, his, leu, pro, thr, ara, gal, lac, mtl, xyl, thi, tsx, rpsL, supE and kdgK) (17), MG1655 (18,19), DM49 (lexA3 derived from AB1157) (20) and ER2566 (lacZ::T7 gene1) (New England Biolabs, Ipswich, MA, USA). The uxaA, dinD and tisAB chromosomal mutants were made in strain BW25113-pKD46 (21) and introduced into AB1157 and MG1655 via T4GT7 transduction (22) using primers uxaAP1/uxaAP2, dinDP1/dinDP2 and tisABP1/tisABP2, respectively (Table 1). In mutant BK4012 (tisAB::kanr) 294 nt of the tisAB gene (position 3851350–3851644 in the E. coli genome, accession nr NC_000913) was replaced with a Kanr cassette. BK4012 have the whole uxaA and the 15 first nucleotides of the dinD antisense regions deleted, leaving the six last amino acids of TisB intact. The SOS box was not deleted in order not to interfere with istR induction.
Table 1.
Oligonucleotides used in this study
 |
Bold bases in TisB1, TisB2, TisB3, tisABuA1 and tisABuA2 nucleotides are mutated. Italic sequences in uxaAP, dinDP and tisABP primers are specific for the kanamycin cassette. T7 promoter sequence in the riboprobes is shown in italic.
Vector pKK232-8 contains a promoterless cat gene allowing for selection of DNA fragments containing promoter activity (23). Plasmid pBK410 was selected from a genomic library and contains a 759-bp fragment from the intergenic region between ilvBL-emrD inserted at the BamHI site of pKK232-8. Expression plasmids pET28b-TisA and pET28b-TisB contains the TisA and TisB open reading frames inserted in the NcoI–BamHI restriction sites of the pET28b(+) vector (Novagen, Madison, WI, USA) using primers TisA F, TisA R, TisB F and TisB R (primers listed in Table 1).
pBK410 mutations M1I, D5stop, K12stop, tisABuA1 and tisABuA2 were made using QuikChange site directed mutagenesis with primers TisB1(M1I) (F and R), TisB2(D5stop) (F and R), TisB3(K12stop) (F and R), tisBuA1 (F and R) and tisABuA2 (F and R), respectively.
Cells were grown in LB media or K-medium and washed in M9 buffer (24).
Generation of lysogens
Lysogenic E. coli was made by infecting the strain with bacteriophage λ and picking and purifying bacteria from the center of a plaque. Lysogeny was confirmed by demonstrating plaque formation on an indicator strain, or demonstrating immunity against reinfection with bacteriophage λ.
Mutagenesis
Exponentially growing bacteria were washed twice with M9 buffer and irradiated with UV light. From the irradiated cells 5 ml were mixed with 5 ml 2 × K-medium and grown overnight. Appropriate dilutions of the culture were plated on ampicillin containing LB agar plates for determining the total number of bacteria and on rifampicin containing LB agar plates (100 µg/ml) for determination of the number of rifR mutants.
Northen blot
RNA was isolated as previously described (25). RNA samples (5 µg) were denatured at 80°C in formaldehyde loading dye (55% formamide, 0.015% SDS, 0.015% Bromophenol Blue, 0.015% Xylene cyanol, 0.3 mM EDTA, 26.8% formaldehyde and 1.5×MOPS) buffer, separated on a 1% agarose, MOPS, formaldehyde gel and transferred to nylon membrane (Hybond N+, Amersham, Little Chalfont, Buckinghamshire, UK) (26).
Radiolabeled RNA probes were synthesized with MAXIscript (Ambion, Austin, TX, USA) (primers for synthesis of T7 template listed in Table 1) and hybridized using ExpressHyb Hybridization Solution (BD biosciences, San Jose, CA, USA). Hybridization signals were visualized using a Typhoon 9410 phosphor imager (Amersham).
Immunoblotting
Overnight cultures of E. coli were diluted 1 : 50 in K-medium and grown for the indicated time periods. Cells were harvested and lysed by plasmolysis (24) and protein extracts were separated on a SDS–polyacrylamide gel (27) and transferred to a PVDF (polyvinylidene difluoride) membrane by electroblotting. The membranes were incubated for 1 h at room temperature with a 1 : 1000 dilution of rabbit anti-LexA serum. The bound antibodies were visualized by incubating with a 1 : 5000 dilution of anti-rabbit IgG(Fc)-AP conjugate (Promega, Madison, WI, USA) and staining with BCIP/NBT (Promega). The LexA blot was scanned with a hp scanjet 4500c and the intensities of the protein bands analysed with the program ImageJ (Rasband,W.S. and ImageJ, U. S. National Institutes of Health, Bethesda, MD, USA, http://rsb.info.nih.gov/ij/, 1997–2007).
Prophage λ reactivation
Strains lysogenic to prophage λ were grown to OD600 of 0.4–0.5 in LB media at 37°C. The cells were pelleted by centrifugation, washed three times in 1 × M9 buffer to remove free bacteriophage, exposed to different UV doses (0–45 J/m2) and spread on LB agar plates with a non-lysogenic indicator strain to measure induction of prophage λ.
