Abstract
RNA thermometers are thermosensors that regulate gene expression by temperature-induced changes in RNA conformation. Naturally occurring RNA thermometers exhibit complex secondary structures which are believed to undergo a series of gradual structural changes in response to temperature shifts. Here, we report the de novo design of considerably simpler RNA thermometers that provide useful RNA-only tools to regulate bacterial gene expression by a shift in the growth temperature. We show that a single small stem-loop structure containing the ribosome binding site is sufficient to construct synthetic RNA thermometers that work efficiently at physiological temperatures. Our data suggest that the thermometers function by a simple melting mechanism and thus provide minimum size on/off switches to experimentally induce or repress gene expression by temperature.
INTRODUCTION
RNA thermometers are RNA-based genetic control systems that sense temperature changes. At low temperatures, the mRNA adopts a conformation that masks the ribosome binding site [Shine–Dalgarno (SD) sequence] within the 5′-untranslated region (5′-UTR) and, in this way, prevents ribosome binding and translation. At elevated temperatures, the RNA secondary structure melts locally, thereby giving the ribosomes access to the ribosome binding site to initiate translation. RNA thermometers differ from riboswitches (1) in that they do not require binding of a ligand (metabolite) to induce the conformational change, but instead, directly respond to temperature (2,3). Biological processes controlled by RNA thermometers include, for example, bacterial pathogenesis (4), heat-shock responses (5–7) and the life cycle of bacteriophages (8). The highly complex RNA secondary structures into which most naturally occurring RNA thermometers can be folded has led to the hypothesis that RNA thermometers may not function as simple on/off switches, but rather represent dimmers (3), in that they may go through a series of distinct conformational changes which in turn may gradually change translation levels with temperature (3,7,9).
Here, we have attempted to construct minimum size synthetic RNA thermometers that are considerably simpler than naturally occurring RNA thermosensors and can be used to induce or repress gene expression by a simple temperature shift. Such thermometers would provide minimum size tools to regulate expression of endogenous genes and transgenes independently of proteinaceous trans-acting factors or chemical inducers. We describe here the design principles for synthetic RNA thermometers and demonstrate that our optimized thermometers are fully functional in vivo as translational regulators at physiological temperatures. Our data show that efficient RNA thermometers can be built from a single small RNA stem-loop structure masking the ribosome binding site, thus providing useful RNA-only tools for the regulation of gene expression in prokaryotes.
MATERIALS AND METHODS
Bacteria
Escherichia coli cells (One Shot® Top10F’, Invitrogen, Karlsruhe, Germany) harboring the various plasmid constructs were grown in Luria–Bertani (LB) medium with ampicillin (100 µg/ml) at 37°C under continuous shaking (180 r.p.m.) unless otherwise stated. To determine the time course of temperature-dependent induction of gene expression, bacteria were precultured at 17°C and, after 2 days, diluted in fresh medium. The growth temperature was then shifted from 17°C to 37°C and samples were taken for protein isolation at the indicated time points. The starting cell density of the cultures for 30 min and 1 h time points was set to OD600 = 0.2, for the 2 h and 3 h time points to OD600 = 0.1 and for all others time points to OD600 = 0.05.
Cloning of 5′-UTR thermosensor elements from the Listeria prfA gene
The oligonucleotides (Table 1) for the assembly of the wild-type (wt) and the T46A-mutated 5′-UTR from the Listeria monocytogenes prfA gene (4) were phosphorylated with T4 polynucleotide kinase (New England Biolabs, Frankfurt/M., Germany) in a reaction containing 150 pmol of each oligonucleotide, 1 mM ATP and 10 U polynucleotide kinase. For annealing of the complementary oligonucleotides, 75 pmol of each phosphorylated oligonucleotides were mixed, incubated at 95°C and slowly cooled down to 35°C. For cloning, vector pBS SK(+) was digested with BamHI and HindIII and ligated to the annealed double-stranded oligonucleotides resulting in plasmids pBS41 (wt prfA 5′-UTR) and pBS42 (T46A mutant version; 4). The gfp coding region together with the 3′-UTR of the plastid rps16 gene was excised as an NcoI and HindIII fragment from a previously constructed expression cassette (10,11) and ligated into pBS41 and pBS42, resulting in vectors pBS43 and pBS44, respectively. The strong tobacco plastid rRNA operon promoter Prrn was amplified with primer pair Prrnfor 5′-TTTTGAGCTCGGTACCCCAAAG-3′ (SacI site underlined) and Prrnrev 5′-TTTTGGATCCGTATCCAAGCGCTTCGTA-3′ (BamHI site underlined) from a plasmid clone (12). The PCR product was cloned as SacI/BamHI into pBS43 and pBS44, resulting in pBS45 and pBS46, respectively.
Table 1.
