Abstract
Although centromere function has been conserved through evolution, apparently no interspecies consensus DNA sequence exists. Instead, centromere DNA may be interconnected through the formation of certain DNA structures creating topological binding sites for centromeric proteins. DNA topoisomerase II is a protein, which is located at centromeres, and enzymatic topoisomerase II activity correlates with centromere activity in human cells. It is therefore possible that topoisomerase II recognizes and interacts with the alpha satellite DNA of human centromeres through an interaction with potential DNA structures formed solely at active centromeres. In the present study, human topoisomerase IIα-mediated cleavage at centromeric DNA sequences was examined in vitro. The investigation has revealed that the enzyme recognizes and cleaves a specific hairpin structure formed by alpha satellite DNA. The topoisomerase introduces a single-stranded break at the hairpin loop in a reaction, where DNA ligation is partly uncoupled from the cleavage reaction. A mutational analysis has revealed, which features of the hairpin are required for topoisomerease IIα-mediated cleavage. Based on this a model is discussed, where topoisomerase II interacts with two hairpins as a mediator of centromere cohesion.
INTRODUCTION
The centromere is the central connection point between sister-chromatids in cell division, and it ensures that the replicated DNA is distributed with one copy to each daughter cell. In higher eukaryotes centromeric DNA generally contains stretches of DNA repeats, and in human cells a number of different repetitive elements are found in connection with the centromeres (1). Among these the alpha satellite DNA repeats may be the only repeat elements, which are found at all conventional human centromeres (2–4). Alpha satellite DNA ranges from around 200 kb per chromosome to several megabases and has a basic repeat unit with an average size of 171 bp (5–7). Comparisons of alpha satellite repeats from a number of primates suggest that they are composed of a variable region and a more conserved region of ∼117 bp and 54 bp, respectively (6). The conserved region contains two regions of dyad symmetry, one of which is made from alternating blocks of homopurine and homopyrimidine elements (8) and has a high A/T content. Dyad symmetries, homopurine and homopyrimidine elements, and a high A/T content seem to be common characteristics of eukaryotic centromeric DNA and of the DNA in the so-called neocentromeres that emerge in non-centromeric DNA to take on centromeric function in the absence of a natural centromere (3,4,8–12).
Although the centromere is an essential element, relatively little is known about how the underlying DNA supports the centromeric function. In most eukaryotic species the exact centromere boundaries are difficult to define and no interspecies consensus of the DNA sequence exists (13–16). Furthermore, even within a single species it has often been impossible to identify sequences absolutely necessary and sufficient for centromere function. Considering also that non-centromeric sequences may take on centromere function and form neocentromeres at times when normal centromeres are lost, and that conventional centromeres are seen losing their function when placed on a DNA element, which contains another functioning centromere (17), it has been obvious to speculate that the centromere identity may lie within the DNA structure more than in its primary sequence, and that this function could be propagated by an epigenetic mechanism, whereby centromeric proteins recognize suitable DNA and bring it into a functional state (3,8).
In line with this, many proteins involved in centromere function are conserved across species (13,18). Topoisomerase II is one of the proteins associated with the centromeres. It is a highly conserved and essential enzyme, which influences the topological conformation of DNA by creating a transient 4-bp staggered double-stranded break in the DNA backbone, through which it can transport another DNA helix (19). This ability explains why the enzyme is required for many aspects of DNA metabolism, including DNA replication, transcription, chromosome condensation/decondensation, as well as sister chromatid separation (19–21). Topoisomerase II associates with centromeric DNA in a cell-cycle-specific manner and concentrates at centromeres as they condensate prior to mitotic cell division (22–24). It is consequently one of the first of the non-constitutive centromeric proteins to locate at the centromeres ahead of mitosis (22). In higher eukaryotes the enzyme is active within a sub-region of the centromeric sequences, which coincides with the location of other centromere proteins (22,23,25,26). Topoisomerase II seems to have a role in cohesion at centromeres, which is regulated by sumoylation of the enzyme (27–30), and more recently, sumoylation of topoisomerase II has been found to direct the enzyme to the inner centromere to allow accurate chromosome separation prior to anaphase onset (31–33). A functional role of topoisomerase II at the centromeres is also strongly suggested by the fact that the activity of the enzyme is restricted to active centromeres at mitosis (25,34). Furthermore, like natural centromeres, neocentromeres, which do not contain any detectable alpha satellite DNA, contain an accumulation of mitotic topoisomerase II activity, whereas topoisomerase II activity is not found at inactive blocks of alpha satellite DNA (34). Centromeric topoisomerase II activity at mitosis thus seems to be connected to centromere activity (34).
To investigate the molecular anatomy underlying the interaction between topoisomerase II and alpha satellite DNA, we have extended our previous in vivo study (34) by this in vitro study using purified human topoisomerase IIα and oligonucleotides representing the conserved part of alpha satellite DNA. The nature of the topoisomerase IIα-mediated cleavage and the effects of different mutations in the alpha satellite DNA sequence on the cleavage were investigated in vitro. In contrast to the general cleavage mechanism of topoisomerase II, we find that the enzyme introduces a single-stranded break at the loop of a hairpin structure formed in the alpha satellite DNA. The cleavage is highly dependent on the finer architecture of the centromeric substrate and bears characteristics of suicidal cleavage.
MATERIALS AND METHODS
Oligonucleotides
All oligonucleotides were obtained from DNA Technology A/S.
