Abstract
We present a simple and novel assay—employing a universal molecular beacon (MB) in the presence of Hg2+—for the detection of single nucleotide polymorphisms (SNPs) based on Hg2+–DNA complexes inducing a conformational change in the MB. The MB (T7-MB) contains a 19-mer loop and a stem of a pair of seven thymidine (T) bases, a carboxyfluorescein (FAM) unit at the 5′-end, and a 4-([4-(dimethylamino)phenyl]azo)benzoic acid (DABCYL) unit at the 3′-end. Upon formation of Hg2+–T7-MB complexes through T–Hg2+–T bonding, the conformation of T7-MB changes from a random coil to a folded structure, leading to a decreased distance between the FAM and DABCYL units and, hence, increased efficiency of fluorescence resonance energy transfer (FRET) between the FAM and DABCYL units, resulting in decreased fluorescence intensity of the MB. In the presence of complementary DNA, double-stranded DNA complexes form (instead of the Hg2+–T7-MB complexes), with FRET between the FAM and DABCYL units occurring to a lesser extent than in the folded structure. Under the optimal conditions (20 nM T7-MB, 20 mM NaCl, 1.0 μM Hg2+, 5.0 mM phosphate buffer solution, pH 7.4), the linear plot of the fluorescence intensity against the concentration of perfectly matched DNA was linear over the range 2–30 nM (R2 = 0.991), with a limit of detection of 0.5 nM at a signal-to-noise ratio of 3. This new probe provides higher selectivity toward DNA than that exhibited by conventional MBs.
INTRODUCTION
The past decade has witnessed the development of many advanced biomolecular recognition probes for highly sensitive and selective detection of DNA molecules (genes) of interest (1–6). One such set of promising probes are single-stranded DNA molecular beacons (DNA-MB) that form hairpin-shaped structures to recognize targeted DNA molecules. To allow the monitoring conformation changes in DNA-MB upon reactions with targeted DNA, a fluorophore and a quencher are covalently conjugated at the termini of each DNA-MB strand. DNA-MBs act as fluorescence resonance energy transfer (FRET)-based switches that are normally in the closed or ‘fluorescence off’ state, but switch to the open or ‘fluorescence on’ state in the presence of target (complimentary) DNA strands (7).
When DNA-MBs are used for the detection of single nucleotide polymorphisms (SNPs), problems associated with their nonspecific binding to DNA-binding proteins and endogenous nuclease degradation occur, leading to false-positive signals and their limited applicability in complex biological samples (8–10). MBs containing nuclease-resistant backbone residues, such as negatively charged phosphorothioates and neutral peptide nucleic acids, have been developed, but they sometimes exhibit toxicity, self-aggregation and nonspecific binding to single-stranded DNA (ss-DNA)-binding protein (SSB) (11–13). To provide high sensitivity and fast hybridization kinetics, hybrid molecular probes consisting of two ss-DNA sequences tethered to two ends of a poly(ethylene glycol) chain have been developed (14). The two ss-DNA sequences are complementary to adjacent areas of a target sequence in such a way that hybridization of the probe with the target brings the 5′- and 3′-ends of the probe in close proximity. Nevertheless, hybrid molecular probes are more difficult to prepare and are more expensive than conventional DNA-MBs.
Probes based on the Hg2+-induced conformational change of a DNA molecule through thymidine (T)–Hg2+–T coordination have been realized for the detection of Hg2+ ions (15–18). A DNA sensor has been employed for the detection of Hg2+ through the enhanced efficiency of FRET as a result of formation of T–Hg2+–T complexes (15). Recently, we presented a simple and rapid colourimetric assay—employing poly-Tn and 13 nm-diameter Au NPs in the presence of salt—for the detection of Hg2+ ions based on Hg2+–DNA complexes inducing the aggregation of Au NPs (17).