RESULTS
Isolation of plasmid pBK410
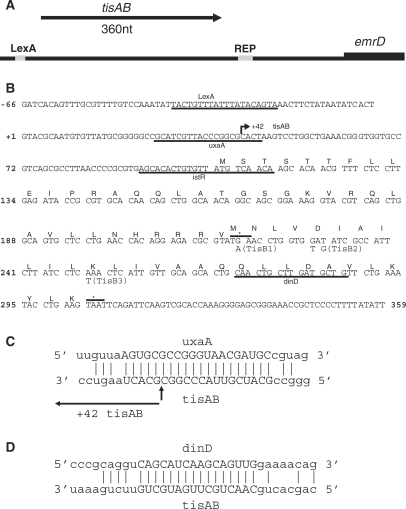
In a search for DNA damage inducible promoters in E. coli, digested chromosomal DNA from AB1157 was ligated in front of the promoter less chloramphenicol gene of promoter probe vector pKK232-8, allowing a positive selection of DNA fragments containing promoters because of the chloramphenicolR phenotype. The library was transformed into wild-type E. coli strain AB1157 and plated on media containing chloramphenicol, or chloramphenicol and 3 mM of the alkylating agent methyl methanesulfonate (MMS). MMS is an inducer of the adaptive response to alkylating agents in E. coli and also a potent inducer of the SOS response. Unexpectedly, one plasmid clone, pBK410, was selected on the basis of conferring MMS sensitivity to AB1157. Sequencing of pBK410 revealed a 759-bp insert from the intergenic region between ilvBL-emrD. This intergenic region has been shown to contain two overlapping sRNAs termed istR-1 and istR-2, and the divergently transcribed SOS-inducible gene termed ysdAB/tisAB (2,15). The pBK410 clone contains the ysdAB/tisAB gene, a single repetitive extragenic palindromic (REP) sequence and the first 37 amino acids of the emrD gene, while the istR genes are not contained in this clone (Figure 1A). A PARALIGN (28) search of the pBK410 insert revealed three sequences repeated in other regions of the E. coli genome, the 20-nt sequence GCATCGTTACCCGGCGCACT is antisense to a sequence at the 3′-end of the uxaA gene (Figure 1C), the 16-nt sequence CAACTGCTTGATGCTG is antisense to a sequence at the 3′-end of the SOS-inducible dinD gene (Figure 1D), and the 22-nt sequence AGCACACTGTGTTATGTCAACA is antisense to the istR sequences. The role of the 22-nt istR repeat in antisense regulation of tisAB has been previously described (15,16).
Figure 1.
Overview and details of the 759-bp pBK410 insert. (A) Direction of transcription of tisAB is shown by arrow and the emrD start is indicated. LexA-binding site and the single repetitive extragenic palindromic (REP) sequence is shaded. (B) Nucleotide sequence of the first 425 bp of the pBK410 insert up to transcriptional stop of tisAB. TisA and TisB open reading frames are labelled. The underlined istR sequence is the complement to the istR-1 and istR-2 sequence, which is described elsewhere (15). Sequence homology to uxaA and dinD is underlined. Numbering is relative to transcriptional startpoint of the 359-nt tisAB RNA. Arrow directed towards right +42 tisAB indicates the cleavage site of the endonucleolytically processed tisAB transcript. TisA and TisB translational stops are indicated with asterisk and line. Base changes leading to mutations TisB1, TisB2 and TisB3 are indicated below the nucleotide sequence.(C) Potential base pairing between uxaA and tisAB RNA is indicated, 20-nt core homology is capitalized. Information on tisAB RNA cleavage is from elsewhere (15). (D) Potential basepairing between dinD and tisAB RNA is indicated, 16-nt core homology is capitalized.
TisAB is overexpressed from plasmid pBK410
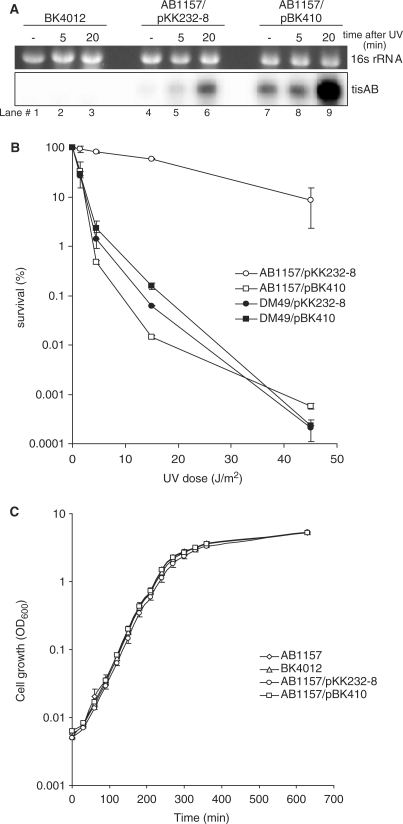
The insert sequence of pBK410 starts 66 nt upstream of the tisAB transcription start (Figure 1B). The functionality of the SOS operator in pBK410 was confirmed by northern analysis using a tisAB specific probe (Figure 2A). Total RNA for northern analysis was isolated from unirradiated and UV irradiated BK4012 (tisAB::kanr) and wild-type AB1157 carrying plasmid pBK410 or the cloning vector pKK232-8. A band corresponding to the 359-nt full-length tisAB transcript is clearly UV inducible in AB1157 carrying the pKK232-8 or pBK410 (Figure 2A, lanes 6 and 9). The band is totally absent in the tisAB deletion mutant BK4012 (tisAB::kanr).
Figure 2.
tisAB expression. (A) Expression of tisAB RNA in BK4012 (tisAB::Kanr), AB1157 transformed with pKK232-8 and AB1157 transformed with pBK410 (tisAB overexpression). Total RNA was extracted from the strains and 5 µg RNA obtained from unexposed cells (–) and cells exposed to UV (50 J/m2) at two different time points (5 and 20 min after UV-exposure) was separated on formaldehyde-1% agarose gel and blotted onto a nylon membrane. The blot was hybridized with a tisAB riboprobe. (B) UV survival was measured by irradiating exponentially growing AB1157 and DM49 (lexA3) transformed with the cloning vector pKK232-8 or plasmid pBK410 (overexpressing tisAB) to different doses of UV in M9 buffer and plating the cells on LB agar plates. Error bars showing the standard deviations are included. (C) 1 : 1000 dilutions of overnight cultures were grown in LB medium for 630 min and OD600 was measured every 30 min.
The northern experiments also illustrate that unirradiated AB1157 expresses significant amounts of tisAB transcript from pBK410 compared to the apparent absence of tisAB signal in AB1157 carrying only the cloning vector, pKK232-8. The level of tisAB transcript expressed from pBK410 in unirradiated AB1157 (Figure 2A, lane 7), is comparable to the fully induced level of tisAB in AB1157 carrying the cloning vector, pKK232-8 (Figure 2A, lane 6). Primer extension and 3′-RACE confirmed the transcriptional start and stop of the tisAB transcript reported earlier (data not shown) (15).