TOP83: TTCAGATAAAAACTAGAAAGAAGCTTT CTGAGAAACTTCTTTGTGTTCTGTGAATTCATCT CACAGAGTTACACCTTTCTTTT
TOP62: TTCAGATAAAAACTAGAAAGAAGCTTT CTGAGAAACTTCTTTGTGTGTTACACCTTTCTTTT
TOP62-G30C: TTCAGATAAAAACTAGAAAGAA GCTTTCTCAGAAACTTCTTTGTGTGTTACACCTT TCTTTT
TOP62-C24A: TTCAGATAAAAACTAGAAAGAAG ATTTCTGAGAAACTTCTTTGTGTGTTACACCTTT CTTTT
TOP62-insG36: TTCAGATAAAAACTAGAAAGAA GCTTTCTGAGAAAGCTTCTTTGTGTGTTACACCT TTCTTTT
TOP62-delC24insG36: TTCAGATAAAAACTAGAA AGAAGTTTCTGAGAAAGCTTCTTTGTGTGTTAC ACCTTTCTTTT
BOT83: AAAAGAAAGGTGTAACTCTGTGAGAT GAATTCACAGAACACAAAGAAGTTTCTCAGAAA GCTTCTTTCTAGTTTTTATCTGAA
BOT62: AAAAGAAAGGTGTAACACACAAAGA AGTTTCTCAGAAAGCTTCTTTCTAGTTTTTATC TGAA
BOT62-delG39insC28: AAAAGAAAGGTGTAACAC ACAAAGAAGCTTTCTCAGAAACTTCTTTCTAGTTTTTATCTGAA
TOP64: TTCAGATAAAAACTAGAAAGAAGCTTT CTTGAAGAAACTTCTTTGTGTGTTACACCTTTCT TTT
TOP66: TTCAGATAAAAACTAGAAAGAAGCTTT CTTTGAAAGAAACTTCTTTGTGTGTTACACCTTT CTTTT
TOP68: TTCAGATAAAAACTAGAAAGAAGCTTT CTTTTGAAAAGAAACTTCTTTGTGTGTTACACCT TTCTTTT
TOP70: TTCAGATAAAAACTAGAAAGAAGCTT TCTTTTTGAAAAAGAAACTTCTTTGTGTGTTACA CCTTTCTTTT
Preparation of cleavage substrates
Most oligonucleotides were purified by preparative polyacrylamide gel-electrophoresis. 3′-end labelling of the single-stranded DNA substrates was performed by incubating 10 pmol oligonucleotide with 20 U of terminal deoxynucleotidyl transferase (TdT) and [α-32P]ddATP in the supplied TdT reaction buffer for 1 h at 37°C. The reaction was terminated by passing the mixture over a Sephadex G-50 column. DNA was finally precipitated and dissolved either in 40 mM Tris–HCl, pH 7.5, 20 mM MgCl2 and 50 mM NaCl, in TE-buffer or in deionized H2O. The labelled DNA was heated to 80°C and cooled to room temperature either in the presence or absence of the complementary oligonucleotide.
Expression and purification of recombinant human topoisomerase IIα
Expression and purification of recombinant human topoisomerase IIα was done as described in (35,36).
Mapping of topoisomerase IIα-mediated cleavage sites
A standard cleavage reaction was set up by incubating ∼4 pmol of topoisomerase IIα with ∼0.1 pmol of labelled substrate in 50 μl of 10 mM Tris–HCl, pH 7.0, 5 mM MgCl2, 60 mM NaCl, 20 μg/ml bovine serum albumin, and 0.1 mM EDTA (cleavage buffer) at 37°C for the indicated time periods. SDS (1% final concentration) was then added to stop the reaction. The samples were subjected to phenol extraction, and the protein-linked cleavage complexes were recovered from the phenol–water interphase as described previously (37). The complexes were subsequently ethanol-precipitated, proteinase K digested (2 mg/ml, 2 h at 45°C) and analysed by electrophoresis on 12% denaturing polyacrylamide gels. Gels were finally dried, and DNA cleavage was visualized by use of phosphor-imaging (BioRad Molecular Imager®FX). The protein fragment covalently linked to the 5′-end of the cleavage products delays the gel migration of the product by one nucleotide, and due to partial proteinase K digestion cleavage products having a longer protein fragment covalently linked can appear, which migrate even slower (37).
Quantification of topoisomerase IIα-mediated cleavage levels
Cleavage reactions of 40 μl were set up with 3′-end labelled TOP83 as described above for the mapping of topoisomerase IIα-mediated cleavage sites and incubated at 37°C for 90 min. When indicated, cleavage reactions included 0.1 mM VM26 (Bristol Myers, Squibb Company), 0.1 mM mAMSA (Parker Davis Corp.), or 1% DMSO. VM26 and mAMSA were dissolved in DMSO. The reactions were stopped by addition of loading buffer (final concentration: 63 mM Tris–HCl, pH 6.8, 1% SDS, 140 mM β-mercaptoethanol, 10% glycerol, bromophenol blue), heated briefly to ∼95°C and cooled down on ice prior to gel-electrophoresis on 8% SDS–polyacrylamide gels. Gels were dried, and cleavage levels were measured by use of phosphor-imaging (BioRad Molecular Imager®FX).
Secondary-structure predictions
Predictions of secondary structures were performed using the program ‘mfold’, which is found online at http://mfold.bioinfo.rpi.edu/cgi-bin/dna-form1.cgi (38). Usually ionic conditions used for the simulations were 80 mM NaCl and 5 mM MgCl2.
RESULTS
Human topoisomerase IIα cleaves primate alpha satellite DNA in vitro
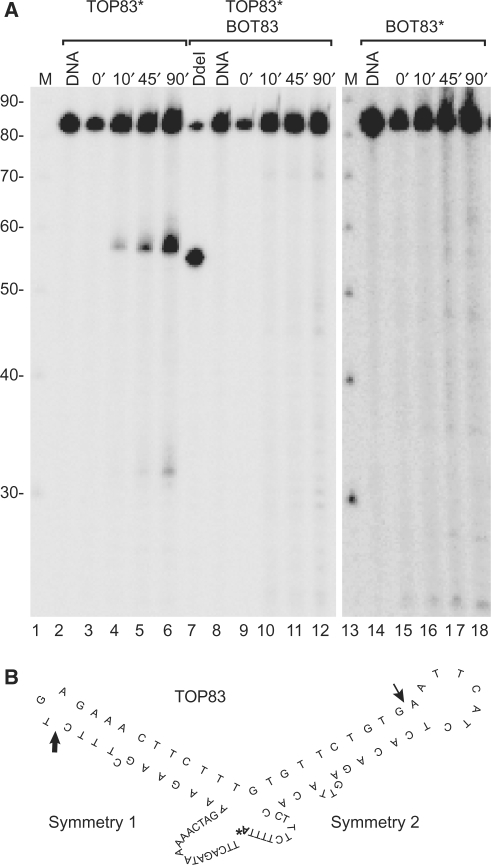
In a previous in vivo study we have demonstrated that topoisomerase II interacts with human alpha satellite DNA in active centromeres, but not in inactive ones (34). To understand how topoisomerase II differentiates between active and inactive alpha satellite DNA we have investigated the interaction between purified human topoisomerase IIα and oligonucleotides representing the conserved part of primate alpha satellite DNA. To this end, two oligonucleotides, TOP83 and BOT83, each representing one strand of the consensus sequence for the most conserved region of the alpha satellite monomer, were constructed. The oligonucleotides contain two regions of dyad symmetry, symmetry 1 and 2, where symmetry 1 comprises a 2-fold dyad symmetry (8). Based on this, the oligonucleotides are predicted to form hairpin structures as shown in Figure 1B for TOP83. The oligonucleotides were incubated with human topoisomerase IIα either separately or as a duplex. To discourage that the dyad symmetry sequences of two identical oligonucleotides should make a partial double-stranded duplex in the reaction mixtures, the oligonucleotides were constructed with 5′- and 3′-ends that were unable to base pair. Furthermore, the substrates were used in low concentrations in the cleavage reaction to favour intra-molecular interactions, thus making hairpin-formation more likely than the formation of partial duplexes. As seen from the cleavage experiment presented in Figure 1A topoisomerase IIα–DNA cleavage complexes are formed when the top strand alone (TOP83) is used as substrate. TOP83 has two cleavage sites, one major site and one minor site, located 56 and 32 nt from the labelled 3′-end, respectively (Figure 1A, lanes 2–6). Interestingly, no comparable cleavage occurred on the complementary bottom strand (BOT83) (lanes 14–18) or on the double-stranded substrate (lanes 8–12).