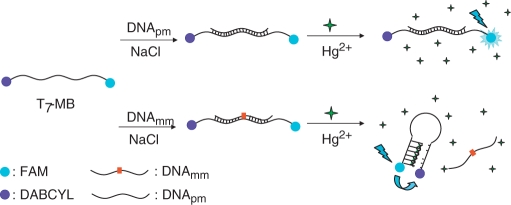
In this article, we present a simple and novel assay—employing T7-MB in the presence of salt and Hg2+—for the detection of SNPs based on Hg2+–DNA complexes inducing a conformational change in T7-MB. The T7-MB contains a stem of a pair of 7-mer T bases that interact with Hg2+ and a loop of 19-mer DNA bases that recognize targeted DNA. According to our previous study (18), for obtaining stable DNA–Hg complexes that allow selective detection of target DNA, the minimum number of T is 14. Therefore, 7-mer bp of Ts in the stem region are necessary in the stem region for providing a proper function. The T7-MB probe contains a donor of carboxyfluorescein (FAM) at the 5′-end, and a quencher of 4-([4-(dimethylamino)phenyl]azo)benzoic acid (DABCYL) at the 3′-end (the sequence of the MB listed in Table 1). The T7-MB is a random-coil structure that changes into a folded structure in the presence of Hg2+ ions through T–Hg2+–T bonding (19–21). As a result of the decreased distance between the donor and quencher, the fluorescence of FAM in the Hg2+–T7-MB complexes becomes weaker because of FRET occurring between the FAM and DABCYL units. When the DNA loop of T7-MB interacts with a targeted DNA more strongly than do the T7 units in the stem with Hg2+, a double-stranded DNA forms, rather than the folded structure. In this case, the FAM and DABCYL units are separated far apart, resulting in FAM fluorescing strongly, as depicted in Scheme 1. We investigated the effect of the Hg2+ concentration on the sensitivity and selectivity of the T7-MB probe, and compared its sensing performance toward SNPs with that of conventional DNA-MBs.
Table 1.
DNA sequences of MBs and trget DNA
| Name | Sequence (5′–3′) |
|---|---|
| T7-MB | FAM-TTTTTTTTCTAAATCACTATGGTCGCTTTTT TT-DABCYL |
| DNA-MB1 | FAM-ACTTAGTTCTAAATCACTATGGTCGCACTA AGT-DABCYL |
| DNA-MB2 | FAM-ACCTAGCTCTAAATCACTATGGTCGCGCTA GGT-DABCYL |
| DNA-MB3 | FAM-GCCGAGCTCTAAATCACTATGGTCGCGCTC GGC-DABCYL |
| DNApm | GCGACCATAGTGATTTAGA |
| DNAmm1 | GCGACCATAATGATTTAGA |
| DNAmm2 | GCGACCATACTGATTTAGA |
| DNAmm3 | GCGACCATATTGATTTAGA |
| DNAmm4 | GCGACCATAGAGATTTAGA |
| DNAmm5 | GCGACCATAGCGATTTAGA |
Bold and underlined letters indicate the mutant bases.
Scheme 1.
Schematic representation of the working principles of the T7-MB.
MATERIALS AND METHODS
Chemicals
Mercury(II) chloride (HgCl2) and magnesium(II) chloride (MgCl2) used in this study were purchased from Aldrich (Milwaukee, WI, USA). Sodium phosphate dibasic anhydrous and sodium phosphate monobasic monohydrate, obtained from J. T. Baker (Phillipsburg, NJ, USA), were used to prepare the phosphate buffer (5.0 mM, pH 7.4). The T7-MB, DNA-MBx (x = 1−3), perfectly matched DNA (DNApm) and mismatched DNA (DNAmmx) (see Table 1 for sequences) were purchased from Integrated DNA Technology, Inc. (Coralville, IA, USA). The sequences in T7-MB and DNA-MBx that do not have any biological targets were randomly designed to provide optimum selectivity toward the target DNAs and hybridization kinetics (4). Milli-Q ultrapure water was used in all experiments.
