Abstract
Understanding the molecular mechanisms that influence transposable element target site preferences is a fundamental challenge in functional and evolutionary genomics. Large-scale transposon insertion projects provide excellent material to study target site preferences in the absence of confounding effects of post-insertion evolutionary change. Growing evidence from a wide variety of prokaryotes and eukaryotes indicates that DNA transposons recognize staggered-cut palindromic target site motifs (TSMs). Here, we use over 10 000 accurately mapped P-element insertions in the Drosophila melanogaster genome to test predictions of the staggered-cut palindromic target site model for DNA transposon insertion. We provide evidence that the P-element targets a 14-bp palindromic motif that can be identified at the primary sequence level, which predicts the local spacing, hotspots and strand orientation of P-element insertions. Intriguingly, we find that the although P-element destroys the complete 14-bp target site upon insertion, the terminal three nucleotides of the P-element inverted repeats complement and restore the original TSM, suggesting a mechanistic link between transposon target sites and their terminal inverted repeats. Finally, we discuss how the staggered-cut palindromic target site model can be used to assess the accuracy of genome mappings for annotated P-element insertions.
INTRODUCTION
Mobile DNA sequences known as transposable elements are naturally occurring mutagenic agents that have been harnessed as experimental tools for genetic analysis in a variety of model organisms (1,2). Of the two major classes of transposable elements that exist—those that transpose directly via a DNA molecule (transposons), and those that transpose indirectly via a RNA intermediate (retrotransposons) (1)—DNA-based transposons have been most widely developed as tools for gene disruption and gene transfer experiments, becoming essential parts of the genetic tool-kit in bacteria (3), fungi (4), plants (5) and animals (2). One of the most advanced transposon systems for genetic analysis is the Drosophila P-element (6), which has been engineered to facilitate a large number of genetic and genomic manipulations including gene disruption, reporter gene analysis, gene and enhancer trapping, misexpression of endogenous genes and the generation of chromosomal aberrations (7).
Because of the widespread utility of the P-element as a tool for Drosophila genetics and genomics, the mechanisms of P-element transposition have been studied intensively over the last 25 years (8). Like many DNA-based transposons, the P-element transposes through a ‘cut and paste’ mechanism that can be divided into two events—excision from the donor site and insertion into a new location in the host genome. Transposition is initiated when the P-element encoded transposase protein forms a tetrameric complex that binds one of the P-element terminal inverted repeats (TIRs) at the donor site (9,10), followed by GTP-dependent synapsis with the other TIR and sequential cleavage of each TIR from the donor site (10,11). The P-element transposase complex then forms a staggered cut of 17-nt at both TIRs (12), exposing the reactive 3′ single-stranded extensions that mediate strand transfer and integration into a new target site (9).
In contrast to donor excision and target site integration, the molecular mechanisms of target site selection for new P-element insertions remain poorly understood. Target site selection at the genomic scale is generally thought to be nonrandom, with P-elements exhibiting a preference for insertion into euchromatic regions (13), a bias towards insertion into 5′-end of genes (14), hotspots for insertion at both the gene (15) and nucleotide (16) levels, and local hopping in the vicinity of donor elements (17). In addition to these factors that suggest the influence of chromatin structure, other studies have reported a role for local DNA sequence/structure in P-element target site selection. Based on a limited sample of only 18 insertions, O’Hare and Rubin (16) first demonstrated that P-elements prefer to insert into an 8-bp GC-rich consensus sequence (GGCCAGAC), which was later confirmed in a expanded sample (n = 61 insertions) by Preston et al. (18). Subsequently, many P-element insertion sites were shown to differ from this consensus sequence (19), and other pregenomic analyses of small samples led to different target motif [e.g. GXTCAGGC, (20)], casting some doubt on the generality of the original target motif reported by O’Hare and Rubin (16). Liao et al. (21) analyzed a much larger set of 1469 P-element insertions sites mapped to partially assembled genome sequences and concluded that ‘although there are base preferences at each position, these are not strong enough to generate a clear consensus sequence’. Instead, these authors argued that the P-element recognizes a 14-bp palindromic structural motif based on a pattern of hydrogen bonding at the target site. More recently, Julian (22) analyzed a sample of 795 P-elements and reported a 14-bp nonpalindromic consensus sequence (ANNGGCCAGACNNT) that extended the GC-rich motif of O’Hare and Rubin (16). These conflicting results have lead us to clarify whether the P-element targets a specific motif and, if so, whether this motif is a palindrome in order to better understand the target site selection of the P-element and other DNA transposons.
The possibility that the P-element targets a palindromic motif is intriguing given the fact that many other DNA transposons in wide variety of organisms, including bacteria, plants, worms, insects and vertebrates, also appear to prefer for palindromic target sequences (Table 1). A palindromic target site recognition model has potential relevance for understanding the mechanisms of transposon integration, since it is consistent with transposase acting as homo-multimeric complex with the target site DNA (9,10,23–26). Additionally, there may be functional connections between palindromic target sites and the TIRs that flank many transposons, which are themselves palindromic sequences. Finally, palindromic target sites are also often observed for retroviruses (27), which use integrase enzymes for integration that share catalytic activity with transposases (26). The palindromic nature of transposon target site recognition is not universally accepted, however, with both palindromic and non-palindromic target site motifs (TSMs) often reported for the same transposon [see conflicting evidence for the P-element (above) or for Tc1 (28,29)]. These discrepancies may have arisen because many pregenomic analyses of transposon insertion site preferences were based on extremely small sample sizes of insertions, natural target sites that have undergone sequence evolution since transposon insertion, or insertions into small artificial target regions (e.g. plasmids) that only allowed a limited exploration of sequence space.
Table 1.
