Abstract
Epitope tagging is a powerful and commonly used approach for studying the physical properties of proteins and their functions and localization in eukaryotic cells. In the case of Saccharomyces cerevisiae, it has been possible to exploit the high efficiency of homologous recombination to tag proteins by modifying their endogenous genes, making it possible to tag virtually every endogenous gene and perform genome-wide proteomics experiments. However, due to the relative inefficiency of homologous recombination in cultured human cells, epitope-tagging approaches have been limited to ectopically expressed transgenes, with the attendant limitations of their nonphysiological transcriptional regulation and levels of expression. To overcome this limitation, a modification and extension of adeno-associated virus-mediated human somatic cell gene targeting technology is described that makes it possible to simply and easily create an endogenous epitope tag in the same way that it is possible to knock out a gene. Using this approach, we have created and validated human cell lines with epitope-tagged alleles of two cancer-related genes in a variety of untransformed and transformed human cell lines. This straightforward approach makes it possible to study the physical and biological properties of endogenous proteins in human cells without the need for specialized antibodies for individual proteins of interest.
INTRODUCTION
Epitope tagging is a powerful approach that makes it possible to perform immunoprecipitation, western blot and immunocytochemistry without the need for specialized antibodies to the protein of interest. In some model organisms such as Saccharomyces cerevisiae, it is possible to epitope tag the endogenous allele of a gene and therefore to study the protein in its natural state. However, in cultured human cells, the inefficiencies of chromosomal modification have limited epitope tagging to the study of ectopically expressed transgenes.
This limitation has important practical repercussions when performing experiments using epitope tags in human cells. First, ectopically expressed transgenes are generally expressed more highly than the endogenous gene (often dramatically so). Second, transgenes generally lack features present in endogenous genes such as a natural promoter, introns, and 5′- and 3′-untranslated regions, all of which contribute to transcriptional and translational regulation. Because of these limitations, results generated using epitope-tagged expression vectors are often criticized as being possible artifacts of overexpression and/or unnatural expression and generally require confirmation with endogenous proteins.
Therefore, it has been desirable to perform epitope tagging on endogenous genes. In the genetically tractable yeast S. cerevisiae, this goal has been realized—virtually every open reading frame has been epitope tagged and studied using proteomics and related approaches (1). However, the limitations of homologous recombination-mediated genetic modification have made the prospect of epitope-tagging endogenous genes in human cells a daunting one at best.
Homologous recombination-based gene targeting in human cells was introduced by Porter and Itzhaki in 1993, and since that time ∼50 different genes (mostly cancer related) have been disrupted by homologous recombination (2,3). Though powerful, this approach has been in limited use because of the perceived technical difficulties in its execution. However, two recent advances have made the creation of modified alleles in human cells more feasible. First, Hirata et al. (4) demonstrated that the use of adeno-associated virus (AAV) vectors for delivery of targeting constructs enhanced the efficiency of gene targeting in human cells by several orders of magnitude. Porteus et al. (5) and Kohli et al. (6) also demonstrated that targeting vectors created in AAV backbones and delivered by infection have reproducibly high efficiencies of gene targeting. Second, Topaloglu et al. (7) recently described a NeoR gene cassette termed a synthetic exon promoter trap (SEPT) that made the creation of promoter trap targeting vectors a simpler and more technically tractable prospect.
Here, we build on these recent advances in human genomic modification and describe a modification of AAV-mediated human somatic cell gene-targeting technique and its application to the creation of epitope tags of endogenous genes in human cells. Importantly, the time needed to create such cell lines is comparable to the time otherwise needed to create and validate polyclonal antibodies to individual endogenous proteins.