Analysis of samples
Aliquots (400 μl) of 5.0 mM phosphate buffer (pH 7.4) containing NaCl (0–250 mM) and MB (20 nM) were maintained at ambient temperature for 10 min. Aliquots (50 μl) of tested DNA (1.0 μM) were added to the solutions, which were then incubated for 30 min. The final ratio of the concentrations of the MB and the tested DNA was 1:5. An aliquot (50 μl) of Hg2+ (0–1.5 μM) was added to each solution, which was then incubated for 2 h prior to fluorescence measurements (Cary Eclipse; Varian, CA, USA) at various temperatures (10–90°C). To evaluate the resistance to endogenous nuclease degradation, aliquots (450 μl) of 5.0 mM phosphate buffer (pH 7.4) containing NaCl (20 mM), MgCl2 (5.0 mM), T7-MB or DNA-MB (20 nM) and Hg2+ (1.0 μM) were maintained at ambient temperature for 2 h. An aliquot (50 μl) of DNase I (final concentration: 5.0 μg/ml) was added to each solution and then the mixtures were subjected to fluorescence measurements after certain periods of time, as indicated in the Results and discussion section. To evaluate the nonspecific binding to SSB, 5.0 mM phosphate buffer (pH 7.4, 450 μl) solutions containing NaCl (20 mM), SSB (100 nM) and T7-MB or DNA-MB (20 nM) were maintained at ambient temperature for 30 min. An aliquot (50 μl) of Hg2+ (1.0 μM) was added to each solution, which was then incubated for 2 h prior to fluorescence measurement.
RESULTS AND DISCUSSION
Sensing behavior
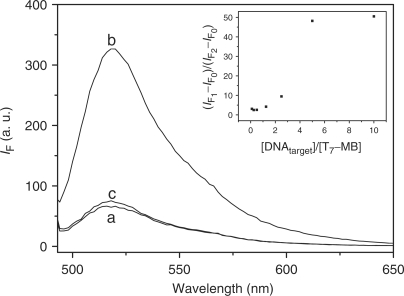
Two aliquots of the T7-MB (20 nM) were separately added to 5.0 mM phosphate buffers containing 20 mM NaCl solution (pH 7.4) in the absence and presence of targeted DNA (DNApm; 100 nM) and then the mixtures were equilibrated for 30 min at ambient temperature. Two aliquots of Hg2+ (final concentration: 1.0 μM) were then added separately to the two mixtures. In the absence of the target DNA, the fluorescence of FAM (excitation wavelength: 475 nm) was low, as indicated in Figure 1 (spectrum a). In the presence of the targeted DNA, the fluorescence (spectrum b) of FAM was higher than that in the absence of the target DNA. These results support the sensing mechanism illustrated in Scheme 1. When using a single base mismatched DNA (DNAmm1) having the sequence listed in Table 1 as a control, the fluorescence of FAM (spectrum c) was only slightly higher than that in the absence of the targeted DNA, suggesting that the T7-MB probe has high specificity toward DNApm. In addition, the selectivity of T7-MB toward DNApm to DNAmm1 increased upon increasing in the ratio of the targeted DNA to T7-MB, and achieved a maximum when the targeted DNA was used in 5-fold excess (inset of Figure 1). The selectivity values of T7-MB (20 nM) toward DNApm over DNAmm1 were 3.0, 3.0, 4.2, 9.5 and 46 when the molar ratios of the target DNA to T7-MB were 0.1, 0.5, 1, 2 and 5, respectively. The selectivity increased upon increasing the concentration of the targeted DNA, because the hybrid structure of T7-MB with DNApm is more stable than that with DNAmm1. The use of four other single base mismatched DNA strands (DNAmm2–5) provided similar results to those obtained using DNAmm1. Furthermore, when using a random DNA sequence (5′-ACCTGGAAGAGTATTGCAA-3′) as a control to test the specificity of our T7-MB, we did not observe any change in the fluorescence. The highly specific nature of our T7-MB probe suggested that it would have great potential for use in SNPs studies.
Figure 1.
Fluorescence spectra of the T7-MB (20 nM) in (a) the absence of target DNA and (b, c) the presence of (b) DNApm (100 nM) and (c) DNAmm1 (100 nM). Inset: the values of (IF1–IF0)/(IF2–IF0) of T7-MB in the presence of DNApm (IF1) and DNAmm1 (IF2), as functions of concentration ratio of DNAtarget to T7-MB. The solution contained 5 mM phosphate buffer (pH 7.4), 1.0 μM Hg2+ and 20 mM NaCl.