Synthetic oligonucleotides used for generating 5′-UTR constructs
| Vector | 5′-UTR | Oligo | Sense and antisense sequences of synthetic oligonucleotides |
|---|---|---|---|
| pBSU0 | U0 | U0_s | 5′-GATCCGAAAAAACTAGTCTGCAGAAGGAGATATACC-3′ |
| U0_as | 5′-CATGGGTATATCTCCTTCTGCAGACTAGTTTTTTCG-3′ | ||
| pBSU1 | U1 | U1_s | 5′-GATCCCACCTTACTAGTCTGCAGAAGGAGATATACC-3′ |
| U1_as | 5′-CATGGGTATATCTCCTTCTGCAGACTAGTAAGGTGG-3′ | ||
| pBSU2 | U2 | U2_s | 5′-GATCCCTCACTTACTAGTCTGCAGAAGGAGATATACC-3′ |
| U2_as | 5′-CATGGGTATATCTCCTTCTGCAGACTAGTAAGTGAGG-3′ | ||
| pBSU3 | U3 | U3_s | 5′-GATCCCTCCATACTAGTCTGCAGAAGGAGATATACC-3′ |
| U3_as | 5′-CATGGGTATATCTCCTTCTGCAGACTAGTATGGAGG-3′ | ||
| pBSU4 | U4 | U4_s | 5′-GATCCATCCTTACTAGTCTGCAGAAGGAGATATACC-3′ |
| U4_as | 5′-CATGGGTATATCTCCTTCTGCAGACTAGTAAGGATG-3′ | ||
| pBSU5 | U5 | U5_s | 5′-GATCCCTCCTAACTAGTCTGCAGAAGGAGATATACC-3′ |
| U5_as | 5′-CATGGGTATATCTCCTTCTGCAGACTAGTTAGGAGG-3′ | ||
| pBSU6 | U6 | U6_s | 5′-GATCCTCTCCTTCACTAGTAAAAAAAAAAAAAAAAAAAAA AAAAAAGGAGATATACC-3′ |
| U6-as | 5′- CATGGGTATATCTCCTTTTTTTTTTTTTTTTTTTTTTTTTT TACTAGTGAAGGAGAG-3′ | ||
| pBSU9 | U9 | U9-s | 5′-GATCCCTCCTTACTAGTCTGCAGAAGGAGATATACC-3′ |
| U9_as | 5′-CATGGGTATATCTCCTTCTGCAGACTAGTAAGGAGG-3′ | ||
| pBSU10 | U10 | U10_s | 5′-GATCCTCTCCTTACTAGTCTGCAGAAGGAGATATACC-3′ |
| U10_as | 5′-CATGGGTATATCTCCTTCTGCAGACTAGTAAGGAGAG-3′ | ||
| pBSU11 | U11 | U11_s | 5′-GATCCCTCCTTCACTAGTCTGCAGAAGGAGATATACC-3′ |
| U11_as | 5′-CATGGGTATATCTCCTTCTGCAGACTAGTGAAGGAGG-3′ | ||
| pBSU12 | U12 | U12_s | 5′-GATCCTCTCCTTCACTAGTCTGCAGAAGGAGATATACC-3′ |
| U12_as | 5′-CATGGGTATATCTCCTTCTGCAGACTAGTGAAGGAGAG-3′ | ||
| pBS45 | prfA-wt | prfA5′-for | 5′-GATCCTGTAAAAAACATCATTTAGCGTGACTTTCTTTCAACA GCTAACAATTGTTGTTACTGCCTA-3′ (BamHI overhang in italics) |
| prfA5′-rev | 5′-ATACCCTAAAAACATTAGGCAGTAACAACAATTGTTAGCT GTTGAAAGAAAGTCACGCTAAATGATGTTTTTTACAG-3′ (overhang complementary to prfA3′-for in bold) | ||
| prfA3′-for | 5′-ATGTTTTTAGGGTATTTTAAAAAAGGGCGATAAAAAACGA TTGGGGGATGAGACATGAACGCTCAAGCCATGG (NcoI site underlined, overhang complementary to prfA5′-rev and prfA5′T46A-rev in bold) | ||
| prfA3′-rev | 5′-AGCTCCATGGCTTGAGCGTTCATGTCTCATCCCCCAATCG TTTTTTATCGCCCTTTTTTAAA-3′ (HindIII overhang in italics, NcoI site underlined) | ||
| pBS46 | prfA-T46A | prfA5′T46A-for | 5′-GATCCTGTAAAAAACATCATTTAGCGTGACTTTCTTTCAACA GCTAACAAATGTTGTTACTGCCTA-3′ (BamHI overhang italic, mutation T46A underlined) |
| prfA5′T46A-rev | 5′-ATACCCTAAAAACATTAGGCAGTAACAACATTTGTTAGCTGTT GAAAGAAAGTCACGCTAAATGATGTTTTTTACAG (overhang complementary to prfA3′-for bold, mutation T46A underlined) |
Design of synthetic 5′-UTR elements containing putative thermosensors
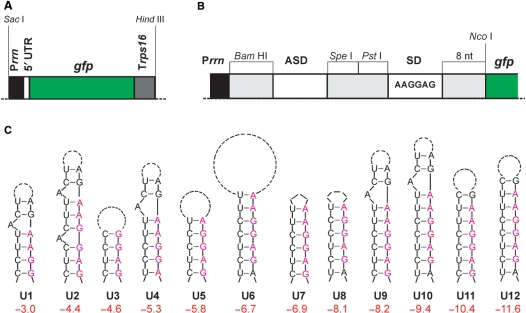
The 5′-UTR elements were designed on the basis of minimal sequence elements (Figure 1B) and contained a BamHI site at the 5′-end followed by an ASD (anti-SD sequence) element and two additional unique restriction sites (SpeI and PstI) allowing insertion and manipulation of the various loop structures. The downstream consensus SD sequence (5′-AAGGAG-3′) was followed by 8-nt spacer derived from the bacteriophage T7 gene 10 leader sequence and a unique NcoI restriction site comprising the translational start codon of gfp. The different synthetic 5′-UTR constructs differ in loop size and/or in the extent of complementarity between ASD and SD (Figure 1C; Table 2). Prediction of the secondary structure formation and calculation of their free energies was performed using the algorithms of the Mfold web server [version 3.2; (13)]. For all constructs, the 5′-UTR sequence excluding the start codon was used as input sequence. The control construct U0 contains no ASD element, but a 6-nt A-stretch instead, thus excluding stable base paring with the SD sequence.
