Abstract
RNA ligases participate in repair, splicing and editing pathways that either reseal broken RNAs or alter their primary structure. Here, we report the characterization of an RNA ligase from the thermophilic archaeon, Methanobacterium thermoautotrophicum. The 381-amino acid Methanobacterium RNA ligase (MthRnl) catalyzes intramolecular ligation of 5′-PO4 single-strand RNA to form a covalently closed circular RNA molecule through ligase-adenylylate and RNA-adenylylate (AppRNA) intermediates. At the optimal temperature of 65°C, AppRNA was predominantly ligated to a circular product. In contrast, at 35°C, phosphodiester bond formation was suppressed and the majority of the AppRNA was deadenylylated. Sedimentation analysis indicates that MthRnl is a homodimer in solution. The C-terminal 127-amino acid segment is required for dimerization, is itself capable of oligomeization and acts in trans to inhibit the ligation activity of native MthRnl. MthRnl can also join single-stranded DNA to form a circular molecule. The lack of specificity for RNA and DNA by MthRnl may exemplify an undifferentiated ancestral stage in the evolution of ATP-dependent ligases.
INTRODUCTION
RNA ligases catalyze the formation of phosphodiester bonds between the 5′-phosphate and 3′-hydroxyl termini of RNA via three sequential nucleotidyltransfer reactions (1). First, the ligase reacts with ATP, forming a covalent ligase–AMP complex with the release of pyrophosphate. In the second step, AMP is transferred from the ligase to the 5′-phosphate terminus of RNA to form adenylylated RNA (AppRNA). Finally, a 3′-hydroxyl group attacks the AppRNA, forming a 5′–3′ phosphodiester linkage and releasing AMP.
There are two families of RNA ligases, Rnl1 and Rnl2, which are distinguished by polynucleotide substrate specificity (2,3). Rnl1 ligases catalyze the joining of broken ends of single-stranded RNA generated by a site-specific RNA endonuclease. Bacteriophage T4Rnl1 functions to repair breaks in the anticodon loop of tRNALys (4). In yeast and plants, tRNA ligase (Trl1) participates in intron splicing (5,6). The intron is cleaved by a site-specific endonuclease that recognizes the fold of the pre-tRNA; Trl1 then joins the two halves of the tRNA. Yeast Trl1 is also responsible for nonspliceosomal splicing of mRNA in the unfolded protein response pathway (7). An Rnl1-type enzyme has been characterized in Autographa californica Baculovirus, although the biological role of this ligase is unknown (8).
The second type of RNA ligase, Rnl2, repairs breaks in double-stranded RNA. While this type of RNA ligase is found in all three phylogenetic domains (3), a biological function is firmly established only for the kinetoplastid RNA ligases (9–11). Kinetoplastid RNA ligases are involved in altering the translational reading frame of mitochondrial mRNAs by the insertion or removal of uridines, directed by a guide RNA sequence. In bacteriophage T4, a second RNA ligase (T4Rnl2) preferentially joins nicks in double-stranded RNA or RNA termini bridged together by a DNA template strand (2,3). Biochemical and structural analysis of T4Rnl2 shows that specificity for RNA is dictated by two terminal ribonucleotides on the 3′-OH side of the nick, while the rest of the nucleotides can be replaced by DNA (2,12).
T4Rnl1 and T4Rnl2 are monomeric proteins composed of two structural domains (2,13,14). The N-terminal adenylyltransferase domains of the enzymes are structurally similar to each other and contain the defining sequence motifs found in the covalent nucleotidyltransferase superfamily (15). Members of this family include ATP-dependent DNA ligases and GTP-dependent mRNA capping enzymes. In contrast, the C-terminal domain of T4Rnl1 and T4Rnl2 are structurally and functionally distinct from each other, as well as from the OB-fold of C-terminal domain found in DNA ligases and mRNA capping enzymes (2,13). Mutational analysis suggests that specificity for RNA is dictated in part by the C-terminal domain. The isolated adenylyltransferase domain of T4Rnl2 can catalyze steps 1 and 3 of the ligation reaction, but is inactive in overall nick-sealing activity and defective in binding to a nicked duplex substrate (14). Residues important for the second step of ligation were mapped within the C-terminal domain of T4Rnl2 (16). In T4Rnl1, removal of the C-terminal domain abolished specificity for tRNA ligation (17). These findings suggest that the C-terminal domain of RNA ligase is important for polynucleotide substrate recognition and specificity.
All archaeal species encode intron-containing tRNAs that are cleaved at a bulge-helix-bulge motif by a splicing endonuclease (18–21). The two halves must be joined enzymatically for the tRNA to function in protein synthesis. Several crenarchaeon pre-rRNAs are known to form circular RNA intermediates during rRNA processing, generated by intramolecular ligation events of two RNA termini (22,23). An intron has been reported in at least one protein-coding gene in the crenarchaea (24,25). The presence of bulge-helix-bulge-like motifs in pre-rRNA and pre-mRNA at the processing sites suggests that the intron sequences are removed by the same splicing endonuclease. While the processes of tRNA end-joining and rRNA circularization have been detected in cell extracts (26–30), the enzymes that catalyze the ligation reactions have not been identified.
A search for polypeptides resembling T4Rnl2 identified candidate RNA ligases from six species of archaea (3). The N-terminal segment of the putative archaeal RNA ligases contained all the defining sequence motifs of the covalent nucleotidyltransferase superfamily. Remarkably, the C-terminal segment bears no resemblance to the primary structure of any known polynucleotide ligases or capping enzymes. These findings raise questions about the evolution and substrate specificity of archaeal ligases.
Here, we characterized the RNA ligase encoded by Methanobacterium thermoautotrophicum ΔH open reading frame MTH1221, which we have named MthRnl. We demonstrated that this 381-amino acid MthRnl is a thermophilic ligase that catalyzes intramolecular ligation of single-stranded RNA through ligase-adenylylate and AppRNA intermediates. MthRnl is also able to circularize single-stranded DNA. MthRnl is a homodimer in solution, and the C-terminal segment is required for dimerization, thermoreactivity and strand-joining activity.