MATERIALS AND METHODS
Creation of pAAV-SEPT-Acceptor and pAAV-TK-Acceptor
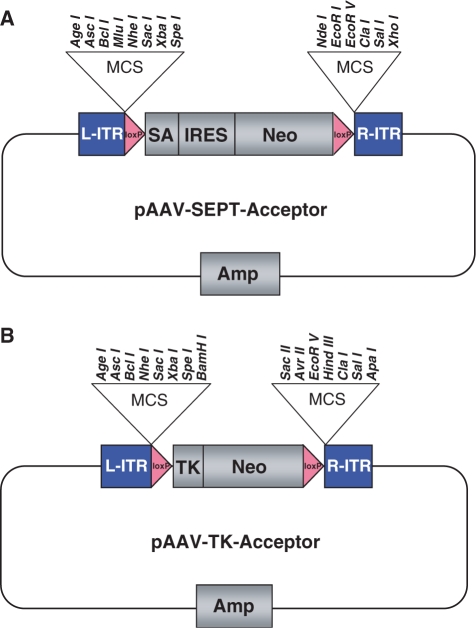
For pAAV-SEPT-Acceptor, the SEPT cassette was PCR amplified using VENT Polymerase (New England Biolabs, Beverly, MA) from the SEPT vector using primers adding consecutive restriction sites 5′ and 3′ to the cassette (7). All primers for this and other recombinant DNA steps were obtained from Integrated DNA Technologies (Coralville, IA, USA) and were PAGE purified. All primer sequences are available from the authors upon request. The 5′ cloning sites (in order 5–3′) were as follows: Not I, Age I, Asc I, Bcl I, Mlu I, Nhe I, Sac I, Xba I and Spe I. The 3′ cloning sites (similarly in order 5–3′) are as follows: Nde I, EcoRI, EcoRV, Cla I, Sal I, Xho I and Not I. The PCR product was then digested with Not I and cloned into the Not I site of pAAV-MCS (Stratagene, La Jolla, CA, USA), an AAV-2-based vector system. The integrity of the resulting construct was confirmed by restriction analysis and sequencing of the insert and junctions. The completed vector and the order of unique sites in the polylinker are depicted in Figure 1A.
Figure 1.
Acceptor vectors for endogenous epitope tagging. pAAV-SEPT-Acceptor (A) and pAAV-TK-Acceptor (B) both contain a FLOXed NeoR gene flanked by polylinkers containing unique, rare-cutting restriction sites chosen to simplify four-way ligations. L-ITR and R-ITR refer to the inverted terminal repeats required for viral packaging. pAAV-SEPT-Acceptor contains a SEPT cassette composed of a splice acceptor (SA), internal ribosome entry site (IRES) and promoterless NeoR gene. pAAV-TK-Acceptor contains a NeoR gene driven by a thymidine kinase (TK) promoter.
For pAAV-TK-Acceptor, the TK-neo cassette was PCR amplified from pMC1-neo-polyA (Stratagene) using primers containing restriction sites (sense primers Sac I, Xba I, Spe I, Bam HI, and antisense primers Sac II, Avr II, Eco RV, HindIII, Cla I) and cloned into the Sac I and Cla I sites of pAAV-SEPT-Acceptor, replacing the SEPT cassette with a TK-neo cassette. The integrity of the resulting construct was confirmed by restriction analysis and sequencing of the insert and junctions. The completed vector and the order of unique sites in the polylinker are depicted in Figure 1B.
PCR-based creation and assembly of epitope-tagging vectors
Homology arms for creation of both PTEN and p53 epitope-tagging vectors were created by PCR from a human genomic DNA template using VENT Polymerase (New England Biolabs) as described by the manufacturer. Homology arms were ∼1 kb in size. In the case of PTEN, the left arm was composed of exon I/intron I, and the right arm was composed of intron I. In the case of p53, the left arm was composed of intron I/exon II/intron II/exon III/intron III, and the right arm was composed of intron III/exon IV/intron IV. The sequence of all PCR primers is available from the authors upon request.
For both vectors, the PCR product composing the left homology arm was digested with Age I and Sac I, and the PCR product composing the right homology arm was digested with Eco RI and Sal I. All enzymes were obtained from New England Biolabs. These left and right arms were then simultaneously cloned into the pAAV-SEPT-Acceptor vector that had been digested with Age I, Sac I, Eco RI and Sal I and treated with Calf Intestinal Alkaline Phosphatase. After electroporation into DH10B cells (Invitrogen), thousands of colonies were obtained, and a subset was tested by whole-cell PCR with a junction-specific primer pair. For the PTEN vector assembly, 9 of 37 (24%) colonies contained plasmids in which the ligation had successfully occurred. For the p53 vector assembly, 19 of 45 (42%) colonies contained the expected ligation event. A subset of positive clones was further confirmed by DNA sequencing; in all cases, the junctions were correct.