Figure 1.
Construction of synthetic RNA thermometers. (A) GFP expression cassette used to test synthetic stem-loop structures for their possible function as RNA thermometer. The structures were built into the 5′-UTR in between the ribosomal RNA operon promoter (Prrn) and the gfp coding region. Trps16: 3′-UTR from the plastid rps16 gene. (B) Enlargement of the 5′-UTR region showing the modular construction of the thermosensors. Restriction sites used to exchange segments of the 5′-UTR are indicated (BamHI, SpeI and PstI). SD denotes the Shine-Dalgarno sequence (ribosome binding site; consensus sequence indicated). ASD sequences designed to base pair with the SD sequence. The start codon for translation of gfp is part of an NcoI restriction site, a 8 nt spacer separates the SD sequence from the start codon. (C) RNA secondary structures tested in this study. The structures are ordered and numbered according to the calculated free energies of the entire 5′-UTR (ΔG in kcal/mol). The number of dashes in the loops corresponds to the number of inter-nucleotide bonds in the loop. Nucleotides belonging to the SD sequence are shown in purple letters when included in the stem. In some constructs, the most stable structure calculated by Mfold included two additional base pairs (formed between 2 nt in the BamHI site and 2 nt in the spacer in between the SD and the start codon), which are separated from the stem-loop shown here by a bulge.
Table 2.
Synthetic 5′-UTR sequences tested for RNA thermometer function
| Control construct |
| U0 5′-GGAUCCGAAAAAACUAGUCUGCAGAAGGAGAUAUACCCaugg-3′ |
| Putative thermoregulators |
| U1 5′-GGAUCCCACCUUACUAGUCUGCAGAAGGAGAUAUACCCaugg-3′ |
| U2 5′-GGAUCCCUCACUUACUAGUCUGCAGAAGGAGAUAUACCCaugg-3′ |
| U3 5′-GGAUCCCUCCAUACUAGUCUGCAGAAGGAGAUAUACCCaugg-3′ |
| U4 5′-GGAUCCAUCCUUACUAGUCUGCAGAAGGAGAUAUACCCaugg-3′ |
| U5 5′-GGAUCCCUCCUAACUAGUCUGCAGAAGGAGAUAUACCCaugg-3′ |
| U6 5′-GGAUCCUCUCCUUCACUAGU(A)25AAGGAGAUAUACCCaugg-3′ |
| U7 5′-GGAUCCCUCCUUCAGAAGGAGAUAUACCCaugg-3′ |
| U8 5′-GGAUCCUCUCCUUAGAAGGAGAUAUACCCaugg-3′ |
| U9 5′-GGAUCCCUCCUUACUAGUCUGCAGAAGGAGAUAUACCCaugg-3′ |
| U10 5′-GGAUCCUCUCCUUACUAGUCUGCAGAAGGAGAUAUACCCaugg-3′ |
| U11 5′-GGAUCCCUCCUUCACUAGUCUGCAGAAGGAGAUAUACCCaugg-3′ |
| U12 5′-GGAUCCUCUCCUUCACUAGUCUGCAGAAGGAGAUAUACCCaugg-3′ |
Restriction sites (BamHI, SpeI, PstI, NcoI) are indicated in italics. ASD and SD region (5′-AAGGAG-3′) are underlined. Complete sequences of the 5′-UTRs are shown (beginning with the transcriptional start site and ending with translational start site). The start codon and the first nucleotide of the coding region (which corresponds to the last nucleotide of the NcoI site) are given in lowercase letters.
All vectors containing synthetic thermosensors are based on vector pBS45 harboring the Prrn – prfA 5′-UTR – gfp – rps16 3′-UTR expression cassette. A shorter version of the Prrn promoter lacking the SD sequence was amplified by PCR using the primer pair Prrn-shortfw (5′-GCTGGAGCTCGGTACCCCAAAG-3′; SacI site underlined) and Prrn-shortrv (5′-GGATCCTCCCAGAAATATAGCC-3′; BamHI site underlined). After cloning of the PCR product into the pCR2.1-TOPO vector (Invitrogen, Karlsruhe, Germany) for control sequencing (MWG-Biotech, Martinsried, Germany), the promoter fragment was excised by digestion with SacI and BamHI and inserted into the similarly cut vector pBS45, resulting in plasmid pBS45_s.