MATERIALS AND METHODS
Cloning, expression and purification of recombinant MthRnl
Methanobacterium thermoautotrophicum ΔH genomic DNA was used as a PCR template to amplify the MTH1221 ORF. Oligonucleotide primers were designed to introduce an NdeI restriction site 5′ of the predicted translation start codon and a BamHI site at the 3′ of the predicted stop codon. A 1.1-kb NdeI–BamHI fragment was inserted into pET16b (EMD Chemicals, Madison, Wisconsin, USA) to generate the plasmid pET-MthRnl. Using this method, the MTH1221 ORF was fused in-frame with a 20-amino acid N-terminal leader peptide containing 10 tandem histidines, under the control of a T7 RNA polymerase promoter. A C-terminal truncation mutant MthRnl (1–253) was constructed by PCR amplification using an antisense primer that introduced translation stop codon in lieu of the codon for Ala254 and a BamHI site immediately 3′ of the new stop codon. An N-terminal truncation mutant MthRnl (255–381) was constructed by PCR amplification using a sense primer that introduced a start codon in lieu of Ala 254 and NdeI site at the new start codon. The PCR products were digested with NdeI and BamHI, and then inserted into pET16b. DNA sequencing confirmed that no alterations in the inserted DNA sequence were introduced during the PCR amplification and cloning procedures.
The pET-MthRnl plasmid was transformed into Escherichia coli strain Rossetta2-(DE3) (EMD Chemicals, Madison, Wisconsin, USA). A 0.5 l culture of E. coli Rossetta2-(DE3)/pET-MthRnl was grown at 37°C in Luria-Bertani medium containing 0.1 mg ampicillin/ml until the A600 reached 0.3. Isopropyl-β-d-thiogalactopyranoside (IPTG) was then added to the culture to a final concentration of 0.3 mM. Incubation was continued at 37°C for 3 h. Cells were harvested by centrifugation and resuspended in 20 ml buffer A [50 mM Tris–HCl (pH 7.5), 250 mM NaCl, 10% sucrose]. Lysozyme was added to a final concentration of 25 µg/ml. The sample was then sonicated for 30 s. Triton X-100 was added to 0.1% final concentration and sonication was repeated. Insoluble material was removed by centrifugation. The soluble extract was then applied to a 1-ml column of Ni-NTA agarose (Qiagen, Valencia, California, USA) that had been equilibrated with buffer A containing 0.1% Triton X-100. The column was washed with the same buffer and then eluted stepwise with buffer B [50 mM Tris–HCl (pH 8.0), 250 mM NaCl, 10% glycerol] containing 0, 50, 100, 200, 500 and 1000 mM imidazole. The polypeptide composition of the column fractions was monitored by SDS–polyacrylamide gel electrophoresis (PAGE). The recombinant MthRnl protein was retained on the column and was recovered predominantly in the 200 mM fraction (∼2 mg of total protein). For expression of MthRnl (1–253) and MthRnl (255–381) proteins, E. coli cells were grown at 37°C until A600 reached 0.4. Cultures were then placed on ice for 10 min, adjusted to 2% ethanol and 0.4 mM IPTG and were further incubated at 17°C for 18 h with constant shaking. Both proteins were purified by Ni-NTA chromatography as described above. Protein concentrations were determined by the Bio-Rad dye-binding assay, using bovine serum albumin (BSA) as the standard.
Adenylyltransferase assay
Standard reaction mixtures (20 µl) containing 50 mM Tris–HCl (pH 8.0), 5 mM dithiothreitol (DTT), 0.5 mM MgCl2, 5 µM [α32P]ATP and 40 pmol of MthRnl were incubated for 5 min at 50°C, unless otherwise specified. The reactions were quenched with 1% SDS and the products were separated by SDS–PAGE. The radiolabeled ligase–AMP adduct was visualized by autoradiography of the dried gel and quantified by scanning the gel with a Storm PhosphorImager.
Ligation assay
Standard reaction mixtures (10 µl) containing 50 mM Tris–HCl (pH 6.5), 0.5 mM MgCl2, 0.5 pmol 32P-labeled RNA or DNA and T4Rnl2 or MthRnl, with or without ATP, were incubated for 30 min at indicated temperatures. Reactions were initiated by adding enzyme and terminated by the addition of 10 µl of 98% formamide/20 mM EDTA. Products were resolved on denaturing 18% polyacrylamide gel containing 7 M urea in 0.5× TBE. The extent of ligation (circular/circular + single stranded) was quantified by scanning the gel with a PhosphorImager.
Glycerol gradient sedimentation
Aliquots (30 µg) of the Ni–agarose preparations of the full-length or truncated MthRnl proteins were mixed with marker proteins (catalase, BSA and cytochrome c) in 0.1 ml of buffer B. The mixtures were layered onto 4.8 ml 15–30% glycerol gradients containing buffer C [50 mM Tris–HCl, (pH 8.0), 150 mM NaCl, 2 mM DTT, 0.05% Triton X-100]. The gradients were centrifuged in a Beckman SW50i rotor at 45 000 r.p.m. for 18 or 23 h at 4°C. Fractions (∼0.2 ml) were collected from the bottoms of the tubes. Aliquots (20 µl) were analyzed by SDS–PAGE. Polypeptides were visualized by staining with Coomassie blue dye. Aliquots (2 µl) of each fraction were assayed for adenylyltransferase activity as described above.
Ligation substrates
Synthetic 24-mer RNA (5′-AUUCCGAUAGUGCGUGUCGCCCUU) and DNA (5′-ATTCCGATAGTGCGTGTCGCCCTT) oligonucleotides were labeled with [γ-32P]ATP at the 5′-end using T4 polynucleotide kinase. Radiolabeled AppRNA was prepared by ligase-mediated AMP transfer of 32P-labeled pRNA in the presence of 1 mM ATP as described (14). Radiolabeled products were then purified by electrophoresis through a nondenaturing polyacrylamide gel.
