Abstract
Friedreich ataxia (FRDA) is caused by hyperexpansion of GAA•TTC repeats located in the first intron of the FXN gene, which inhibits transcription leading to the deficiency of frataxin. The FXN gene is an excellent target for therapeutic intervention since (i) 98% of patients carry the same type of mutation, (ii) the mutation is intronic, thus leaving the FXN coding sequence unaffected and (iii) heterozygous GAA•TTC expansion carriers with ∼50% decrease of the frataxin are asymptomatic. The discovery of therapeutic strategies for FRDA is hampered by a lack of appropriate molecular models of the disease. Herein, we present the development of a new cell line as a molecular model of FRDA by inserting 560 GAA•TTC repeats into an intron of a GFP reporter minigene. The GFP_(GAA•TTC)560 minigene recapitulates the molecular hallmarks of the mutated FXN gene, i.e. inhibition of transcription of the reporter gene, decreased levels of the reporter protein and hypoacetylation and hypermethylation of histones in the vicinity of the repeats. Additionally, selected histone deacetylase inhibitors, known to stimulate the FXN gene expression, increase the expression of the GFP_(GAA•TTC)560 reporter. This FRDA model can be adapted to high-throughput analyses in a search for new therapeutics for the disease.
INTRODUCTION
Friedreich ataxia (FRDA), a severe autosomal recessive neurodegenerative disease, is the most frequent inherited ataxia with a prevalence of one in 30 000–50 000 Caucasians (1,2). FRDA is caused by hyperexpansion of GAA•TTC repeats in the first intron of the FXN gene. Normal alleles contain <30 triplets while disease-causing expanded FXN alleles have from 66 to ∼1700 GAA•TTC repeats (1,3,4). The elongated GAA•TTC sequence suppresses the expression of the FXN gene, causing a deficiency of frataxin that leads to a serious imbalance in mitochondrial iron metabolism (5). The amount of frataxin detected in FRDA patient cells varies between 5% and 30% of the frataxin level found in unaffected individuals (1,6–8). Since the coding sequence of the FXN gene in FRDA patients does not carry mutations, alleviating the transcriptional block imposed by the trinucleotide repeats is an attractive target for therapeutic intervention. Importantly, asymptomatic, heterozygous carriers of the GAA•TTC expansion have ∼40–50% of normal levels of FXN mRNA and protein concentrations (8,9). Hence, an increase in frataxin expression to levels found in carriers or even a modest enhancement of frataxin production may have a therapeutic effect.
Two major mechanisms of transcription inhibition by long GAA•TTC repeats have been postulated (10–12). First, the expanded GAA•TTC repeats can adopt non-B DNA structures such as triplexes, bitriplexes and/or sticky DNA (13–17) as well as stable DNA•RNA hybrid conformations (18–20). These structures can affect many aspects of DNA metabolism such as replication, recombination and genome stability (12). Long tracts of GAA•TTC were demonstrated to inhibit transcription in vitro and in cell cultures (14,18,19,21). Second, recent studies suggest that FXN gene silencing is induced by expanded GAA•TTC repeats via chromatin modifications that are characteristic of heterochromatin (9,22–24). Heterochromatin hallmarks such as reduced level of histone H3 and H4 acetylation accompanied by an increased trimethylation of lysine 9 in histone H3, were especially apparent immediately downstream and upstream of an expanded repeat tract, whereas the FXN promoter did not show significant chromatin alterations (9,23–25).
A limited number of molecules that reverse transcriptional silencing of the FXN have been described (10,11,26). Research in the field of FRDA therapeutics is substantially hampered by a lack of appropriate models of expanded GAA•TTC alleles for screening compound libraries or approved drugs. Current methods of monitoring changes in frataxin mRNA and protein expression, based predominantly on quantitative PCR and western blots, are laborious and inefficient, especially for high-throughput applications. Additionally, low levels of the FXN mRNA and frataxin present in the FRDA cells make the quantitative analyses difficult and error-prone.
Several human lymphoblast cell lines, derived from FRDA patients and the expansion carriers, are currently available. Studies related to pathogenesis of FRDA have also been conducted using primary lymphocytes derived from patients as well as using RNAi-induced frataxin knockdown cells (9,27,28). Although existing cell-based models are valuable for discovering new aspects of FRDA pathogenesis and for evaluating the efficacy of pre-selected compounds that act on the endogenous FXN gene, they are not adequate for high-throughput analyses. Two reporter FRDA cell lines designed for compound screening have been described (29,30). Due to either short GAA•TTC repeats and/or their location within the reporter construct, neither of these cell lines fully recapitulates the molecular defects of endogenous expanded FXN genes.
To accelerate the discovery of new FRDA therapeutics and to study the molecular pathways involved in repeat-induced gene silencing, we designed, constructed and validated a cell line containing a GFP reporter minigene with an intronic (GAA•TTC)560 tract. This reporter construct recapitulates many of the characteristics of the endogenous expanded FXN gene such as reduced mRNA and protein levels, patterns of chromatin modifications, and repeat instability. Expression of the minigene is stimulated by compounds known to increase levels of frataxin in cells from FRDA patients. This molecular model of FRDA can be utilized in high-throughput screening of large compound libraries in a search for new pharmacological agents with potential therapeutic benefits in FRDA.
MATERIALS AND METHODS
Construction of the GFP_(GAA•TTC) cell lines
pRW5656, a derivative of pcDNA5/FRT/TO (Invitrogen), was constructed by cloning the GFP gene containing the adenovirus exon (Ad2) from pGFP-Ad2_wt (31) into the NotI/KpnI sites of the vector. Subsequently, a polylinker, containing Bsu36I and BssHII recognition sequences, was cloned into the XcmI site of the intron followed by removal of the Ad2 exon by PmlI and EcoRV digestion, creating pGFP_Int.
pRW3823 (13) was a source of the (GAA•TTC)270 tract while the longest sequence (560 GAA•TTC repeats) was obtained using PCR from genomic DNA isolated from the GM16210 cell line (NIGMS Human Genetic Cell Repository at The Coriell Institute for Medical Research, Camden, NJ, USA). Long GAA•TTC tracts were amplified as described (1). A cell line harboring (GAA•TTC)70 repeats was a product of spontaneous repeat deletion, which occurred most likely during integration of a plasmid containing 560 repeats into the HEK293Flp-InT-Rex cells (Invitrogen). The plasmid DNA as well as the PCR product were cleaved by Bsu36I and BssHII endonucleases (recognition sites are present in the sequences flanking the repeats in the intron 1 of the FXN gene) and ligated into the Bsu36I/BssHII cleaved pGFP_Int. Plasmids containing full-length GAA•TTC repeats as determined using polyacrylamide gel electrophoresis of the excised inserts were site specifically integrated into the genome of HEK293Flp-InT-Rex cells (Invitrogen) creating four cell lines containing the GFP minigene: GFP_(GAA•TTC)0, GFP_(GAA•TTC)70, GFP_(GAA•TTC)270 and GFP_(GAA•TTC)560.