Effect of Hg2+ concentration
The sensing capability of our T7-MB probe for DNA depends on the interplay of the complexes formed between T7 and Hg2+ and between the DNA sequence in the loop and the tested DNA. Thus, we expected that the specificity and sensitivity of our T7-MB probe would depend on the concentration of Hg2+, because it affects the amount of Hg2+–T7-MB complex formed. We investigated the effect of Hg2+ at various concentrations (0–1.5 μM) on the fluorescence of the FAM unit in the T7-MB in the absence of tested DNA. Upon increasing concentration of Hg2+ in the presence of 20 nM T7-MB (Figure 2A, closed square), the fluorescence of FAM initially decreased rapidly (from 0 to 0.5 μM) and then decreased more gradually (from 0.5 to 1.5 μM). This result suggests that the folded DNA structure was more stable in the presence of higher concentrations of Hg2+. To support this hypothesis, we conducted melting temperature measurements; here, we define Tm as the temperature at which the fluorescence of FAM reaches 50% of its original value. Upon increasing the temperature, the fluorescence intensity increased as a result of breaking the T–Hg2+–T bonds (Figure 2B). Upon increasing the Hg2+ concentration, the value of Tm increased, reaching a plateau at the concentration of Hg2+ of 1.0 μM (inset to Figure 2B).
Figure 2.
(A) Plots of (closed square) the fluorescence intensity at 518 nm of T7-MB (20 nM) and (open square) the values of (IF1– IF0)/(IF2–IF0) of T7-MB in the presence of DNApm (IF1) and DNAmm1 (IF2), both as functions of the concentration of Hg2+ (0–1.5 μM). (B) Fluorescence spectra of the T7-MB (20 nM) recorded a various temperatures. Inset: plot of the value of Tm of T7-MB as a function of the concentration of Hg2+ (0–1.5 μM). Other conditions were the same as those described inFigure 1.
The results in Figure 2 suggest that the concentration of Hg2+ is an important factor determining the specificity of the T7-MB. Thus, to determine the optimal Hg2+ concentration under the tested conditions, we plotted (IF1 – IF0)/(IF2 – IF0) against the Hg2+ concentration, where IF0, IF1, and IF2 are the fluorescence intensities of the FAM unit in T7-MB in the absence of the targeted DNA and in the presence of DNApm and DNAmm1, respectively. A higher value of this ratio indicates better specificity of the T7-MB probe toward DNApm over DNAmm. Figure 2A (open square) indicates that the ratio was maximized at an Hg2+ concentration of 1.0 μM; at higher concentrations (e.g. 10 μM), the T7-MB prefers to complex with Hg2+, reducing its ability to recognize its target DNA. In addition, the temperature also affected the specificity of the T7-MB. The specificity of the T7-MB probe toward DNApm over DNAmm achieved a plateau at ambient temperature (25–30°C). At higher temperature, the T–Hg2+–T bonds were broken as a result of decreasing the specificity (Figure S1). Thus, the optimal conditions—providing the highest specificity of the T7-MB toward its target DNA—involved the use of 20 nM T7-MB in 5.0 mM phosphate buffer (pH 7.4) containing 1.0 μM Hg2+ and 20 mM NaCl at ambient temperature.
Next, we separately investigated the kinetics of forming folded structures of the T7-MB with and without targeted DNA in the presence of Hg2+. The fluorescence intensity of the T7-MB decreased immediately once Hg2+ was added. However, the fluorescence intensities took 1.5 and 2.0 h to achieve constant values in the presence of DNApm and DNAmm1, respectively (Figure S2). Figure S2 reveals that the folded rate of the T7-MB with DNAmm1 was slower than that with DNApm. The kinetics of this probe is slow, because some undesired Hg-oligonucleotide complexes may be kinetically preferred formed, especially in the case of DNAmm1 (20). Based on these kinetics, we employed an equilibrium time of 2.0 h in the following experiments.
