Abstract
Porphyromonas gingivalis HmuY is a putative heme-binding lipoprotein associated with the outer membrane. It is part of an operon together with a gene encoding an outer-membrane hemin utilization receptor (HmuR) and four uncharacterized genes. A similar operon organization was found in Bacteroides fragilis and B. thetaiotaomicron, with the former containing an additional HmuY homologue encoded upstream of the hmuR-like gene. In P. gingivalis cultured under heme-limited conditions, a ~1-kb hmuY transcript was produced at high levels along with some ~3.5 and ~9-kb transcripts. Compared with the parental strain, mutants deficient in hmuY or hmuR or hmuY–hmuR gene function grew more slowly and bound lower amounts of hemin and hemoglobin. Significantly, they grew more slowly or were unable to grow when human serum was used as the sole iron/heme source. Analysis of the hmu promoter showed that it is regulated by iron. The HmuY protein normally occurs as a homodimer, but in the presence of hemin it may form tetramers. These results show that HmuY may be the first reported member of a new class of proteins in Porphyromonas and Bacteroides species involved in heme utilization, a function being exerted in conjunction with HmuR, an outer-membrane heme transporter.
Keywords: Porphyromonas gingivalis, HmuR, Heme outer-membrane receptor, HmuY, Heme-binding protein, Heme uptake, Lipoprotein, His-tag
Introduction
Periodontitis is an inflammatory disease involving microbial, genetic, immunological, and environmental factors. Porphyromonas gingivalis is a black-pigmented, Gram-negative anaerobic bacterium which has been implicated as a major etiological agent in the development of chronic periodontitis. Proliferation of P. gingivalis in the gingival pocket is dependent on the acquisition of iron and heme (Olczak et al. 2005). Within the human host, heme-binding proteins are a major source of these growth factors essential for the invading organisms. In vitro, P. gingivalis can acquire heme from various hemoproteins, including hemoglobin, hemoglobin–haptoglobin, and hemin bound to hemopexin or to serum albumin, indicating that this bacterium has a mechanism for removing heme from the host heme-containing proteins (reviewed in Genco and Dixon 2001; Potempa et al. 2003; Olczak et al. 2005). To acquire iron and heme, P. gingivalis employs hemagglutinins, proteases (particularly gingipains), lipoproteins, and specific outer-membrane receptors (Nelson et al. 2003; Potempa et al. 2003; Olczak et al. 2005). P. gingivalis is capable of storing heme on its cell surface and transporting the entire heme molecule into the cell. We identified an outer-membrane hemin utilization receptor (HmuR) in P. gingivalis involved in the acquisition of both free hemin and heme bound to hemoproteins (Simpson et al. 1999, 2000). The role of the hmuR gene in heme accumulation has been extensively studied through mutant construction and biochemical analysis of the protein (Simpson et al. 2000, 2004; Olczak et al. 2001; Liu et al. 2006; Olczak 2006).
The hmuR gene in the P. gingivalis A7436 strain is located in an operon with a hmuY gene and four other genes, so far uncharacterized (Simpson et al. 2000). An operon with identical structure was found in P. gingivalis W83 genome (Nelson et al. 2003; Lewis et al. 2006). In contrast, a different organization of the hmu operon was described in strains ATCC 53977, 381, and W50 (Karunakaran et al. 1997). In these strains, instead of the entire hmuR gene, a truncated version, termed hemR, was reported to be in association with part of a prtT gene. In the ATCC 53977 strain, the open reading frame (ORF) preceding the hemR gene (Karunakaran et al. 1997) was shown to encode a protein identical to HmuY (Nelson et al. 2003; Simpson et al. 2000). While we were working on this subject, recent data (Lewis et al. 2006; Ong et al. unpublished data) demonstrated that in the P. gingivalis W83 and W50 strains the previously identified HmuY protein was longer than previously reported (Simpson et al. 1999, 2000; Karunakaran et al. 1997; LANL and TIGR databases). The deduced amino-acid sequence analysis of full-length HmuY suggests that it is a putative membrane-associated lipoprotein. HmuY shows no sequence similarity to proteins deposited in databases except for peptides derived from a 30 kDa (a heated protein showing a molecular mass of 24 kDa) hemin-binding envelope protein described earlier by Kim et al. (1996) in the P. gingivalis 381 strain. Recently, we reported that truncated recombinant HmuY protein bound hemin and ATP in vitro (Olczak et al. 2006). Our preliminary analysis also showed that HmuY may be functional in the form of dimers/oligomers. Interestingly, the N-terminus of mature HmuY is identical to a protein purified from the P. gingivalis envelope (Mihara and Holt 1993a, b; Mihara et al. 1993). The protein, designated as FAF (fibroblast-activating factor), was identified in P. gingivalis W50, W83, and ATCC 33277, but not in the ATCC 25285 strain. It exerted a significant proliferation-stimulating eVect on normal human gingival fibroblasts and displayed functional similarity to several human-derived growth factors. The authors also found that FAF possessed a weak phosphatase activity, which may affect bone resorption.