Figure 1.
Topoisomerase IIα introduces a single-stranded break at the loop of a hairpin structure formed by the conserved part of centromeric alpha satellite DNA. (A) Topoisomerase IIα-mediated cleavage of the 3′-end labelled substrates, TOP83 (lanes 2–6), TOP83 hybridized to BOT83 (lanes 8–12), or BOT83 (lanes 14–18). The asterisk indicates radioactive labelling of the substrate. The incubation time for the cleavage reaction is indicated above the individual lanes. Lanes 1 and 13, DNA marker increasing in steps of 10 bases; lanes 2, 8 and 14, DNA controls without topoisomerase IIα; lane 7, digestion of the labelled duplex substrate with the restriction enzyme DdeI. Migration of the cleavage fragments is retarded with one nucleotide due to a small peptide remaining covalently linked to the DNA after proteinase K treatment. The smear above the cleavage bands represents cleavage products having a longer protein fragment covalently linked due to partial proteinase K digestion (37). (B) Predicted secondary structure of TOP83, the major and minor cleavage sites are indicated by a thick and a thin arrow, respectively. The radioactive nucleotide added to the substrate in the labelling reaction is indicated by a bold ‘A’ with an asterisk.
The observed cleavage sites coincide with the centre of the dyad symmetries and correspond to the loop part of the hairpin structures. The strongest site is observed within symmetry 1 and the weaker site within symmetry 2 as schematically illustrated (Figure 1B). The results thus suggest that topoisomerase IIα recognizes a hairpin structure in alpha satellite DNA as opposed to standard B-form DNA. Hairpin structures have earlier been demonstrated to be among the preferred substrates for topoisomerase II (39–42,44).
When DNA is incubated with topoisomerase II, a cleavage/religation equilibrium is normally established within seconds, where the level of cleavage complexes that can be trapped by SDS is constant once the equilibrium is reached (19). As seen from the time-course experiment presented in Figure 1, cleavage complexes accumulate with time when topoisomerase IIα is incubated with TOP83. This indicates that the enzyme to some extent allows an uncoupling of the cleavage and religation reactions on this substrate, a characteristic of topoisomerase II-mediated suicidal cleavage, where uncoupling takes place due to a loss of positioning of the two DNA termini for religation (37,43).
Topoisomerase IIα-mediated cleavage of alpha satellite DNA in vitro is stimulated by VM26 and inhibited by mAMSA
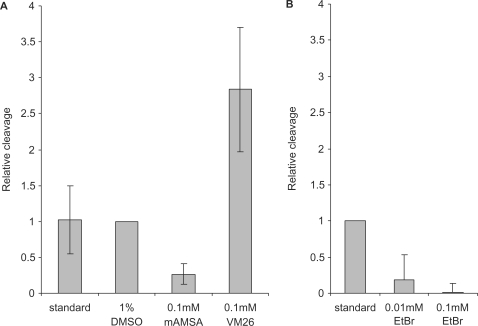
Topoisomerase II activity in the centromeric regions of human chromosomes has been detected by the use of the topoisomerase II-poisons VM26 and VP16, both belonging to the epidophyllotoxins (11,25,26,34). These drugs stabilize topoisomerase II–DNA cleavage complexes by inhibiting the DNA religation reaction (45). To examine the effect of this drug type on topoisomerase IIα-mediated cleavage of alpha satellite DNA in our reaction design, cleavage reactions were performed in the presence of VM26. As shown in Figure 2A, VM26 stimulates topoisomerase IIα-mediated cleavage of alpha satellite DNA, as it does when normal duplex DNA is used as substrate (45). The fact that more cleavage complexes are trapped in the presence of VM26 demonstrates that some DNA religation must take place upon cleavage of alpha satellite DNA in the absence of drug, so religation is not completely uncoupled from cleavage.
Figure 2.
Topoisomerase IIα-mediated cleavage of the centromeric hairpin is differentially affected by VM26 and mAMSA. (A) Topoisomerase IIα-mediated cleavage of TOP83 in the absence or presence of the indicated concentrations of VM26 and mAMSA. Experiments with DMSO were used as reference, as VM26 and mAMSA were dissolved in DMSO. ‘Standard’ denotes experiments without DMSO and drugs. The results are the means +/– SD of four independent experiments. (B) Topoisomerase IIα-mediated cleavage of TOP83 in the absence or presence of the indicated concentrations of ethidium bromide (EtBr). Cleavage experiments were performed as described in (A). Cleavage levels are relative to cleavage levels obtained in the absence of EtBr. The results are the means +/– SD of two independent experiments.
Like VM26, mAMSA is often used for stimulating topoisomerase II-mediated cleavage by inhibiting enzyme-mediated DNA religation (46). However, mAMSA is a DNA intercalator (47), and as such mAMSA may perturb hairpin structure and in this way inhibit topoisomerase IIα-mediated cleavage in the present setup. When mAMSA was included in the in vitro cleavage reaction, cleavage was reduced to about 25% of the level observed in the absence of drug. Cleavage inhibition was similarly observed when the DNA intercalator, ethidium bromide (EtBr), was included in the cleavage reaction (Figure 2B). Like EtBr, mAMSA probably inhibits cleavage due to a perturbation of the hairpin structure. Thus, the result strongly supports that a specific hairpin is formed in the conserved part of alpha satellite DNA in vitro and that this structure is essential for cleavage by topoisomerase IIα.