Sensitivity and specificity
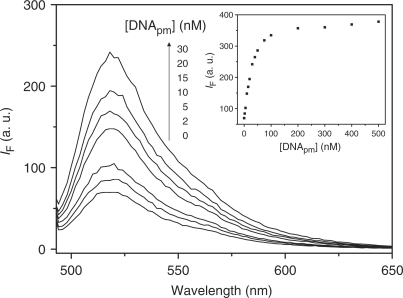
We investigated the sensitivity of the T7-MB at different concentrations toward DNApm. Figure 3 indicates that the fluorescence intensity increased upon increasing the concentration of DNApm when using 20 nM T7-MB. We obtained a linear response (R2 = 0.991) of the fluorescence intensity against the concentration of DNApm over the range 2–30 nM, (inset to Figure 3), with a limit of detection of 0.5 nM at a signal-to-noise ratio of 3. The LODs of DNApm by using T7-MB at the concentrations of 10.0 and 50.0 nM were 0.48 and 1.20 nM, respectively. High concentration of T7-MB probe produced high background fluorescence intensity, leading to decreases in the sensitivity. When using low concentrations (<20 nM) of T7-MB, poor selectivity toward DNApm is problematic. Relative to other existing methods for the detection of DNA using DNA-MBs (the optimum conditions as shown in Figure S3), the T7-MB probe provides at least a 3-fold improvement in sensitivity. The relative standard deviation for quantitation of DNA using the T7-MB probe was <0.8%.
Figure 3.
Fluorescence spectra of T7-MB (20 nM) recorded at various concentrations of DNApm. Inset: plot of the fluorescence intensity at 518 nm of T7-MB (20 nM) as a function of the concentration of DNApm. Other conditions were the same as those described in Figure 1.
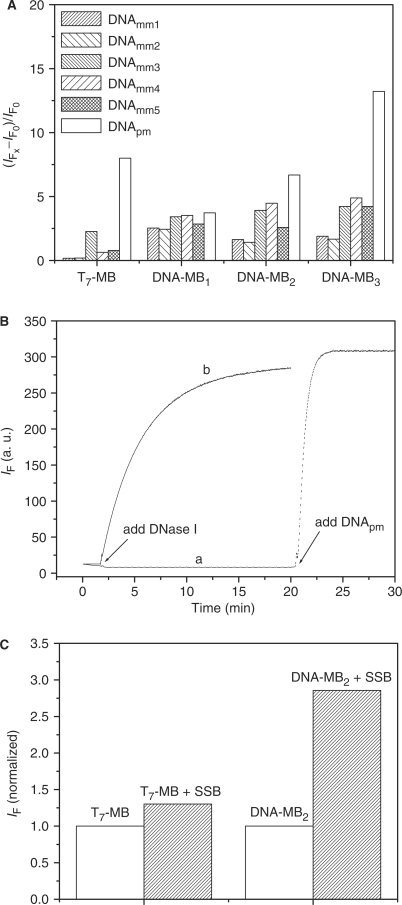
To compare the present system to a conventional DNA-MB probe for the study of SNPs, we employed the two systems separately for the detection of DNApm and five mismatched strands DNAmm1–5. Because the stability of DNA-MBx probe depends on the GC content in the stem, three different DNA-MBx probe (x = 1–3; no Hg2+) as listed in Table 1 were chosen. The performances of the four MB probes were evaluated according to the values of (IF–IF0)/IF0, where IF0 is the florescence intensity of the FAM in T7-MB or DNA-MB in the absence of target DNA and IF values are those in the presence of DNApm or DNAmm1–5, respectively. Figure 4A reveals that our T7-MB probe exhibits enhanced specificity over the conventional DNA-MBx under the optimal conditions (20 nM T7-MB in the presence of 1.0 μM Hg2+ or DNA-MBx (x = 1–3), 20 mM NaCl and 5.0 mM phosphate buffer solution, pH 7.4 at 35°C). We further conducted similar experiments under physiological conditions (150 mM NaCl, 5.0 mM KCl, 1.0 mM MgCl2, 1.0 mM CaCl2 and 25 mM Tris–HCl buffer solution, pH 7.4). The specificity values of T7-MB and DNA-MBx (x = 1–3) toward DNApm over DNAmm1 were 69-fold for the T7-MB probe (20 nM in the presence of 100 μM Hg2+), and 1.0-, 1.1- and 1.2-fold for DNA-MBx (20 nM; x = 1–3), respectively. We also compared the stabilities of the T7-MB and DNA-MB2 probes in the presence of the endonuclease DNase I (Figure 4B). The DNA-MB2 degraded rapidly once DNase I was added, whereas the T7-MB remained unaffected for at least 20 min under otherwise identical conditions. After 2 h, at least 50% of the T7-MB in the presence of Hg2+ remained in its folded structure, based on changes in the fluorescence