The aim of this study was to further characterize the role of HmuY and HmuR in heme acquisition by P. gingivalis. Our results not only complement and clarify existing findings, but also further our understanding of the nature of HmuY protein. We suggest that HmuY may be the first reported member of a new class of lipoproteins involved in heme utilization, a function being exerted in conjunction with the outer-membrane receptor HmuR.
Materials and methods
Nucleotide and amino-acid sequence analyses
Sequence data for P. gingivalis, Bacteroides fragilis, B. thetaiotaomicron, Prevotella intermedia, Treponema denticola, and Aggregatibacter actinomycetemcomitans were obtained from The Institute for Genomic Research (http://tigr.org) and Los Alamos National Laboratory (http://oralgen.lanl.gov). Primary sequences of all proteins were obtained from the Swiss Protein Data Bank (Bairoch and Boeekmann 1991). Multiple sequence alignments of HmuY and its related sequences were carried out by the CLUS-TAL X program (Jeanmougin et al. 1998) with default parameters, the multiple alignments being manually adjusted if necessary. Prediction of lipoproteins and signal peptides in Gram-negative bacteria was performed using LipoP 1.0 Server (http://www.cbs.dtu.dk/services/LipoP) (Juncker et al. 2003). Protein sequences were compared using BLASTP program (http://www.ncbi.nlm.nih.gov/BLAST/) (Altschul et al. 1997). Secondary structure prediction was carried out using the SOPMA program of Network Protein Sequence Analysis (http://npsa-pbil.ibcp.fr/cgi-bin/secpred_sopma.pl) (Geourjon and Deleage 1995).
Bacterial strains and growth conditions
Porphyromonas gingivalis wild-type strains A7436, W83, and 381 were maintained as described previously (Simpson et al. 2000) on anaerobic blood agar plates (ABA, Graso). WS1 (hmuR::ermF), TO1 (hmuY::tetQ), and TO2 (hmuY::tetQ, hmuR::ermF) mutant strains (Table 1S) were grown on ABA plates supplemented with 1 µg/ml erythromycin alone, 1 µg/µl tetracycline alone, and both 1 µg/µl erythromycin and 1 µg/µl tetracycline, respectively. TO3 and TO4 in-frame hmuY deletion mutant strains (Table 1S) were cultured on ABA plates supplemented with 1 µg/ml erythromycin. Escherichia coli TOP10 and ER2566 strains were maintained in Luria–Bertani (LB) medium (Sigma) and incubated at 37°C. The E. coli K-12 EB53 strain was cultured in LB or M9 medium supplemented with 2 µg/ml δ-aminolevulinic acid (ALA; Sigma).