Palindromic transposon target site sequences are common across all major kingdoms of life
| Transposon | Length of TSD (bp) | TSM | Taxon | Reference |
|---|---|---|---|---|
| IS231A | 11 | GGGNNNNNCCC | Bacteria | (59) |
| IS630 | 2 | CTAG | Bacteria | (60) |
| IS903 | 9 | WTTYANNNNNNNNNTRAAW | Bacteria | (25,61) |
| Tn3/IS3000 | 5 | TWNTAWTANWA | Bacteriaa | (24,62,63) |
| Tn4652 | 5 | GTAWTAC | Bacteria | (64) |
| Tn5/IS50 | 9 | AGNTYWRANCT | Bacteria | (65) |
| Tn10 | 9 | GNNGCTNAGCNNC | Bacteria | (23,44) |
| Ac/Ds | 8 | CTTATAAG | Plant | (5,66) |
| Mu | 9 | CCTNNNNNNNNNAGG | Plant | (45,67) |
| Tc1 | 2 | CAYATATRTG | Worm | (28,29) |
| Tc3 | 2 | AWATATWT | Worm | (29) |
| Tc5 | 3 | MYTNARK | Worm | (29) |
| Hermes | 8 | GTGNNCAC | Insect | (68) |
| Hobo | 8 | GTTTAAAC | Insect | (69) |
| Minos | 2 | ATATATAT | Insect | (70) |
| Mos | 2 | AATATATATT | Insectb | (71) |
| P-element | 8 | ATRGTCCGGACWAT | Insect | This study; (21) |
| SB | 2 | RCAYATATRTGY | Vertebrate | (72–74) |
Note that the length of the target site motif is often longer than the TSD, indicated in bold. IUPAC ambiguity codes are as follows: N = A/C/G/T, W = A/T, Y = C/T, R = A/G, M = A/C, K = G/T.
aData are for a bacterial transposon mobilized in a fungal genomic background.
bData are for an insect transposon mobilized in a worm genomic background.
To understand transposon target site selection properly, it is necessary to investigate large sample of target sites in their in vivo genomic context immediately following insertion. Large-scale transposon insertion projects, such as the P-element gene disruption projects in Drosophila melanogaster (30–35) provide excellent functional genomic data to study models of target site selection for DNA transposons. Here, we analyze a sample of over 10 000 reliably mapped P-element insertions and provide evidence that the P-element prefers a staggered-cut palindromic target motif that can be identified at the primary sequence level. Moreover, we show that the local spacing, hotspots and strand orientation of P-element insertions across the genome support a palindromic insertion site model for transposon target site selection. These results have important implications for understanding the structure inverted repeat DNA transposons and their mechanisms of transposition, as well as for the analysis of artificial and natural transposon insertions in genome sequences.
MATERIALS AND METHODS
P-element insertion sites were obtained from release 5.6 of the D. melanogaster genome annotation (36). The majority of these data are from large-scale transposon insertion projects (30–35) with additional insertions curated from literature. Data manipulation was conducted in custom PERL (version 5.8.6) programs using BioPERL (version 1.3) (37) modules. Data and statistical analysis was performed in the R programming language (version 2.6.2) (http://cran.r-project.org/). In reality, P-element insertions occur between adjacent nucleotides in the genome and therefore should be annotated in genome sequences on inter-base coordinates. However, annotations in FlyBase are on base coordinates and therefore P-element insertion sites are represented differently on the positive and negative strands (i.e. at the base after the insertion site on the positive strand and at the base before the insertion site on the negative strand). To make coordinate systems comparable on the positive and negative strands for analysis of distances between P-element insertions, we added 1 bp to the coordinates of insertions on the negative strand, but retained the annotated coordinate in Supplementary Files 1 and 3.
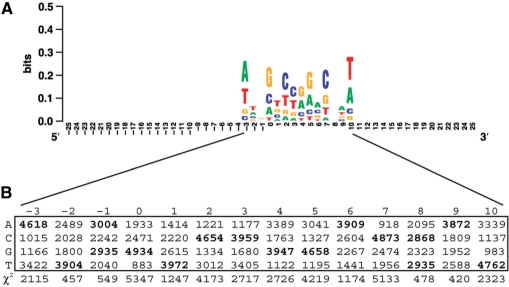
To determine if the P-element targets a specific motif at the primary sequence level, we generated sequence logos (38) from sets of aligned P-element insertion sites. Insertions at the same coordinate on the same strand were collapsed to create sets of nonredundant insertion sites. To do this, we extracted a 51-bp window centered around each insertion site (−25 and +25 from the insertion site) and used Weblogo (version 2.8.2) (39) with the following options (c -k 1 -w 15 -h 5 -Y -B 0.5 -n -s -25 -T 0.1 -b). Logos were created for both positive and negative stand insertions for each ‘family’ of P-elements generated from distinct insertion screens. Since sequence logos measure the information content and not the statistical significance of a motif, we tested of each position in the motif for deviation from expected genome-wide base composition using a χ2-test.
To measure the match of individual insertion sites to the putative P-element TSM, a position frequency matrix (PFM) was generated from a nonredundant set of aligned P-element insertions sites. Insertions on the negative strand were reverse complemented before including into the initial PFM and scoring, thus all sites in our model are oriented relative to the positive strand. Since no significant differences were observed between nucleotide frequencies at complementary positions (e.g. positions 1 and 14; seven χ2-tests, all P > 0.04), we averaged frequencies of complementary nucleotides at corresponding positions around the plane of reverse-complement symmetry (e.g. positions 1 and 14) to construct our final PFM for scoring target sites. This palindromic PFM was used to score individual insertion sites using PATSER (version 3b.5) (40) with the following parameters: -A a:t 0.29 c:g 0.21 -d2 -R. For each insertion site, we evaluated the match to PFM by calculating a log-likelihood ‘motif score’ for the distinct target sites that would give rise to that insertion site on the positive and negative strands. In addition, for each target site we calculated (i) a ‘half-site score’ by assessing the match of the 5′ and 3′ half of the target site to first seven columns of the 14-bp PFM, and (ii) a ‘palindrome score’ that ranges from zero to seven, with a score of one given to each pair of corresponding positions in the palindrome that had complementary nucleotides and a score of zero given for noncomplementary nucleotides.
RESULTS
To ensure large enough sample sizes and reliable genome mappings for our analysis of P-element target site preferences, we restricted our analysis to four families of P-element (GT1, SUPor-P, EPgy2 and XP) from the D. melanogaster Release 5.6 genome annotation that were obtained from large-scale screens that were localized to precise sequence coordinates using inverse PCR after completion of the D. melanogaster genome sequence (33,35). These families of P-elements each had a large number of insertions (>500) with a high proportion of insertions mapped to a single base pair (>90%) and mapped to a specific strand (>90%). Preliminary analyses showed that inclusion of data from other P-element screens generated systematic biases in subsequent analyses because of inaccurate genome mappings (see Discussion section). Table 2 summarizes characteristics of genome mappings for 10 860 insertions from the four P-element families analyzed in this study.
Table 2.