Synthetic 5′-UTR sequences were inserted into the BamHI/NcoI-digested pBS45_s as annealed synthetic oligonucleotides with BamHI and NcoI overhangs (Tables 1 and 2). The control U0 (in vector pBSU0) and the putative thermoregulative 5′-UTR elements U1 (vector pBSU1), U2 (vector pBSU2), U3 (vector pBSU3), U4 (vector pBSU4), U5 (vector pBSU5), U6 (vector pBSU6), U9 (vector pBSU9), U10 (vector pBSU10), U11 (vector pBSU11) and U12 (vector pBSU12) were obtained from synthetic DNA oligonucleotides (Metabion International AG, Martinsried, Germany; Table 1) by annealing single-stranded sense and antisense oligonucleotides with 5′-BamHI and 3′-NcoI overhangs. To this end, sense and antisense oligonucleotides were pairwise annealed by mixing the two oligonucleotides in water (150 pmol each) in a volume of 20 µl, heating at 95°C for 5 min and slowly cooling down to 25°C in a standard heating block. pBSU7 (containing 5′-UTR element U7) and pBSU8 (containing 5′-UTR element U8) were obtained after religation of SpeI/PstI restricted and mung bean nuclease treated (1 U enzyme per microgram DNA, 30-min incubation at 30°C; New England Biolabs) vectors pBSU11 and pBSU10, respectively. The sequences of all plasmid clones were confirmed by control sequencing using the M13 reverse primer (MWG-Biotech).
RNA isolation and northern blot analysis
Total cellular RNA was isolated from E. coli grown at different temperatures using the peqGOLD TriFast reagent (Peqlab GmbH, Erlangen, Germany) following the manufacturer's instructions. To remove contaminating DNA, DNase I treatment (1 U recombinant RNase-free DNase I per microgram RNA; Roche, Mannheim, Germany) was performed at 37°C for 30 min in a buffer containing 10 mM MgCl2, 10 mM Tris–HCl pH 7.5 and 1 mM EDTA followed by phenol–chloroform–isoamylalcohol (25:24:1) extraction and ethanol precipitation. For northern blot analysis, total RNA samples were separated on 1% formaldehyde containing agarose gels and blotted onto Hybond nylon membranes (GE Healthcare, Freiburg, Germany). Hybridizations were performed at 65°C in Rapid-Hyb buffer (GE Healthcare) according to the manufacturer's protocol.
A 16S rRNA-specific probe (467 bp) was produced by PCR amplification of E. coli genomic DNA with primers P16Srrn-F (5′-CAAGCGGTGGAGCATGTGG-3′) and (P16Srrn-R 5′-GGCGGTGTGTACAAGGCCC-3′). PCR was performed in an Eppendorf thermocycler according to standard protocols (1 min at 95°C, 45 s at 52°C–62°C, 30 s at 72°C, 32 cycles). A gfp-specific probe was generated by excising the entire coding region with NcoI and XbaI from a plasmid clone. Fragments were purified by agarose gel electrophoresis following extraction of the excised gel slices using the GFX™ PCR (DNA and Gel Band Purification) kit (GE Healthcare). Hybridization probes were labeled by random priming with radioactive [α-32P]dCTP (GE Healthcare).
Protein isolation and immunoblotting
Transformed E. coli strains were grown at different temperatures (17°C, 22°C, 30°C and 37°C) until they reached the end of the exponential phase (OD600 around 1.0). Prior to protein isolation, bacterial cultures were brought to identical ODs (OD600 = 0.4). Bacterial pellets were frozen in liquid nitrogen and then dissolved in 400 µl lysis buffer (50 mM HEPES, 300 mM NaCl, 0.5% SDS, pH 8.0). After 30 min incubation on ice, cells were disrupted by sonication (amplitude 10%, 15 s; Sonifier®, W-250 D, G. Heinemann Ultraschall und Labortechnik, Schwäbisch Gmünd, Germany) and the lysate was denatured at 95°C for 3 min. The BCA protein assay (Pierce, Rockford, IL, USA) was used for protein quantification. For western blotting, samples of 1 µg total soluble protein were separated by electrophoresis in denaturating SDS–polyacrylamide gels and transferred to Hybond P PVDF membranes (GE Healthcare) using a semidry blotting system (SEDEC-M, PeqLab, Erlangen, Germany) and standard transfer buffer (25 mM Tris–HCl, 192 mM glycine, pH 8.3). Immunobiochemical detection was carried out with a GFP-specific primary monoclonal mouse antibody (JL-8; Clontech, Mountain View, CA, USA) using the ECL Plus™ detection system (GE Healthcare).
β-Galactosidase assays
The E. coli lacZ gene encoding the β-galactosidase reporter enzyme was amplified from E. coli strain K12 with primers PLacZ5′NcoI (5′-AACCATGGCCATGATTACGGATTC-3′) and PLacZ3′XbaI (5′-AATCTAGATTACCATTCGCCATTCAG-3′) introducing NcoI and XbaI restriction sites at the 5′-end and 3′-end of the product, respectively (restriction sites underlined in primer sequences). In this way, the gfp coding region in the pBSU clones could be directly replaced by lacZ as NcoI/XbaI fragment. The cloning vector pBluescript SK(+) (Stratagene, Amsterdam, Netherlands) carrying the lacZ gene under control of the original lacZ promoter was included as an independent constitutive control using IPTG (80 µg/ml LB medium) as an inducer for expression.