Construction and isolation of P. gingivalis hmuY and hmuY–hmuR mutants
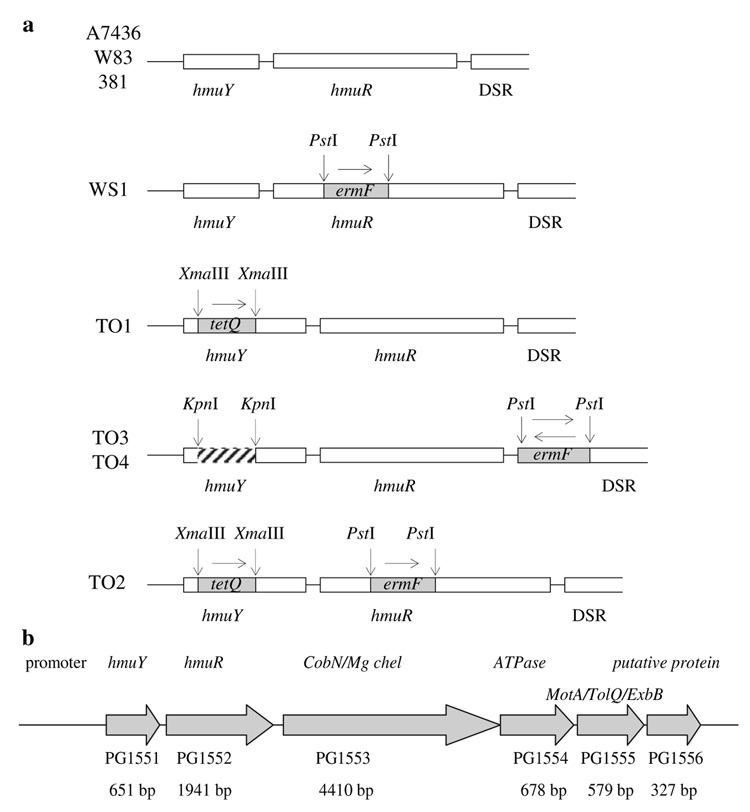
To construct insertional mutants (Fig. 1a), the hmuY gene amplified from P. gingivalis A7436 genomic DNA was cloned into the BamHI and HindIII restriction sites of the pGEM-3Zf(−) plasmid (Promega). All primers used in this study are listed in Table 2S. Then a B. fragilis tetQ encoding a tetracycline-resistance gene was cut out from pWS1 plasmid (Simpson et al. 2000) and cloned into the native XmaIII restriction site of the hmuY gene, resulting in pTOY18 plasmid (Table 1S). The plasmid was linearized using the NdeI restriction enzyme and introduced into P. gingivalis A7436 wild-type strain (to construct the single hmuY mutant TO1) or into the WS1 mutant strain (to construct the double hmuY–hmuR mutant TO2) by electro-poration (Simpson et al. 2000). To construct the in-frame hmuY deletion mutants TO3 and TO4, 100 bp of the 3′ end of hmuY gene and 201 bp of the 5′ end of hmuY gene together with the entire hmuR gene were amplified. The respective primers added the SacI and KpnI and the KpnI and SalI restriction sites, respectively. The PCR products were digested with KpnI and ligated. Then the hmuY gene lacking its internal 300 bp together with the entire hmuR gene was amplified using ligation mixture as a template, digested with the SacI and SalI restriction enzymes, and cloned into the pGEM-3Zf(–) plasmid containing the downstream region of hmuR gene (DSR) and ermF encoding the erythromycin-resistance gene (Rasmussen et al. 1986) cloned between hmuR and DSR (Fig. 1a, b) as described previously (Liu et al. 2006). The ermF cassette was cloned in both directions, resulting in pTOY19 and pTOY20 plasmids (Table 1S). All constructs and homologous recombination between the constructs and the chromosomal DNA of P. gingivalis were verified by PCR and sequencing analyses (data not shown).
Fig. 1.
Construction of P. gingivalis mutant strains (a) and the physical organization of the hemin-uptake gene cluster (hmu) of P. gingivalis (b)
Phenotypic analysis of P. gingivalis mutants
The mutants’ growth in the presence of hemin, hemoglobin, or human serum as sole iron/heme sources was characterized as described previously (Simpson et al. 2000; Liu et al. 2006). To determine if the P. gingivalis mutant strains were affected in their ability to bind different iron sources, the binding of hemin or hemoglobin to the cells was examined using a spectrophotometric assay (Simpson et al. 2000; Liu et al. 2006).
Northern blotting
Total RNA was isolated according to the manufacturer’s protocol (A&A Biotechnology) from the P. gingivalis A7436 and W83 wild-type and mutant strains grown in Schaedler broth (SB), basal medium supplemented with 160 µM dipirydyl (BMD), and BM supplemented with 1.5 µM hemin or 5% human serum. Cell samples were collected from 12-h cultures. A 220-bp PCR product (amplified by primers listed in Table 2S) was used for in vitro transcription of the hmuY-specific RNA probe using DIG Northern Starter Kit (Roche). RNA (1.5 µg per lane) was separated in a formaldehyde agarose gel, transferred onto a nylon membrane (Brightstar-Plus; Ambion), and incubated with the probe labeled with digoxygenin as recommended by the manufacturer (Roche). To check the RNA purity and integrity, separate agarose gel was stained with ethidium bromide.