RESULTS
Purification and adenylyltransferase activity of MthRnl
We produced the 45-kDa MthRnl as a His10-tagged fusion protein in E. coli and purified the recombinant protein by Ni–agarose chromatography (Figure 1A). The initial ligation step involves formation of a covalent enzyme–AMP intermediate (EpA). EpA formation by MthRnl was detected with high sensitivity and specificity by label transfer from [α-32P]ATP to the enzyme (Figure 1B). Note that the lower molecular weight species associated with adenylyltransferase activity was recovered in the peak Ni–agarose fraction, suggesting that some MthRnl was proteolyzed during expression in E. coli. The native size of the MthRnl was determined by sedimentation through a 15–30% glycerol gradient (Figure 1C). A single activity peak (Figure 1C) coincided with MthRnl polypeptide (data not shown). An S value of 6.1 was determined relative to protein standards sedimented in a parallel gradient, suggesting that MthRnl is a homodimer in solution.
Figure 1.
Purification and adenylyltransferase activity. (A) Purification of MthRnl. Aliquots (10 µl) of the Ni–agarose preparation of His-tagged MthRnl were resolved by SDS–PAGE followed by Coomassie blue staining. The MthRnl polypeptide is denoted by an arrow. The positions and sizes (kilodalton) of marker polypeptides are indicated on the left. (B) Adenylyltransferase activity. Aliquots (1 µl) of indicated Ni–agarose fractions were assayed for adenylyltransferase activity. A 32P-labeled ligase–AMP intermediate, denoted by an asterisk, was visualized by autoradiography of the dried gel. (C) Sedimentation analysis. MthRnl was centrifuged in a 15–30% glycerol gradient as described in Materials and methods section. Aliquots (1 µl) of the gradient fractions indicated were assayed for adenylyltransferase activity, gauged by the signal intensity of the radiolabeled MthRnl polypeptide (PSL, photo stimulatable luminescence). The peaks of the marker proteins, catalase (11.3 S), BSA (4.3 S) and cytochrome c (1.8 S), are indicated.
The adenylyltransferase activity was optimal between 55°C to 65°C and declined as the temperature was elevated to 85°C (Figure S1A). The extent of EpA formation increased in proportion with the amount of input protein (Figure S1B). EpA formation depended on a divalent cation cofactor. This requirement was satisfied by 2 mM magnesium, manganese, cobalt and, to a lesser extent, by calcium and zinc (Figure S1C). The activity was optimal between 1–2 mM MnCl2 and 0.5–1 mM MgCl2 (data not shown). EpA formation increased as a function of ATP concentration and reached saturation near 10 µM ATP (Figure S1D). Half saturation was achieved at ∼1 µM ATP. Based on the molar amount of AMP transfer versus the amount of ligase added, we estimated that ∼2% of input enzyme molecules were converted to 32P-EpA at saturating ATP. The remaining enzyme preparation likely consisted of preadenylylated enzyme.
Ligation activity
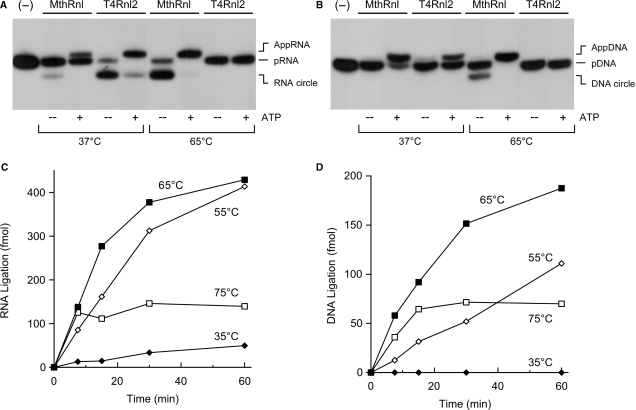
We assayed the ligation activity of MthRnl using 32P-labeled 24-mer RNA (pRNA), at either 37°C or 65°C, in the presence or absence of ATP. At 37°C in the absence of ATP, MthRnl formed trace amount of circular RNA product, which migrated ∼2-nt faster than the input pRNA (Figure 2A). When the reaction temperature was elevated to 65°C, MthRnl converted most of input 24-mer strand to a circular RNA. Inclusion of 1 mM ATP in the reaction suppressed the formation of ligated circle, and accumulated AppRNA, which migrated 1-nt slower than the input pRNA. As the ATP concentration was increased, a transition from the circular product to AppRNA was observed, with a midpoint at ∼50 µM ATP (Figure S2A). As previously noted for T4Rnl2, the excess ATP reacts with a free ligase and dissociate prematurely from AppRNA (31). The adenylylated ligase is unable to bind to and seal the AppRNA strand.
Figure 2.
MthRnl can circularize single-stranded RNA and single-stranded DNA at elevated temperature. (A) RNA ligation. Standard ligation reactions containing 5 pmol MthRnl or T4Rnl2, with or without 1 mM ATP, were incubated for 30 min at either 37°C or 65°C as indicated. Reaction products were resolved by denaturing PAGE. An autoradiogram of the gel is shown. Position of pRNA, AppRNA and circularized RNA (RNA circle) are indicated. A control reaction, lacking enzyme, is shown in the lane indicated by (−). (B) DNA ligation. Identical to (A) except that the substrate for ligation was 32P-labeled 24-mer DNA (pDNA). Reaction products, AppDNA and circularized DNA (cDNA) are indicated. (C) Kinetics and temperature-dependency. A reaction mixture (50 µl) containing 50 mM Tris–HCl (pH 6.5), 0.5 mM MgCl2, 2.5 pmol pRNA and 10 pmol MthRnl was incubated at either 35°C, 55°C, 65°C or 75°C. Aliquots (10 µl) were withdrawn at the times indicated and quenched immediately with formamide–EDTA. The level of circular RNA product is plotted as a function of incubation time. (D) Identical to (C) except that the substrate for ligation was 24-mer DNA.
We asked whether MthRnl is capable of joining 32P-labeled DNA oligonucleotide (pDNA) (Figure 2B). In the presence of ATP at 65°C, MthRnl readily transferred the AMP to pDNA to form adenylylated DNA (AppDNA). In the absence of ATP, MthRnl converted pDNA into a DNA circle. In contrast, T4Rnl2 is unable to circularize the DNA strand. Furthermore, MthRnl was capable of circularizing chimeric 24-mer pDNA–RNA oligonucleotides (12 nt of deoxyribonucleotides at 5′-PO4 followed by 12 nt ribonucleotides at the 3′-OH), or 24-mer pRNA–DNA (12 nt ribonucleotides at 5′-PO4 followed by 12 nt deoxyribonucleotides at the 3′-OH) (data not shown). These results indicate that MthRnl does not discriminate between RNA and DNA for phosphodiester bond formation.