UV sensitization of wild-type E. coli by overexpression of tisAB
Several of the SOS-inducible genes in E. coli have a direct role in DNA repair, and mutants of SOS genes are often more sensitive to UV irradiation. To investigate a potential role of the tisAB gene in repair of UV induced DNA lesions, we tested the BK4012 (tisAB::kanr) mutant for UV resistance. We found no detectable difference in UV survival of BK4012 relative to AB1157, suggesting that the tisAB gene does not have a direct role in repair of UV mediated DNA damage (data not shown).
Survival experiments showed that the UV sensitivity of AB1157 carrying pBK410 was similar to the UV sensitivity of the lexA3 strain DM49 (Figure 2B), which is unable to induce the SOS response due to defective LexA cleavage (20). In addition to the tisAB gene, plasmid pBK410 also contains a repetitive extragenic palindromic (REP) sequence and the first 37 aa of the emrD coding sequence (Figure 1A). To investigate if the presence of the repetitive extragenic palindromic sequence and the emrD sequence in pBK410 affects the UV sensitisation of AB1157, plasmid pBK411 was generated. In plasmid pBK411 the repetitive extragenic palindromic sequence and emrD sequences downstream of the tisAB terminator were removed and the ability of pBK411 to confer sensitivity to AB1157 was measured. Plasmids pBK410 and pBK411 were found to confer the same UV sensitivity to AB1157 (data not shown). Plasmid pBK410 expresses significant levels of tisAB from uninduced cells at a level comparable to fully induced AB1157 carrying only the cloning vector pKK232-8. To examine the toxicity of tisAB expression from plasmid pBK410 we compared the growth of AB1157 carrying pBK410 or pKK232-8 and the tisAB deletion mutant BK4012. The growth curve shown in Figure 2C clearly demonstrates that uninduced expression of tisAB from pBK410 is not toxic. The data suggest that the reduction in UV resistance of AB1157 result from overexpression of the plasmid encoded tisAB gene product, and that a chromosomal deletion of tisAB does not affect any cellular functions contributing to UV survival.
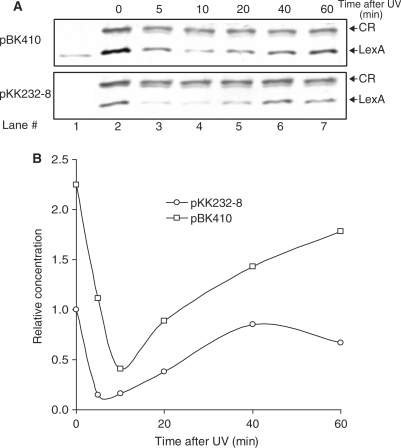
Overexpression of tisAB leads to higher levels of LexA
Activation of the SOS regulon depends on cleavage of the LexA repressor, in which the level of LexA before induction and the kinetics of LexA cleavage after induction is well characterized (1,29). The DM49 (lexA3) like UV sensitivity of wild-type AB1157 carrying pBK410 could be explained by a delay in SOS induction caused by higher levels of tisAB. We therefore tested the effect of tisAB overexpression on LexA repressor cleavage in UV irradiated cells. The level of LexA in protein extracts from strain AB1157 carrying either pBK410 or the cloning vector pKK232-8 were followed for 60 min after UV irradiation by immunoblotting with LexA antibody (Figure 3A). The level of uncleaved LexA repressor in unirradiated cells (Figure 3A, lane 2) is higher in AB1157 in the presence of pBK410 compared to AB1157 carrying only pKK232-8. A scan of the LexA immunoblot suggests that the level of LexA repressor in AB1157 is approximately twice the amount in cells carrying pBK410 compared to normal cells (Figure 3B). Figure 3A and B suggest that in cells carrying pBK410 the amount of LexA repressor 5 min after irradiation is comparable to the basal level of LexA in unirradiated cells carrying only pKK232-8, indicating a 5-min delay of SOS induction due to tisAB overexpression. Both the kinetics of LexA degradation after UV irradiation and the accumulation of LexA 10–20 min after UV irradiation appears to be parallel in cells carrying pBK410 compared to normal cells (Figure 3A and B), and indicates that only the endpoint levels of LexA is higher. Our immunoblotting experiments could not detect any differences in the amount of crossreacting (CR) proteins appearing on the western blot, suggesting that the effect from pBK410 is specific to LexA (Figure 3A). The results demonstrate that the constitutive overexpression of tisAB from pBK410 elevates the level of LexA in uninduced AB1157 resulting in higher LexA levels throughout the induction period and a shorter phase of full induction.
Figure 3.
Immunoblotting and quantitation of LexA repressor in AB1157 transformed with pKK232-8 (cloning vector) or pBK410 (tisAB overexpression). (A) Reaction mixtures containing 25 μg plasmolysed protein extracts from unexposed and cells exposed to UV for indicated times, separated by SDS–polyacrylamide gel electrophoresis, electroblotted to PVDF membrane and visualized by BCIP/NBT via rabbit anti-LexA serum. Lane 1 contains 3 ng of purified LexA protein. CR indicates a cross-reacting protein of unknown origin, shown here for normalization. (B) The bands in (A) were scanned and quantified as indicated in Materials and methods section. Intensities of LexA were normalized against the intensity of the cross-reacting protein (CR). All values are relative to LexA in uninduced AB1157/pKK232-8.
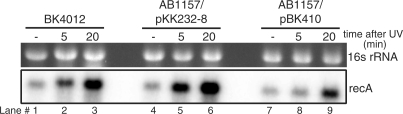
Delayed induction of recA mRNA by overexpression of tisAB
To determine if inhibition of LexA cleavage by overexpression of tisAB also influences the induction of other SOS-inducible genes, we measured the induction of recA mRNA in AB1157 in the presence of plasmid pBK410. The recA mRNA level indicates the kinetics and amount of induction of the RecA protein, which is a key recombinational and SOS regulatory protein. Exponentially growing cultures of BK4012 (tisAB::kanr) and AB1157 carrying either pBK410 or the cloning vector pKK232-8, were irradiated with 50 J/m2 and the amount of recA mRNA was determined prior to, and 5 and 20 min after irradiation by northern hybridization against a strand specific recA probe (Figure 4). The level of tisAB RNA was also determined by rehybridizing the northern blot with a probe against tisAB. As shown in Figure 4, recA mRNA is rapidly induced both in AB1157 and in BK4012 (tisAB::kanr), reaching a relatively high level after 5 min and is further increased after 20 min. In contrast, induction of recA mRNA after 5 min is barely detectable in AB1157 when pBK410 is present (Figure 4, lane 8). The uninduced level of recA mRNA also appears to be lower in the presence of pBK410 (Figure 4, lane 7). The result implies that overexpression of tisAB from plasmid pBK410 delays recA mRNA induction.