Summary of reliably mapped P-element insertions in the Release 5.6 Flybase genome annotation
| P-element family name | Number of insertions | Number mapped to 1 bp (%) | Number mapped to +/− strand (%) | Number on + strand (%) |
|---|---|---|---|---|
| GT1 | 556 | 531 (95.50) | 496 (93.4) | 260 (52.42) |
| SUPor-P | 2297 | 2288 (99.61) | 2134 (93.27) | 1065 (49.91) |
| EPgy2 | 3496 | 3473 (99.34) | 3258 (93.81) | 1630 (50.03) |
| XP | 5311 | 4974 (93.65) | 4972 (99.96) | 2479 (49.86) |
| Total | 11 660 | 11 266 (96.62) | 10 860 (96.40) | 5434 (50.04) |
Numbers reported include redundant insertions in the same insertion site.
The P-element targets a 14-bp palindromic motif
We constructed separate sequence logos for insertions on the positive and negative strands for insertions that were mapped to a single base pair for each family. For all four families, we observed the same palindromic TSM for insertions mapped either to the plus or minus strands (Figure 1, Supplementary File 2). The similarity in TSM for the different families suggests that the local target site preferences is intrinsic to the P-element and is not family or screen dependent. Therefore, we pooled insertions for EPgy2, GT1, SUPor-P and XP into one sample for all subsequent the analyses of P-element insertion preferences. These four families include a total of 10 860 insertions located in 10 221 nonredundant insertion sites in the D. melanogaster euchromatin.
Figure 1.
The P-element targets a 14-bp palindromic TSM. (A) Sequence logo depicting the relative base usage for a 51 bp window centered around 10 221 P-element insertion sites. The insertion site on the positive strand is just before position zero, and the insertion site on the negative strand is just after position seven. Insertions on the minus strand have been reverse complemented before being included in the alignment. The Y-axis is in bit (log base 2) units of the usage of bases in the motif relative to the random expectation of equal frequency. (B) Table of base usage in the 14-bp TSM and χ2 statistics testing the null hypothesis that base usage at each position of the motif is random under the genome-wide background base composition in D. melanogaster. All positions deviate significantly from random base usage (3 df, P < 2.2 × 10−16 for all 14 motif positions).
Alignment of these 10 221 high-quality P-element insertion sites in the D. melanogaster genome revealed an optimal 14-bp palindromic target motif with the consensus sequence ATRGTCCGGACWAT (Figure 1). This 14-bp palindromic TSM for the P-element is consistent with the 14-bp palindromic hydrogen bonding pattern reported for an independent set of insertions from the EP screen (21), but differs from the originally reported 8-bp nonpalindromic P-element TSM [GGCCAGAC, (16)]. When oriented with respect to insertion sites on the positive strand, the center of the TSM is offset to the right of the insertion site (position 0), starting at position −3 and extending to position +10, since the P-element endonuclease makes a staggered cut with an 8-nt 3′ overhang upon integration. The central 8 nt of this motif represent the target site duplication (TSD) generated by P-element upon integration (16). The lowest information content positions in the motif directly flanking the core TSD base pairs where the P-element endonuclease cleaves DNA, and the highest information content site are at the termini of the motif (positions −3 and 10). In contrast to previous work (21), we find strong statistical support for a clear consensus sequence: all columns in the 14-bp motif deviate significantly from the overall base composition of the D. melanogaster genome sequence (A = T = 29%, G = C = 21%; 14 χ2-tests, 3 df, all P < 2.2 × 10−16) (Figure 1). We note that an important property of this staggered-cut palindromic TSM model is that each target site has two distinct insertion sites, one each on the positive and negative strands.
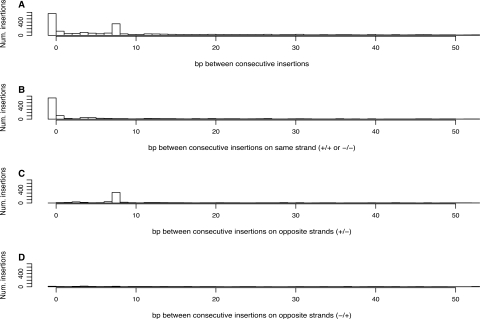
The palindromic target site model predicts nonrandom local spacing of P-element insertions
Under a model of a palindromic target site with a staggered cut, we reasoned that there would be hotspot target sites in the genome, into which multiple P-elements integrate either in the same insertion site on the same strand, or into the two different insertion sites on opposite strands. If such ‘opposite-strand’ hotspot target sites exist in the genome, they are predicted to have a characteristic pattern of local spacing of 8 bp between consecutive insertions, with one insertion on the positive strand followed by the next insertion on the negative strand. Figure 2 shows the distribution of distances between consecutive P-element insertions for all insertions, and for consecutive insertions on the same strand (+/+ and −/−) or opposite strands (+/− or −/+). The local spacing between P-element insertions shows a clear tendency for the P-element to insert with either a distance of zero or 8-bp apart (Figure 2A). Consistent with the prediction of the palindromic target site model, the excess of 0-bp distances are only found between consecutive insertions on the same strand (+/+ or −/−) (Figure 2B), while the excess of 8-bp distances are found only between consecutive insertions on the +/− opposite strand configuration (Figure 2C) but not the −/+ opposite strand configuration (Figure 2D).
Figure 2.
Nonrandom local spacing reveals two types of P-element insertion hotspot. (A) Distances, in base pairs (bp), between all consecutive P-element insertions in the genome. (B) Distances between consecutive P-element insertions on the same strand (+/+ or −/−), showing same-strand hotspots at a distance of 0 bp. (C and D) Distances between consecutive P-element insertions on opposite strands (+/− or −/+), showing opposite-strand hotspots at a distance of 8 bp. Note that the x-axis has been truncated at 50 bp in all three panels for clarity.
The excess of 0-bp distances on the same strand is consistent with previous findings that the P-element often inserts into the exact same base pair in the genome (16,33,41,42). However, in contrast to previous reports that suggested insertion can occur in either strand at the same nucleotide (16,41), we find that the overwhelming majority (375/392, 95.6%) of insertion sites with more than one insertion at the same nucleotide occur on the same strand. A tendency for P-element inserts to be spaced 8-bp apart on opposite strands has not been reported previously, and is uniquely predicted under the 14-bp palindromic target site model for P-element integration, but not under a model of random target site selection. These results also reveal that there are in fact two types of hotspot target sites for P-element insertion at the nucleotide level (i) those that have multiple insertions into the exact same coordinate on the same strand and (ii) those that have multiple insertions into sites spaced exactly 8-bp apart on opposite strands in the +/− configuration. Moreover, the relative proportions of insertions into the two types of hotspot target sites (655 same strand: 351 opposite strand) are consistent with random strand integration, which are expected to occur in a 2:1 same-strand:opposite-strand ratio if the strand at a hotspot is chosen randomly (binomial test, P = 0.299).