The rates of pRNA and pDNA circularization by MthRnl at various temperatures are shown in Figure 2C and D. The ligation activities were optimal at 65°C for both pRNA and pDNA circularization, but declined significantly as the temperature decreased to 35°C. While the enzyme was initially active at 75°C, the yield of circular products ceased after a 10-min incubation. Approximately 0.5 pmol pRNA or 0.2 pmol pDNA can be circularized per picomole of MthRnl in 30 min. The rate of ligation represents a single turnover reaction by the preadenylylated enzyme, as ATP was omitted from the reaction to detect overall ligation.
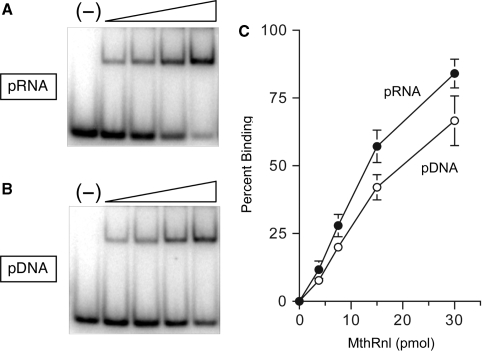
A native gel mobility shift assay was used to compare the binding of MthRnl to pRNA and pDNA (Figure 3). Binding reactions were performed in the absence of metal to preclude the conversion of substrate to product during incubation. Incubating MthRnl with pRNA or pDNA resulted in formation of a discrete ligase-oligonucleotide complex, which migrated slower than the free substrate. The apparent dissociation constant, calculated as described by Riggs et al. (32), was ∼1.5 µM for pRNA binding. MthRnl also forms complex with pDNA with comparable binding affinity (Kd = ∼ 2 µM). These results further support the finding that MthRnl do not discriminate between DNA and RNA.
Figure 3.
Nucleic acid binding. (A) RNA binding. Reaction mixtures (10 µl) contained 50 mM Tris–HCl (pH 8), 5% glycerol, 1 pmol pRNA and 3.8, 7.5, 15, or 30 pmol MthRnl (from left to right within each titration series). A control reaction, lacking enzyme, is shown in the lane indicated by (−). After incubation for 5 min at 22°C, products were analyzed by native PAGE and visualized by autoradiography. (B) Identical to (A) except that the substrate for ligation was pDNA. (C) The extent of pRNA binding and pDNA binding by MthRnl. Percentage of protein–RNA complex (pRNA) and protein–DNA complex (pDNA) formed was plotted as a function of input protein. The data shown represent the average of three separate binding experiments. Standard error bars are included for each datum point.
Further characterization of MthRnl ligation activity was performed using pRNA substrate. Ligation activity required a divalent cation cofactor (Figure S2B). Activity was optimal between 0.2 and 2 mM magnesium or manganese (data not shown). Cobalt supported ligation, whereas zinc failed to promote formation of AppRNA (Figure S2B). Trace amounts of AppRNA were detected with copper. In the presence of calcium, MthRnl accumulated AppRNA, suggesting that calcium cannot support the third step of the reaction. The effect of pH on circularization of pRNA was examined over a pH range of 4.0–9.0 in the absence of ATP (data not shown). The yield of circular product was optimal between pH 5.5 and 7.5. Reducing the pH to 5.0 resulted in accumulation of AppRNA, and suppression of the intramolecular ligation reaction. Further reduction of the pH below 4.5 abolished all activities. Activity also declined gradually at alkaline pH levels, such that the yield of circular product at pH 9.0 was 20% of that at pH 7.0, without accumulating AppRNA. Similar pH effects were found previously for T4 and KVP40 Rnl2 (3,31). The standard ligation reaction contained 25 mM NaCl from the enzyme solution. Activity was reduced by 20% in the presence of 100 mM NaCl (data not shown) and by ∼50% in 200 mM NaCl.
Phosphodiester formation at a preadenylylated RNA
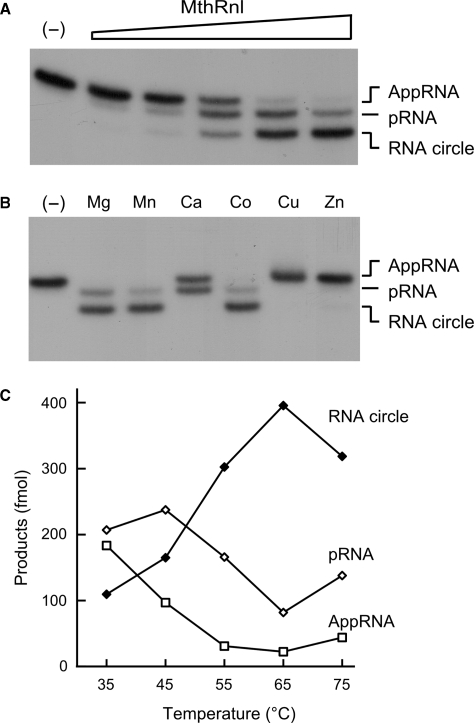
We examined step 3 of the ligation reaction in isolation using preadenylylated RNA substrate. In the presence of magnesium without added ATP, formation of a phosphodiester bond was proportional to the amount of input MthRnl at 65°C (Figure 4A). MthRnl can also deadenylylate AppRNA, by a reversal of step 2 of the ligation pathway. This is evidenced by the appearance of pRNA, which migrates between AppRNA and circular RNA. At saturating concentrations of MthRnl, ∼80% of the input AppRNA substrate was converted to ligated circles, whereas the rest was deadenylylated to pRNA.
Figure 4.