RT-PCR
Total RNA was isolated as described above. Contaminating DNA was eliminated by on-column DNAse digestion with an RNAse-free DNAse kit (Macherey-Nagel) and the absence of DNA was verified by PCR (data not shown). Reverse transcriptase reaction (RT) was performed using 3 µg of RNA, random hexamers, and Superscript III reverse transcriptase according to the protocol (Invitrogen). Then PCR amplification was carried out for 24 (hmuY), 29 (hmuR), 39 (hmuY–hmuR), or 18 (16S rRNA) cycles in a reaction mixture containing primers specific to the respective sequences (Table 2S) and 1 µl of the RT reaction sample as a template. P. gingivalis-specific 16S rRNA, which is constitutively expressed, was used as a control.
Complementation analysis
An E. coli hemA mutant with an enzymatic lesion in the heme biosynthesis pathway was used to characterize heme utilization through the hmuY–hmuR system by complementation of the biosynthesis defect. For this purpose, HmuY alone, HmuR alone, or both HmuY and HmuR were expressed under their native promoter in the E. coli K-12 EB53 strain (Eberspacher and Braun 1980). Regions containing the hmu promoter and hmuY gene or the promoter and both hmuY and hmuR genes were amplified from the genomic DNA and ligated into the XbaI and HindIII or XbaI and KpnI restriction sites of the pTriEx-4 vector (Novagen), respectively, resulting in pTOYP1 and pTOYP2 plasmids (Table 2S). To construct the plasmid containing the hmu promoter and the hmuR gene, the promoter region was amplified and cloned into the XbaI and EcoRI restriction sites of the pTriEx-4 vector, resulting in the pTOYP plasmid. Then the hmuR gene was amplified and cloned into the EcoRI and KpnI restriction sites of the pTOYP plasmid, resulting in pTOYP3 plasmid (Table 1S). The transformants were selected on LB plates supplemented with 100 µg/ml ampicillin and the resultant plasmids were verified by PCR and sequencing analyses (data not shown). Finally, the E. coli EB mutant strain was transformed with the pTOYP, pTOYP1, pTOYP2, or pTOYP3 plasmids and grown on LB or M9 plates supplemented with 2 µg/ml ALA and 100 µg/ml ampicillin at 37°C.
To test the utilization of different iron sources in a liquid medium, E. coli cells were cultured in LB or M9 medium supplemented with ALA to an OD600 of about 0.6. Then the cells were starved in LB or M9 medium without ALA for 5 h. These cultures served as the inoculum into LB or M9 medium alone or media supplemented with 160 µM dipirydyl and hemin, hemoglobin, or ALA. Cultures (initial OD660 ~0.1) were incubated and the OD was measured after 8 h. Growth on different iron sources was also tested by spreading 100 µl of the starved cultures on LB or M9 agar plates or plates with 160 µM dipirydyl. Then paper disks loaded with 10 µl of ALA (0.2 mg/ml) and varying concentrations of hemoglobin (1–50 µM) or hemin (1–500 µM) were placed and the plates were incubated for 1–3 days at 37°C. To test for spontaneous revertants, all E. coli transformants were grown on LB or M9 plates without ALA.
Transcriptional fusions
To determine hmu promoter activity, constructs containing the entire promoter region or the promoter region fused inframe to a lacZ gene were amplified on P. gingivalis A7436 genomic DNA and pUC18 plasmid, respectively. First the PCR product amplified on genomic DNA was digested and cloned into the XbaI and XhoI restriction sites of the pTriEx-4 vector, resulting in the pTOYPA1 plasmid (Table 1S). Then the lacZ gene was amplified and cloned into the pTOYPA1 plasmid within the XhoI restriction site, resulting in pTOYPA2 plasmid. E. coli cells transformed with the respective plasmids were grown overnight in 10 ml of LB medium. This culture served as an inoculum (1 × 108 cells) to 25 ml of LB medium supplemented with 150 µM dipirydyl (low-iron conditions) or LB supplemented with 100 µM FeCl3 (high-iron conditions) and the bacteria were grown for 8 h. β-galactosidase activity was measured after sonication of the cells by the method of Miller (1972) with o-nitrophenyl-β-d-galactopyranoside (ONPG; 3 mg/ml) as a substrate, and the resultant enzymatic activity was expressed in Miller units.