Correct integrants were selected using 200 μg/ml hygromycin according to the manufacturer's recommendations. Individual hygromycin-resistant colonies were isolated using cloning discs (Fisher), expanded and analyzed for repeat size and GFP expression level. All constructs were sequenced prior to as well as after establishing the stable cell lines.
Cell culture
HEK293Flp-InT-Rex cells were cultured in DMEM medium (Sigma) supplemented with 10% FBS, 2 mM l-glutamine, 100 U each of penicillin and streptomycin, 200 µg/ml hygromycin B and 5 µg/ml of blastocidin (Invitrogen). Cells were grown at 37°C in 5% CO2. Transfection of HEK293Flp-InT-Rex cells was carried out using Lipofectamine2000 (Invitrogen) in OptiMEM medium (Invitrogen) according to the supplier's instructions. GM16210 (FRDA, (GAA•TTC)580/580), GM15850 (FRDA, (GAA•TTC)650/1030) and GM15851 (unaffected control) lymphoblast cells were propagated in RPMI 1640 medium with 2 mM l-glutamine, 15% FBS and 100 U of penicillin and streptomycin at 37°C in 5% CO2.
PCR and qRT-PCR
Amplification of long GAA•TTC repeats was carried out using previously described primers (2500F, 629) and conditions (1). PCR products were cleaved using appropriate restriction endonucleases and purified using agarose gel electrophoresis (Qiagen).
The length of the GAA•TTC tract integrated into the HEK293Flp-InT-Rex cell lines was analyzed using intronic primers 5′CTTCCCTTTACACAACGTTTGGGTT3′ and 5′GTACTGTTTGGATTCAGTGAGGGACT3′. The level of GFP expression was determined using quantitative reverse transcription PCR (qRT-PCR) with primers complementary to exons 1 and 2 of this gene (5′GCGACGTAAACGGCCACAAGTT3′; 5′ATGCCCTTCAGCTCGATGCGGT3′). The same primer pair was used for analysis of the splicing of the GFP intron. QRT-PCR was carried out using a Stratagene Mx3005P system and Brilliant® II SYBR® Green QPCR Master Mix (Stratagene). In the qRT-PCR experiments, GAPDH was used for normalization (primers 5′GAAGGTGAAGGTCGGAGTC3′ and 5′GAAGATGGTGATGGGATTTC3′). All qRT-PCR analyses were carried under the identical conditions of 95°C for 20 s, 55°C for 30 s and 72°C for 1 min; 40 cycles. Reverse transcription PCR to analyze splicing of the GFP mRNA was modified by extending the elongation step of the reaction to 5 min. Total RNA was isolated using PureLink™ Micro-to-Midi™ Total RNA Purification System (Invitrogen). Genomic DNA was extracted with a GFX Genomic DNA Purification Kit (GE Healthcare).
Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described (9). For each immunoprecipitation experiment, the amount of lysate corresponding to 25–50 μg of total DNA was incubated with one of the following antibodies: anti-H3K9ac (07-352), anti-H3K14ac (07-353), anti-H4K5ac (07-327), anti-H4K8ac (07-328) and anti-H3K9me3 (17-625). All antibodies were from Upstate Biotechnology/Millipore. Samples were quantitated in triplicate by quantitative PCR (qPCR) using the standard curve method. The primers used in this study were: for the region upstream of the GAA•TTC repeats in the GFP intron 5′AATAGCCTCCTGACCACAGATCCTT3′ and 5′CCATGTGACATCTAGCCCGCA3′; for the region downstream of the GAA•TTC repeats in the GFP intron, 5′ACAGCAAAGACTGGAGCAACCCAT3′ and 5′CCCCATGAGAACCCACAGTGTT3′; and, for GAPDH, 5′CACCGTCAAGGCTGAGAACG3′ and 5′ATACCCAAGGGAGCCACACC3′. Statistical calculations were conducted using SigmaPlot 2000.
RESULTS
Construction of a reporter minigene containing long, intronic GAA•TTC repeats
Since expanded GAA•TTC repeats are located in intron 1 of the FXN gene, ∼1.5 kb downstream of the first exon, we constructed a set of reporter minigenes based on a eukaryotic variant of the GFP gene, with different numbers of GAA•TTC repeats in an artificial intron derived from the rat Pem1 gene (31) (Figure 1). Repeats present in GFP intron 1 are located 1.2 kb from the exon/intron junction. The minigenes were integrated by site-specific recombination into the genome of the HEK293Flp-InT-Rex cell line (see Materials and methods section). The use of identical sites of integration for different constructs allows for direct comparison between cell lines, eliminating any potential bias resulting from random integration events in different chromosomal contexts.
Figure 1.
Schematic diagram of the FXN and GFP genes containing intronic GAA·TTC repeats. (A) Structure of the FXN gene (5′-region) indicating the position of the trinucleotide repeats within intron 1. Expansions longer than (GAA•TTC)66 repeats lead to FRDA. Bu and Bh designate recognition sites for Bsu36I and BssHII restriction enzymes, respectively. These endonucleases were used to clone GAA•TTC tracts into the GFP intron. (B) Structures of the GFP_(GAA•TTC) minigenes. The GFP gene, expressed under control of the CMV promoter and the tetracycline operator/repressor (TetO2), was divided into two exons separated by a 1.7-kb intron. The GAA•TTC tracts of different lengths (0, 70, 270 and 560 repeats) were cloned into the Bsu36I and BssHII restriction sites 1.2 kb from the 5′-end of the intron 1. These minigenes were site-specifically integrated into the HEK293Flp-InT-Rex cells creating a set of GFP_(GAA•TTC) reporter cell lines harboring different numbers of GAA•TTC repeats.
Four cell lines were created that differ in the number of repeats present in the intron of the GFP gene: GFP_(GAA•TTC)560, GFP_(GAA•TTC)270, GFP_(GAA•TTC)70 and a control cell line without repeats (GFP_(GAA•TTC)0). In all cases, the expression of the GFP minigene is under the control of the tetracycline inducible CMV promoter, with the tetracycline repressor encoded in the genome of the HEK293Flp-InT-Rex cells (Figure 1).