To determine temperature inducibility of lacZ expression, E. coli cultures were grown at different temperatures and bacterial pellets were frozen in liquid nitrogen. After solubilization of the bacterial pellet in buffer H (100 mM HEPES/KOH, pH 7.0; 150 mM NaCl; 2 mM MgCl2; 1% BSA), 0.1% SDS and 0.1% chloroform were added to complete cell disruption. For colorimetric assays, 125 µl of O-nitrophenyl-β-d-galactopyranoside (O-NPG; 4 mg/ml in buffer H), a synthetic substrate for β-galactosidase, were added to the lysate and samples were incubated at 30°C for 5 h. After centrifugation, extinction of the supernatant was measured at 405 nm in a spectrophotometer (Ultrospec 3100 pro, GE Healthcare).
RESULTS
Design of synthetic RNA thermometers
We reasoned that, if RNA thermometers indeed function by the proposed conceptually simple melting mechanism (3,4,9), it should be possible to design them de novo. We further assumed that, if the unmasking of the ribosome binding site is the only requirement for the thermometer to work, much less complicated RNA secondary structures should suffice, provided that their melting temperatures can be adjusted precisely and tightly enough to the relevant physiological temperature range. To test these hypotheses, we sought to construct minimum RNA thermometers that consist only of a single stem-loop structure containing the ribosome binding site.
In order to be able to test a variety of different structures, we constructed a reporter gene cassette for expression in E. coli (Figure 1A). To minimize the possibility that host factors interfere with translational regulation of the reporter, the gene cassette was constructed entirely from synthetic (heterologous) sequences and did not contain any bacterial sequence. The reporter gene was the gfp gene from the jellyfish Aequorea victoria (14) and the expression elements were derived from the tobacco chloroplast (plastid) genome: the promoter from the ribosomal RNA operon (Prrn) and the 3′-untranslated region (3′-UTR) from the gene for the ribosomal protein S16 (rps16; Figure 1A). As the plastid genome is of prokaryotic origin, these elements function well in eubacteria (15). The consensus SD sequence 5′-AAGGAG-3′ served as ribosome binding site (Figure 1B). Upstream of the SD, three unique restriction site were integrated to allow for insertion of short sequence stretches that, in part, are complementary to the SD and thus would mask the ribosome binding site in an RNA secondary structure that potentially could function as an RNA thermometer (Figure 1B and C; Tables 1 and 2). These sequences are referred to as ASD sequences (Figure 1B). To avoid formation of complex RNA secondary structures within the 5′-UTR, the length of the 5′-UTR was reduced to a minimum: transcription initiated at the BamHI site, immediately upstream of the ASD, and only 8 nt were present between the SD and the translational start codon of gfp (Figure 1B), a distance considered to be in the optimal range for efficient initiation of protein biosynthesis in E. coli. Figure 1C provides an overview of the putative RNA secondary structures tested for their thermometer properties in this study. For simplicity, the constructs are numbered here consecutively according to their calculated free energies (rather than the order in which they were constructed and experimentally tested; cf. Figure 2). In addition, a control construct U0 was produced, in which no ASD was present (Tables 1 and 2).
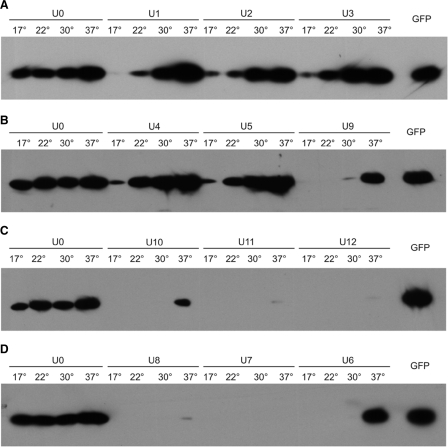
Figure 2.
Test of temperature dependence of GFP expression controlled by synthetically designed RNA thermometers. One microgram of total soluble protein was loaded in all lanes. To allow for quantitative comparisons, the temperature series for the control construct U0 as well as a sample of purified GFP protein (20 ng) are included in all western blots. (A) Constructs with weak RNA secondary structures. (B) Three constructs with secondary structures of intermediate strength. (C) Constructs with relatively stable RNA secondary structures. (D) Three constructs with free energies adjusted to theoretical values between those of U5 and U9.