Figure 4.
Expression of recA mRNA in BK4012 (tisAB::Kanr), AB1157 and AB1157 transformed with pBK410 (tisAB overexpression). Total RNA was extracted from the strains and 5 µg RNA obtained from unexposed cells (–) and cells exposed to UV (50 J/m2) at two different time points (5 and 20 min after UV-exposure) was separated on formaldehyde-1% agarose gel and blotted onto a nylon membrane. The blot was hybridized with a recA riboprobe.
Overexpression of tisAB inhibits UV induction of prophage λ
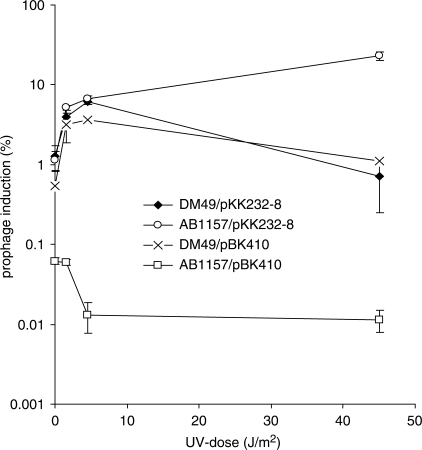
Induction of prophage λ in lysogenic E. coli cells requires RecA coprotease activity for cleavage of the λ CI repressor in a manner similar to LexA cleavage (30,31). To test if overexpression of tisAB from pBK410 inhibits induction of prophage λ we made λ-lysogens of AB1157 and DM49 (lexA3) and transformed the strains with either pBK410 or the cloning vector pKK232-8. The lysogenic strains were irradiated with UV and appropriate dilutions plated on AB1157 indicator strain to measure induction of prophage λ (Figure 5). Wild-type AB1157 in the presence of pBK410 shows a 20-time lower proportion of lysogenic cells spontaneously inducing prophage λ compared to AB1157 carrying only the cloning vector (0.06% and 1.2%, respectively) (Figure 5). In the presence of pBK410 and after UV irradiation the proportion of lysogenic AB1157 inducing λ decreases approximately six times. Induction of prophage λ in DM49 (lexA3) appears to be unaffected by the presence of pBK410. Lysogenic DM49 gave rise to small plaques during prophage λ induction while AB1157 produced bigger plaques. The size of the plaques formed by AB1157 in the presence of pBK410 was similar to those formed by AB1157 carrying the cloning vector, implying that the relatively low number of cells inducing λ in AB1157 carrying pBK410 exhibited wild-type kinetics of λ-induction. It thus appears that tisAB inhibits UV induction of prophage λ supporting a role for tisAB in regulation of the SOS response.
Figure 5.
Inhibition of prophage λ induction. Lysogenic AB1157 (λ) and DM49 (lexA3) (λ) were grown in K-medium to mid-exponential phase, washed, resuspended in M9 buffer and exposed to UV as indicated. The datapoints shown is an average of three experiments. Error bars showing the standard deviations are included.
Overexpression of tisAB inhibits cell filamentation
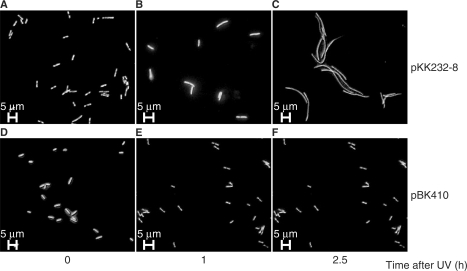
After SOS induction E. coli form long filaments as a consequence of activation of the septation inhibitor SulA (32). To test if overexpression of tisAB from plasmid pBK410 affected filamentation, exponentially growing cultures of AB1157 carrying pBK410 or the cloning vector pKK232-8 were UV irradiated with 50 J/m2 and the cells were examined at intervals with fluorescent microscopy to determine the degree of filamentation (Figure 6). In the presence of the cloning vector, strain AB1157 displayed short filaments 1 h after UV exposure and produced long filaments after 2.5 h (Figure 6B and C). AB1157 carrying pBK410 was not generating filaments at all after UV exposure (Figure 6E and F), indicating a role of tisAB in modulation of filamentation via regulation of the SOS response.
Figure 6.
Fluorescence microscopy of UV exposed (50 J/m2) cells carrying the cloning vector pKK232-8 or plasmid pBK410 (tisAB overexpression) followed by growth for 1 and 2.5 h. Cells were grown in K-medium and stained with acridine orange. (A) AB1157/pKK232-8, unexposed; (B) AB1157/pKK232-8, 1 h after UV; (C) AB1157/pKK232-8, 2.5 h after UV; (D) AB1157/pBK410, unexposed; (E) AB1157/pBK410, 1 h after UV; (F) AB1157/pBK410, 2.5 h after UV.
Moderate decrease in UV mutagenesis by the presence of pBK410
An important SOS response is the increase in mutation frequency as a result of the action of error-prone translesion polymerases, PolIV (dinB/P) and PolV (umuDC), which are both induced as part of the SOS response (1). To examine the influence of plasmid pBK410 on SOS mutagenesis in wild type E. coli, we UV irradiated AB1157 carrying the pBK410 and compared the frequency of rifR forming colonies with AB1157 carrying the cloning vector pKK232-8. The results summarized in Table 2 indicate that among the surviving fraction of AB1157 carrying pBK410 there appears to be a moderate reduction in UV-induced mutation frequency compared to AB1157 carrying only the vector.