Ligation of preadenylylated RNA substrate. (A) Protein titration. Reaction mixtures (10 µl) containing 50 mM Tris–HCl (pH 6.5), 0.5 mM MgCl2, 0.5 pmol AppRNA and 0.5, 1, 2, 4, 8 pmol of MthRnl (proceeding from left to right within each titration series) were incubated for 30 min at 65°C. A control reaction, lacking enzyme, is shown in the lane indicated by a (–). An autoradiogram of the gel is shown. Positions of pRNA, AppRNA and RNA circle are indicated. (B) Metal specificity. Reaction mixtures (10 µl) containing 50 mM Tris–HCl (pH 6.5), 0.5 pmol AppRNA, 5 pmol MthRnl and either 0.5 mM MgCl2, MnCl2, CaCl2, CoCl2, CuSO2 or ZnSO2, were incubated for 30 min at 65°C. Control reaction without divalent cation is indicated by (−). (C) Temperature dependency. Reaction mixtures (10 µl) containing 50 mM Tris–HCl (pH 6.5), 0.5 mM MgCl2, 0.5 pmol AppRNA, 5 pmol MthRnl were incubated for 15 min at either 35°C, 45°C, 55°C, 65°C and 75°C. Level of AppRNA, RNA circle and deadenylylated pRNA, are plotted as functions of temperature.
The extent of phosphodiester bond formation (step 3) and deadenylylation (the reverse of step 2) of AppRNA was analyzed at various temperatures (Figure 4C). Similar to what was found for the overall ligation reaction, the optimum temperature for phosphodiester bond formation was 65°C. At lower temperatures, however, the reverse reaction is favored. This is shown by the shift in the distribution of pRNA and ligated circles. At 35°C, nearly 40% of the input AppRNA was deadenylylated to form pRNA, whereas only 20% was ligated to circular RNA. Thus, under an optimum temperature, MthRnl promotes the reaction forward in the direction of phosphodiester bond formation and against deadenylylation.
Circularization and deadenylylation of AppRNA requires the presence of a divalent cation (Figure 4B). This requirement can be satisfied by 2 mM magnesium, manganese or cobalt, but not by copper or zinc. We note that a reaction with calcium failed to convert the AppRNA into a circular product. Instead, 50% of the input AppRNA was deadenylylated to pRNA. This result is in agreement with the effect of calcium in overall ligation reaction (Figure S2B). MthRnl displayed a strong step 3 arrest in the presence of calcium that resulted in accumulation of the AppRNA intermediate.
Deletion analysis
To determine how the C-terminal segment contributes to MthRnl function, the protein was split into two segments, N-terminal MthRnl (1–253) and C-terminal MthRnl (255–381). These truncated proteins were produced in E. coli as His10-tag fusions and were purified from soluble lysates by Ni–agarose chromatography. SDS–PAGE analysis revealed proteins of expected sizes (Figure 5A).
Figure 5.
Characterization of MthRnl (1–253) and MthRnl (255–381). (A) Purification. Aliquots (4 µg) of Ni–agarose preparations of full-length MthRnl, MthRnl (1–253), and MthRnl (255–381) were analyzed by SDS–PAGE followed by Coomassie blue staining. The positions and sizes (in kilodalton) of marker proteins are indicated on the left. (B) EpA formation. Aliquots (0.2 µg) of purified proteins in (A) were assayed for adenylyltransferase activity with 0.16 µM [α-32P]ATP at 50°C. EpA complex was visualized by autoradiography of the dried gel. (C) Effect of temperature on MthRnl (1–253)–AMP formation. Standard adenylyltransferase reaction mixtures contained 250 pmol of MthRnl (1–253). Reaction was incubated at indicated temperature. Autoradiography of the dried gel is shown. (D) Effect of temperature on MthRnl (1–253) ligation. Standard ligation reactions containing 20 pmol MthRnl (1–253) was incubated at indicated temperature in the absence of ATP. Control reaction with 10 pmol of full-length MthRnl incubated at 75°C is indicated by (+). (E) RNA binding. Reaction mixtures contained 50 mM Tris–HCl (pH 8), 2% glycerol, 1 pmol pRNA and either 7.5, 15 and 30 pmol of MthRnl(1–253) or MthRnl (255–381). Control reactions with 30 pmol of full-length MthRnl or without protein are indicated by (+) and (−), respectively.
MthRnl (1–253) was defective in overall ligation but retained the ability to form EpA intermediate, bind to pRNA and transfer AMP to pRNA, albeit less efficiently than wild-type ligase (Figure 5). Deletion of C-terminal segment from MthRnl altered the thermoreactivity of the enzyme. The EpA and AppRNA formation was optimum at 25°C and 37°C, respectively (Figure 5C and D). The extent of EpA formation by MthRnl (1–253) at 25°C was 10% of the native MthRnl at 50°C (Figures S1B and S3B). MthRnl (1–253) accumulated AppRNA in the composite ligation assay, either in the presence or absence of ATP (Figure 5D and data not shown), and was unable to convert isolated AppRNA to a circular RNA in the temperature range of 25–75°C (data not shown). Binding of MthRnl (1–253) to pRNA was reduced to ∼30% seen with the full-length protein at equivalent protein concentrations (Figure S3C). The extent of AppRNA formation by MthRnl (1–253) at 25°C was <10% of the native enzyme at 65°C (Figure S3D). The 30-kDa MthRnl (1–253) sedimented as a single discrete component between BSA and cytochrome c with an observed sedimentation coefficient of 3.6, suggesting that MthRnl (1–253) is a monomer (Figure 6A).
Figure 6.
Sedimentation profile of MthRnl (1–253) and MthRnl (255–381). (A) Sample containing MthRnl (1–253) and marker proteins were centrifuged in a 15–30% glycerol gradient at 45 000g for 23 h at 4°C. Aliquots of the gradient fractions were resolved by SDS–PAGE followed by Coomassie blue staining. The glycerol gradient fraction numbers are specified above each lane. The identities of the polypeptides are indicated. (B) Sample containing MthRnl (255–381) and marker proteins were centrifuged in glycerol gradient. Distribution of MthRnl (1–253) and MthRnl (255–381) was unaffected by the marker proteins in the gradient (not shown).
As expected, MthRnl (255–381) was inert for EpA formation (Figure 5B). MthRnl (255–381) failed to form a detectable protein–pRNA complex (Figure 5E) and did not support overall ligation or AppRNA circularization (data not shown). Mixing MthRnl (1–253) and MthRnl (255–381) together did not restore ligation activity (data not shown). Sedimentation analysis revealed that 17-kDa MthRnl (255–381) self-associates to form a high-molecular weight oligomeric species with S values ranging from 7.6 to 12.4 (Figure 6B). We conclude that the 253-amino acid segment at the N-terminal is capable of ligase-adenylylation and RNA-adenylylation. The 127-amino acid segment at the C-terminal has an intrinsic ability to self-associate and this region is required for the phosphodiester bond formation and dimerization.