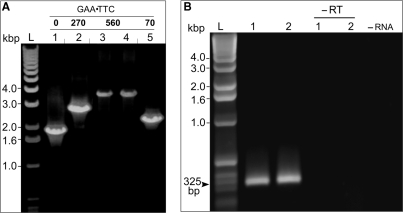
Long GAA•TTC repeats are inherently unstable during propagation in prokaryotic cells (32–35). Even short-term culturing of cells containing plasmids with tracts longer than ∼(GAA•TTC)100 repeats leads to significant deletions within the repetitive sequence (32,33). In our experiments, 40–50% of plasmids designed to contain the (GAA•TTC)270 insert and >90% of plasmids intended to harbor the (GAA•TTC)560 tract resulted in deleted repeats after a short culture (∼15 generations) in Escherichia coli HB101. However, the remaining 10% of the undeleted plasmid (in the case of the (GAA•TTC)560 tract) was sufficient for site-specific integration into the HEK293Flp-InT-Rex genome (Supplementary Figure 1). The size of GAA•TTC repeats in genome-integrated minigenes was determined using PCR. In the majority of cases, no changes in repeat size were observed during the integration process. We detected only one spontaneous, large deletion (from 560 to 70 GAA•TTC repeats) in the 15 individual clones we analyzed (Figure 2A, lane 5).
Figure 2.
Integration of the GFP_(GAA•TTC) minigenes into the HEK293Flp-InT-Rex cells. (A) PCR analyses of lengths of the GAA•TTC repeats in the minigenes integrated into the HEK293Flp-InT-Rex cells. (B) Analyses of splicing of the GFP mRNA using cDNA PCR. The splicing product of the minigene containing 560 GAA•TTC repeats (lane 1) is identical to the splicing product of the GFP_(GAA•TTC)0 minigene (lane 2). Both the size and sequence analyses confirmed the correct splicing pattern of the GFP mRNA exons. Control reactions were conducted without reverse transcriptase (–RT) or RNA template (–RNA).
Prior studies suggest that the expanded GAA•TTC repeats in the intron 1 of the FXN gene do not influence the splicing of the FXN pre-mRNA (18). On the other hand, long trinucleotide repeating sequences, including GAA•TTC repeats, are known to affect splicing and pre-mRNA processing when inserted into the intron of reporter minigenes (36,37). Therefore, we analyzed whether the GFP_(GAA•TTC)560 mRNA is properly processed. Reverse transcription PCR showed a single 325-bp band corresponding to the predicted spliced product between exons 1 and 2 of the GFP gene (Figure 2B). Using PCR conditions allowing for amplification of long cDNA fragments, we were unable to detect any additional mRNA resulting from aberrant or incomplete splicing of the GAA•TTC containing GFP intron (e.g. mRNA with nonspliced intron; data not shown). These studies demonstrate that long GAA•TTC repeats can be integrated into the genome of human cells and, when located in an intron, these repeats do not interfere with splicing pattern of our GFP reporter minigene. We cannot entirely exclude a possibility that potential aberrant splicing products are unstable, thus beyond our detection threshold.
Stable transmission of the long GAA•TTC repeats
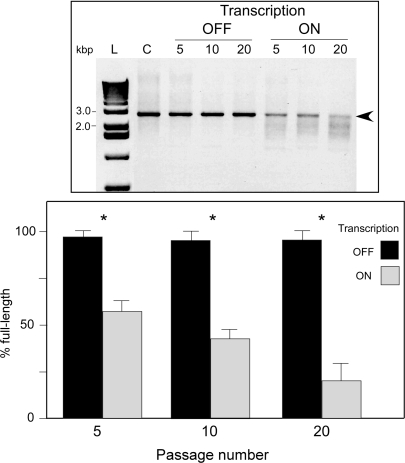
Long GAA•TTC tracts have been shown to exhibit both intergenerational and somatic instability in human cells (38–40) which can impose a serious challenge to generate a reliable cellular model of any repeat expansion disorder. Since transcription through repeats is one of the most potent inducers of their instability (41–44), we analyzed whether transcription can influence the stability of the (GAA•TTC)560 tract. We cultured the GFP_(GAA•TTC)560 cell line for 5, 10 and 20 passages (one passage ≈ 2.3 population doublings) in the presence (transcription on) and in the absence (transcription off) of tetracycline (0.1 μg/ml). Repeat length analyses revealed a very strong effect of transcription on the GAA•TTC tract instability (Figure 3). Similar to cultured human lymphocytes, a strong bias for contractions was observed (38). Culturing the cells for only five passages resulted in >50% reduction of the amount of the full-length (GAA•TTC)560 insert (Figure 3). After 20 passages, <20% of the PCR products contained GAA•TTC tract of the original length. However, we did not observe significant (GAA•TTC)560 tract instability in the absence of transcription (Figure 3). Although transcription of the GFP_(GAA•TTC)560 minigene is significantly reduced compared to the GFP_(GAA•TTC)0 control construct (see below), these results demonstrate that even relatively low levels of transcription strongly stimulate the instability of long GAA•TTC repeats. Therefore, to maintain stability of the GFP_(GAA•TTC)560 minigene, we blocked reporter gene expression by culturing in the absence of tetracycline.
Figure 3.
Effect of transcription on instability of the (GAA•TTC)560 tract in GFP_(GAA•TTC)560 minigene. Cell line harboring GFP_(GAA•TTC)560 minigene was cultured for 5, 10 and 20 passages (12, 24 and 48 population doublings) in the presence or absence of tetracycline (0.1 µg/ml). Repeat region was amplified using PCR and the lost of the full-length (GAA•TTC)560 band (indicated by arrowhead) quantitated. Asterisks indicate statistically significant differences (P < 0.01).
Long GAA•TTC repeats induce transcriptional silencing of the reporter gene
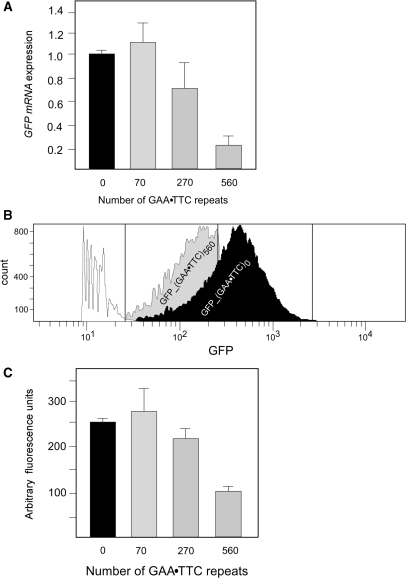
To determine whether GAA•TTC repeats located in the intron of our reporter minigene effectively silence its expression, we used quantitative RT-PCR to measure the level of GFP mRNA in the GFP_(GAA•TTC)70, GFP_(GAA•TTC)270 and GFP_(GAA•TTC)560 cell lines relatively to the GFP mRNA detected in GFP_(GAA•TTC)0 cells. The longest tract of (GAA•TTC)560 led to 4- to 5-fold reduction of the GFP mRNA (Figure 4A). The expression of GFP_(GAA•TTC)270 was only mildly affected (∼25% decrease as compared to the GFP_(GAA•TTC)0). Insertion of 70 intronic GAA•TTC repeats had no effect on the minigene transcription level (Figure 4A). We also analyzed the effects of the GAA•TTC repeat tracts on GFP levels measuring fluorescence using a plate reader and by FACS (Figure 4B and C). The GFP_(GAA•TTC)560 cell line exhibited 2.5-fold less fluorescence than cells harboring the GFP_(GAA•TTC)0. In contrast, GFP fluorescence detected in the cells expressing minigenes containing 70 and 270 GAA•TTC repeats was not statistically different from the fluorescence observed in the control GFP_(GAA•TTC)0 cells.