Site-directed mutagenesis for addition of FLAG epitopes
Site-directed mutagenesis for the addition of FLAG epitope tags to the left homology arms of the PTEN and p53 vectors was performed using the Quikchange Kit (Stratagene) with PAGE-purified oligonucleotides designed to add an in-frame FLAG tag (IDT). A total of 22% and 42% of colonies tested by PCR and sequencing for the PTEN and p53 vectors, respectively, contained the desired 24-nt FLAG insertion (GAT/TAC/AAG/GAT/GAC/GAC/GAT/AAG encoding DYKDDDDK).
Tissue culture and identification of epitope-tagged clones
Transient stocks of AAV-2 virions were created by cotransfection of 293T cells with epitope-tagging vectors together with pAAV-RC (Stratagene) and pHELPER (Stratagene) using FUGENE 6 (Roche, Indianapolis, IN, USA) as previously described (8). Two days after transfection, media was aspirated and cell monolayers were scraped into 1 ml PBS and subjected to four cycles of freeze/thaw (consisting of 10 min freeze in a dry-ice ethanol bath and 10 min thaw in a 37°C water bath, vortexing after each thaw). The lysate was then clarified by centrifugation at 12 000 r.p.m. for 10 min in a benchtop microfuge to remove cell debris, and the virus-containing supernatant was aliquoted and stored at −80°C. No further viral purification or concentration was performed.
These virus stocks were titered by qPCR as described (9). The titer of the FLAG-p53 epitope-tagging vector was 4.0 × 1010 genome copies/ml, and the titer of the FLAG-PTEN epitope-tagging vector was 4.9 × 1010 genome copies/ml. Hundred microliters of virus was then used to infect cells in T25 tissue culture flask, and cells were passaged at limiting dilution into 96-well plates in the presence of G418 1 day after infection. The concentrations of G418 used were as follows: 0.4 mg/ml for BJ–hTERT and RPE–hTERT cells, 0.6 mg/ml for HCT116 cells and 1.0 mg/ml for LN229 and A172 cells. Individual G418-resistant clones were expanded and used for the preparation of genomic DNA using standard techniques. Clones were tested for homologous integration of the targeting vector using a primer pair specific for the targeted allele. PCR products from clones with homologous integration of the targeting vector were then sequenced to determine whether the FLAG epitope had been inserted into the genome of each cell line. All primer sequences used in these steps are available from the authors upon request.
Once individual clones were identified in this way, they were infected with a Cre-expressing adenovirus as previously described (8). Individual clones were expanded by limiting dilution and tested for the restoration of G418-sensitivity.
Preparation of protein lysates, affinity purification and western blot
Protein lysates to be used directly for western blot were prepared in RIPA buffer. Nuclear and cytoplasmic lysates used for FLAG purification were prepared using a modification of Dignam's nondetergent lysis method (10,11). Protein concentrations were quantified using the bicinchoninic assay (Pierce, Rockford, IL, USA).
For FLAG affinity purification, α-FLAG M2 beads (SIGMA, St Louis, MO, USA) were washed once with TBS, then resuspended in nuclear or cytoplasmic fractions derived from parental or epitope-tagged cells, and rotated at 4° for 1 h. Beads were then washed three times in TBS, packed into a Poly-Prep chromatography Column (Biorad, Hercules, CA) and bound proteins were eluted with 100 ng/μl FLAG peptide in three 1-ml fractions. Fractions were concentrated by TCA precipitation, resuspended in sample buffer and separated by SDS–PAGE.
Western blot was performed using standard techniques. Primary antibodies used were as follows: PTEN clone 6H2.1 (Cascade Biosciences, Winchester, MA, USA), p53 clone DO-1 (Calbiochem, San Diego, CA, USA), FLAG polyclonal F7425 (SIGMA) and HAUSP A300-033A (Bethyl Labs, Montgomery, TX, USA).