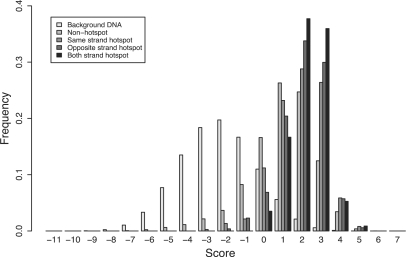
The palindromic target site model predicts hotspots for P-element insertion
If the palindromic motif in Figure 1 is a biologically meaningful representation of P-element target site preferences, we predict (i) that the observed P-element target sites should match the 14-bp motif better than background DNA sequences in the genome, and (ii) that hotspot target sites should match the 14-bp motif better than nonhotspot target sites. As found for the 14-bp hydrogen bonding pattern in (21), P-element target sites have significantly higher scoring matches to the palindromic TSM relative to the distribution of scores for all possible target sites in the genome (Mann–Whitney U-test, P < 2.2 × 10−16) (Figure 3). We extend this finding to show that hotspot target sites for P-element insertion have better motif scores than nonhotspot target sites. This is true for all hotspot types: the 375 same-strand hotspot target sites, the 221 opposite-strand hotspots, and the 98 target sites that are hotspots by both criteria, all match the palindromic TSM better than the 9208 target sites that are hit only once (Mann–Whitney U-tests, P < 2.2 × 10−16). In general, we observe that the rank order of median motif scores for the four classes of target sites is: nonhotspots < same-strand hotspots < opposite-strand hotspots < both-strand hotspots.
Figure 3.
The 14-bp palindromic TSM discriminates P-element insertion sites, hotspots and background DNA. Shown are the distributions of log-likelihood scores of the 14-bp palindromic TSM relative to random background base composition for nontarget site background DNA, nonhotspot target sites with one insertion and hotspot target sites with more than one insertion. See main text for details on different types of hotspots.
The palindromic nature of the TSM raises the question of whether hotspots for P-element insertion might be influenced by whether a target site is simply a good palindrome or specifically a good match to the target site sequence. For example, if complementary substitutions occurred at corresponding positions (e.g. 1 and 14) of an optimal target site, the target site would remain a perfect palindrome but deviate by two substitutions from the optimal target motif. To evaluate whether hotspots are more influenced by match to the target sequence or palindromicity, we tested for associations between the number of insertions per target site with motif score and/or palindrome score. In this analysis, we pooled all insertions from either same-strand and/or opposite-strand hotspots into the same target site giving a dataset of 9902 nonredundant target sites. We found a highly significant positive correlation of number of hits per target site with motif score (Spearman's correlation, ρ = 0.154; P < 2.2 × 10−16) and weak positive correlation with palindrome score (Spearman's correlation, ρ = 0.029; P = 0.003). We also evaluated the partial correlation of each score since motif score and palindrome score are also positively correlated with each other (Spearman's correlation, ρ = 0.216; P < 2.2 × 10−16), and found that the motif score, given the palindrome score, remains significantly associated with the number of hits per target site (Spearman's partial correlation, ρ = 0.151, P < 2.2 × 10−16), but not the converse (Spearman's partial correlation, ρ = −0.0038; P = 0.70). These results indicate that the match to the optimal target motif is more important in determining the frequency of P-element insertion than being a good palindrome.
No strand bias for P-element insertion
Because of the base pair complementarity of double-stranded DNA, matches to any palindromic motif should be distributed equally on both strands of the genome sequence, regardless of the motif sequence, genome-wide base composition or degree of mismatch allowed to the optimal motif. As expected under the palindromic insertion model, roughly equal proportions of P-elements insert into the positive and negative strands for all reliable mapped families of P-element (Table 2). Slight differences from the expected 50%:50% ratio for a particular family is consistent with a small degree of experimental or computational error in strand mapping. Across all families, we find that 5434 of the 10 860 (50.04%) P-element insertions that are mapped to a single base pair are found on the positive strand, which is not statistically different from the expected proportion of 50% (binomial test, P = 0.9464). The lack of strand bias for the P-element is consistent with previous results showing that the distribution of insertion sites for the Caenorhabditis elegans Tc1 transposon is the same on the positive and negative strands (43).
Evidence against sequential half-site recognition of palindromic target sites
As noted previously, matches to a palindromic motif score equally on both DNA strands, which raise the question: given a match to a full target site, how does the P-element determine which of the two possible strands to insert into if matches to the whole motif are equivalent on both strands? As has been suggested previously for other transposons (8,23–25), the existence of a palindromic TSM for the P-element is consistent with the action of a homo-multimeric transposase complex recognizing the target site. Biochemical evidence suggests that the P-element transposase acts in a tetrameric complex during donor excision (9,10), and therefore it is plausible that a multimeric complex is retained in the transposome during target integration. Under this model, we reasoned that the choice of strand might be mediated by sequential recognition of the half sites by protomers of the multimeric transposome complex, which could lead to one strand of the DNA providing a better match to the 7-bp half-site motif. For example, if the first protomer recognized the better half site, this could coordinate the transposome complex and lead to integration on the strand with the higher 5′ half-site score. This scenario would allow for symmetry breaking of the full 14-bp palindromic target motif, and provide a mechanism for predictable strand selection. Since only 47 of all possible 414 14-bp sequences (0.006%) are perfect palindromes, the vast majority of possible target sites break perfect palindrome symmetry and lead to a clear strand prediction.
To test if the half-site recognition model predicts the strand of P-element integration, we evaluated the difference in scores between the 5′ and 3′ half of each insertion site. We found no difference in the half-site scores that would support a model of half-site symmetry breaking through monomer recognition, with 49.9% (5105/10 221) of insertion sites having a better 5′ half-site score and 49.7% (5090/10 221) having a better 3′ half-site score, and only 0.2% (26/10 221) having equivalent 5′ and 3′ half-site scores. Thus, we conclude that the mechanism of P-element strand selection is inconsistent with a sequential half-site recognition model, but is consistent with simultaneous multimer recognition and random strand integration. The inability to find evidence for predictable strand integration supports the unbiased genome-wide strand mappings and the relative proportions of same- and opposite-strand hotspots reported above. Together, these results are consistent with a model of random strand selection during target site integration, which parallels the random choice of which termini is chosen first in the P-element donor excision reaction (11).