Figure 4.
Long intronic GAA•TTC repeats inhibit GFP expression. (A) Results of the qRT-PCR analysis of the GFP mRNA expression in four cell lines harboring 70, 270 and 560 repeats relative to the GFP_(GAA•TTC)0 cell line without GAA•TTC repeats in the GFP gene. (B) Analysis of the GFP expression using fluorescence activated cell sorter (FACS). Approximately 50 000 cells for each cell line were analyzed. HEK293Flp-InT-Rex cells (white) were used as a control. (C) Analysis of the GFP expression using a fluorescence plate reader. Data are collected from duplicate analyses of two 96-well plates per cell line.
In summary, we demonstrated that (GAA•TTC)560 located in the intron of the reporter gene significantly inhibits the expression of the minigene. The level of transcriptional silencing observed for the GFP_(GAA•TTC)560 reporter is similar to the transcriptional inhibition found in FRDA patients. These results show that long intronic GAA•TTC repeats, isolated from their natural chromosomal and FXN gene contexts, are sufficient to induce molecular consequences typical of the GAA•TTC expansions in the FRDA cells.
Gene silencing is induced by heterochromatin-specific histone modifications
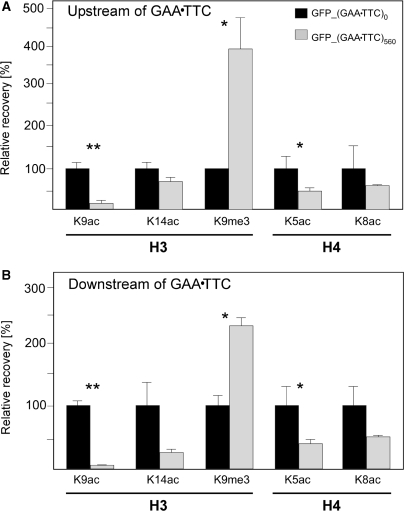
Since transcriptional silencing of pathological FXN alleles is associated with pronounced changes in the acetylation and methylation of histones present in the vicinity of expanded GAA•TTC repeats (9,23,24), to validate our reporter construct, we analyzed histone modifications in the regions immediately upstream and downstream of the GAA•TTC tracts in the GFP_(GAA•TTC)0 and GFP_(GAA•TTC)560 cell lines. ChIP followed by qPCR (Figure 5) revealed that acetylation of histone H3 lysine 9 and 14 (H3K9 and H3K14) in the region immediately upstream of the GAA•TTC repeats was decreased in the GFP_(GAA•TTC)560 cells compared to the cells without the repeats. Similarly, levels of acetylation of H4K5 and H4K8 were reduced in the vicinity of long repeats. In addition, changes in H3K9 acetylation were accompanied by a 3-fold increase in trimethylation of K9 (Figure 5A). Analogous chromatin changes were detected in the region downstream of the GAA•TTC repeats (Figure 5B). Thus, posttranslational histone modifications observed in the vicinity of the GAA•TTC repeats in GFP_(GAA•TTC)560 minigene are consistent with chromatin changes observed in the FXN gene in FRDA cells.
Figure 5.
Histone modifications induced by long GAA•TTC repeats. Chromatin modifications were analyzed in the GFP_(GAA•TTC)560 and GFP_(GAA•TTC)0 cell lines using the ChIP assay. Antibodies specific for acetylation and trimethylation of different lysine residues in human histones H3 and H4 were used. The relative recovery was determined using qPCR in relation to the GAPDH using primer pairs for regions immediately upstream (A) and downstream (B) of the GAA•TTC repeats. Recovery of the GFP_(GAA•TTC)0 for each antibody was set to the value of 100. Error bars indicate standard deviation from at least two independent ChIP experiments quantitated in triplicate. Asterisks indicate statistically significant differences (*P < 0.05 and **P < 0.005).
GFP_(GAA•TTC)560 minigene expression is enhanced by specific inducers of FXN transcription
A number of compounds have been shown to relieve the transcriptional silencing of the mutated FXN gene in FRDA cells, including histone deacetylase inhibitors (HDACi) (9). We analyzed the effects of different compounds on the expression of the GFP_(GAA•TTC)560 and GFP_(GAA•TTC)0 minigenes. For comparison, the influence of these compounds on FXN expression was determined in the human lymphoblast cell lines GM16210 and GM15850 which were derived from FRDA patients as well as in a control cell line GM15851 derived from an unaffected individual. Four HDAC inhibitors, HDACi 106 [N1-(2-aminophenyl)-N7-p-tolylheptanediamide (25), HDACi 4b (N1-(2-aminophenyl)-N7-phenylheptanediamide, (9)], the hydroxamic acids oxamflatin and SAHA (suberoylanilide hydroxamic acid) significantly increased (P < 0.05) the expression of the GFP_(GAA•TTC)560 reporter ∼1.4- to 2.5-fold (Table 1). Also, these compounds induced expression of the FXN gene harboring expanded GAA•TTC repeats in the two cell lines derived from FRDA patients (P < 0.05). The two benzamides HDACi 106 and 4b which had the most positive effect in FRDA cells also demonstrated the largest increases in expression of the GFP_(GAA•TTC)560 minigene. Importantly, with the exception of SAHA, stimulation of expression was specific to genes harboring GAA•TTC repeats, since these compounds did not affect expression of the GFP_(GAA•TTC)0 control minigene or expression of the FXN gene in the control GM15851 cell line (data not shown). Four other compounds tested, splitomicin, resveratrol, nicotinamide and scriptaid had no influence on the expression of the GFP_(GAA•TTC)560 and GFP_(GAA•TTC)0 minigenes or on FXN gene transcription. Hence, similarities in the response to different compounds, strongly support a common mechanism of transcriptional silencing of expanded FXN genes and the GFP_(GAA•TTC)560 minigene.
Table 1.