Mass spectrometry
Protein sequence analysis was performed at the Harvard Mass Spectrometry and Proteomics Resource Laboratory by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (µLC/MS/MS) on a Thermo LTQ-Orbitrap mass spectrometer. These MS/MS spectra were then correlated with known sequences (12,13).
RESULTS
Universal acceptor vectors for single-ligation assembly of AAV-based targeting vectors
One factor limiting the ease and speed of human somatic cell gene-targeting projects has been the technical challenges inherent to vector assembly. Several different approaches have been tried in an effort to simplify this process. For example, we and others have described approaches that exploit the high rate of homologous recombination in S. cerevisiae to build targeting vectors with the needed junctions without the need for conveniently located restriction sites (14). Kohli et al. (6) have employed a nested PCR strategy for the creation of targeting vectors with only a single-ligation step. However, these and other related strategies have had the disadvantages of either requiring multiple consecutive assembly steps and/or being insufficiently robust.
In an effort to remedy these difficulties, universal AAV-2 acceptor vectors were created that make it possible to perform vector assembly in a single ligation (Figure 1A and B, described in detail in Materials and Methods section). These acceptor vectors contain polylinkers with multiple unique rare-cutting restriction sites flanking a FLOXed NeoR gene, making it possible to subclone PCR-generated homology arms into the acceptor vector simultaneously.
Two such acceptor vectors were created, which were identical except that one contained a TK-neo gene for creation of so-called ‘promoter containing’ targeting vectors and the other contained a SEPT-neo gene for creation of promoter trap targeting vectors. Of note, the SEPT cassette contains a splice acceptor followed by an IRES-neo and was recently described by Topaloglu et al. (7). The specific cloning strategy employed for creation of these vectors is described in detail in Materials and Methods section.
A generalizable approach for the creation of human epitope-tagging vectors and its application to p53
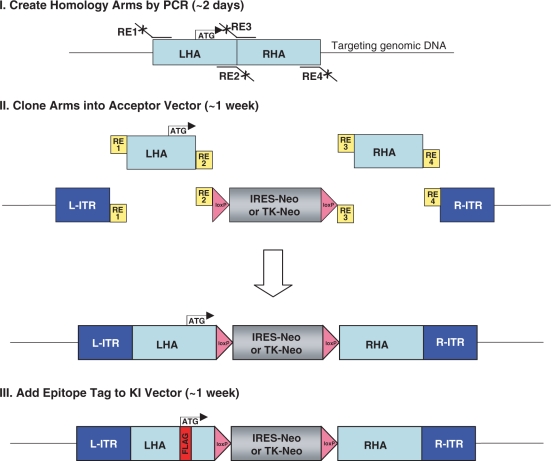
These new acceptor vectors made it theoretically possible to build targeting vectors in a single-ligation step. To do this, left and right homology arms were created by PCR from a human genomic DNA template, and then simultaneously cloned into the polylinkers that had been built into the acceptor vectors. This was successfully performed for p53 in ∼9 days, as described in Materials and Methods section and depicted in Figure 2 (Steps I and II). Importantly, this ligation step was extremely robust, and the expected recombinant plasmid was present in 42% of all tested bacterial colonies.
Figure 2.
Scheme for creation of epitope-tagging vectors. Step I: left and right homology arms (LHA and RHA) are created by PCR from a human genomic DNA template using primers tailed with restriction sites needed for subsequent cloning. Step II: the PCR products are digested and cloned simultaneously into either pAAV-SEPT-Acceptor or pAAV-TK-Acceptor that had been digested with the same enzymes and treated with calf intestinal alkaline phosphatase. Step III: the desired epitope tag is added to the LHA by site-directed mutagenesis.