Experimental testing of synthetic RNA thermometers
All constructs were transformed into E. coli and tested for temperature responsiveness of GFP expression to assess their possible functionality as RNA thermometer. To this end, the bacterial strains were grown at four different physiological temperatures: 17°C, 22°C, 30°C and 37°C followed by protein extraction and determination of GFP protein amounts by western blotting (Figure 2). The control construct U0 showed nearly constant GFP accumulation over the tested temperature range. However, a slight increase of GFP production with temperature was reproducibly noticed, which is most likely due to transcription and translation being somewhat more efficient at higher temperatures. Constructs harboring structures of potentially low melting stability (U1, U2 and U3; Figure 1C) displayed a significantly more pronounced temperature-dependent GFP accumulation than U0, in that GFP accumulation was very low at 17°C and 22°C (Figure 2A). This suggests that, at low growth temperatures, the ribosome binding sites may not be fully accessible and indicates that the stability range of these RNA structures allows some temperature-dependent melting to occur in vivo. We next tested a series of structures with higher stability (U4, U5 and U9; Figure 1C). While the temperature-dependent upregulation of GFP expression in U4 and U5 was not significantly different from U1, U2 and U3 (Figure 2A and B), construct U9 displayed a much clearer inducibility (Figure 2B). We, therefore, designed a series of constructs with potentially higher stabilities than U9 (U10, U11 and U12; Figure 1C) and another series with slightly lower calculated free energies (U6, U7 and U8; Figure 1C). As subtle differences in stabilities cannot be achieved with simply changing the size of the stem of the structure (Figure 1C), we modified the size of the loop as an additional parameter. Increase of the loop size has slightly destabilizing effects and, in combination with the number of base pairs (and the GC content) of the stem, almost any desired theoretical stability value can be obtained (Figure 1C). The two constructs with the potentially strongest RNA secondary structures (U11 and U12; Figure 1C) showed very little GFP accumulation at 37°C, suggesting that the structures might be too stable to be efficiently melted. U10 showed a similar temperature inducibility as construct U9 (Figure 2C), but, consistent with its higher stability, did not reach similar GFP accumulation levels. Interestingly, the series with slightly weaker structures (U6, U7 and U8; Figure 1C) displayed strong differences in GFP accumulation levels. Although all three constructs showed a clear temperature-dependent GFP accumulation, the western blots had to be strongly overexposed to detect the GFP protein in the 37°C samples from the U7 and U8 constructs (Figure 2D and data not shown). In contrast, GFP expression at 37°C in U6 was similarly high as in the U0 control, suggesting that the U6 structure can be efficiently melted at 37°C. Importantly, no detectable GFP accumulation occurred at 17°C, 22°C and 30°C, indicating that U6 represents a rather tight temperature-dependent switch. Comparison with a purified GFP standard revealed that GFP accumulated in U6 bacterial strains to at least 1% of the total soluble protein (Figure 2D).
To test whether the temperature-dependent GFP protein accumulation in our best performing RNA thermometer U6 was indeed due to translational regulation and not caused by high temperature-induced stabilization of the gfp mRNA, we compared mRNA accumulation at all four growth temperatures by RNA gel blot analyses. No significant temperature-dependent change in RNA accumulation was seen (Figure 3A), suggesting that the observed temperature inducibility of GFP accumulation is due to translational regulation. This indicates that the designed simple stem-loop structures may indeed function as RNA thermometers in vivo.
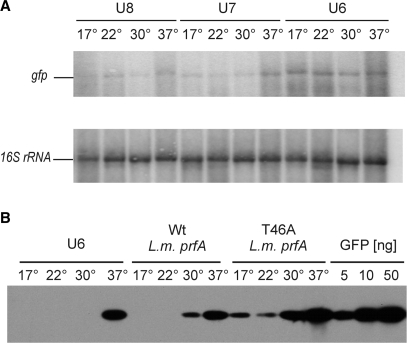
Figure 3.
Analysis of temperature dependence of RNA and protein accumulation in E. coli strains harboring gfp constructs under the control of RNA thermometers. (A) Accumulation of gfp mRNA is independent of the growth temperature. Data are shown for the best performing structure U6 and two other constructs, U7 and U8. After hybridization to a gfp-specific probe, the blot was stripped and rehybridized to a 16S rRNA probe as loading control. (B) Comparison of the efficacy of the synthetic RNA thermometer U6 with the natural thermometer in the 5′-UTR of the prfA gene from L. monocytogenes. T46A is a mutant variant of the prfA thermometer carrying a point mutation that slightly destabilizes the SD-masking secondary structure. For quantitative comparison, a dilution series of purified recombinant GFP protein is also shown.
Comparison of synthetic and natural RNA thermometers
As our designed synthetic thermometers are considerably smaller and structurally simpler than all natural RNA thermometers described so far, we next wanted to compare their background expression and inducibility with a well-studied natural thermometer. The prfA gene of the pathogenic bacterium L. monocytogenes harbors an RNA thermometer in its 5′-UTR (4). Melting of a secondary structure masking the ribosome binding site causes the PrfA protein, a transcriptional activator controlling key virulence genes, to accumulate at 37°C to much higher levels than at 30°C. A point mutation (T46A) weakening the SD-containing stem-loop structure was shown to make expression at lower temperatures more leaky (4), consistent with the proposed melting mechanism. We integrated both the wt prfA thermometer and the T46A variant into our expression cassette and compared it with our best performing synthetically designed RNA thermometer, U6. Interestingly, U6 performed even better than the prfA thermometer in that temperature inducibility was stronger and background expression at 17–30°C was virtually absent (Figure 3B).
Properties of synthetic RNA thermometers as inducible gene expression systems
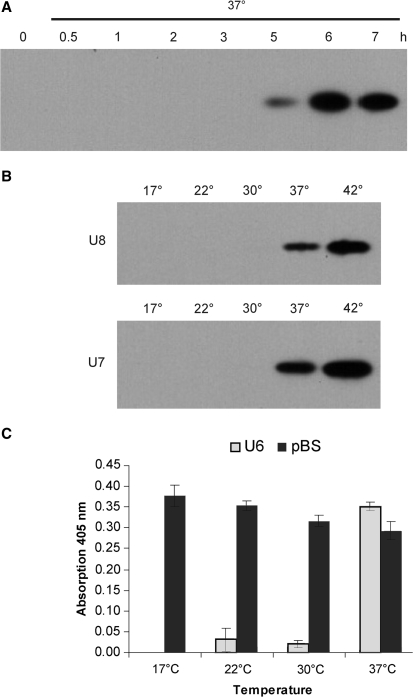
To determine the time course of temperature-dependent protein accumulation under control of the U6 thermometer, we shifted bacterial cultures from 17°C to 37°C and followed the accumulation of GFP protein. The steady-state level of protein accumulation was reached after ∼6 h (Figure 4A), consistent with the notion that full induction at the protein level not only requires melting of the thermometer to occur, but also translation to proceed for some time, before the steady state of GFP accumulation is reached. How quickly the steady state is reached is negatively correlated to protein stability. As GFP is a compactly folded and hence relatively stable protein, it is unsurprising that this process takes a few hours.