Effect of selected compounds on the GFP_(GAA·TTC)560 minigene and FXN gene expression
| Compound | Concentration | Increase of GFP_(GAA·TTC)560 expression (%) | SD (%) | Effect on FXN level in FRDA cell lines (%) | SDa (%) |
|---|---|---|---|---|---|
| DMSO | 0.1% | 100 | 8 | 100 | 7 |
| *HDACi 106b | 10 μM | 243 | 11 | 265 | 19 |
| *HDACi 4bb | 10 μM | 194 | 9 | 251 | 18 |
| *Oxamflatin | 1 μM | 193 | 8 | 154 | 18 |
| *SAHAb | 2.5 μM | 139 | 23 | 168 | 14 |
| Splitomicin | 20 μM | 103 | 6 | 91 | 11 |
| Resveratrol | 20 μM | 99 | 2 | 94 | 19 |
| Nicotinamide | 2 mM | 89 | 9 | 106 | 17 |
| Scriptaid | 1 μM | 77 | 21 | 93 | 25 |
| **Trichostatin Ab | 0.05 μM | 85 | 6 | 124 | 11 |
GFP expression was measured using a fluorescence plate reader. Levels of the FXN mRNA in GM16210 and GM15850 cell lines were determined using qRT-PCR. Changes of expression are presented relative to the effect of 0.1% DMSO.
aHigh SD values result from the inclusion of qRT-PCR data obtained using two different FRDA cell lines.
bThe influence of these compounds on FXN expression and/or frataxin levels was reported previously.
*Compounds significantly (P < 0.05) stimulating expression of both the GFP_(GAA•TTC)560 minigene and the mutated FXN gene.
**Trichostatin A had an inhibitory effect on the GFP_(GAA•TTC)560 minigene expression (P < 0.05), while it did not significantly affect FXN transcription in the lymphoblast cell lines.
DISCUSSION
A molecular model of FRDA was constructed, characterized and validated that recapitulates many of the molecular hallmarks of the mutated FXN allele. Also, this model was adapted for high-throughput studies related to the GAA•TTC repeat-mediated transcriptional silencing and drug discovery. Similar to the FXN gene, long GAA•TTC repeats inserted into the intron of the GFP gene significantly inhibit the expression of this reporter at both the mRNA and protein levels, as inferred from decrease in fluorescence. Analogous to the FRDA cells, silencing is linked to epigenetic changes in the vicinity of the expanded repeats. Moreover, the chromatin modifications observed in cells carrying the GFP_(GAA•TTC)560 minigene are strikingly similar to the posttranslational histone modifications found in the FXN gene harboring an expanded GAA•TTC tract. Finally, we demonstrated that the GAA•TTC-induced transcriptional silencing can be partially alleviated by the same compounds shown to stimulate FXN expression in human cell lines.
Long pathogenic GAA•TTC repeats, isolated from their natural chromosomal context of the FXN gene, are capable of inducing posttranslational changes in chromatin and efficiently inhibit expression of the reporter gene. Hence, the GAA•TTC silencing effect appears to be independent of the endogenous exonic/intronic sequences and a promoter and is attributed to the GAA•TTC repeats per se.
Pattern of chromatin modifications observed in our model cell line harboring 560 GAA•TTC repeats is characteristic for all FRDA cells and animal model systems analyzed so far (9,23–25). Although the extent of histone modifications observed in human lymphoblast cell lines, primary lymphocytes from FRDA patients and the GFP_(GAA•TTC)560 minigene differ, hypoacetylation of H3K9, H3K14, H4K5 and H4K8 as well as increased H3K9 trimethylation was consistently detected in the vicinity of the expanded repeats (9). Additionally, similar histone modifications were detected in human brain tissue from two FRDA patients, particularly downstream of the GAA•TTC tract (23). The epigenetic changes were also observed in brain tissues from transgenic mouse harboring mutated FXN gene; however, the level of hypoacetylation observed in the FRDA mouse was lower than in human samples, most likely due to the relatively short GAA•TTC tracts (∼200 repeats) (23). Despite of a significant difference in the size of GAA•TTC repeats between transgenic mouse and human FRDA samples, decreased acetylation and increased trimethylation of the H3K9 were consistently found in both systems.
The GAA•TTC repeats are one of the most abundant microsatellites found in the human and mouse genomes (45–48). More than 13 000 GAA•TTC tracts spanning on average 74 bp/Mb have been identified in the human genome with multiple loci containing polymorphic tracts longer than 100 bp (46–48). Length polymorphism of the repeating sequences plays an important role in regulation of expression and protein function providing a source of both quantitative and qualitative phenotypic variation (49–51). The repeat sequences located in a different chromosomal context may act as gene expression modifiers via length-dependent epigenetic DNA and histone modifications (52). Relatively short GAA•TTC repeats, inserted randomly into the mouse genome, confer variegation of a transgene expression (22). Other trinucleotide repeating sequences such as CTG•CAG and CGG•CCG, which are expanded in certain human neurological diseases (53), can alter chromatin status. Moderately expanded (<200 copies) CGG•CCG repeats in the 5′-UTR of the fragile X mental retardation gene (FMR1) may facilitate its increased transcription in patients with fragile X-associated tremor/ataxia syndrome (FXTAS) (54,55). On the other hand, large CGG•CCG expansions in this gene (>200 copies) are associated with histone hypoacetylation and methylation of the adjacent CpG islands resulting in transcriptional silencing of this gene (56–58). Large expansions of the CTG•CAG repeats in the congenital form of myotonic dystrophy type I are also associated with heterochromatin formation and CpG island methylation (59). Expansions of both CTG•CAG and CGG•CCG repeats also influence positioning and stability of the nucleosomes in vitro and in vivo (60–63).
The GAA•TTC repeats belong to the large group of polypurine•polypyrimidine (R•Y) sequences that are greatly overrepresented in the human genome (64). Recent studies showed that nearly 3000 R•Y tracts longer than 100 bp and more than 800 R•Y sequences longer that 250 bp exist in the human genome (64). The R•Y tracts, including GAA•TTC repeats, have a high propensity to form triplex DNA structures (12,65,66). These noncanonical DNA conformations can trigger changes in gene expression either directly, by blocking the progression of the transcriptional apparatus, or indirectly, via the posttranslational chromatin modifications (9,18,19,67).
Results of analyses conducted in vitro and in prokaryotic cells indicated that tracts of 30–100 GAA•TTC repeats significantly inhibit transcription through the repeat region (14,18–21,67). In our molecular model of FRDA, a tract of 270 GAA•TTC repeats did not considerably inhibit the expression of the reporter minigene (∼75 and 85% of the GFP_(GAA•TTC)0 protein and mRNA levels, respectively). This result is consistent with studies conducted in FRDA patients and mouse models (68), thus indicating that mammalian models of the GAA•TTC-induced transcriptional silencing are more relevant to the mechanism of the disease than in vitro or prokaryotic systems. Homozygous knock-in Frda230/230GAA mice expressed 66–83% of the wild-type levels of frataxin (68), which is similar to the amount of GFP expressed in the GFP_(GAA•TTC)270 cells. As expected, insertion of a substantially longer tract of the (GAA•TTC)560 repeats significantly reduced (∼45 and 20% of the GFP_(GAA•TTC)0 fluorescence and mRNA levels, respectively) the expression of the reporter gene as compared to the GFP_(GAA•TTC)0 cells.