Table 2.
Mutation frequencies in cells carrying the cloning vector pKK232-8 and plasmid pBK410 (tisAB overexpression)
| Strain | Plasmid | UV (J/m2) | Survival (%) | Mutation frequency (10−7) |
|---|---|---|---|---|
| AB1157 | pBK410 | 0 | 100 | 0.28 |
| 5 | 7 | 3.9 | ||
| 15 | 0.005 | 9.1 | ||
| pKK232-8 | 0 | 100 | 0.56 | |
| 5 | 80 | 7.1 | ||
| 15 | 36 | 23.0 | ||
| DM49 (lexA3) | pBK410 | 0 | 100 | 0.33 |
| 5 | 12 | 2.6 | ||
| 15 | 4.6 | 6.3 | ||
| pKK232-8 | 0 | 100 | 0.33 | |
| 5 | 13 | 1.5 | ||
| 15 | 2.4 | 3.5 |
The role of the YsdA/TisA and YsdB/TisB open reading frames
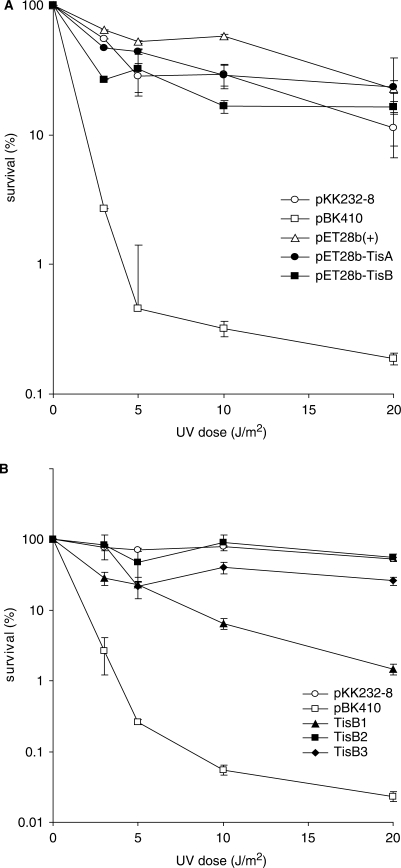
The extreme UV sensitivity plasmid pBK410 inflicts on wild-type E. coli provides a sensitive assay to investigate the functional role of different parts of the tisAB gene. An obvious candidate for the anti-SOS phenotype we observe from plasmid pBK410 is the TisB protein which is translated from +42 tisAB RNA in vitro (16). To study the effect of the two open reading frames, TisA and TisB, in sensitizing E. coli to UV, we cloned TisA and TisB into the T7 polymerase regulated expression vector pET28b(+). The two clones where transformed into the T7 polymerase expressing strain ER2566 and sensitivity to increasing doses of UV was measured. ER2566 transformed with pBK410 was included as a control, and the results in Figure 7A demonstrate that strain ER2566 carrying pBK410 display UV sensitivity comparable to AB1157 carrying pBK410 (Figure 2B). Moreover, ER2566 carrying pET28b-TisA or pET28b-TisB is not significantly more sensitive to UV exposure than the vector control (Figure 7A). We also observed that after UV irradiation ER2566 carrying the pET28b-TisA and pET28b-TisB constructs formed colonies with similar size to ER2566 carrying only the cloning vector pET28b(+). The lack of UV sensitization from the TisB expression construct can be explained by the special way TisB is translated (16), and to further investigate the role of TisB we constructed mutations TisB1, TisB2 and TisB3 (Figure 1B) in the pBK410 plasmid and assayed for UV sensitivity. Figure 7B shows that mutations TisB2 (D5stop, GAT to TAG) and TisB3 (K12stop, AAA to TAA) eliminates the sensitizing phenotype from pBK410 thereby confirming the role of TisB in the phenotype (15). However, the pBK410 mutation TisB1 (M1I, ATG to ATA) which was made in order to mutate the TisB start-codon without destroying the overlapping TisA stop-codon still confers substantial UV sensitivity to AB1157 (Figure 7B). A northern experiment confirmed that these mutant plasmids produced similar amounts of tisAB transcript (data not shown). The results show that the role of TisB translation in vivo necessitates a closer examination.
Figure 7.
UV survival of TisA and TisB expression plasmids (A) and TisB mutants of pBK410 (B). (A) Cloning of TisA and TisB in expression plasmid pET28b(+) is described in Materials and methods section. Cells were grown to exponentially phase in K-medium added 1mM IPTG for induction. Survival was measured by irradiating exponentially growing ER2566 transformed with pKK232-8, pBK410 (tisAB overexpression), pET28b(+), pET28b(+)-TisA and pET28b(+)-TisB to different doses of UV in M9 buffer and plating the cells on LB agar plates. The points shown is average of three experiments, error bars showing standard deviations are included. (B) UV survival of AB1157 transformed with pBK410 and the TisB mutated derivatives TisB1(M1I), TisB2(D5stop) and TisB3(K12stop). Cells were grown to exponentially phase in K-medium, washed in M9, irradiated with different doses of UV and plated on LB agar.