intensity. This behavior arose mainly because the folded structure of the T7-MB is more stable than the random-coil structure of the DNA-MBx. We finally compared the resistance of the T7-MB and DNA-MB2 probes toward nonspecific binding proteins. DNA-MBx are subjected to nonspecific binding to SSB. Binding of the DNA-MB2 to SSB caused it to remain in a randomly coiled structure, leading to a false-positive signal (Figure 4C). For simplicity, we normalized the fluorescence intensities of the two MBs in the presence of SSB to their respective values in the absence of SSB. Interestingly, our results reveal that the T7-MB was barely affected after the addition of excess SSB, indicating that this probe is superior to conventional MBs for detecting target DNA strands within biological samples containing high amounts of SSB. Table 2 compares our present approach with four popular approaches [conventional DNA-MB, locked nucleic acid (LNA)-MB, superquenchers-MB and hybrid-MB] to SNPs study with respect to detection limit, specificity and resistance to SSB and nuclease digestion. The specificity of our method is superior to the other four methods. The sensitivity of our approach is comparable to those of superquenchers-MB and hybrid-MB approaches, and is better than those of conventional DNA-MB and LNA-MB approaches. Like our approach, LNA-MB and hybrid-MB resist to the binding of SSB and nuclease digestion. However, the LNA-MB and hybrid-MB are more difficult and expensive to prepare. Nevertheless, the use of toxic Hg2+ ions, albeit in small amounts, in our probe system is a disadvantageous feature. This disadvantage can be overcome by using different DNA sequences that respond to the presence of lower-toxicity metal ions such as Ag+ and K+ ions (22–27).
Figure 4.
(A) Fluorescence enhancements of T7-MB and DNA-MBx (20 nM) in the presence of DNAmm1, DNAmm2, DNAmm3, DNAmm4, DNAmm5 and DNApm. The final concentration ratios of the T7-MB and DNA-MBx to the tested DNA were 1:5. The fluorescence measurements of T7-MB and DNA-MBx were at ambient temperature and 35°C, respectively. (B) Digestion of (a) T7-MB and (b) DNA-MB2 (20 nM) by DNase I (5.0 μg/ml) in the presence of 5.0 mM MgCl2. (C) Responses of the two MBs toward the presence of SSB. The final ratio of the concentrations of MB and SSB was 1:5. Other conditions were the same as those described in Figure 1.
Table 2.
Comparison of SNPs studies using T7-MB and other four different approaches
| Type of MB | Detection limit of the DNApm (nM) | Specificitya | Resistance |
||
|---|---|---|---|---|---|
| SSB | Nuclease | Reference | |||
| Conventional DNA-MB | 1.5 | 7.0 | No | No | In this study |
| T7-MB | 0.5 | 69 | Yes | Yes | In this study |
| LNA-MB | 10 | 10 | Yes | Yes | Wang, L., et al. (6) |
| Superquenchers- MB | 0.1 | 30 | No | No | Yang, C.J., et al.(7) |
| Hybrid-MB | 0.8 | 25 | Yes | Yes | Yang, C.J., et al. (14) |
aSpecificity: (IF1–IF0)/(IF2–IF0) where IF0, IF1 and IF2 are the fluorescence intensities of the fluorophore units in the MBs without the targeted DNA and with DNApm and DNAmm, respectively.