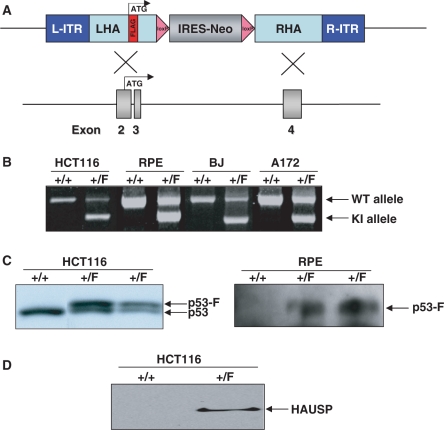
Since the left arm of the targeting vector was designed to contain the initiating methionine codon, it was possible to add an epitope tag to the sequences encoding the amino-terminus of the encoded protein. Of note, a comparison of myc, HA, FLAG and V5 epitope tags showed that FLAG provides the best combination of efficiency and specificity for immunoprecipitation (Kim,J.S. and Waldman,T. unpublished data). As such, FLAG epitopes were employed in the epitope-tagging vectors described herein. The details of the mutagenesis reaction for insertion of FLAG are described in Materials and methods section, and the scheme is depicted in Figure 2 (Step III). Of note, this reaction was extremely robust and efficient (42% of the resulting bacterial colonies contained the FLAG insertion), and took ∼1 week to perform. The resulting p53 epitope-tagging vector and the targeting event it is designed to perform are depicted in Figure 3A.
Figure 3.
Epitope tagging of p53 in human cells. (A) The FLAG-p53 epitope-tagging vector is designed to recombine into intron I/exon II/intron II/exon III/intron III of human p53, adding a FLAG epitope immediately following the initiating methionine of p53 in exon II. Subsequent Cre-mediated recombination removes the IRES-NeoR gene, leaving behind FLAG coding sequences in exon II and a single LoxP site in intron III (not shown). (B) PCR-based identification of epitope-tagged alleles. Depicted are PCR results from HCT116, RPE, BJ and A172 epitope-tagged cells. +/+ refers to homozygosity for the unmodified parental allele, whereas +/F refers to derivatives in which one allele has been modified by the addition of an in-frame amino-terminus FLAG epitope. (C) Western blot-based confirmation of epitope-tagged alleles. The left panel depicts a direct western blot using p53 antibodies on lysates from parental HCT116 cells (+/+) and two independently-derived heterozygous (+/F) FLAG-p53 epitope-tagged clones. The addition of a FLAG epitope increases the size of the encoded protein by ∼1 kDa. The right panel depicts a FLAG IP/FLAG western blot from parental RPE cells (+/+) and two independently derived epitope-tagged derivatives (+/F). (D) Identification of the p53-interacting protein HAUSP by immunoprecipitation. Nuclear extracts (125 μg) from parental HCT116 cells (+/+) and epitope-tagged derivatives (+/F) were incubated with α-FLAG M2 agarose beads followed by FLAG-peptide elution, TCA precipitation, SDS–PAGE and HAUSP western blot.
Creation of human cells with endogenous FLAG-tagged alleles of p53
Once the p53 epitope-tagging vector had been created, it was packaged into AAV virions and used to infect human cells. The four recipient cell lines chosen all harbor wild-type p53 genes and include both immortalized primary cells [hTERT-immortalized retinal pigment epithelial (RPE) cells and hTERT-immortalized BJ fibroblasts] and transformed cell lines derived from two different tumor types (HCT116 colon cancer cells and A172 glioblastoma multiforme cells). Individual G418-resistant colonies were obtained and tested by PCR for the presence of homologous integration of the epitope-tagging vector (Figure 3B). The efficiencies of targeted integration were as follows: HCT116—14%, BJ–hTERT—40%, RPE–hTERT—43%, A172—30%. PCR sequencing demonstrated that all the PCR-positive clones had undergone the desired modification in which a FLAG tag had been inserted in-frame immediately after the initiating methionine of the endogenous gene. Positive clones were infected with a Cre-expressing adenovirus as described in Materials and Methods section to remove the FLOXed NeoR gene and restore the targeted allele to its natural configuration.