Overexpression of tisAB mediates SOS-induced cleavage of the uxaA mRNA
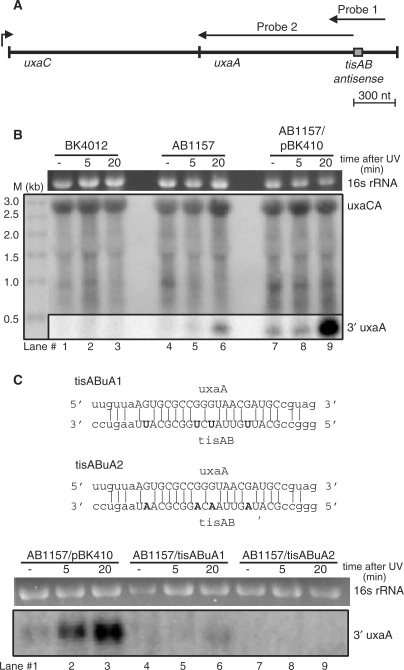
The presence of both uxaA and dinD antisense sequence in the tisAB transcript suggest antisense regulation between the tisAB-uxaA and tisAB-dinD transcripts. The size of the core tisAB-uxaA antisense region is 20 nt (Figure 1C). Notably, tisAB RNA isolated from stationary phase is cleaved in the tisAB-uxaA antisense region (15). The uxaA gene is the last gene in an operon with uxaC, forming an uxaCA mRNA of 2900 nt, where the tisAB-uxaA antisense region is 300 nt upstream of a terminator sequence in the 3′-end of uxaA (Figure 8A). To determine whether the uxaA mRNA is cleaved in the antisense region, total RNA was isolated from unirradiated and UV irradiated exponentially growing cultures of BK4012 (tisAB::Kanr) and AB1157 carrying either pBK410 or the cloning vector pKK232-8. A 400 nt strand specific RNA probe (probe 1, Figure 8A) spanning 200 nt to each side of the tisAB antisense region of uxaA was hybridized to a northern blot of isolated total RNA (Figure 8B, lower panel). As shown in Figure 8B, lane 9, in the presence of pBK410 a band corresponding in size to a 300 nt 3′-cleavage product of uxaA is clearly present in AB1157 20 min after UV irradiation. A weaker band with similar size is also visible in AB1157 carrying the cloning vector pKK232-8 (Figure 8B, lane 6). The 300 nt band is totally absent in the tisAB::kanr mutant BK4012. The 400-nt probe detected only a faint full-length uxaCA mRNA of 2900 nt (data not shown), therefore the blot was rehybridized to a second probe (probe 2, Figure 8A) covering the first 1100 nt of uxaA. This probe produced a signal corresponding to the full-length uxaCA transcript, confirming the faint band detected with the 400 nt. To further examine the role of tisAB on uxaA mRNA processing we made two pBK410 mutant plasmids (tisABuA1 and tisABuA2) with base substitutions in the tisAB-uxaA antisense sequence (Figure 8C). The mutant plasmid, tisABuA1, contained four C to U substitutions while the other plasmid, tisABuA2, contained C to A substitutions in the same positions. The northern experiment demonstrates that the ability to process uxaA mRNA is totally abolished with the tisABuA2 plasmid (lanes 7–9) and strongly impaired with the tisABuA1 construct (lanes 4–6), indicating that GA basepairing in the antisense region disrupt interference completely while GU basepairing partly maintain uxaA mRNA processing. It thus appears that SOS-induced cleavage of uxaA mRNA in the 3′ antisense region is dependent on tisAB expression.
Figure 8.
Analysis of tisAB mediated uxaA cleavage. (A) Map of uxaCA operon and position of northern probes and antisense region. Drawing is to scale. (B) Expression and cleavage of uxaA mRNA in BK4012 (tisAB::Kanr), AB1157 and AB1157 transformed with pBK410 (overexpressing tisAB). Total RNA was extracted from the three isogenic strains and 5 µg RNA obtained from unexposed cells (–) and cells exposed to UV (50 J/m2) at two different time points (5 and 20 min after UV exposure) was separated on formaldehyde-1% agarose gel and blotted onto a nylon membrane. The blot was hybridized with a 400 nt uxaA riboprobe (Probe 1) covering the uxaA/tisAB antisense sequence (lower panel), and rehybridized with a 1100 nt uxaA riboprobe (Probe 2) covering only the sequence upstream of the antisense sequence. Size marker: 4 µg Millennium Markers-Formamide (Ambion). (C) Sequence alignments indicating base changes of the pBK410 derived mutants tisABuA1 and tisABuA2 followed by cleavage of uxaA mRNA in AB1157 transformed with pBK410 and the antisense mutants tisABuA1 and tisABuA2. Northern was carried out as in (B) except the blot was hybridized only with the 400-nt probe (Probe 1).
Delayed UV sensitization of the uxaA, dinD double mutant after overexpression of tisAB
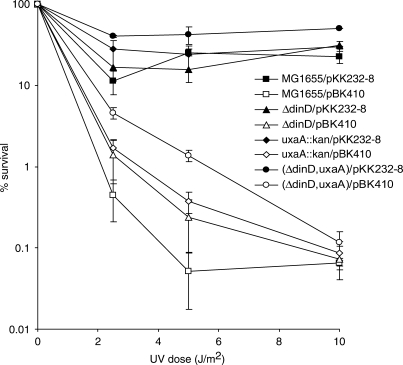
The tisAB-dependent processing of uxaA mRNA raises the reciprocal question if uxaA mRNA is able to mediate an antisense effect on tisAB activity. In order to investigate any effect of the uxaA and dinD antisense sequences on UV sensitivity conferred by tisAB overexpression we performed survival assays in uxaA and dinD deletion mutants. The UV sensitivity of isogenic MG1655, BK5060 (uxaA::kanr), BK5131(ΔdinD) and BK5065 (ΔdinD, uxaA::kanr) carrying plasmid pBK410 or pKK232-8 suggests that pBK410 is less able to confer sensitivity to BK5065 (ΔdinD, uxaA::Kanr) compared to MG1655 when irradiated with 2 and 5 J/m2 (Figure 9). The results shows that MG1655 carrying pBK410 reaches a lower plateau of UV sensitivity at around 0.1% survival after irradiation with 5 J/m2 while BK5065 (ΔdinD, uxaA::Kanr) carrying pBK410 is less sensitive at this UV dose. The results indicate that UV sensitivity conferred by overexpression of tisAB is partially dependent on both the uxaA and dinD genes.
Figure 9.
Effect of uxaA and dinD mutations on UV survival of cells carrying overexpressing tisAB (pBK410). Cells were grown in K-medium to exponential phase, washed in M9, UV irradiated with indicated doses and plated on LB agar plates.
DISCUSSION
In this study we have demonstrated that overexpression of tisAB in wild type E. coli interferes with SOS induction. Overexpression of tisAB to nontoxic levels leads to higher amount of LexA repressor both in unirradiated AB1157 and throughout the induction period causing a delay in recA mRNA induction. AB1157 overexpressing tisAB were sensitive to UV, displayed lower level of spontaneous and induced prophage λ induction, and were lacking cell filamentation. In addition we found that expression of tisAB is necessary for cleavage of the galacturonate catabolism specific uxaA mRNA.