DISCUSSION
Understanding the mechanisms that control transposable element insertion and persistence in genomic DNA is a fundamental challenge in genome biology. Here, we have used patterns of P-element insertion in the D. melanogaster genome to provide evidence for a staggered-cut palindromic target site recognition model for DNA transposon insertion, which has implications for both evolutionary and functional genomics. Consistent with other previous large-scale analyses by Liao et al. (21), we have found that the P-element targets a 14-bp palindromic TSM. We find evidence that the palindromic motif has a clear consensus sequence, whereas Liao et al. (21) argued that it is a structural motif based on patterns of hydrogen bonds. As structure and sequence are intimately related at the DNA sequence level, we make no claim about which of these factors is causal. We have also shown that the local, nonrandom pattern of P-element spacing is uniquely predicted by the palindromic TSM, and that match to the TSM is a better predictor of P-element insertion frequency than palindromicity itself. We have further shown that there is no local or genome-wide strand bias for P-element insertion, consistent with a model of random strand integration. We conclude that staggered-cut palindromic target site model is a sufficient to explain the insertion preferences of the D. melanogaster P-element and, together with the widespread occurrence of staggered-cut palindromic target sites in disparate taxa, suggest that this model may apply generally to other cut-and-paste DNA transposons as well.
Our main findings are unlikely to be affected by systematic biases in our dataset since we have chosen to analyze large families of P-element insertions that have hallmarks of being accurately mapped to genome coordinates. However, some of our results, such as the relatively small difference in the score distribution of hotspot and nonhotspot target sites or the relatively low correlation of the motif score with the number of insertions per target site, can in part be explained by the fact that many P-element screens aimed to create nonredundant set of insertions for each gene in the genome (30,31,33). Thus, many additional target sites in our dataset are actually hotspots for P-element insertion but are observed to have only a single insertion. Despite this bias, the partitioning of all target sites into hotspot and nonhotspot sites is conservative with respect to the null hypothesis that there is no difference between these categories in their similarity to the TSM. We also note that since the P-element TSM is constructed from a nonredundant set of insertions, an increase in the score of same-strand hotspots is not biased by multiple insertions at the same target site being represented multiply in the motif alignment. However, because opposite-strand hotspots were an unexpected result of our analysis, we did not consider these insertions as redundant in our original set of insertion sites. Thus, insertions from opposite-strand hotspots are represented multiply in the motif alignment, and are expected to be biased towards higher match scores, as observed. Nevertheless, the same-strand hotspot results clearly demonstrate that the palindromic motif has explanatory power to discriminate P-element target sites from background and to discriminate hotspots from nonhotspot target sites.
Implications of the staggered-cut palindromic transposon target site model
Our analysis of the P-element target site preferences, along with a growing body of evidence from other DNA transposons (Table 1), suggests a general model for target site selection. The main feature of this model is that the optimal target site is a palindromic sequence/structural motif, which contains within it a staggered cut that is smaller than the length of the full target motif. This model has several implications including that (i) sequences flanking the TSD are important for the target site selection (44,45), (ii) the target site is not the same as the TSD and (iii) each target site has two distinct insertion sites on the positive and negative strands. Furthermore, since a palindromic motif is distributed equally on the positive and negative strand across the genome, (iv) DNA transposons are expected to insert with equal frequency on both strands. This last property of the palindromic target site model justifies the null hypothesis of studies that attempt to infer the postinsertion effects of natural selection from biases in transposon orientation in genome sequences (46,47). Importantly, we do not claim that all cut-and-paste DNA transposons use a staggered-cut palindromic target site that conforms to this model, rather that this model may represent the general ancestral mechanism with exceptions viewed as derived evolutionary states. For example, under one transposition pathway, the Tn7 transposon has strong insertion preferences for a nonpalindromic target site (CCCCGCT) adjacent to its recognition sequence attTn7 (48). However, unlike most transposons that encode a single transpose gene, Tn7 is unusual in that it encodes multiple transpose proteins that work in hetero-multimeric complexes that vary according to different transposition pathways (48). Other DNA transposons such as piggyBac recognize an invariant staggered-cut palindromic target site sequence (TTAA), which does not depend on any flanking DNA sequences. This exception may be explained by the facts that piggyBac uses a divergent transposase that shares little sequence similarity with other DDE transposases (49) and uses an unusual DNA synthesis-independent mechanism of target site integration (50).
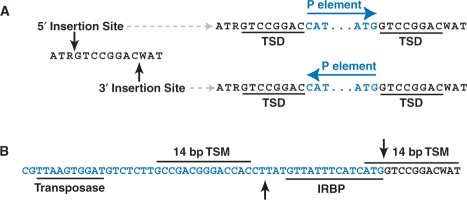
One key feature of the palindromic target site model we propose is that transposon integration destroys the original target site, leaving the TSD on both ends of the transposon, but only the 5′ flanking nucleotides at the 5′-end, and vice versa. In the case of the P-element target site, the central 8 bp is duplicated plus 3 bp of the target site nucleotides on either the 5′- and 3′-ends. Intriguingly, we observe that the terminal 3 bp flanking the TSD at the 5′ (ATR…) and 3′ (…WAT) end of the target site motif exhibit sequence complementary to the terminal 3 bp of the 3′ (…ATG) and 5′ (CAT…) ends, respectively, of the P-element TIRs (Figure 4A). Thus, although P-element integration destroys the complete 14-bp motif, the first 3 nt of the TIR sequences inserted into the target site by the P-element complement the missing part of the target motif at both ends of the insertion. Because of the palindromic nature of the TIRs, complementation occurs regardless of whether the 5′ or 3′ insertion site is used and the P-element is consequently orientated in the 5′ or 3′ direction.
Figure 4.
Model of P-element sequences in the context of the palindromic target site. Genomic sequences are shown in black, P-element sequences are shown in blue and cut sites for transposase activity are shown as black arrowheads. (A) The terminal three nucleotides of the P-element inverted repeats restore and complement the optimal target sequences flanking the TSD. Specifically, the terminal 3 bp flanking the TSD at the 5′ (ATR…) and 3′ (…WAT) end of the TSM are complementary to the terminal 3 bp of the 3′ (…ATG) and 5′ (CAT…) ends, respectively, of the P-element TIRs. Note that this occurs on both ends of the P-element regardless of whether the 5′ or 3′ insertion site is used and the resulting orientation of the P-element insertion. (B) TSMs in the P-element terminal repeat and the target site flank the 17-bp staggered cut sites for donor excision. Shown also are the positions of binding sites for transposase and the IRBP. Only the 3′ terminus of the P-element is shown for clarity, and a similar configuration exists in inverted orientation at the 5′ terminus.