Validation of human cells with epitope-tagged p53 genes
HCT116 and RPE–hTERT cells in which the endogenous p53 gene had been modified via the addition of an amino-terminus FLAG were then validated by immunoprecipitation and western blot with antibodies to p53 and FLAG. As depicted in Figure 3C, parental HCT116 cells had a single molecular weight species of p53 protein, whereas heterozygous epitope-tagged cells had equimolar amounts of two molecular weight species—the endogenous protein, and slightly larger protein reflecting the increased molecular weight caused by the addition of the FLAG tag. Similarly, IP/western blots performed with FLAG antibodies demonstrated the presence of FLAG-p53 protein in epitope-tagged RPE–hTERT cells but not in parental cells (Figure 3C).
Herpesvirus-associated ubiquitin-specific protease (HAUSP) is a recently discovered p53-binding protein that regulates its stability (15). To determine if epitope-tagged cells could be employed to confirm this interaction in HCT116 cells with endogenous proteins, nuclear lysates were prepared from parental and p53 epitope-tagged cells, immunoprecipitated using a FLAG affinity matrix and interrogated by western blot with antibodies to HAUSP. As depicted in Figure 3D, endogenous HAUSP clearly interacted with endogenous epitope-tagged p53 in HCT116 cells, demonstrating the functional integrity of the epitope-tagged allele of p53 and providing proof-of-principle for the use of cells with endogenous epitope-tagged alleles for the confirmation of endogenous protein–protein interactions.
Creation, validation and application of human cells with epitope-tagged PTEN genes
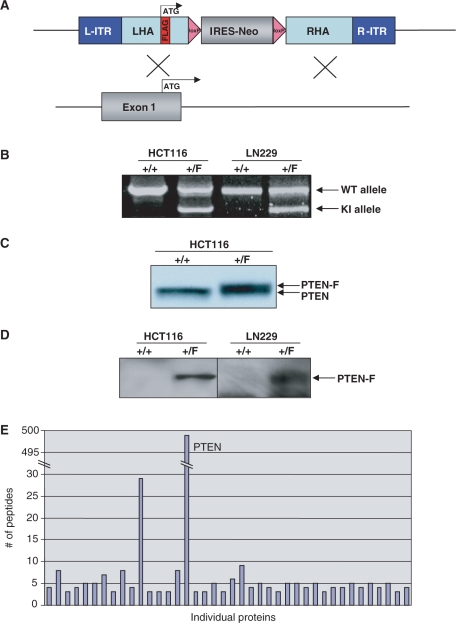
To further demonstrate the generality of the approach, an epitope-tagging vector designed to add a FLAG epitope tag to the amino terminus of the PTEN tumor suppressor gene was created (Figure 4A). The vector was packaged into AAV virions and used to infect HCT116 colon cancer cells and LN229 glioblastoma multiforme cells, both of which harbor wild-type PTEN genes. Individual G418-resistant colonies were obtained and tested by PCR for the presence of homologous integration of the vectors containing the epitope tag (Figure 4B). The efficiencies of targeted integration were as follows: HCT116—25%, LN229—4%. Positive clones were infected with a Cre-expressing adenovirus to remove the FLOXed NeoR gene and restore the targeted allele to its natural configuration.
Figure 4.
Epitope tagging of PTEN in human cells. (A) The FLAG-PTEN epitope-tagging vector is designed to recombine into exon I/intron I of human PTEN, adding a FLAG epitope immediately following the initiating methionine of PTEN in exon I. Subsequent Cre-mediated recombination would remove the IRES-NeoR gene, leaving behind FLAG-coding sequences in exon I and a single LoxP site in intron I (not shown). (B) PCR-based identification of epitope-tagged alleles. Depicted are PCR results from HCT116 and LN229 PTEN epitope-tagged cells. +/+ refers to homozygosity for the unmodified parental allele, whereas +/F refers to derivatives in which one allele has been modified by the additional of an in-frame amino-terminus FLAG epitope. (C) Western blot-based confirmation of epitope-tagged alleles. Depicted is a direct western blot with PTEN antibodies on lysates from parental (+/+) HCT116 cells and a heterozygous (+/F) FLAG-PTEN epitope-tagged clone. The addition of a FLAG epitope increases the size of the encoded protein by ∼1 kDa. (D) The left panel depicts a FLAG IP/FLAG western blot from parental HCT116 cells (+/+) and a FLAG-PTEN epitope-tagged derivative (+/F). The right panel depicts a FLAG IP/FLAG western blot from parental LN229 cells (+/+) and a FLAG-PTEN epitope-tagged derivative (+/F). (E) Mass spectrometry. FLAG immunoprecipitates from parental HCT116 cells and epitope-tagged derivatives were separated by SDS–PAGE and subjected by mass spectrometry, as described in Materials and Methods section. Depicted are the peptide counts from individual proteins that were present in immunoprecipitates from epitope-tagged cells but not in immunoprecipitates from parental cells.