Topoisomerase IIα-mediated cleavage is determined by the finer architecture of the hairpin formed in alpha satellite DNA
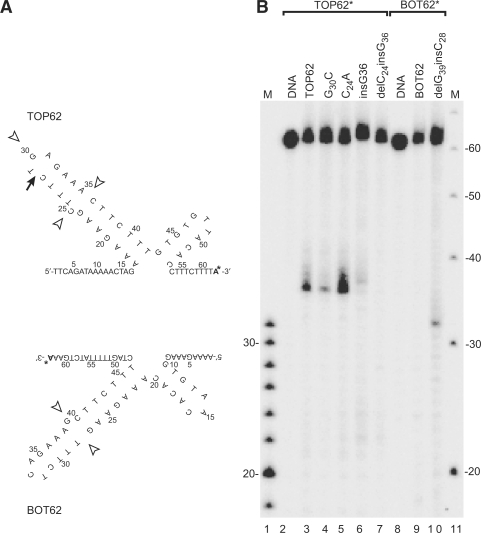
The finding that topoisomerase IIα recognizes and cleaves hairpin structures formed in alpha satellite DNA leaves unexplained why we only observe cleavage in the top strand (TOP83) and not in the bottom strand (BOT83). The two oligonucleotides are very similar in the region containing the cleavage sites, and both are expected to form hairpin structures equally well. However, the differences between the two oligonucleotides must somehow determine the differences in enzyme activity. The deviations between TOP83 and BOT83 in the region around the major cleavage site are, firstly, the base at the top of the hairpin loop, which is a G in TOP83 and a C in BOT83. Secondly, the unpaired base looping out from the middle of the stem is a C in TOP83 and a G in BOT83; and thirdly, the orientation of the unpaired base is different between TOP83 and BOT83. To investigate the importance of each of these differences for the specific interaction of topoisomerase IIα with the two oligonucleotides, variant oligonucleotides were designed, containing mutations at these specific positions. To simplify the analysis we first eliminated symmetry 2 from the substrates, resulting in TOP62 and BOT62 (Figure 3A). This did not influence the cleavage pattern of symmetry 1, as cleavage still occurs at the same position, in this case 35 nt from the labelled 3′-end (compare Figure 3B, lanes 3 and 9 with lanes 6 and 18 in Figure 1A). Therefore, TOP62 and BOT62 were used for the mutational analysis.
Figure 3.
Topoisomerase IIα-mediated cleavage of the centromeric hairpin is determined by an unpaired base in the stem of the hairpin structure. (A) Hairpin structures of the oligonucleotides TOP62 and BOT62, which are identical to TOP83 and BOT83, respectively, except for the lack of most of symmetry 2. The arrow indicates the position of topoisomerase IIα-mediated cleavage; open arrowheads represent positions involved in the mutational analysis shown in (B). The radioactive nucleotide added to the substrate in the labelling reaction is indicated by a bold ‘A’ with an asterisk. (B) Topoisomerase IIα-mediated cleavage of mutated forms of TOP62 and BOT62. The incubation time was 90 min in all cleavage experiments. Migration of the cleavage fragments is retarded with one nucleotide due to a small peptide remaining covalently linked to the DNA after proteinase K treatment. The smear above the cleavage bands represents cleavage products having a longer protein fragment covalently linked due to partial proteinase K digestion (37). The specific mutations are indicated above the lanes. Lane 1, DNA marker increasing in steps of 2 bases; lane 11, DNA marker increasing in steps of 10 bases.
Changing the G at the loop of the hairpin in TOP62 to a C decreased, but did not abolish topoisomerase IIα-mediated cleavage of TOP62 (Figure 3B, lane 4).
To investigate the importance of the unpaired base at the stem of the hairpin, an experiment was first set up with a substrate in which a pairing G was inserted opposite the unpaired C. This resulted in a significant decrease in DNA cleavage (Figure 3B, lane 6), demonstrating that the unpaired base in TOP62 has a stimulatory effect on topoisomerase IIα-mediated cleavage. BOT62 also contains an unpaired base. In this case it is a G, which is located on the 3′-side of the stem. To determine why this substrate is not recognized by topoisomerase IIα, the structure of BOT62 was mimicked by removing the unpaired C in TOP62 and replacing it with an unpaired G at the opposite side of the stem. This abolished topoisomerase IIα-mediated cleavage completely (Figure 3B, lane 7). In agreement with this, removing the unpaired G in BOT62 and inserting a C at the opposite position, thus mimicking TOP62, resulted in cleavage of BOT62 at the expected position, 31 nt from the labelled 3′-end (Figure 3B, lane 10). Based on these results it was concluded that topoisomerase IIα-mediated cleavage of symmetry 1 is dependent upon the orientation of the unpaired base. Experiments were next set up to investigate whether the nature of the unpaired base is important for cleavage. To this end, a substrate was created in which the unpaired C was changed into an A. This base change actually increased cleavage of the substrate (Figure 3B, lane 5).
In conclusion, although the base at the hairpin loop affects topoisomerase IIα-mediated cleavage of the hairpin structure, what is most important for cleavage is the existence and orientation of the unpaired base in the stem of the hairpin.
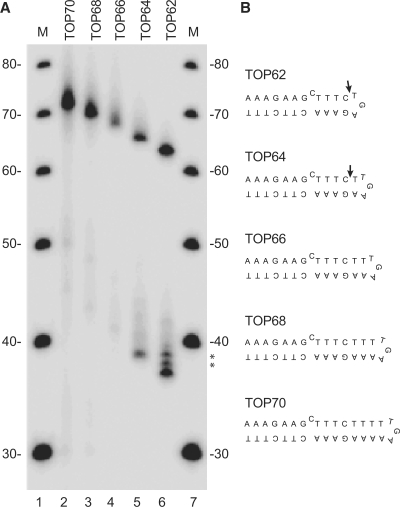
The minimal duplex substrate that gives optimal cleavage with eukaryotic topoisomerase II is a 28-bp DNA fragment (48) and in agreement with earlier footprinting studies (49–51), a recent structure of the central cleavage/religation domain of yeast topoisomerase II in complex with DNA has revealed that the bound DNA is bent with ∼26 bp of the DNA helix being within van der Waals distance of the enzyme (52). The hairpin cleaved in the present study is only half the length of the duplex normally bound in the enzyme. Thus, to learn more about how the enzyme interacts with this substrate, oligonucleotides were designed in which the length of the stem was increased by inserting extra base pairs between the unpaired base and the loop (Figure 4B). Incubation of topoisomerase IIα with the smallest of these substrates (TOP64, one additional base pair between the unpaired base and the loop) resulted in cleavage of the oligonucleotide (Figure 4A, lane 5). The cleavage site was again located 4 nt downstream of the unpaired base as seen with TOP62 (lane 6), so TOP64 extends deeper into the enzyme, resulting in the production of a 5 bases long 5′ overhang upon cleavage. It thus seems that the unpaired base defines the cleavage position. Surprisingly, increasing the distance between the unpaired base and the hairpin loop further practically abolished cleavage of the substrate (lanes 2–4).