To further evaluate the role of the C-terminal region in dimerization, we asked whether MthRnl (255–381) can potentially associate with MthRnl and alter the ligation activity. Full-length MthRnl was mixed with increasing amounts of MthRnl (255–381) protein and then assayed for ligation activity in the presence and absence of ATP. Addition of MthRnl (255–381) inhibits the overall ligation of native enzyme to circularize pRNA (Figure 7A). At 1:1 molar ratio, the activity was reduced to 60% of the activity of MthRnl alone. Activity progressively declined as concentration of MthRnl (255–381) was increased. MthRnl (255–381) had little or no effect on ligase-adenylylation or sealing preformed AppRNA (Figure 7B and D), but elicited a concentration-dependent inhibition on RNA adenylylation (Figure 7C). The inhibitory effect was specific to MthRnl, as MthRnl (255–381) had little or no effect on overall ligation or AppRNA formation by T4Rnl2. The most likely explanation for these observation is that inhibition is due to hetrodimerization between MthRnl and MthRnl (255–381). MthRnl (255–381) may disrupt MthRnl homodimerization and block the RNA adenylylation step of the ligation pathway.
Figure 7.
Inhibition of MthRnl ligation activity by MthRnl (255–381). (A) Effect on overall ligation. Ten picomole of MthRnl was preincubated with indicated amount of MthRnl (255–381) for 5 min at 65°C followed by standard ligation assay in the absence of ATP. (B) Effect on EpA formation. Five picomole of MthRnl was preincubated with indicated amount of MthRnl (255–381), followed by standard adenylyltransferase assay. (C) Effect on pRNA adenylylation. Identical to (A) except the ligation reaction contained 100 µM ATP. (D) Effect on AppRNA-sealing activity. Identical to (A) except that ligation reaction contained 0.5 pmol of AppRNA in the absence of ATP. Percent product were normalized to the control level in the absence of MthRnl (255–381) (defined as 1.0) and then plotted as a function of added MthRnl (255–381) (closed circle). The data shown represent the average of three separate experiments. Standard error bar are included for each datum point. For a control reaction in (A) and (C), 10 pmol of T4Rnl2 was preincubated with indicated amount of MthRnl (255–381) for 5 min at 25°C prior to ligation assay at 25°C as described (3). Percent product were normalized to the control level in the absence of T4Rnl2 and then plotted as a function of added MthRnl (255–381) (open circle).
DISCUSSION
Here, we show that archaea possess a strand-joining enzyme that can catalyze intramolecular ligation of single-stranded RNA. Recombinant MthRnl reacts with ATP to form a covalent ligase–AMP complex, then transfers AMP to pRNA to form an AppRNA intermediate and finally seals AppRNA to form a circular RNA product. All three steps of the ligation reactions require a divalent cation as a cofactor. We also determined that calcium can support AppRNA formation, but does not support phosphodiester bond formation, which is a property shared with Methanobacterium DNA ligase (33). The optimal temperature for strand-joining activity was 65°C, which is also the growth temperature of M. thermoautotrophicum. Using the AppRNA intermediate as a substrate, we showed that MthRnl, at its optimal temperature, favors phosphodiester bond formation to promote the ligation reaction forward to completion. Under suboptimal temperatures, however, deadenylylation of AppRNA was the preferred reaction. These results suggest that MthRnl may alter its protein conformation during the transition from step 2 and step 3 reactions. Remodeling of active site in transition of step 2 to step 3 catalysis has been suggested by the structures of T4Rnl2 (2).
A defining property of MthRnl is the apparent lack of specificity to seal a polynucleotide substrate. MthRnl forms a stable complex with both pRNA and pDNA and the rate of circularization was comparable. The substrate specificity of MthRnl differs from that of T4Rnl2 and Deinococcus ligase, which both discriminate between RNA and DNA (12,34). MthRnl more closely resembles T4Rnl1 and RM378 Rnl1, both of which are able to circularize DNA but less efficiently than RNA (35–37). Our preliminary experiments show that MthRnl was unable to join a nick on a double-stranded DNA substrate (Gu,H., Yoshinari,S. and Ho,C.K., unpublished data).
Another distinct feature of MthRnl is its ability to self-associate to form a homodimer. Archaeal RNA ligase is the only polynucleotide ligase known to homodimerize. RNA ligases from phages (T4Rnl1, T4Rnl2 and KVP 40) and Deinococcus (DraRnl) are monomeric (3,13,31,34) as are most DNA ligases. In this study, we showed that the C-terminal segment (residues 255–381) is required for homodimerization and is itself capable of oligomerization. While this article was in preparation, the van Tilbeurgh group reported a crystal structure of Pyrococcus abyssi RNA ligase (an ortholog of MthRnl) in complex with AMPPNP (38). Pyrococcus RNA ligase adopts a homodimeric structure with an N-terminal nucleotidyltransferase and C-terminal dimerization domains. The dimerization interface is formed by a four-helix bundle, from residues 245 to 313 in the Pyrococcus RNA ligase (equivalent of MthRnl residues 243–310). Additional contacts are found at the C-terminal end of the protein, which protrudes into the active sites of the partner molecules. Our results are in agreement with the structure of Pyrococcus RNA ligase in that the C-terminal domain of archaea ligase is responsible for dimerization.
Based on our deletion analysis, dimerization likely contributes to the strand-joining activity of MthRnl. Removal of the C-terminal segment significantly diminished the ligase-adenylylation and RNA-adenylylation steps, and completely abolished phosphodiester bond formation. The isolated C-terminal domain can suppress the RNA-adenylylation step in trans. In light of the structure of Pyrococcus RNA ligase, we envision that C-terminal portion of the partner molecule makes direct contact with RNA or active site residues to promote strand-joining activity. Our preliminary experiments show that deletion of 15 amino acid from the C-terminal end severely reduces the MthRnl activity (Gu,H. and Ho,C.K., unpublished data). We speculate that isolated C-terminal domain associates with the monomeric enzyme, but is incapable of inducing the conformation changes necessary to catalyze the RNA-adenylylation step. Alternatively, the N-terminal catalytic domains need to be brought together as a dimer to promote the step 2 reaction. Further mutational analyses should define resides that are involved in dimerization, to address how homodimerization contributes to ligation activity.