According to the present model for SOS induction (1), the enhanced basal level of LexA repressor in unirradiated AB1157 carrying pBK410 could explain the 5 min delay in recA mRNA induction (Figure 4). The significance of the delayed recA induction could be severe given the important role of the RecA protein in recovery after UV irradiation. The lack of UV induced cell filamentation in AB1157 carrying pBK410 (Figure 6F) also suggests that the higher levels of LexA leads to insufficient induction of SulA, the SOS-inducible inhibitor of septation (33).
Our data demonstrated that UV-induced mutagenesis in AB1157 appears to be normal in the surviving fraction of irradiated cells carrying pBK410. Further, the plaques formed by lysogenic AB1157 carrying pBK410 displayed a size similar to wild-type cells. Most likely, a subpopulation of surviving cells behaves as wild-type cells, and does not express the anti-SOS phenotype.
An important question regarding the inhibition of SOS functions by overexpression of tisAB is what functional part of the tisAB gene is responsible for the inhibitory effect. An obvious candidate is the TisB peptide which has been reported to encode an SOS-inducible toxin (15,16). However, our protein expression constructs containing the TisB or TisA reading frames under transcriptional control of an IPTG-inducible promoter failed to confer cell death (Figure 2C) or to produce detectable UV sensitivity to the host, as compared to plasmid pBK410 (Figure 7A). Assuming that the TisB peptide is the component mediating inhibition of SOS functions, the lack of phenotype from pET28 constructs could simply be explained by a lack of TisB expression from this system. It has been shown in vitro that TisB translation is effective only from the +42 transcript of tisAB, suggesting that the RNA structure and context could be important for TisB translation. In addition, our observation that the TisB1(M1I) mutation in pBK410 still confers considerable UV sensitivity to wild-type E. coli (Figure 7B) suggests that the role of TisB translation in vivo needs to be further investigated. However, our data indicate that the actual function of the tisAB gene under normal levels of induction is not a toxin that kills cells under stress, but a function inhibiting or regulating activities important during SOS induction. Several steps in the sequential events leading to SOS induction could be affected by tisAB. Overexpression of tisAB could directly or indirectly affect the cleavage or binding properties of LexA repressor and λ cI repressor leading to higher basal levels of the proteins. By example, a change in internal pH affects LexA affinity for operators (34). The RecA filament or the RecA protein itself could be inhibited by tisAB overexpression analogous to the function of DinI and RecX proteins (9). The key to understand the target of tisAB appears to rest in how and if TisB is translated in vivo and the possible target and location of the protein.
The presence of antisense sequences to both the uxaA gene and the SOS-inducible dinD gene (Figure 1C and D) suggests additional complexity in the way tisAB mediates its anti-SOS effect and also the way tisAB itself is regulated. Typically, antisense RNA sequences affect RNA stability or translation of mRNA (35). We were able to demonstrate that tisAB expression affects processing of uxaA mRNA by mediating cleavage of the uxaA transcript, giving rise to a 300 nt cleavage fragment from the 3′-end of uxaA. Since cleavage occurs in the coding region of uxaA it will result in inactivation of the uxaA mRNA, and possibly having a polar effect on the upstream uxaC on the same mRNA. The uxaA gene is specific for galacturonate catabolism in E. coli, and the UxaA enzyme (altronate dehydratase) mediates formation of 2-keto-3-deoxy-gluconate (KDG) from galacturonate which is further phosphorylated to a substrate for the Eda enzyme in the Entner–Doudoroff pathway (14). The Eda enzyme is known to play a role in the SOS response, as the Eda protein is induced by DNA-damaging agents in an RecA-dependent manner, and it is hypothesized that Eda is necessary for recovery of respiration following the SOS response (13). We may speculate if overexpression of tisAB reduces the level of UxaA and the amount of KDG formed, thereby inhibiting the cells’ ability to recover respiration following SOS induction.
We also investigated the opposite possibility that uxaA mRNA via the uxaA/tisAB antisense region had a role in generating +42 tisAB RNA. In our hands the level of +42 tisAB RNA from UV exposed log phase cells were found to be below detection level in acrylamide northern experiments (data not shown). However, in agreement with Vogel et al. (15) the +42 tisAB cleavage product was present in northern experiments with RNA obtained from stationary phase cells exposed to UV (data not shown). Further, wild-type and uxaA cells showed the same level + 42 tisAB RNA after UV exposure indicating that uxaA mRNA may not promote cleavage of tisAB. Nevertheless, these experiments are not excluding that uxaA modulate processing of tisAB RNA. In fact we may speculate if uxaA mRNA inhibit tisAB cleavage under growth conditions in which uxaA is upregulated to avoid an SOS inhibitory effect of the +42 tisAB transcript.
We were not able to demonstrate any tisAB-mediated degradation of dinD mRNA, similar to uxaA mRNA degradation. It is intriguing that tisAB and dinD, both being induced by the SOS response, has sequences with potential of mediating interaction between the two RNA transcripts. The DinD protein function is not known, the gene is dispensable for growth, and the only phenotype known of dinD is an recA-dependent cold sensitivity of the dinD mutation pcsA68 (36). It should also be noted that SOS-mediated induction of β-galactosidase from the dinD2::Mud(ApR,lac) mutant is delayed (37), suggesting a possible role for DinD in SOS induction.
FUNDING
Norwegian Research Council (FUGE-CAMST); and the Anders Jahres Foundation for Medical Research. Funding for open access publication charge: Rikshospitalet, Oslo, Norway.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Marie Aslaksen Røed for technical assistance and James Booth for critical reading of the manuscript.