Figure 4.
Topoisomerase IIα-mediated cleavage of the centromeric hairpin is inhibited if the distance between the unpaired base and the hairpin loop is increased. (A) Topoisomerase IIα-mediated cleavage of TOP62 derived substrates having 1, 2, 3 or 4 extra base pairs inserted between the unpaired base and the hairpin loop as indicated above the lanes. Lanes 1 and 7, DNA marker increasing in steps of 10 bases. The asterisks indicate cleavage products in lane 6, which migrate slower due to partial proteinase K digestion. The substrates are schematically presented in (B). The arrows indicate the cleavage site.
The finding may be explained by considering that topoisomerase II is a dimer, where each of the two enzyme subunits is responsible for cleavage of one DNA strand, resulting in a double-stranded break with two protruding 5′-ends of four bases. Thus, if the enzyme in our assay binds two hairpins located as ‘kissing-loops’ (Figure 5A), they will add up to the size of a standard minimum substrate and the single-stranded cleavage observed in the hairpin loop will actually be part of a double-stranded break with the second cleavage site being at the same position in an identical hairpin bound by the other subunit. The ‘kissing-loop’ model is consistent with the observation that only TOP62 and TOP64, but not the longer hairpins are cleaved, as these two substrates probably are the only ones, which in a loop-to-loop configuration give a reasonably spacing between the two cleavage sites for the enzyme to perform double-stranded cleavage, while presenting the unpaired base in the stem of the hairpin in the appropriate position.
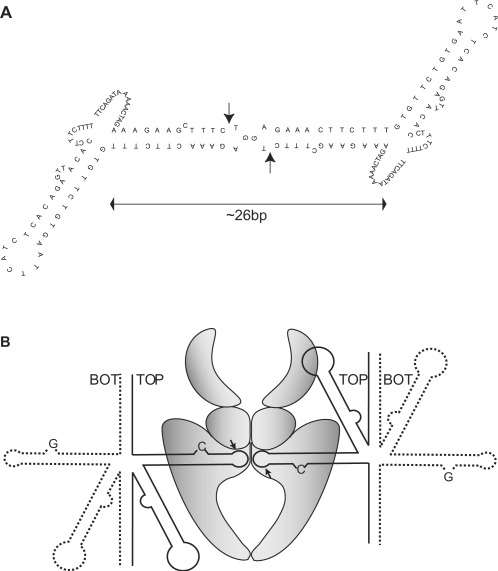
Figure 5.
‘Kissing-loop’ model of two centromeric DNA hairpins bound to topoisomerase IIα. (A) Two DNA hairpins located loop-to-loop will fulfil the requirement of the enzyme for binding ∼26 bp of DNA. The ‘kissing-loops’ mimic a standard DNA duplex allowing human topoisomerase IIα to introduce a ‘double-stranded break’ as indicated by the arrows. (B) Simultaneous topoisomerase IIα-mediated cleavage of two ‘top strand’ hairpins may provide cohesion of sister-chromatids in the centromeric region. Since the enzyme only binds top strand hairpins, the two hairpins held together and cleaved by topoisomerase IIα must originate from two different alpha satellite monomers. If the two monomers belong to two-sister chromatids, the enzyme will bind the sister-chromatids together until DNA religation has taken place.
DISCUSSION
The aim of the present work has been to elucidate the possible connection between chromosomal centromeres and DNA topoisomerase II, which are both essential for cell propagation. Our present study of topoisomerase IIα-mediated cleavage of centromeric DNA sequences in vitro has revealed that the enzyme only interacts with one of the two complementary hairpins formed in the conserved part of the primate alpha satellite DNA repeat. The enzyme introduces a single-stranded break at the loop of the hairpin, where cleavage is guided by an unpaired base in the stem of the loop.
Mechanistic considerations concerning topoisomerase IIα-mediated cleavage of centromeric hairpin structures
The observed cleavage in alpha satellite DNA is interesting from a mechanistic point of view in that cleavage occurs in a hairpin structure rather than in normal duplex DNA. Our findings demonstrate that the existence and orientation of an unpaired base in the stem of the hairpin is important for the cleavage event and that the base determines the cleavage position. This suggests that the unpaired base interacts with a particular contact point inside the active site of the enzyme, which would explain why we consistently see cleavage four bases distal to the position of this base. The unpaired base thus serves to position the hairpin correctly within the active site of the enzyme. Topoisomerase II has previously been found to recognize and cleave DNA hairpins (44), though cleavage in this case was observed at the base of the hairpin stems. Furthermore, strong topoisomerase II recognition sequences have been shown by NMR to form hairpin structures in vitro, where the normal topoisomerase II cleavage site is located near the top of the hairpin, in or just next to the hairpin loop, as seen with the hairpins in the present study (39–42).