Figure 4.
Characteristics of synthetic RNA thermometers. (A) Time course of GFP accumulation under control of the U6 secondary structure after induction by a temperature shift from 17°C to 37°C. (B) Temperature dependence of GFP protein accumulation in bacterial strains harboring the U7 and U8 thermometers. Higher GFP accumulation at 42°C suggests more efficient melting of the RNA secondary structures. (C) U6 confers temperature dependence of lacZ expression. A bacterial strain harboring a pBluescript vector carrying the lacZ gene under control of its own promoter and grown in the presence of the inductor IPTG served as constitutive control. Hydrolysis of O-NPG, a synthetic substrate of the β-galactosidase, was measured spectroscopically. Data represent the means of three independent biological replicas. The standard deviation is indicated.
U7 and U8, two constructs with slightly higher stabilities than the best performing construct U6 (Figure 1C), also showed temperature inducibility, but yielded much lower GFP protein accumulation at 37°C than U6 (Figure 2D and data not shown). We reasoned that, if our designed RNA thermometers indeed function by the proposed temperature-dependent melting mechanism, the U7 and U8 thermometers should function more efficiently at temperatures higher than 37°C. To test this hypothesis, we comparatively analyzed the temperature-dependent GFP accumulation at 37°C and 42°C. Both U7 and U8 showed significantly higher protein accumulation at 42°C than at 37°C (Figure 4B) indicating that melting of the RNA secondary structure may be more efficient at 42°C and providing additional evidence for thermometer function being dependent on the stability of the designed stem-loop structure. This suggests that RNA thermometers can be optimized for any desired shift temperature by adjusting the stability of the stem-loop structure acting as molecular switch.
Finally, we wanted to exclude the possibility that additional, fortuitously forming secondary structures within the gfp coding region contribute to the thermometer function of our synthetically designed thermosensors. To this end, we replaced the gfp coding region with the coding region of lacZ, a reporter gene encoding the easily assayable enzyme β-galactosidase, and tested the U6 stem-loop for conferring temperature dependency onto β-galactosidase accumulation. Indeed, expression of β-galactosidase under control of the U6 structure was strongly temperature dependent and highly induced at 37°C (Figure 4C) confirming that the function of the synthetic RNA thermometers is independent of the coding region and most probably exclusively mediated by the short stem-loop containing 5′-UTR.
DISCUSSION
In the course of this work, we have developed synthetic RNA thermometers suitable to translationally regulate gene expression in bacteria at physiological temperatures. Our results show that a single small stem-loop structure is sufficient to construct an efficient RNA thermosensor suggesting that the mechanism of RNA thermometers is considerably simpler than generally assumed and may not require complex RNA secondary structures, helical stacking, intramolecular docking or intricate tertiary interactions. This is consistent with a recent report demonstrating that a natural RNA thermometer from Salmonella enterica could be reduced to a single stem-loop and yet retained temperature responsiveness (16). Our data also demonstrate that synthetic RNA thermometers can be exploited as simple on/off switches to induce or repress gene expression in response to temperature (3,9). While the induction mode can be used to induce metabolic pathways by temperature or overexpress a protein of interest, the repression mode can be used, for example, as a reverse genetics tool to study the functions of essential genes (i.e. genes that cannot be knocked-out) by gradually downregulating their expression and following the phenotypic and physiological effects.
Our two best performing RNA thermometers (U6 and U9; Figure 2) trigger expression to similarly high levels at 37°C, but differ considerably in their secondary structural properties. While U6 has a large loop and a perfectly matched stem, U9 has a small loop and a single bulged-out nucleotide in the stem, which was introduced as a stability-reducing element. The finding that very different structures can be similarly effective as RNA switches suggests that widely different structural elements (bulges, mismatches, loop size) and their combinations can be explored to optimize RNA thermometers for the desired switch temperature.
The thermometers described here were optimized for best performance at 37°C, the optimum growth temperature of E. coli. Thermometers optimized for any other temperature close to 37°C can probably most easily be produced by keeping the stem structures of one of our best performing thermometers (U6, U9, U10) and making subtle adjustments to the loop size (Figure 1C). This is because, unlike alterations in the stem, changes in the number of nucleotides in the loop by one or a few nucleotides have only slightly stabilizing or destabilizing effects on the RNA structure. Thus, it should be possible to design a thermometer optimized for any desired inducing temperature by simply testing a series of constructs with different loop sizes. As both the loop sequence and the closing base pair can contribute the stability of a stem–loop, such sequence modifications may also be worthwhile testing when trying to optimize a thermometer for a new switch temperature.