Complementation and restoration of the destroyed P-element target site suggests a mechanistic link between staggered-cut palindromic target sites and the structure of the TIR transposons, specifically involving the terminal nucleotides of the TIRs. In the case of the P-element, biochemical evidence shows a close association between the P-element transposase and the last two P-element nucleotides during donor excision (9). Moreover, a special role for terminal nucleotides in the P-element TIRs may explain the strong conservation of only the first 3 nt of the TIRs among P-element family members in insects and vertebrates (51), and the widespread conservation of the first and last two nucleotides (5′-CA…TG-3′) across diverse transposon families (52,53). The possibility that P-element sequences may complement and restore their target sites may also explain why P-elements continue to favor a target site even if when there is a preexisting insertion (17,34,54,55), effectively allowing a nonlethal insertion to regenerate a ‘safe-haven’ for other insertions. Moreover, since the P-element TIRs provide the optimal sequences in the restored TSM, P-element insertion is expected to always improve the original TSM and make it more likely to be a hotspot. Finally, multiple insertions into the same target site are predicted to be in inverted orientation and separated by exactly eight bp, as has been demonstrated for the unstable singedweak allele (41). The singedweak allele is also hypermutable and undergoes reversion by precise excision of one P-element or the other at a high rate (41), and thus the recurrent targeting to safe-haven hotspots may increase subsequent rates of P-element remobilization.
The potential significance of target site complementation by the P-element termini is strengthened by the fact that high scoring sites for the TSM are found at positions 20–33 and 2875–2888 of the 5′ and 3′ P-element terminal inverted repeats, respectively, just internal to the inverted repeat binding protein (IRBP) site (56) (Figure 4B). These sites are in the upper 25th percentile of the distribution of target site scores, and are likely to be bona fide target sites since genetic evidence has revealed that a hotspot for P-element insertion exists at bp 19–26 of the P-element itself (57). Since the P-element termini each carry one target site and the target site is duplicated and complemented at each end after insertion, four high scoring target sites (two at each end) are available for transposase activity at a donor site, which is fully consistent with the action of a tetrameric complex during donor excision (9,10). Additionally, the two high scoring TSMs at each end of the integrated P-element closely flank the 17-bp staggered cut sites (Figure 4B), suggesting that the transposase complex may be coordinated to its cut sites during donor excision by the two TSMs.
More practically, the destruction of the full target site on integration requires analysis of preintegration (not postintegration) target sequences to determine true transposon target site preferences. Additionally, if terminal TIR sequences can partially complement postintegration target sites, it may be difficult to determine whether sequences at the termini of a single transposon insertion are part of the TIR or the target site. This issue was raised previously for the Tc3 transposon, and resolved by changing the sequences of target sites (58). In fact, this ambiguity between TIRs and staggered-cut palindromic target sites may underscore the functional connections between the TIR structures of transposons and their target site sequences.
The palindromic target site model can confirm annotated transposon insertion sites
Although a palindromic target site model cannot predict the strand of a transposon insertion given a target site, it can be used to confirm the strand of an insertion site given its correctly annotated location in the genome. This is because under a staggered-cut palindromic model, transposons do not insert into the center of their target site. Therefore, an insertion at a given nucleotide position in the genome is generated by two different target sites on the positive and negative strand that can have different motif scores. To demonstrate the utility of this property of our model, we scored each P-element insertion site in our dataset at the two potential target sites on either strand that would give rise to an insertion at this nucleotide to see if the higher scoring target site confirms the annotated strand in FlyBase. Remarkably, we found that the top-scoring strand under our palindromic motif model confirms the annotated strand for 90.4% (9243/10 221) of P-element insertion sites in our dataset, confirming the high quality genome mappings of the four families analyzed here. The inability to perfectly confirm the annotated strand P-element insertion given its location is consistent with a probabilistic mechanism for the choice of P-element strand integration and/or some residual error in the genome mappings in our dataset.
In contrast, we confirmed the strand for only 67.1% (3823/5694) of the remaining insertion sites mapped to a single base pair from other P-element screens omitted from our analysis (Supplementary File 3). We interpret this result to indicate that upwards of 20% of these P-elements from other families may have incorrect strand or coordinate mappings in D. melanogaster genome annotation, and is the primary reason these families not analyzed here. These errors likely arise from multiple sources, as shown by differences in the sequence logos (Supplementary File 4) and the rate of strand confirmation in the three most abundant P-element families not included in our study—EP, GawB and LacW (31–33). For example, the GawB insertions show the correct sequence logo on the positive strand, but the logo appears to be shifted by 1 nt on the negative strand, indicating a subtle difference in coordinate systems on the positive and negative strands. This is also reflected in the fact that we confirmed positive strand insertions at the same rate (91%, 976/1072) as accurately mapped families above, but we confirmed negative strand insertions at a much lower rate (66%, 681/1031). In contrast, the EP family logos show much reduced information content, a shift in logos for both positive and negative strand insertions, and a lower rate of strand confirmation on the positive strand (53.6%, 543/1012) than on the negative strand (72.9%, 621/852). The lacW family appears to be the most poorly mapped set of insertions with nearly all insertions (86%, 508/589) mapped to the positive strand, logos that do not resemble any of the other families, and low rates of strand confirmation on both the positive and negative strands. The degree of these potential errors in coordinate or strand mapping are unknown but could have important consequences for use of these P-element collections by Drosophila researchers, including the misexpression of the incorrect neighboring locus for EP-elements mapped to the incorrect strand.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The University of Manchester.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sam Griffiths-Jones, Stefan Roberts and members of the Bergman Lab for stimulating discussion throughout the project; and Don Rio, Roger Hoskins, Hugo Bellen and two anonymous reviewers for helpful comments on the article.