When topoisomerase II interacts with a duplex DNA substrate it introduces a double-stranded break, but the cleavage observed in the alpha satellite DNA hairpin is apparently a single-stranded break resulting from cleavage by only one of the subunits in the dimeric enzyme. Earlier studies have revealed that cleavage by the two enzyme subunits occurs sequentially and can be fully uncoupled, thus resulting in topoisomerase II-mediated single-stranded breaks (53,54). Uncoupling of the two subunits has been observed under specific reaction conditions, where Ca++ is substituting Mg++ as the divalent cation in the reaction mixture. Subunit uncoupling would seem the natural outcome if one subunit lacked a DNA substrate. In case the enzyme binds a single hairpin, an elongation of the hairpin stem will be expected to favour cleavage due to more appropriate substrate binding. However, extensions of the hairpin stem length by more than 1 bp abolished cleavage (Figure 4). In particular, it is striking that cleavage did not occur with a hairpin having a 4-bp extension, which in principle should make the hairpin long enough to accommodate a double-stranded break. The observed hairpin cleavage pattern may be explained by a simultaneous interaction of the enzyme with two identical hairpins, which will fulfil the substrate requirements for both subunits. Thus, we believe that topoisomerase IIα interacts with two hairpins located as ‘kissing-loops’ in the active site of the enzyme as presented in Figure 5A. The ‘kissing-loops’ will mimic a normal duplex with enzyme–DNA contacts throughout the whole DNA-binding pocket of the enzyme, and both enzyme subunits will be able to introduce a break, resulting in a double-stranded break. In support of this, the distance between the two break points, when either TOP62 or TOP64 are used as substrate, approximately matches the distance between the two break points in a normal 4-bp staggered topoisomerase II-mediated double-stranded break. In further support of the ‘kissing-loop’ model, a guanine is preferred over a cytosine at the top of the hairpin loop (Figure 3). In the model these two bases come into close contact, and pairing between the two guanines may occur (55).
According to the nature of topoisomerase II-mediated cleavage, the sizes of the cleaved hairpins as well as the cleavage position will make sure that the subunit binding the hairpin is not the one cleaving it. An interesting feature of the ‘kissing-loop’ model is therefore that each subunit of the enzyme after cleavage will be covalently attached to the hairpin bound by the other subunit. Such an enzyme–DNA interaction will make the normal DNA strand-passage activity of the enzyme impossible, since no gap will exist in the DNA upon cleavage. Instead, the two subunits of the topoisomerase II enzyme become interlinked via a DNA-bridge, and as a consequence the two DNA molecules are bound to each other through a protein-bridge.
The centromeric DNA hairpins may represent a distinct class of topoisomerase II targets used in centromere cohesion
The observed topoisomerase IIα-mediated cleavage of hairpins formed in centromeric alpha satellite DNA in vitro may mimic in vivo events since centromeres are believed to be defined by structure rather than primary DNA sequence, and since topoisomerase II cleaves centromeres in vivo. The possibility that centromeric sequences form secondary structures including hairpins, has been suggested for budding yeast, Drosophila and human (8,56–60), and the fact that topoisomerase II cleaves active but not inactive centromeres could be an indication that only active centromeres contain the right structures for topoisomerase II recognition (25,34).
The mapping of topoisomerase II-mediated cleavage sites at centromeres in vivo has been done in the presence of drugs that stabilize the topoisomerase II–DNA cleavage-complex (11,25,26). In the present study, VM26 was found to stimulate the cleavage of the centromeric alpha satellite repeats in vitro as it does in vivo. In contrast, mAMSA inhibited cleavage (Figure 2A), suggesting that the hairpin structures are sensitive to mAMSA intercalation. Only a few studies have examined topoisomerase II-localization or cleavage at the centromeres in the presence of mAMSA in vivo. The results of these experiments are not consistent, probably partly due to differences in the drug concentrations employed (23,26). However, in cells where the localization of topoisomerase II at centromeres seems to be lost upon mAMSA-treatment, the enzyme still remains at chromosome arms (23). This indicates that the topoisomerase II populations at the chromosome axis and at the centromeres interact differently with DNA. Thus, at the chromosomal arms topoisomerase II may interact with normal B-form DNA and cleavage complexes will accumulate in the presence of mAMSA, but at the centromeres a fraction of the enzymes may recognize and interact with specific DNA structures that are removed by mAMSA treatment. The differential interaction of topoisomerase II with the DNA at the two different locations could be mediated and/or regulated by topoisomerase II enzymes bearing different post-translational modifications.
It has been suggested that a subpopulation of the cellular topoisomerase II plays a role for sister chromatid cohesion in the centromeric region (28). If the hairpin cleavage observed in the present study in vitro is indeed a reflection of the in vivo situation, it may be connected to a cohesion role of topoisomerase II at centromeres. If the enzyme simultaneously binds and cleaves two hairpins from alpha satellite repeats belonging to two sister chromatids, the binding may very well function as a kind of cohesion as illustrated in the model presented in Figure 5B. The covalent bonds formed between the two enzyme subunits and the DNA and the strong connection between the two topoisomerase II subunits may be ideal for the purpose of cohesion. Topoisomerase II is probably the only enzyme able to make such covalent attachment to two different DNA-strands at the same time. Interestingly, the potential existence of alternative systems for linking certain regions of repeated DNA in a cohesin-independent manner has also been proposed by others (20).
In Saccharomyces cerevisiae sumoylation of topoisomerase II has been found to be necessary for the separation of sister chromatid centromeres, and it has been suggested that the modified enzyme regulates a DNA structure, which is required for centromeric cohesion (28,32). Interestingly, the decatenation activity of topoisomerase II is not increased by sumoylation (27), indicating that the potential cohesion function of the enzyme is not associated with its ability to (de-)catenate DNA. The data are in line with the hairpin-to-hairpin cohesion model presented here. Thus, topoisomerase II may have at least two functions during cell division; a decatenation function, which ensures correct segregation of sister chromatids, and a cohesion function, which assists in holding sister-centromeres together (28,61–63). Interestingly, the findings presented here suggest that these two functions of the enzyme may be carried out in two completely different ways.
FUNDING
Danish Cancer Society Grant (DP05009); the Danish Medical and Natural Science Research Counsils; the Danish Agency for Science, Technology and Innovation; and the Novo Nordisk Foundation. Funding for open access charge: The Danish Cancer Society Grant DP05009.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Vibe Oestergaard for purification of human topoisomerase IIα.