The cloning and stable maintenance of long GAA•TTC repeats is a crucial step in the construction of reporter genes containing expanded repeating sequences. Tracts longer than 100 GAA•TTC repeats frequently undergo rapid deletions, particularly in plasmids cultivated in prokaryotic hosts during cloning procedures (19,32,33). Interestingly, (GAA•TTC)560 tracts were unstable even in the context of genomic DNA. A weak stimulation of transcription through the GAA•TTC repeats led to the destabilization of the repeats, whereas inhibition of transcription resulted in complete stabilization of the repeats. While transcription has been shown to stimulate CTG•CAG, GAA•TTC and CGG•CCG repeat instabilities in prokaryotic systems (33,41,42), the effect of transcription was demonstrated only for CTG•CAG repeats in eukaryotic cells (43,44). Transcription-induced GAA•TTC repeat instability observed in our model system has important implications for the progressive somatic GAA•TTC repeat instability observed in FRDA patients (39,40,69). Transcription has been postulated recently to be a major factor influencing CTG•CAG repeat instability in nondividing neuronal cells (43,44). Perhaps the same mechanism is responsible for somatic GAA•TTC instabilities observed in dorsal root ganglia (DRG) cells of the FRDA patients (39).
Studies on the molecular pathogenesis and potential therapeutic strategies for FRDA are generally conducted using lymphoblast cell lines and primary cells derived from FRDA patients. In addition, mouse models are employed to elucidate various aspects of the FRDA development. These model systems are invaluable in the analyses of selected, specific processes related to the disease etiology. On the other hand, they cannot be used in the comprehensive, high-throughput screens in search for new treatment strategies. High-throughput cell-based approaches require assays that can be quantitatively monitored using rapid and sensitive reporter systems.
Two research groups created FRDA reporter cell lines in the search for compounds capable of alleviating GAA•TTC-induced silencing. Sarsero et al. (29) generated an in-frame fusion between human FXN gene and the GFP gene. This construct, however, does not contain expanded GAA•TTC repeats, which is the hallmark of the FRDA pathogenesis and a primary genetic defect leading to the disease. The fusion reporter harbors only 6 GAA•TTC repeats and therefore it only allows for the identification of molecules that act on the wild-type promoter and not on the repeats. A second reporter cell line generated by Hebert and coworkers (30) used fragments of the FXN gene containing 15 and 148 GAA•TTC repeats fused to the GFP reporter gene. In this construct, the GAA•TTC repeats were located in the 5′-UTR of the reporter; consequently, the inhibition of the reporter gene expression likely results from the interference with translation rather than with transcription.
No effective treatment is available for FRDA at the present time. Histone deacetylase inhibitors are currently the most promising compounds for targeting the FXN gene silencing (9,25). We demonstrated that HDACIs, effective in induction of the frataxin expression, are also capable of stimulation of the GFP_(GAA•TTC)560 minigene expression. Hence, this molecular model of FRDA can be utilized to analyze large collections of compounds, in a high-throughput setting, to discover new candidate drugs capable of alleviating the GAA•TTC-mediated transcriptional repression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (ES11347 to R.D.W.); Friedreich's Ataxia Research Alliance (to R.D.W.); Seek a Miracle (to R.D.W.); the Robert A. Welch Foundation (to R.D.W.); Friedreich's Ataxia Research Alliance (to M.N.); National Ataxia Foundation (to M.N.); National Institutes of Health (R21NS055781 to J.M.G.); Friedreich's Ataxia Research Alliance (to J.M.G.); National Ataxia Foundation (postdoctoral support for E.S.). Funding for open access charge: Friedreich's Ataxia Research Alliance and National Ataxia Foundation.
Conflict of interest statement. None declared
Supplementary Material
REFERENCES
- 1.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 2.Pandolfo M. Friedreich's ataxia. In: Wells RD, Ashizawa T, editors. Genetic Instabilities and Neurological Diseases. 2nd edn. San Diego, CA: Elsevier-Academic Press; 2006. pp. 277–296. [Google Scholar]
- 3.Cossee M, Schmitt M, Campuzano V, Reutenauer L, Moutou C, Mandel JL, Koenig M. Evolution of the Friedreich's ataxia trinucleotide repeat expansion: founder effect and premutations. Proc. Natl Acad. Sci. USA. 1997;94:7452–7457. doi: 10.1073/pnas.94.14.7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montermini L, Andermann E, Labuda M, Richter A, Pandolfo M, Cavalcanti F, Pianese L, Iodice L, Farina G, Monticelli A, et al. The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum. Mol. Genet. 1997;6:1261–1266. doi: 10.1093/hmg/6.8.1261. [DOI] [PubMed] [Google Scholar]
- 5.Pandolfo M. Iron metabolism and mitochondrial abnormalities in Friedreich ataxia. Blood Cells Mol. Dis. 2002;29:536–547. doi: 10.1006/bcmd.2002.0591. [DOI] [PubMed] [Google Scholar]
- 6.Campuzano V, Montermini L, Lutz Y, Cova L, Hindelang C, Jiralerspong S, Trottier Y, Kish SJ, Faucheux B, Trouillas P, et al. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 7.Montermini L, Richter A, Morgan K, Justice CM, Julien D, Castellotti B, Mercier J, Poirier J, Capozzoli F, Bouchard JP, et al. Phenotypic variability in Friedreich ataxia: role of the associated GAA triplet repeat expansion. Ann. Neurol. 1997;41:675–682. doi: 10.1002/ana.410410518. [DOI] [PubMed] [Google Scholar]
- 8.Pianese L, Turano M, Lo Casale MS, De Biase I, Giacchetti M, Monticelli A, Criscuolo C, Filla A, Cocozza S. Real time PCR quantification of frataxin mRNA in the peripheral blood leucocytes of Friedreich ataxia patients and carriers. J. Neurol. Neurosurg. Psychiatry. 2004;75:1061–1063. doi: 10.1136/jnnp.2003.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat. Chem. Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 10.Gottesfeld JM. Small molecules affecting transcription in Friedreich ataxia. Pharmacol. Ther. 2007;116:236–248. doi: 10.1016/j.pharmthera.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebert MD. Targeting the gene in Friedreich ataxia. Biochimie. 2008;90:1131–1139. doi: 10.1016/j.biochi.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Wells RD. DNA triplexes and Friedreich ataxia. FASEB J. 2008;22:1625–1634. doi: 10.1096/fj.07-097857. [DOI] [PubMed] [Google Scholar]