REFERENCES
- 1.Craig NL. Mobile DNA II. Washington, DC: ASM Press; 2002. [Google Scholar]
- 2.Mates L, Izsvak Z, Ivics Z. Technology transfer from worms and flies to vertebrates: transposition-based genome manipulations and their future perspectives. Genome Biol. 2007;8(Suppl. 1):S1. doi: 10.1186/gb-2007-8-s1-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Venter JC. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 4.Ross-Macdonald P, Coelho PS, Roemer T, Agarwal S, Kumar A, Jansen R, Cheung KH, Sheehan A, Symoniatis D, Umansky L, et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- 5.Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, Akiyama K, Kamiya A, Ito T, Shinozaki K. A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 2004;37:897–905. doi: 10.1111/j.1365.313x.2004.02009.x. [DOI] [PubMed] [Google Scholar]
- 6.Rubin GM, Kidwell MG, Bingham PM. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell. 1982;29:987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- 7.Ryder E, Russell S. Transposable elements as tools for genomics and genetics in Drosophila. Brief Funct. Genomic Proteomic. 2003;2:57–71. doi: 10.1093/bfgp/2.1.57. [DOI] [PubMed] [Google Scholar]
- 8.Rio DC. In: Mobile DNA II. Craig N, editor. Washington, DC: ASM Press; 2002. pp. 484–518. [Google Scholar]
- 9.Beall EL, Rio DC. Transposase makes critical contacts with, and is stimulated by, single-stranded DNA at the P element termini in vitro. EMBO J. 1998;17:2122–2136. doi: 10.1093/emboj/17.7.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang M, Cecconi C, Bustamante C, Rio DC. Analysis of P element transposase protein-DNA interactions during the early stages of transposition. J. Biol. Chem. 2007;282:29002–29012. doi: 10.1074/jbc.M704106200. [DOI] [PubMed] [Google Scholar]
- 11.Tang M, Cecconi C, Kim H, Bustamante C, Rio DC. Guanosine triphosphate acts as a cofactor to promote assembly of initial P-element transposase-DNA synaptic complexes. Genes Dev. 2005;19:1422–1425. doi: 10.1101/gad.1317605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beall EL, Rio DC. Drosophila P-element transposase is a novel site-specific endonuclease. Genes Dev. 1997;11:2137–2151. doi: 10.1101/gad.11.16.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berg CA, Spradling AC. Studies on the rate and site-specificity of P element transposition. Genetics. 1991;127:515–524. doi: 10.1093/genetics/127.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelley MR, Kidd S, Berg RL, Young MW. Restriction of P-element insertions at the Notch locus of Drosophila melanogaster. Mol. Cell Biol. 1987;7:1545–1548. doi: 10.1128/mcb.7.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green MM. Genetic instability in Drosophila melanogaster: de novo induction of putative insertion mutations. Proc. Natl Acad. Sci. USA. 1977;74:3490–3493. doi: 10.1073/pnas.74.8.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Hare K, Rubin GM. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983;34:25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- 17.Tower J, Karpen GH, Craig N, Spradling AC. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preston CR, Sved JA, Engels WR. Flanking duplications and deletions associated with P-induced male recombination in Drosophila. Genetics. 1996;144:1623–1638. doi: 10.1093/genetics/144.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrell J, Modolell J. The Drosophila extramacrochaetae locus, an antagonist of proneural genes that, like these genes, encodes a helix-loop-helix protein. Cell. 1990;61:39–48. doi: 10.1016/0092-8674(90)90213-x. [DOI] [PubMed] [Google Scholar]
- 20.Bellen HJ, Kooyer S, D’Evelyn D, Pearlman J. The Drosophila couch potato protein is expressed in nuclei of peripheral neuronal precursors and shows homology to RNA-binding proteins. Genes Dev. 1992;6:2125–2136. doi: 10.1101/gad.6.11.2125. [DOI] [PubMed] [Google Scholar]
- 21.Liao GC, Rehm EJ, Rubin GM. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2000;97:3347–3351. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Julian AM. M.Sc. Thesis, College Station: Texas A&M University Press; 2003. Use of bioinformatics to investigate and analyze transposable element insertions in the genomes of Caenorhabditis elegans and Drosophila melanogaster and into the target plasmid PGDV1. [Google Scholar]
- 23.Halling SM, Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982;28:155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- 24.Davies CJ, Hutchison C.A., III Insertion site specificity of the transposon Tn3. Nucleic Acids Res. 1995;23:507–514. doi: 10.1093/nar/23.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu WY, Derbyshire KM. Target choice and orientation preference of the insertion sequence IS903. J. Bacteriol. 1998;180:3039–3048. doi: 10.1128/jb.180.12.3039-3048.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haren L, Ton-Hoang B, Chandler M. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 1999;53:245–281. doi: 10.1146/annurev.micro.53.1.245. [DOI] [PubMed] [Google Scholar]
- 27.Wu X, Li Y, Crise B, Burgess SM, Munroe DJ. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J. Virol. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korswagen HC, Durbin RM, Smits MT, Plasterk RH. Transposon Tc1-derived, sequence-tagged sites in Caenorhabditis elegans as markers for gene mapping. Proc. Natl Acad. Sci. USA. 1996;93:14680–14685. doi: 10.1073/pnas.93.25.14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preclin V, Martin E, Segalat L. Target sequences of Tc1, Tc3 and Tc5 transposons of Caenorhabditis elegans. Genet. Res. 2003;82:85–88. doi: 10.1017/s0016672303006359. [DOI] [PubMed] [Google Scholar]
- 30.Spradling AC, Stern DM, Kiss I, Roote J, Laverty T, Rubin GM. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc. Natl Acad. Sci. USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi S, Ito K, Sado Y, Taniguchi M, Akimoto A, Takeuchi H, Aigaki T, Matsuzaki F, Nakagoshi H, Tanimura T, et al. GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis. 2002;34:58–61. doi: 10.1002/gene.10137. [DOI] [PubMed] [Google Scholar]
- 33.Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryder E, Blows F, Ashburner M, Bautista-Llacer R, Coulson D, Drummond J, Webster J, Gubb D, Gunton N, Johnson G, et al. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004;167:797–813. doi: 10.1534/genetics.104.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 36.Drysdale RA, Crosby MA. FlyBase: genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 41.Roiha H, Rubin GM, O’Hare K. P element insertions and rearrangements at the singed locus of Drosophila melanogaster. Genetics. 1988;119:75–83. doi: 10.1093/genetics/119.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shilova VY, Garbuz DG, Myasyankina EN, Chen B, Evgen’ev MB, Feder ME, Zatsepina OG. Remarkable site specificity of local transposition into the Hsp70 promoter of Drosophila melanogaster. Genetics. 2006;173:809–820. doi: 10.1534/genetics.105.053959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Luenen HG, Plasterk RH. Target site choice of the related transposable elements Tc1 and Tc3 of Caenorhabditis elegans. Nucleic Acids Res. 1994;22:262–269. doi: 10.1093/nar/22.3.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bender J, Kleckner N. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. Proc. Natl Acad. Sci. USA. 1992;89:7996–8000. doi: 10.1073/pnas.89.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dietrich CR, Cui F, Packila ML, Li J, Ashlock DA, Nikolau BJ, Schnable PS. Maize Mu transposons are targeted to the 5′ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics. 2002;160:697–716. doi: 10.1093/genetics/160.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 47.Cutter AD, Good JM, Pappas CT, Saunders MA, Starrett DM, Wheeler TJ. Transposable element orientation bias in the Drosophila melanogaster genome. J. Mol. Evol. 2005;61:733–741. doi: 10.1007/s00239-004-0243-0. [DOI] [PubMed] [Google Scholar]