REFERENCES
- 1.Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC. Human centromeric DNAs. Hum. Genet. 1997;100:291–304. doi: 10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- 2.Choo KH. The Centromere. New York: Oxford University Press Inc; 1997. [Google Scholar]
- 3.Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 4.Murphy TD, Karpen GH. Centromeres take flight: alpha satellite and the quest for the human centromere. Cell. 1998;93:317–320. doi: 10.1016/s0092-8674(00)81158-7. [DOI] [PubMed] [Google Scholar]
- 5.Choo KH, Vissel B, Nagy A, Earle E, Kalitsis P. A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 1991;19:1179–1182. doi: 10.1093/nar/19.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koch JE, Kolvraa S, Petersen KB, Gregersen N, Bolund L. Oligonucleotide-priming methods for the chromosome-specific labelling of alpha satellite DNA in situ. Chromosoma. 1989;98:259–265. doi: 10.1007/BF00327311. [DOI] [PubMed] [Google Scholar]
- 7.Waye JS, Willard HF. Chromosome-specific alpha satellite DNA: nucleotide sequence analysis of the 2.0 kilobasepair repeat from the human X chromosome. Nucleic Acids Res. 1985;13:2731–2743. doi: 10.1093/nar/13.8.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koch J. Neocentromeres and alpha satellite: a proposed structural code for functional human centromere DNA. Hum. Mol. Genet. 2000;9:149–154. doi: 10.1093/hmg/9.2.149. [DOI] [PubMed] [Google Scholar]
- 9.Alonso A, Mahmood R, Li S, Cheung F, Yoda K, Warburton PE. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum. Mol. Genet. 2003;12:2711–2721. doi: 10.1093/hmg/ddg282. [DOI] [PubMed] [Google Scholar]
- 10.Craig JM, Wong LH, Lo AW, Earle E, Choo KH. Centromeric chromatin pliability and memory at a human neocentromere. EMBO J. 2003;22:2495–2504. doi: 10.1093/emboj/cdg232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kas E, Laemmli UK. In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J. 1992;11:705–716. doi: 10.1002/j.1460-2075.1992.tb05103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lo AW, Magliano DJ, Sibson MC, Kalitsis P, Craig JM, Choo KH. A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 2001;11:448–457. doi: 10.1101/gr.167601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukagawa T. Assembly of kinetochores in vertebrate cells. Exp. Cell Res. 2004;296:21–27. doi: 10.1016/j.yexcr.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Henikoff S. Near the edge of a chromosome's “black hole”. Trends Genet. 2002;18:165–167. doi: 10.1016/s0168-9525(01)02622-1. [DOI] [PubMed] [Google Scholar]
- 15.Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- 16.Tyler-Smith C, Floridia G. Many paths to the top of the mountain: diverse evolutionary solutions to centromere structure. Cell. 2000;102:5–8. doi: 10.1016/s0092-8674(00)00004-0. [DOI] [PubMed] [Google Scholar]
- 17.Choo KH. Centromerization. Trends Cell. Biol. 2000;10:182–188. doi: 10.1016/s0962-8924(00)01739-6. [DOI] [PubMed] [Google Scholar]
- 18.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 19.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 20.Porter AC, Farr CJ. Topoisomerase II: untangling its contribution at the centromere. Chromosome Res. 2004;12:569–583. doi: 10.1023/B:CHRO.0000036608.91085.d1. [DOI] [PubMed] [Google Scholar]
- 21.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 22.Rattner JB, Hendzel MJ, Furbee CS, Muller MT, Bazett-Jones DP. Topoisomerase II alpha is associated with the mammalian centromere in a cell cycle- and species-specific manner and is required for proper centromere/kinetochore structure. J. Cell. Biol. 1996;134:1097–1107. doi: 10.1083/jcb.134.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sumner AT. The distribution of topoisomerase II on mammalian chromosomes. Chromosome Res. 1996;4:5–14. doi: 10.1007/BF02254938. [DOI] [PubMed] [Google Scholar]
- 24.Taagepera S, Rao PN, Drake FH, Gorbsky GJ. DNA topoisomerase II alpha is the major chromosome protein recognized by the mitotic phosphoprotein antibody MPM-2. Proc. Natl Acad. Sci. USA. 1993;90:8407–8411. doi: 10.1073/pnas.90.18.8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Floridia G, Zatterale A, Zuffardi O, Tyler-Smith C. Mapping of a human centromere onto the DNA by topoisomerase II cleavage. EMBO Rep. 2000;1:489–493. doi: 10.1093/embo-reports/kvd110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spence JM, Critcher R, Ebersole TA, Valdivia MM, Earnshaw WC, Fukagawa T, Farr CJ. Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha-satellite array. EMBO J. 2002;21:5269–5280. doi: 10.1093/emboj/cdf511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azuma Y, Arnaoutov A, Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. J. Cell Biol. 2003;163:477–487. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- 29.Diaz-Martinez LA, Gimenez-Abian JF, Clarke DJ. Chromosome cohesion - rings, knots, orcs and fellowship. J. Cell Sci. 2008;121:2107–2114. doi: 10.1242/jcs.029132. [DOI] [PubMed] [Google Scholar]
- 30.Vagnarelli P, Morrison C, Dodson H, Sonoda E, Takeda S, Earnshaw WC. Analysis of Scc1-deficient cells defines a key metaphase role of vertebrate cohesin in linking sister kinetochores. EMBO Rep. 2004;5:167–171. doi: 10.1038/sj.embor.7400077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azuma Y, Arnaoutov A, Anan T, Dasso M. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 2005;24:2172–2182. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, Shuai K, Grosschedl R, van Deursen JM. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi Y, Yong-Gonzalez V, Kikuchi Y, Strunnikov A. SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics. 2006;172:783–794. doi: 10.1534/genetics.105.047167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen CL, Wandall A, Kjeldsen E, Mielke C, Koch J. Active, but not inactive, human centromeres display topoisomerase II activity in vivo. Chromosome Res. 2002;10:305–312. doi: 10.1023/a:1016571825025. [DOI] [PubMed] [Google Scholar]