The physical and biochemical properties of MthRnl raise interesting questions about the kinds of reactions that MthRnl catalyzes in vivo. Archaea needs RNA ligase to perform tRNA splicing. Introns are also found in pre-rRNA as well as pre-mRNA in some crenarchaea (22,23,25). These RNAs contain bulge-helix-bulge-like motifs at the intron–exon boundary, which are cleaved symmetrically by the splicing endonuclease to produce single-stranded overhangs at both ends of exon and intron. Evidence suggests that joining of exons and circularization of introns take place concurrently (22,28). We speculate that homodimeric structure of archaeal RNA ligase may facilitate both ligation events simultaneously.
Studies of tRNA end-joining reaction in Haloferax volcanii cell extracts suggests that the mechanism of tRNA ligation is likely different from that used by yeast and plants. In yeast and plants, the intrinsic end-healing activity of Trl1 first converts the 2′-, 3′-cyclic phosphate and 5′-OH ends, generated by endonuclease cleavage, into 2′-PO4/3′-OH and phosphorylates 5′-OH by GTP-dependent kinase (5). The 3′-OH and 5′-PO4 are subsequently joined to yield a 3′–5′ phosphodiester bond at the spliced junction. The end-joining activity of Trl1 is similar to the ligation activity of MthRnl, in that they both require a divalent cation and ATP for joining the 3′-OH and 5′-PO4 ends. However, in H. volcanii, tRNA end-joining reaction does not require a divalent cation, and the 2′-, 3′-cyclic phosphate and 5′-OH are joined by a single-step reaction to form a 3′–5′phosphodiester linkage (28,30). These findings raise the possibility that an RNA ligase other than MthRnl may be responsible for tRNA ligation. Indeed the MtRnl-like enzyme is not ubiquitous in all archaea species. Of the 48 archaeal genomes that have been published, a credible homologue of MthRnl is found in 29 species. It is possible that some archaea species may have two different types of RNA ligases, similar to mammalian cells where both yeast/plant-type and H. volcanii-type ligation activities have been detected (39–41). Alternatively, different archaea species may have different types of RNA ligases.
Ho et al. (3) proposed that members of the covalent nucleotidyltransferase superfamily evolved by fusion of common Rnl2-like catalytic domain to a variable domain to provide biological specificity for RNA processing and DNA repair. The archaeal RNA ligases apparently fused the N-terminal catalytic domain to a novel C-terminal domain that can mediate homodimerization. The lack of specificity for polynucleotide substrate (RNA versus DNA) by MthRnl might exemplify an undifferentiated ancestral stage in the evolution of ATP-dependent ligases. We hypothesize that the C-terminal region of archaeal RNA ligases may also contribute to the selectivity for certain types of RNA or DNA, or promote interactions with other proteins that are involved in nucleic acid repair.
FUNDING
University at Buffalo Interdisciplinary Research Development Fund (UB2020). Funding for open access charge: Department of Biological Sciences, SUNY at Buffalo.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Stewart Shuman (Sloan Kettering Institute) for Methanobacterium genomic DNA and for helpful discussion and advice; and Dr Paul Gollnick and Dr Michael Yu (SUNY Buffalo) for critically reading the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
REFERENCES
- 1.Uhlenbeck OC, Gumport RI. T4 RNA Ligase. In: Boyer PD, editor. The Enzymes. Vol. 15, Academic Press: New York; 1982. pp. 31–58. [Google Scholar]
- 2.Nandakumar J, Shuman S, Lima CD. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell. 2006;127:71–84. doi: 10.1016/j.cell.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 3.Ho CK, Shuman S. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc. Natl Acad. Sci. USA. 2002;99:12709–12714. doi: 10.1073/pnas.192184699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amitsur M, Levitz R, Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abelson J, Trotta CR, Li H. tRNA splicing. J. Biol. Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 6.Englert M, Beier H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005;33:388–399. doi: 10.1093/nar/gki174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 8.Martins A, Shuman S. Characterization of a baculovirus enzyme with RNA ligase, polynucleotide 5'-kinase, and polynucleotide 3'-phosphatase activities. J. Biol. Chem. 2004;279:18220–18231. doi: 10.1074/jbc.M313386200. [DOI] [PubMed] [Google Scholar]
- 9.McManus MT, Shimamura M, Grams J, Hajduk SL. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA. 2001;7:167–175. doi: 10.1017/s1355838201002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rusché LN, Huang CE, Piller KJ, Hemann M, Wirtz E, Sollner-Webb B. The two RNA ligases of the Trypanosoma brucei RNA editing complex: cloning the essential band IV gene and identifying the band V gene. Mol. Cell Biol. 2001;21:979–989. doi: 10.1128/MCB.21.4.979-989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Wirtz E, Salavati R, Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- 12.Nandakumar J, Shuman S. How an RNA ligase discriminates RNA versus DNA damage. Mol. Cell. 2004;16:211–221. doi: 10.1016/j.molcel.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 13.El Omari K, Ren J, Bird LE, Bona MK, Klarmann G, LeGrice SF, Stammers DK. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J. Biol. Chem. 2006;281:1573–1579. doi: 10.1074/jbc.M509658200. [DOI] [PubMed] [Google Scholar]