CONCLUSIONS
We have developed a new sensing strategy for SNPs study using T7-MB probe in the presence of Hg2+. This new approach is simple, sensitive, selective and cost-effective for studying SNPs. The T7-MB probe in the presence of Hg2+ has greater resistance toward nuclease digestion and undergoes less nonspecific binding with SSB. When compared with the conventional MB approaches, the T7-MB probe provides a greater specificity toward perfect-matched DNA over mismatched DNA and is more stable in the presence of high concentrations of salt. When SNPs study under physiological conditions is needed, the stability and specificity of the T7-MB probe can be further improved by carefully controlling Hg2+ concentrations and/or the stem length. The superior characteristics of the T7-MB probe show its great potential for use in SNPs studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Council of Taiwan (NSC 96-2627-M-002-013 and NSC 96-2627-M-002-014); National Taiwan University for PDF support (96R0066-37 to Y.-W.L.). Funding for open access publication charge: National Science Council of Taiwan (NSC 96-2627-M-002-013 and NSC 96-2627-M-002-014).
Conflict of interest statement. None declared.
REFERENCES
- 1.Peng X, Greenberg MM. Facile SNP detection using bifunctional cross-linking oligonucleotide probes. Nucleic Acids Res. 2008;36:e31. doi: 10.1093/nar/gkn052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolpashchikov DM. Split DNA enzyme for visual single nucleotide polymorphism typing. J. Am. Chem. Soc. 2008;130:2934–2935. doi: 10.1021/ja711192e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grossmann TN, Roglin L, Seitz O. Triplex molecular beacons as modular probes for DNA detection. Angew. Chem. Int. Ed. Engl. 2007;46:5223–5225. doi: 10.1002/anie.200700289. [DOI] [PubMed] [Google Scholar]
- 4.Kim Y, Yang CJ, Tan W. Superior structure stability and selectivity of hairpin nucleic acid probes with an L-DNA stem. Nucleic Acids Res. 2007;35:7279–7287. doi: 10.1093/nar/gkm771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Chu X, Liu Y, Jiang JH, He Z, Zhang Z, Shen G, Yu RQ. A colorimetric method for point mutation detection using high-fidelity DNA ligase. Nucleic Acids Res. 2005;33:e168. doi: 10.1093/nar/gni163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Yang CJ, Medley CD, Benner SA, Tan W. Locked nucleic acid molecular beacons. J. Am. Chem. Soc. 2005;127:15664–15665. doi: 10.1021/ja052498g. [DOI] [PubMed] [Google Scholar]
- 7.Yang CJ, Lin H, Tan W. Molecular assembly of superquenchers in signaling molecular interactions. J. Am. Chem. Soc. 2005;127:12772–12773. doi: 10.1021/ja053482t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher TL, Terhorst T, Cao X, Wagner RW. Intracellular disposition and metabolism of fluorescently labeled unmodified and modified oligonucleotides microinjected into mammalian cells. Nucleic Acids Res. 1993;21:3857–3865. doi: 10.1093/nar/21.16.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leonetti JP, Mechti N, Degols G, Gagnor C, Lebleu B. Intracellular distribution of microinjected antisense oligonucleotides. Proc. Natl Acad. Sci. USA. 1991;88:2702–2706. doi: 10.1073/pnas.88.7.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uchiyama H, Hirano K, Kashiwasake-Jibu M, Taira K. Detection of undergraded oligonucleotides in vivo by fluorescence resonance energy transfer. J. Biol. Chem. 1996;271:380–384. doi: 10.1074/jbc.271.1.380. [DOI] [PubMed] [Google Scholar]