HCT116 and LN229 cells in which the endogenous PTEN gene had been modified via the addition of an amino-terminus FLAG were then validated by immunoprecipitation and western blot with antibodies to PTEN and FLAG. As depicted in Figure 4C, parental HCT116 cells had a single molecular weight species of PTEN protein, whereas heterozygous epitope-tagged cells had equimolar amounts of two molecular weight species—the endogenous protein, and slightly larger protein reflecting the increased molecular weight caused by the addition of the FLAG tag. Similarly, IP/western blots performed with FLAG antibodies demonstrated the presence of FLAG-PTEN protein in epitope-tagged HCT116 cells and LN229 cells but not in parental cells (Figure 4D).
To provide proof-of-principle that cells with endogenous epitope tags could be valuable for mass spectrometric analysis of protein complexes, cytoplasmic lysates from parental and epitope-tagged cells were affinity purified using a FLAG affinity matrix. After concentration by TCA precipitation, eluents were separated by SDS–PAGE, stained with Coomassie Brilliant Blue (not shown) and gel lanes excised for comparative mass spectrometry as described in Materials and Methods section. The majority of proteins identified in this analysis were present in both samples, and correspond to well known contaminants of FLAG purification (not shown). However, 498 peptides corresponding to PTEN were present in the epitope-tagged cells, and none in the parental cells. The peptide counts for individual proteins present exclusively in FLAG-PTEN cells are depicted in Figure 4E. Interestingly, peptides corresponding to several cellular proteins other than PTEN were uniquely present in FLAG immunoprecipitates from the epitope-tagged cells. These peptides undoubtedly correspond to a combination of bona-fide PTEN interacting proteins and false positives; their confirmation and validation will be described elsewhere.
DISCUSSION
There are a number of possible uses for the stable cell lines harboring endogenous epitope-tagged genes. First, the endogenous epitope-tagged cell lines will likely prove valuable for purification of endogenous proteins and the identification of novel protein complexes. Second, cells harboring epitope-tagged endogenous alleles will be useful for validating novel protein–protein interactions discovered using other approaches (such as yeast two hybrid) when sensitive and specific antibodies to the components of the complex are unavailable. Third, such stable cell lines may prove valuable in industry for the purification of medically useful therapeutic proteins in situations where ectopically expressed proteins are either inactive or insufficiently active due to altered stoichiometry and other issues.
The creation of endogenous epitope tags is a particularly powerful use of human somatic cell gene-targeting technology. One important limitation to the use of gene targeting for the creation of complete knockouts in human cells has been the need for sequential targeting of multiple alleles. As such, attempts to create homozygous deletions of genes in human cells have been limited to the few known immortal human cell lines that are diploid (or near-diploid). In contrast, the successful application of endogenous epitope tagging only requires that a single allele be modified, making it possible to create epitope tags in virtually any diploid or aneuploid human cell line that can be cultured, infected with AAV and cloned. In fact, of the cells used in this study, RPE–hTERT, BJ–hTERT and HCT116 are near-diploid, whereas A172 and LN229 are aneuploid.
During preparation of this manuscript, Zhang et al. (16) described a related approach for the creation of endogenous epitope tags in human cells and their application to CHIP/CHIP studies. The approach they describe is different from that described here in that the epitope tag was prebuilt into the acceptor vector and as such simplifies creation of epitope-tagging vectors by a single step. However, one disadvantage of the vector architecture described by Zhang et al. is that epitope tag must be placed in the carboxyl terminus of the encoded protein. Furthermore, the vector system described herein provides the option of using the promoter trap architecture, leading to substantially higher efficiencies of targeted integration than those reported by Zhang et al. The next generation of epitope-tagging vector systems will likely use a combination of the approaches described by Zhang et al. and those described herein, and will also likely employ dual tags to avoid the contaminants common to single tag purifications.