- 13.Sakamoto N, Chastain PD, Parniewski P, Ohshima K, Pandolfo M, Griffith JD, Wells RD. Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich's ataxia. Mol. Cell. 1999;3:465–475. doi: 10.1016/s1097-2765(00)80474-8. [DOI] [PubMed] [Google Scholar]
- 14.Sakamoto N, Ohshima K, Montermini L, Pandolfo M, Wells RD. Sticky DNA, a self-associated complex formed at long GAA.TTC repeats in intron 1 of the Frataxin gene, inhibits transcription. J. Biol. Chem. 2001;276:27171–27177. doi: 10.1074/jbc.M101879200. [DOI] [PubMed] [Google Scholar]
- 15.Vetcher AA, Napierala M, Iyer RR, Chastain PD, Griffith JD, Wells RD. Sticky DNA, a long GAA.GAA.TTC triplex that is formed intramolecularly, in the sequence of intron 1 of the frataxin gene. J. Biol. Chem. 2002;277:39217–39227. doi: 10.1074/jbc.M205209200. [DOI] [PubMed] [Google Scholar]
- 16.Vetcher AA, Napierala M, Wells RD. Sticky DNA: effect of the polypurine.polypyrimidine sequence. J. Biol. Chem. 2002;277:39228–39234. doi: 10.1074/jbc.M205210200. [DOI] [PubMed] [Google Scholar]
- 17.Son LS, Bacolla A, Wells RD. Sticky DNA: in vivo formation in E. coli and in vitro association of long GAA*TTC tracts to generate two independent supercoiled domains. J. Mol. Biol. 2006;360:267–284. doi: 10.1016/j.jmb.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 18.Bidichandani SI, Ashizawa T, Patel PI. The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am. J. Hum. Genet. 1998;62:111–121. doi: 10.1086/301680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohshima K, Montermini L, Wells RD, Pandolfo M. Inhibitory effects of expanded GAA•TTC triplet repeats from Intron I of the Friedreich ataxia gene on transcription and replication in vivo. J. Biol. Chem. 1998;275:14588–14595. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- 20.Grabczyk E, Mancuso M, Sammarco MC. A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 2007;35:5351–5359. doi: 10.1093/nar/gkm589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krasilnikova MM, Kireeva ML, Petrovic V, Knijnikova N, Kashlev M, Mirkin SM. Effects of Friedreich's ataxia (GAA)n*(TTC)n repeats on RNA synthesis and stability. Nucleic Acids Res. 2007;35:1075–1084. doi: 10.1093/nar/gkl1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–913. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 23.Al-Mahdawi S, Pinto RM, Ismail O, Varshney D, Lymperi S, Sandi C, Trabzuni D, Pook M. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum. Mol. Genet. 2008;17:735–746. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
- 24.Greene E, Mahishi L, Entezam A, Kumari D, Usdin K. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res. 2007;35:3383–3390. doi: 10.1093/nar/gkm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rai M, Soragni E, Jenssen K, Burnett R, Herman D, Coppola G, Geschwind DH, Gottesfeld JM, Pandolfo M. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS ONE. 2008;3:e1958. doi: 10.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson RB. Experimental therapeutics for Friedreich's ataxia. In: Wells RD, Ashizawa T, editors. Genetic Instabilities and Neurological Diseases. 2nd edn. San Diego, CA: Elsevier-Academic Press; 2006. pp. 297–303. [Google Scholar]
- 27.Napoli E, Morin D, Bernhardt R, Buckpitt A, Cortopassi G. Hemin rescues adrenodoxin, heme a and cytochrome oxidase activity in frataxin-deficient oligodendroglioma cells. Biochim. Biophys. Acta. 2007;1772:773–780. doi: 10.1016/j.bbadis.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Tan G, Napoli E, Taroni F, Cortopassi G. Decreased expression of genes involved in sulfur amino acid metabolism in frataxin-deficient cells. Hum. Mol. Genet. 2003;12:1699–1711. doi: 10.1093/hmg/ddg187. [DOI] [PubMed] [Google Scholar]
- 29.Sarsero JP, Holloway TP, Li L, McLenachan S, Fowler KJ, Bertoncello I, Voullaire L, Gazeas S, Ioannou PA. Evaluation of an FRDA-EGFP genomic reporter assay in transgenic mice. Mamm. Genome. 2005;16:228–241. doi: 10.1007/s00335-004-3021-9. [DOI] [PubMed] [Google Scholar]
- 30.Grant L, Sun J, Xu H, Subramony SH, Chaires JB, Hebert MD. Rational selection of small molecules that increase transcription through the GAA repeats found in Friedreich's ataxia. FEBS Lett. 2006;580:5399–5405. doi: 10.1016/j.febslet.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc. Natl Acad. Sci. USA. 2004;101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakamoto N, Larson JE, Iyer RR, Montermini L, Pandolfo M, Wells RD. GGA*TCC interrupted triplets in long GAA*TTC repeats inhibit the formation of triplex and sticky DNA structures, alleviate transcription inhibition, and reduce genetic instabilities. J. Biol. Chem. 2001;276:27178–27187. doi: 10.1074/jbc.M101852200. [DOI] [PubMed] [Google Scholar]
- 33.Napierala M, Bacolla A, Wells RD. Increased negative superhelical density in vivo enhances the genetic instability of triplet repeat sequences. J. Biol. Chem. 2005;280:37366–37376. doi: 10.1074/jbc.M508065200. [DOI] [PubMed] [Google Scholar]
- 34.Pollard LM, Sharma R, Gomez M, Shah S, Delatycki MB, Pianese L, Monticelli A, Keats BJ, Bidichandani SI. Replication-mediated instability of the GAA triplet repeat mutation in Friedreich ataxia. Nucleic Acids Res. 2004;32:5962–5971. doi: 10.1093/nar/gkh933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollard LM, Bourn RL, Bidichandani SI. Repair of DNA double-strand breaks within the (GAA*TTC)n sequence results in frequent deletion of the triplet-repeat sequence. Nucleic Acids Res. 2008;36:489–500. doi: 10.1093/nar/gkm1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorbunova V, Seluanov A, Dion V, Sandor Z, Meservy JL, Wilson JH. Selectable system for monitoring the instability of CTG.CAG triplet repeats in mammalian cells. Mol. Cell. Biol. 2003;23:4485–4493. doi: 10.1128/MCB.23.13.4485-4493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baralle M, Pastor T, Bussani E, Pagani F. Influence of Friedreich ataxia GAA noncoding repeat expansions on pre-mRNA processing. Am. J. Hum. Genet. 2008;83:77–88. doi: 10.1016/j.ajhg.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma R, Bhatti S, Gomez M, Clark RM, Murray C, Ashizawa T, Bidichandani SI. The GAA triplet-repeat sequence in Friedreich ataxia shows a high level of somatic instability in vivo, with a significant predilection for large contractions. Hum. Mol. Genet. 2002;11:2175–2187. doi: 10.1093/hmg/11.18.2175. [DOI] [PubMed] [Google Scholar]