- 11.Braasch DA, Corey DR. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn H, Demidov VV, Coull JM, Fiandaca MJ, Gildea BD, Frank-Kamenetskii MD. Hybridization of DNA and PNA molecular beacons to single-stranded and double-stranded DNA targets. J. Am. Chem. Soc. 2002;124:1097–1103. doi: 10.1021/ja0041324. [DOI] [PubMed] [Google Scholar]
- 13.Egholm M, Buchardt O, Nielsen PE, Berg RH. Peptide nucleic acids (PNA). Oligonucleotide analogues with an achiral peptide backbone. J. Am. Chem. Soc. 1992;114:1895–1897. [Google Scholar]
- 14.Yang CJ, Martinez K, Lin H, Tan W. Hybrid molecular probe for nucleic acid analysis in biological samples. J. Am. Chem. Soc. 2006;128:9986–9987. doi: 10.1021/ja0618346. [DOI] [PubMed] [Google Scholar]
- 15.Ono A, Togashi H. Highly selective oligonucleotide-based sensor for mercury(II) in aqueous solutions. Angew. Chem. Int. Ed. Engl. 2004;43:4300–4302. doi: 10.1002/anie.200454172. [DOI] [PubMed] [Google Scholar]
- 16.Xue X, Wang F, Liu X. One-step, room temperature, colorimetric detection of mercury (Hg2+) using DNA/nanoparticle conjugates. J. Am. Chem. Soc. 2008;130:3244–3245. doi: 10.1021/ja076716c. [DOI] [PubMed] [Google Scholar]
- 17.Liu C-W, Hsieh Y-T, Huang C-C, Chang H-T. Detection of mercury(II) based on Hg2+—DNA complexes inducing the aggregation of gold nanoparticles. Chem. Commun. 2008:2242–2244. doi: 10.1039/b719856f. [DOI] [PubMed] [Google Scholar]
- 18.Chiang C-K, Huang C-C, Liu C-W, Chang H-T. Oligonucleotide-based fluorescence probe for sensitive and selective detection of Mercury(II) in aqueous solution. Anal. Chem. 2008;80:3716–3721. doi: 10.1021/ac800142k. [DOI] [PubMed] [Google Scholar]
- 19.Tanake Y, Yamaguchi H, Oda S, Kondo T, Nomura M, Kojima C, Ono A. NMR spectroscopic study of a DNA duplex with mercury-mediated T-T base pairs. Nucleosides, Nucleotides Nucleic Acids. 2006;25:613–624. doi: 10.1080/15257770600686154. [DOI] [PubMed] [Google Scholar]
- 20.Miyake Y, Togashi H, Tashiro M, Yamaguchi H, Oda S, Kudo M, Tanake Y, Kondo Y, Sawa R, Fujimoto T, et al. MercuryII-mediated formation of thymine-HgII-thymine base pairs in DNA duplexes. J. Am. Chem. Soc. 2006;128:2172–2173. doi: 10.1021/ja056354d. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka Y, Oda S, Yamaguchi H, Kondo Y, Kijima C, Ono A. 15N-15N J-coupling across HgII: direct observation of HgII-mediated T-T base pairs in DNA duplex. J. Am. Chem. Soc. 2007;129:244–245. doi: 10.1021/ja065552h. [DOI] [PubMed] [Google Scholar]
- 22.Ono A, Miyake Y. Highly selective binding of metal ions to thymine-thymine and cytosine–cytosine base pairs in DNA duplexes. Nucleic Acids Symp. Ser. 2001;1:227–228. doi: 10.1093/nass/3.1.227. [DOI] [PubMed] [Google Scholar]
- 23.Matsuda S, Romesberg FE. Optimization of interstrand hydrophobic packing interactionswithin unnatural DNA base pairs. J. Am. Chem. Soc. 2004;126:14419–14427. doi: 10.1021/ja047291m. [DOI] [PubMed] [Google Scholar]
- 24.Atwell S, Meggers E, Spraggon G, Schultz PG. Structure of a copper-mediated base pair in DNA. J. Am. Chem. Soc. 2001;123:12364–12367. doi: 10.1021/ja011822e. [DOI] [PubMed] [Google Scholar]
- 25.Meggers E, Holland PL, Tolman WB, Romesberg FE, Schultz PG. A novel copper-mediated DNA base pair. J. Am. Chem. Soc. 2000;122:10714–10715. [Google Scholar]
- 26.Tanaka K, Tengeiji A, Kato T, Toyama N, Shiro M, Shionoya M. Efficient incorporation of a copper hydroxypyridone based pair in DNA. J. Am. Chem. Soc. 2002;124:12494–12498. doi: 10.1021/ja027175o. [DOI] [PubMed] [Google Scholar]
- 27.Huang C-C, Chang H-T. Aptamer-based fluorescence sensor for rapid detection of potassium ions in urine. Chem. Commun. 2008:1461–1463. doi: 10.1039/b718752a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.