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and mutagenesis. Am. Soc. Microbiol. 2006:463–612. [Google Scholar]
- 2.Fernandez De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 3.Hertman I, Luria SE. Transduction studies on the role of a rec+ gene in the ultraviolet induction of prophage lambda. J. Mol. Biol. 1967;23:117–133. doi: 10.1016/s0022-2836(67)80021-4. [DOI] [PubMed] [Google Scholar]
- 4.Brooks K, Clark AJ. Behavior of lambda bacteriophage in a recombination deficienct strain of Escherichia coli. J. Virol. 1967;1:283–293. doi: 10.1128/jvi.1.2.283-293.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sassanfar M, Roberts JW. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 6.Bagdasarian M, Bailone A, Bagdasarian MM, Manning PA, Lurz R, Timmis KN, Devoret R. An inhibitor of SOS induction, specified by a plasmid locus in Escherichia coli. Proc. Natl Acad. Sci. USA. 1986;83:5723–5726. doi: 10.1073/pnas.83.15.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutreix M, Backman A, Celerier J, Bagdasarian MM, Sommer S, Bailone A, Devoret R, Bagdasarian M. Identification of psiB genes of plasmids F and R6-5. Molecular basis for psiB enhanced expression in plasmid R6-5. Nucleic Acids Res. 1988;16:10669–10679. doi: 10.1093/nar/16.22.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golub E, Bailone A, Devoret R. A gene encoding an SOS inhibitor is present in different conjugative plasmids. J. Bacteriol. 1988;170:4392–4394. doi: 10.1128/jb.170.9.4392-4394.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lusetti SL, Drees JC, Stohl EA, Seifert HS, Cox MM. The DinI and RecX proteins are competing modulators of RecA function. J. Biol. Chem. 2004;279:55073–55079. doi: 10.1074/jbc.M410371200. [DOI] [PubMed] [Google Scholar]
- 10.Yasuda T, Nagata T, Ohmori H. Multicopy suppressors of the cold-sensitive phenotype of the pcsA68 (dinD68) mutation in Escherichia coli. J. Bacteriol. 1996;178:3854–3859. doi: 10.1128/jb.178.13.3854-3859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stohl EA, Brockman JP, Burkle KL, Morimatsu K, Kowalczykowski SC, Seifert HS. Escherichia coli RecX inhibits RecA recombinase and coprotease activities in vitro and in vivo. J. Biol. Chem. 2003;278:2278–2285. doi: 10.1074/jbc.M210496200. [DOI] [PubMed] [Google Scholar]
- 12.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cayrol C, Petit C, Raynaud B, Capdevielle J, Guillemot JC, Defais M. Recovery of respiration following the SOS response of Escherichia coli requires RecA-mediated induction of 2-keto-4-hydroxyglutarate aldolase. Proc. Natl Acad. Sci. USA. 1995;92:11806–11809. doi: 10.1073/pnas.92.25.11806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peekhaus N, Conway T. What's for dinner?: Entner-Doudoroff metabolism in Escherichia coli. J. Bacteriol. 1998;180:3495–3502. doi: 10.1128/jb.180.14.3495-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogel J, Argaman L, Wagner EG, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Darfeuille F, Unoson C, Vogel J, Wagner EG. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell. 2007;26:381–392. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Dewitt SK, Adelberg EA. The occurrence of a genetic transposition in a strain of Escherichia coli. Genetics. 1962;47:577–585. doi: 10.1093/genetics/47.5.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guyer MS, Reed RR, Steitz JA, Low KB. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb. Symp. Quant. Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- 19.Blattner FR, Plunkett G., III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 20.Mount DW, Low KB, Edmiston SJ. Dominant mutations (lex) in Escherichia coli K-12 which affect radiation sensitivity and frequency of ultraviolet lght-induced mutations. J. Bacteriol. 1972;112:886–893. doi: 10.1128/jb.112.2.886-893.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson GG, Young KY, Edlin GJ, Konigsberg W. High-frequency generalised transduction by bacteriophage T4. Nature. 1979;280:80–82. doi: 10.1038/280080a0. [DOI] [PubMed] [Google Scholar]
- 23.Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984;27:151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- 24.Seeberg E, Strike P. Excision repair of ultraviolet-irradiated deoxyribonucleic acid in plasmolyzed cells of Escherichia coli. J. Bacteriol. 1976;125:787–795. doi: 10.1128/jb.125.3.787-795.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saetrom P, Sneve R, Kristiansen KI, Snove O., Jr, Grunfeld T, Rognes T, Seeberg E. Predicting non-coding RNA genes in Escherichia coli with boosted genetic programming. Nucleic Acids Res. 2005;33:3263–3270. doi: 10.1093/nar/gki644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chomczynski P. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal. Biochem. 1992;201:134–139. doi: 10.1016/0003-2697(92)90185-a. [DOI] [PubMed] [Google Scholar]
- 27.Schagger H, von JG. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 28.Rognes T. ParAlign: a parallel sequence alignment algorithm for rapid and sensitive database searches. Nucleic Acids Res. 2001;29:1647–1652. doi: 10.1093/nar/29.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Little JW. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie. 1991;73:411–421. doi: 10.1016/0300-9084(91)90108-d. [DOI] [PubMed] [Google Scholar]
- 30.Craig NL, Roberts JW. E. coli recA protein-directed cleavage of phage lambda repressor requires polynucleotide. Nature. 1980;283:26–30. doi: 10.1038/283026a0. [DOI] [PubMed] [Google Scholar]
- 31.Little JW. Autodigestion of lexA and phage lambda repressors. Proc. Natl Acad. Sci. USA. 1984;81:1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huisman O, D’Ari R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature. 1981;290:797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- 33.Gottesman S, Maurizi MR. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol. Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dri A-M, Moreau PL. Control of the LexA regulon by pH: evidence for a reversible inactivation of the LexA repressor during the growth cycle of Escherichia coli. Mol. Microbiol. 1994;12:621–629. doi: 10.1111/j.1365-2958.1994.tb01049.x. [DOI] [PubMed] [Google Scholar]
- 35.Wagner EG, Simons RW. Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 36.Ohmori H, Saito M, Yasuda T, Nagata T, Fujii T, Wachi M, Nagai K. The pcsA gene is identical to dinD in Escherichia coli. J. Bacteriol. 1995;177:156–165. doi: 10.1128/jb.177.1.156-165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenyon CJ, Walker GC. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc. Natl Acad. Sci. USA. 1980;77:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]