It should be noted that, although there is some correlation between the theoretically calculated stabilities of the tested RNA secondary structures and their performance as RNA thermometers, this correlation is not strict. Constructs U7 and U8 represent the most notable exception in that their calculated stabilities are in between those of U6 and U9 (two of the best performing constructs; Figure 1C), but yet they yield much lower GFP levels at 37°C than U6 and U9 (Figure 2). Although Mfold, the algorithm used to calculate the stabilities of the structures (13), usually provides good estimates, the stabilities in vivo may depend on a number of incalculable factors, including the ionic milieu and electrostatic interactions with other macromolecules. It thus cannot be excluded that U7 and U8 are considerably more stable in vivo than theoretically predicted. However, a perhaps more likely explanation is that U9 and U10, two well-performing constructs with calculated higher stabilities than U7 and U8, harbor an unpaired nucleotide in the stem (Figure 1C), which could be responsible for their better meltability. Finally, it must also be borne in mind that, in addition to sheer structural stability, also the kinetics of RNA folding can have a significant impact on the efficiency of translation initiation (17).
While the exact contribution of loop size and/or bulges to overall stability may be difficult to calculate reliably, all constructs that have the same predicted basic structure show a good correlation between the calculated stabilities of the secondary structures and their performance as thermosensors. For example, constructs U4 and U10 are derivatives of the well-performing thermosensor U9 (Figure 2B) in that they have identical loop size (and sequence) and the same basic stem structure (Figure 1C). Compared to U9, U10 carries a stabilizing mutation, i.e. an extra base pair in the stem. This results in loss of inducibility at 30°C and less efficient inducibility at 37°C (Figure 2B and C). In contrast, U4 carries a destabilizing mutation, i.e. a deletion of 1 bp from the stem. As expected, this leads to a substantial increase in leakiness of the thermometer in that significant expression occurs already at 17°C and 22°C (Figure 2B). Likewise, introduction of a mismatch into the stem (as in construct U2) destabilizes the structure and increases leakiness at low temperatures (Figure 2A). These correlations support the assumption that the predicted secondary structures of our designed RNA thermometers indeed form in vivo.
As all eubacteria use a SD sequence-based mechanism of translation initiation (18,19), the RNA thermometers described here should be universally applicable. Modifications may be necessary to adjust the thermometers to the optimum growth temperature of a given bacterial species, but this should be easily achievable using the design criteria outlined here. As the RNA structure masking the ribosome binding site is apparently the only component required to confer the regulatory properties, no prior knowledge about endogenous gene regulation systems in the bacterial species of interest is required.
In summary, the synthetic thermometers described here should provide superb minimum size tools to induce or repress gene expression by a simple temperature shift. Moreover, they represent RNA-only tools that function independently of protein factors, do not require the addition of chemical inducers and thus should be widely applicable.
FUNDING
Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 2.Storz G. An RNA thermometer. Genes Dev. 1999;13:633–636. doi: 10.1101/gad.13.6.633. [DOI] [PubMed] [Google Scholar]
- 3.Narberhaus F, Waldminghaus T, Chowdhury S. RNA thermometers. FEMS Microbiol. Rev. 2006;30:3–16. doi: 10.1111/j.1574-6976.2005.004.x. [DOI] [PubMed] [Google Scholar]
- 4.Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 5.Morita MT, Tanaka Y, Kodama TS, Kyogoku Y, Yanagi H, Yura T. Translational induction of heat shock transcription factor sigma32: evidence for a built-in RNA thermosensor. Genes Dev. 1999;13:655–665. doi: 10.1101/gad.13.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nocker A, Hausherr T, Balsiger S, Krstulovic NP, Hennecke H, Narberhaus F. A mRNA-based thermosensor controls expression of rhizobial heat shock genes. Nucleic Acids Res. 2001;29:4800–4807. doi: 10.1093/nar/29.23.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhury S, Maris C, Allain F.H.-T, Narberhaus F. Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 2006;25:2487–2497. doi: 10.1038/sj.emboj.7601128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altuvia S, Oppenheim AB. Translational regulatory signals within the coding region of the bacteriophage lambda cIII gene. J. Bacteriol. 1986;167:415–419. doi: 10.1128/jb.167.1.415-419.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wurbs D, Ruf S, Bock R. Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. Plant J. 2007;49:276–288. doi: 10.1111/j.1365-313X.2006.02960.x. [DOI] [PubMed] [Google Scholar]
- 11.Ruf S, Karcher D, Bock R. Determining the transgene containment level provided by chloroplast transformation. Proc. Natl Acad. Sci. USA. 2007;104:6998–7002. doi: 10.1073/pnas.0700008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl Acad. Sci. USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 15.Bock R. Transgenic chloroplasts in basic research and plant biotechnology. J. Mol. Biol. 2001;312:425–438. doi: 10.1006/jmbi.2001.4960. [DOI] [PubMed] [Google Scholar]
- 16.Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 2007;65:413–424. doi: 10.1111/j.1365-2958.2007.05794.x. [DOI] [PubMed] [Google Scholar]
- 17.Poot RA, Tsareva NV, Boni IV, van Duin J. RNA folding kinetics regulates translation of phage MS2 maturation gene. Proc. Natl Acad. Sci. USA. 1997;94:10110–10115. doi: 10.1073/pnas.94.19.10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy JEG, Brimacombe R. Prokaryotic translation initiation: the interactive pathway leading to initiation. Trends Genet. 1994;10:402–407. doi: 10.1016/0168-9525(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 19.Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]