- 14.Ho CK, Wang LK, Lima CD, Shuman S. Structure and mechanism of RNA ligase. Structure. 2004;12:327–339. doi: 10.1016/j.str.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Shuman S, Lima CD. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Nandakumar J, Ho CK, Lima CD, Shuman S. RNA substrate specificity and structure-guided mutational analysis of bacteriophage T4 RNA ligase 2. J. Biol. Chem. 2004;279:31337–31347. doi: 10.1074/jbc.M402394200. [DOI] [PubMed] [Google Scholar]
- 17.Wang LK, Nandakumar J, Schwer B, Shuman S. The C-terminal domain of T4 RNA ligase 1 confers specificity for tRNA repair. RNA. 2007;13:1235–1244. doi: 10.1261/rna.591807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belfort M, Weiner A. Another bridge between kingdoms: tRNA splicing in archaea and eukaryotes. Cell. 1997;89:1003–1006. doi: 10.1016/s0092-8674(00)80287-1. [DOI] [PubMed] [Google Scholar]
- 19.Lykke-Andersen J, Aagaard C, Semionenkov M, Garrett RA. Archaeal introns: splicing, intercellular mobility and evolution. Trends Biochem. Sci. 1997;22:326–331. doi: 10.1016/s0968-0004(97)01113-4. [DOI] [PubMed] [Google Scholar]
- 20.Marck C, Grosjean H. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: evolutionary implications. RNA. 2003;9:1516–1531. doi: 10.1261/rna.5132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kjems J, Garrett RA. Ribosomal RNA introns in archaea and evidence for RNA conformational changes associated with splicing. Proc. Natl Acad. Sci. USA. 1991;88:439–443. doi: 10.1073/pnas.88.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burggraf S, Larsen N, Woese CR, Stetter KO. An intron within the 16S ribosomal RNA gene of the archaeon Pyrobaculum aerophilum. Proc. Natl Acad. Sci. USA. 1993;90:2547–2550. doi: 10.1073/pnas.90.6.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe Y, Yokobori S, Inaba T, Yamagishi A, Oshima T, Kawarabayasi Y, Kikuchi H, Kita K. Introns in protein-coding genes in Archaea. FEBS Lett. 2002;510:27–30. doi: 10.1016/s0014-5793(01)03219-7. [DOI] [PubMed] [Google Scholar]
- 25.Yoshinari S, Itoh T, Hallam SJ, DeLong EF, Yokobori S, Yamagishi A, Oshima T, Kita K, Watanabe Y. Archaeal pre-mRNA splicing: a connection to hetero-oligomeric splicing endonuclease. Biochem. Biophys. Res. Commun. 2006;346:1024–1032. doi: 10.1016/j.bbrc.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 26.Armbruster DW, Daniels CJ. Splicing of intron-containing tRNATrp by the archaeon Haloferax volcanii occurs independent of mature tRNA structure. J. Biol. Chem. 1997;272:19758–19762. doi: 10.1074/jbc.272.32.19758. [DOI] [PubMed] [Google Scholar]
- 27.Kjems J, Garrett RA. Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell. 1988;54:693–703. doi: 10.1016/s0092-8674(88)80014-x. [DOI] [PubMed] [Google Scholar]
- 28.Salgia SR, Singh SK, Gurha P, Gupta R. Two reactions of Haloferax volcanii RNA splicing enzymes: joining of exons and circularization of introns. RNA. 2003;9:319–330. doi: 10.1261/rna.2118203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomes I, Gupta R. RNA splicing ligase activity in the archaeon Haloferax volcanii. Biochem. Biophys. Res. Commun. 1997;237:588–594. doi: 10.1006/bbrc.1997.7193. [DOI] [PubMed] [Google Scholar]
- 30.Zofallova L, Guo Y, Gupta R. Junction phosphate is derived from the precursor in the tRNA spliced by the archaeon Haloferax volcanii cell extract. RNA. 2000;6:1019–1030. doi: 10.1017/s1355838200000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin SM, Ho CK, Miller ES, Shuman S. Characterization of bacteriophage KVP40 and T4 RNA ligase 2. Virology. 2004;319:141–151. doi: 10.1016/j.virol.2003.10.037. [DOI] [PubMed] [Google Scholar]
- 32.Riggs AD, Newby RF, Bourgeois S. lac repressor–operator interaction. II. Effect of galactosides and other ligands. J. Mol. Biol. 1970;51:303–314. doi: 10.1016/0022-2836(70)90144-0. [DOI] [PubMed] [Google Scholar]
- 33.Sriskanda V, Kelman Z, Hurwitz J, Shuman S. Characterization of an ATP-dependent DNA ligase from the thermophilic archaeon Methanobacterium thermoautotrophicum. Nucleic Acids Res. 2000;28:2221–2228. doi: 10.1093/nar/28.11.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martins A, Shuman S. An RNA ligase from Deinococcus radiodurans. J. Biol. Chem. 2004;279:50654–50661. doi: 10.1074/jbc.M407657200. [DOI] [PubMed] [Google Scholar]
- 35.Blondal T, Hjorleifsdottir SH, Fridjonsson OF, Aevarsson A, Skirnisdottir S, Hermannsdottir AG, Hreggvidsson GO, Smith AV, Kristjansson JK. Discovery and characterization of a thermostable bacteriophage RNA ligase homologous to T4 RNA ligase 1. Nucleic Acids Res. 2003;31:7247–7254. doi: 10.1093/nar/gkg914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCoy MI, Gumport RI. T4 ribonucleic acid ligase joins single-strand oligo(deoxyribonucleotides) Biochemistry. 1980;19:635–642. doi: 10.1021/bi00545a005. [DOI] [PubMed] [Google Scholar]
- 37.Sugino A, Snoper TJ, Cozzarelli NR. Bacteriophage T4 RNA ligase. Reaction intermediates and interaction of substrates. J. Biol. Chem. 1977;252:1732–1738. [PubMed] [Google Scholar]
- 38.Brooks MA, Meslet-Cladiere L, Graille M, Kuhn J, Blondeau K, Myllykallio H, van Tilbeurgh H. The structure of an archaeal homodimeric ligase which has RNA circularization activity. Protein Sci. 2008;17:1336–1345. doi: 10.1110/ps.035493.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Filipowicz W, Shatkin AJ. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell. 1983;32:547–557. doi: 10.1016/0092-8674(83)90474-9. [DOI] [PubMed] [Google Scholar]
- 40.Laski FA, Fire AZ, RajBhandary UL, Sharp PA. Characterization of tRNA precursor splicing in mammalian extracts. J. Biol. Chem. 1983;258:11974–11980. [PubMed] [Google Scholar]
- 41.Zillmann M, Gorovsky MA, Phizicky EM. Conserved mechanism of tRNA splicing in eukaryotes. Mol. Cell Biol. 1991;11:5410–5416. doi: 10.1128/mcb.11.11.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.