- 48.Peters JE, Craig NL. Tn7: smarter than we thought. Nat. Rev. Mol. Cell Biol. 2001;2:806–814. doi: 10.1038/35099006. [DOI] [PubMed] [Google Scholar]
- 49.Sarkar A, Sim C, Hong YS, Hogan JR, Fraser MJ, Robertson HM, Collins FH. Molecular evolutionary analysis of the widespread piggyBac transposon family and related “domesticated” sequences. Mol. Genet. Genomics. 2003;270:173–180. doi: 10.1007/s00438-003-0909-0. [DOI] [PubMed] [Google Scholar]
- 50.Mitra R, Fain-Thornton J, Craig NL. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008;27:1097–1109. doi: 10.1038/emboj.2008.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammer SE, Strehl S, Hagemann S. Homologs of Drosophila P transposons were mobile in zebrafish but have been domesticated in a common ancestor of chicken and human. Mol. Biol. Evol. 2005;22:833–844. doi: 10.1093/molbev/msi068. [DOI] [PubMed] [Google Scholar]
- 52.Collins JJ, Anderson P. The Tc5 family of transposable elements in Caenorhabditis elegans. Genetics. 1994;137:771–781. doi: 10.1093/genetics/137.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee I, Harshey RM. Patterns of sequence conservation at termini of long terminal repeat (LTR) retrotransposons and DNA transposons in the human genome: lessons from phage Mu. Nucleic Acids Res. 2003;31:4531–4540. doi: 10.1093/nar/gkg499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang P, Spradling AC. Efficient and dispersed local P element transposition from Drosophila females. Genetics. 1993;133:361–373. doi: 10.1093/genetics/133.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Timakov B, Liu X, Turgut I, Zhang P. Timing and targeting of P-element local transposition in the male germline cells of Drosophila melanogaster. Genetics. 2002;160:1011–1022. doi: 10.1093/genetics/160.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rio DC, Rubin GM. Identification and purification of a Drosophila protein that binds to the terminal 31-base-pair inverted repeats of the P transposable element. Proc. Natl Acad. Sci. USA. 1988;85:8929–8933. doi: 10.1073/pnas.85.23.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eggleston WB. Ph.D. Thesis, Madison: University of Wisconsin Press; 1990. P element transposition and excision in Drosophila: interactions between elements. [Google Scholar]
- 58.van Luenen HG, Colloms SD, Plasterk RH. The mechanism of transposition of Tc3 in C. elegans. Cell. 1994;79:293–301. doi: 10.1016/0092-8674(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 59.Hallet B, Rezsohazy R, Mahillon J, Delcour J. IS231A insertion specificity: consensus sequence and DNA bending at the target site. Mol. Microbiol. 1994;14:131–139. doi: 10.1111/j.1365-2958.1994.tb01273.x. [DOI] [PubMed] [Google Scholar]
- 60.Tenzen T, Ohtsubo E. Preferential transposition of an IS630-associated composite transposon to TA in the 5′-CTAG-3′ sequence. J. Bacteriol. 1991;173:6207–6212. doi: 10.1128/jb.173.19.6207-6212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu WY, Thompson W, Lawrence CE, Derbyshire KM. Anatomy of a preferred target site for the bacterial insertion sequence IS903. J. Mol. Biol. 2001;306:403–416. doi: 10.1006/jmbi.2000.4421. [DOI] [PubMed] [Google Scholar]
- 62.Kumar A, Seringhaus M, Biery MC, Sarnovsky RJ, Umansky L, Piccirillo S, Heidtman M, Cheung KH, Dobry CJ, Gerstein MB, et al. Large-scale mutagenesis of the yeast genome using a Tn7-derived multipurpose transposon. Genome Res. 2004;14:1975–1986. doi: 10.1101/gr.2875304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seringhaus M, Kumar A, Hartigan J, Snyder M, Gerstein M. Genomic analysis of insertion behavior and target specificity of mini-Tn7 and Tn3 transposons in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:e57. doi: 10.1093/nar/gkl184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kivistik PA, Kivisaar M, Horak R. Target site selection of Pseudomonas putida transposon Tn4652. J. Bacteriol. 2007;189:3918–3921. doi: 10.1128/JB.01863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goryshin IY, Miller JA, Kil YV, Lanzov VA, Reznikoff WS. Tn5/IS50 target recognition. Proc. Natl Acad. Sci. USA. 1998;95:10716–10721. doi: 10.1073/pnas.95.18.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ito T, Motohashi R, Kuromori T, Noutoshi Y, Seki M, Kamiya A, Mizukado S, Sakurai T, Shinozaki K. A resource of 5,814 dissociation transposon-tagged and sequence-indexed lines of Arabidopsis transposed from start loci on chromosome 5. Plant Cell Physiol. 2005;46:1149–1153. doi: 10.1093/pcp/pci112. [DOI] [PubMed] [Google Scholar]
- 67.Fernandes J, Dong Q, Schneider B, Morrow DJ, Nan GL, Brendel V, Walbot V. Genome-wide mutagenesis of Zea mays L. using RescueMu transposons. Genome Biol. 2004;5:R82. doi: 10.1186/gb-2004-5-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guimond N, Bideshi DK, Pinkerton AC, Atkinson PW, O’Brochta DA. Patterns of Hermes transposition in Drosophila melanogaster. Mol. Genet. Genomics. 2003;268:779–790. doi: 10.1007/s00438-002-0800-4. [DOI] [PubMed] [Google Scholar]
- 69.O’Brochta DA, Warren WD, Saville KJ, Atkinson PW. Interplasmid transposition of Drosophila hobo elements in non-drosophilid insects. Mol. Gen. Genet. 1994;244:9–14. doi: 10.1007/BF00280181. [DOI] [PubMed] [Google Scholar]
- 70.Metaxakis A, Oehler S, Klinakis A, Savakis C. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics. 2005;171:571–581. doi: 10.1534/genetics.105.041848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Granger L, Martin E, Segalat L. Mos as a tool for genome-wide insertional mutagenesis in Caenorhabditis elegans: results of a pilot study. Nucleic Acids Res. 2004;32:e117. doi: 10.1093/nar/gnh111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vigdal TJ, Kaufman CD, Izsvak Z, Voytas DF, Ivics Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 2002;323:441–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- 73.Carlson CM, Dupuy AJ, Fritz S, Roberg-Perez KJ, Fletcher CF, Largaespada DA. Transposon mutagenesis of the mouse germline. Genetics. 2003;165:243–256. doi: 10.1093/genetics/165.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.