- 35.Oestergaard VH, Bjergbaek L, Skouboe C, Giangiacomo L, Knudsen BR, Andersen AH. The transducer domain is important for clamp operation in human DNA topoisomerase IIalpha. J. Biol. Chem. 2004;279:1684–1691. doi: 10.1074/jbc.M309624200. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Thyssen A, Westergaard O, Andersen AH. Position-specific effect of ribonucleotides on the cleavage activity of human topoisomerase II. Nucleic Acids Res. 2000;28:4815–4821. doi: 10.1093/nar/28.24.4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersen AH, Sorensen BS, Christiansen K, Svejstrup JQ, Lund K, Westergaard O. Studies of the topoisomerase II-mediated cleavage and religation reactions by use of a suicidal double-stranded DNA substrate. J. Biol. Chem. 1991;266:9203–9210. [PubMed] [Google Scholar]
- 38.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amir-Aslani A, Mauffret O, Bittoun P, Sourgen F, Monnot M, Lescot E, Fermandjian S. Hairpins in a DNA site for topoisomerase II studied by 1H- and 31P-NMR. Nucleic Acids Res. 1995;23:3850–3857. doi: 10.1093/nar/23.19.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amir-Aslani A, Mauffret O, Sourgen F, Neplaz S, Maroun RG, Lescot E, Tevanian G, Fermandjian S. The hairpin structure of a topoisomerase II site DNA strand analyzed by combined NMR and energy minimization methods. J. Mol. Biol. 1996;263:776–788. doi: 10.1006/jmbi.1996.0615. [DOI] [PubMed] [Google Scholar]
- 41.El Amri C, Mauffret O, Monnot M, Tevanian G, Lescot E, Porumb H, Fermandjian S. A DNA hairpin with a single residue loop closed by a strongly distorted Watson-Crick G x C base-pair. J. Mol. Biol. 1999;294:427–442. doi: 10.1006/jmbi.1999.3270. [DOI] [PubMed] [Google Scholar]
- 42.Mauffret O, Amir-Aslani A, Maroun RG, Monnot M, Lescot E, Fermandjian S. Comparative structural analysis by [1H,31P]-NMR and restrained molecular dynamics of two DNA hairpins from a strong DNA topoisomerase II cleavage site. J. Mol. Biol. 1998;283:643–655. doi: 10.1006/jmbi.1998.2095. [DOI] [PubMed] [Google Scholar]
- 43.Gale KC, Osheroff N. Uncoupling the DNA cleavage and religation activities of topoisomerase II with a single-stranded nucleic acid substrate: evidence for an active enzyme-cleaved DNA intermediate. Biochemistry. 1990;29:9538–9545. doi: 10.1021/bi00493a007. [DOI] [PubMed] [Google Scholar]
- 44.Froelich-Ammon SJ, Gale KC, Osheroff N. Site-specific cleavage of a DNA hairpin by topoisomerase II. DNA secondary structure as a determinant of enzyme recognition/cleavage. J. Biol. Chem. 1994;269:7719–7725. [PubMed] [Google Scholar]
- 45.Burden DA, Osheroff N. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998;1400:139–154. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- 46.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:221–253. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 47.Capranico G, Binaschi M. DNA sequence selectivity of topoisomerases and topoisomerase poisons. Biochim. Biophys. Acta. 1998;1400:185–194. doi: 10.1016/s0167-4781(98)00135-3. [DOI] [PubMed] [Google Scholar]
- 48.Lund K, Andersen AH, Christiansen K, Svejstrup JQ, Westergaard O. Minimal DNA requirement for topoisomerase II-mediated cleavage in vitro. J. Biol. Chem. 1990;265:13856–13863. [PubMed] [Google Scholar]
- 49.Lee MP, Sander M, Hsieh T. Nuclease protection by Drosophila DNA topoisomerase II. Enzyme/DNA contacts at the strong topoisomerase II cleavage sites. J. Biol. Chem. 1989;264:21779–21787. [PubMed] [Google Scholar]
- 50.Spitzner JR, Muller MT. A consensus sequence for cleavage by vertebrate DNA topoisomerase II. Nucleic Acids Res. 1988;16:5533–5556. doi: 10.1093/nar/16.12.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomsen B, Bendixen C, Lund K, Andersen AH, Sorensen BS, Westergaard O. Characterization of the interaction between topoisomerase II and DNA by transcriptional footprinting. J. Mol. Biol. 1990;215:237–244. doi: 10.1016/S0022-2836(05)80342-0. [DOI] [PubMed] [Google Scholar]
- 52.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 53.Andersen AH, Christiansen K, Zechiedrich EL, Jensen PS, Osheroff N, Westergaard O. Strand specificity of the topoisomerase II mediated double-stranded DNA cleavage reaction. Biochemistry. 1989;28:6237–6244. doi: 10.1021/bi00441a015. [DOI] [PubMed] [Google Scholar]
- 54.Zechiedrich EL, Christiansen K, Andersen AH, Westergaard O, Osheroff N. Double-stranded DNA cleavage/religation reaction of eukaryotic topoisomerase II: evidence for a nicked DNA intermediate. Biochemistry. 1989;28:6229–6236. doi: 10.1021/bi00441a014. [DOI] [PubMed] [Google Scholar]
- 55.Skelly JV, Edwards KJ, Jenkins TC, Neidle S. Crystal structure of an oligonucleotide duplex containing G.G base pairs: influence of mispairing on DNA backbone conformation. Proc. Natl Acad. Sci. USA. 1993;90:804–808. doi: 10.1073/pnas.90.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gallego J, Golden EB, Stanley DE, Reid BR. The folding of centromeric DNA strands into intercalated structures: a physicochemical and computational study. J. Mol. Biol. 1999;285:1039–1052. doi: 10.1006/jmbi.1998.2334. [DOI] [PubMed] [Google Scholar]
- 57.Nonin-Lecomte S, Leroy JL. Structure of a C-rich strand fragment of the human centromeric satellite III: a pH-dependent intercalation topology. J. Mol. Biol. 2001;309:491–506. doi: 10.1006/jmbi.2001.4679. [DOI] [PubMed] [Google Scholar]
- 58.Ortiz-Lombardia M, Cortes A, Huertas D, Eritja R, Azorin F. Tandem 5′-GA:GA-3′ mismatches account for the high stability of the fold-back structures formed by the centromeric Drosophila dodeca- satellite. J. Mol. Biol. 1998;277:757–762. doi: 10.1006/jmbi.1998.1646. [DOI] [PubMed] [Google Scholar]
- 59.Tal M, Shimron F, Yagil G. Unwound regions in yeast centromere IV DNA. J. Mol. Biol. 1994;243:179–189. doi: 10.1006/jmbi.1994.1645. [DOI] [PubMed] [Google Scholar]
- 60.Zhu L, Chou SH, Reid BR. A single G-to-C change causes human centromere TGGAA repeats to fold back into hairpins. Proc. Natl Acad. Sci. USA. 1996;93:12159–12164. doi: 10.1073/pnas.93.22.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DiNardo S, Voelkel K, Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc. Natl Acad. Sci. USA. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Downes CS, Mullinger AM, Johnson RT. Inhibitors of DNA topoisomerase II prevent chromatid separation in mammalian cells but do not prevent exit from mitosis. Proc. Natl Acad. Sci. USA. 1991;88:8895–8899. doi: 10.1073/pnas.88.20.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]