In summary, here, we describe and implement a generalizable approach that makes it possible to create epitope tags of endogenous genes in human cells in the same time frame that would otherwise be needed to create and validate polyclonal antibodies. Further implementation and refinement of this approach may ultimately make it possible to pursue large-scale proteomics approaches in human cells in the same way as they are currently being pursued in more genetically tractable model organisms.
FUNDING
National Institutes of Health (R01CA11569, T32CA009686 to C.B.); American Cancer Society (RSG-06-191-01). Funding for open access charge: R01CA11569.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 2.Porter ACG, Itzhaki JE. Gene targeting in human somatic cells: complete inactivation of an interferon-inducible gene. Eur. J. Biochem. 1993;218:273–278. doi: 10.1111/j.1432-1033.1993.tb18375.x. [DOI] [PubMed] [Google Scholar]
- 3.Rago C, Vogelstein B, Bunz F. Genetic knockouts and knockins in human somatic cells. Nat. Protoc. 2007;2:2734–2746. doi: 10.1038/nprot.2007.408. [DOI] [PubMed] [Google Scholar]
- 4.Hirata R, Chamberlain J, Dong R, Russell DW. Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat. Biotechnol. 2002;20:735–738. doi: 10.1038/nbt0702-735. [DOI] [PubMed] [Google Scholar]
- 5.Porteus MH, Cathomen T, Weitzman MD, Baltimore D. Efficient gene targeting mediated by adeno-associated virus and DNA double strand breaks. Mol. Cell Biol. 2003;23:3558–3565. doi: 10.1128/MCB.23.10.3558-3565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B. Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses. Nucleic Acids Res. 2004;32:e3. doi: 10.1093/nar/gnh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Topaloglu O, Hurley PJ, Yildirim O, Civin CI, Bunz F. Improved methods for the generation of human gene knockout and knockin cell lines. Nucleic Acids Res. 2005;33:e158. doi: 10.1093/nar/gni160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JS, Lee C, Bonifant CL, Ressom H, Waldman T. Activation of p53-dependent growth suppression in human cells by mutations in PTEN or PIK3CA. Mol. Cell Biol. 2007;27:662–677. doi: 10.1128/MCB.00537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veldwijk MR, Topally J, Laufs S, Hengge UR, Wenz F, Zeller WJ, Fruehauf S. Development and optimization of a real-time quantitative PCR-based method for the titration of AAV-2 vector stocks. Mol. Ther. 2002;6:272–278. doi: 10.1006/mthe.2002.0659. [DOI] [PubMed] [Google Scholar]
- 10.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung SY, Malovannaya A, Wei J, O’Malley B, Qin J. Proteomic analysis of stead-state nuclear hormone receptor coactivator complexes. Mol. Endocrinol. 2006;19:2451–2465. doi: 10.1210/me.2004-0476. [DOI] [PubMed] [Google Scholar]
- 12.Eng JK, McCormick AL, Yates J.R., III. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 13.Chittum HS, Lane WS, Carlson BA, Roller PP, Lung FT, Lee BJ, Hatfield DL. Rabbit β-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry. 1998;37:10866–10870. doi: 10.1021/bi981042r. [DOI] [PubMed] [Google Scholar]
- 14.Lee C, Kim JS, Waldman T. PTEN gene targeting reveals a radiation-induced size checkpoint in human cancer cells. Cancer Res. 2004;64:6906–6914. doi: 10.1158/0008-5472.CAN-04-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Guo C, Chen Y, Shulha HP, Schnetz MP, LaFramboise T, Bartels CF, Markowitz S, Weng Z, Scacheri PC, et al. Epitope tagging of endogenous proteins for genome-wide ChIP-chip studies. Nat. Methods. 2008;5:163–165. doi: 10.1038/nmeth1170. [DOI] [PMC free article] [PubMed] [Google Scholar]