- 39.De Biase I, Rasmussen A, Endres D, Al-Mahdawi S, Monticelli A, Cocozza S, Pook M, Bidichandani SI. Progressive GAA expansions in dorsal root ganglia of Friedreich's ataxia patients. Ann. Neurol. 2007;61:55–60. doi: 10.1002/ana.21052. [DOI] [PubMed] [Google Scholar]
- 40.De Biase I, Rasmussen A, Monticelli A, Al-Mahdawi S, Pook M, Cocozza S, Bidichandani SI. Somatic instability of the expanded GAA triplet-repeat sequence in Friedreich ataxia progresses throughout life. Genomics. 2007;90:1–5. doi: 10.1016/j.ygeno.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 41.Mochmann LH, Wells RD. Transcription influences the types of deletion and expansion products in an orientation-dependent manner from GAC*GTC repeats. Nucleic Acids Res. 2004;32:4469–4479. doi: 10.1093/nar/gkh787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowater RP, Jaworski A, Larson JE, Parniewski P, Wells RD. Transcription increases the deletion frequency of long CTG.CAG triplet repeats from plasmids in Escherichia coli. Nucleic Acids Res. 1997;25:2861–2868. doi: 10.1093/nar/25.14.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 44.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Astolfi P, Bellizzi D, Sgaramella V. Frequency and coverage of trinucleotide repeats in eukaryotes. Gene. 2003;317:117–125. doi: 10.1016/s0378-1119(03)00659-0. [DOI] [PubMed] [Google Scholar]
- 46.Clark RM, Bhaskar SS, Miyahara M, Dalgliesh GL, Bidichandani SI. Expansion of GAA trinucleotide repeats in mammals. Genomics. 2006;87:57–67. doi: 10.1016/j.ygeno.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Subramanian S, Madgula VM, George R, Mishra RK, Pandit MW, Kumar CS, Singh L. Triplet repeats in human genome: distribution and their association with genes and other genomic regions. Bioinformatics. 2003;19:549–552. doi: 10.1093/bioinformatics/btg029. [DOI] [PubMed] [Google Scholar]
- 48.Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22:253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 50.Hammock EA, Young LJ. Microsatellite instability generates diversity in brain and sociobehavioral traits. Science. 2005;308:1630–1634. doi: 10.1126/science.1111427. [DOI] [PubMed] [Google Scholar]
- 51.Fondon JW, 3rd, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc. Natl Acad. Sci. USA. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dorer DR, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- 53.Wells RD, Ashizawa T. Genetic Instabilities and Neurological Diseases. 2nd edn. San Diego, CA: Elsevier-Academic Press; 2006. [Google Scholar]
- 54.Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, Hagerman PJ. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hagerman RJ, Hagerman PJ. The fragile X premutation: into the phenotypic fold. Curr. Opin. Genet. Dev. 2002;12:278–283. doi: 10.1016/s0959-437x(02)00299-x. [DOI] [PubMed] [Google Scholar]
- 56.Chiurazzi P, Pomponi MG, Pietrobono R, Bakker CE, Neri G, Oostra BA. Synergistic effect of histone hyperacetylation and DNA demethylation in the reactivation of the FMR1 gene. Hum. Mol. Genet. 1999;8:2317–2323. doi: 10.1093/hmg/8.12.2317. [DOI] [PubMed] [Google Scholar]
- 57.Pietrobono R, Pomponi MG, Tabolacci E, Oostra B, Chiurazzi P, Neri G. Quantitative analysis of DNA demethylation and transcriptional reactivation of the FMR1 gene in fragile X cells treated with 5-azadeoxycytidine. Nucleic Acids Res. 2002;30:3278–3285. doi: 10.1093/nar/gkf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coffee B, Zhang F, Warren ST, Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat. Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- 59.Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 60.Wang YH, Amirhaeri S, Kang S, Wells RD, Griffith JD. Preferential nucleosome assembly at DNA triplet repeats from the myotonic dystrophy gene. Science. 1994;265:669–671. doi: 10.1126/science.8036515. [DOI] [PubMed] [Google Scholar]
- 61.Wang YH, Gellibolian R, Shimizu M, Wells RD, Griffith J. Long CCG triplet repeat blocks exclude nucleosomes: a possible mechanism for the nature of fragile sites in chromosomes. J. Mol. Biol. 1996;263:511–516. doi: 10.1006/jmbi.1996.0593. [DOI] [PubMed] [Google Scholar]
- 62.Wang YH, Griffith J. Expanded CTG triplet blocks from the myotonic dystrophy gene create the strongest known natural nucleosome positioning elements. Genomics. 1995;25:570–573. doi: 10.1016/0888-7543(95)80061-p. [DOI] [PubMed] [Google Scholar]
- 63.Wang YH, Griffith J. Methylation of expanded CCG triplet repeat DNA from fragile X syndrome patients enhances nucleosome exclusion. J. Biol. Chem. 1996;271:22937–22940. [PubMed] [Google Scholar]
- 64.Bacolla A, Collins JR, Gold B, Chuzhanova N, Yi M, Stephens RM, Stefanov S, Olsh A, Jakupciak JP, Dean M, et al. Long homopurine*homopyrimidine sequences are characteristic of genes expressed in brain and the pseudoautosomal region. Nucleic Acids Res. 2006;34:2663–2675. doi: 10.1093/nar/gkl354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sinden RR. DNA Structure and Function. San Diego, CA: Academic Press; 1994. [Google Scholar]
- 66.Bissler JJ. Triplex DNA and human disease. Front. Biosci. 2007;12:4536–4546. doi: 10.2741/2408. [DOI] [PubMed] [Google Scholar]
- 67.Grabczyk E, Usdin K. The GAA.TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miranda CJ, Santos MM, Ohshima K, Smith J, Li L, Bunting M, Cossee M, Koenig M, Sequeiros J, Kaplan J, et al. Frataxin knockin mouse. FEBS Lett. 2002;512:291–297. doi: 10.1016/s0014-5793(02)02251-2. [DOI] [PubMed] [Google Scholar]
- 69.Bidichandani SI, Purandare SM, Taylor EE, Gumin G, Machkhas H, Harati Y, Gibbs RA, Ashizawa T, Patel PI. Somatic sequence variation at the Friedreich ataxia locus includes complete contraction of the expanded GAA triplet repeat, significant length variation in serially passaged lymphoblasts and enhanced mutagenesis in the flanking sequence. Hum. Mol. Genet. 1999;8:2425–2436. doi: 10.1093/hmg/8.13.2425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.