Expression of HmuY and HmuR in E. coli cells and HmuY purification
The entire full-length version of the hmuY gene, the gene lacking the 21-amino-acid sequence encoding the predicted signal peptide and the lipidation Cys residue, the gene lacking the predicted signal peptide, the lipidation Cys residue and four additional amino acids (GKKK), and the entire hmuR gene were amplified on P. gingivalis A7436 genomic DNA and cloned into the respective NcoI and KpnI or XhoI restriction sites of the pTriEx-4 vector (all primers are listed in Table 2S). The resultant constructs (pCB, pDB, pFAF, and pHMUR, respectively; Table 1S) were used to overexpress HmuY protein variants containing the C-terminal fusion protein (HSV-His-tag) or HmuR protein containing the C-terminal His-tag. Recombinant HmuY protein variants not associated with the outer membrane were expressed and purified as described previously (Olczak et al. 2006) with slight modifications. Binding to metal affinity beads (TALON; Clontech) was performed at pH 6.8, and the protein was eluted with 150 mM imidazol in 20 mM sodium phosphate buffer, pH 6.8, containing 300 mM NaCl and 10% glycerol. To express HmuY without a fusion protein, the respective hmuY region was cloned into the NcoI and HindIII restriction sites of the pTriEx-4 vector, resulting in the pHMUY11 plasmid (Table 1S). The protein was expressed as described previously (Olczak et al. 2006) and purified using ion-exchange chromatography (DEAE-Sephacel; Amersham Pharmacia) in 50 mM Tris/HCl buVer, pH 7.6. The unbound fraction containing HmuY was further re-purified using gel filtration chromatography (Sephadex G-75; Amersham Pharmacia) in 20 mM sodium phosphate buffer, pH 7.4, containing 250 mM NaCl. The samples were dialyzed against PBS containing 1% glycerol and finally against PBS or 20 mM sodium phosphate buffer, pH 7.4, containing 20 mM NaCl.
UV–vis and CD spectra
Hemin solution preparation and examination of hemin binding to recombinant HmuY by UV–vis spectra were performed as described previously (Olczak et al. 2006). The CD spectra were recorded on a Jasco J-715 spectrophotometer at 20°C using a 10-mm-path-length cell with a 5 µM HmuY solution in 20 mM sodium phosphate buffer, pH 7.4, containing 20 mM NaCl. Data points were collected every 0.2 nm from 205 to 265 nm at a speed of 100 nm/min. The baseline was corrected by subtracting the spectra of the buffer remaining after dialysis measured under identical conditions. The signal-to-noise ratio was improved by accumulating five scans. The spectra are reported in terms of mean residue molar ellipticity [θ] (deg cm2 dmol−1). The formula used to calculate the mean residue ellipticity was: [θ] = (θ × 100 × MW)/(c × l × NA), where [θ] is the mean residue ellipticity, θ the experimental ellipticity in mdeg, MW the molecular weight of the protein in Daltons, c the protein concentration in mg/ml, l the cuvette’s path length in cm, and NA the number of amino acids of the protein.
Analytical gel filtration chromatography
Analytical gel filtration chromatography was performed using a Superdex 200 HR 10/30 column (Amersham Pharmacia) coupled to an FPLC system (Amersham Pharmacia). The column was equilibrated with 20 mM phosphate buffer, pH 7.4, containing 250 mM NaCl. Sample volumes of 200 µl (130 µg of protein) were injected onto the column and eluted at a flow rate of 0.5 ml/min. Proteins were followed by absorption measurements at 280 nm. Molecular masses of protein peaks were estimated relative to those of standard proteins: ribonuclease A (13.7 kDa), chymotrypsinogen (25 kDa), ovalbumin (43 kDa), bovine serum albumin (67 kDa), and catalase (232 kDa). The void volume was determined using blue dextran.
SDS-PAGE and Western blotting
The total protein concentration was determined using the modified Bradford method (Roti-Nanoquant, Roth) (Zor and Selinger 1996). Recombinant proteins were analyzed by SDS-PAGE or PAGE followed by staining with Coomassie Brilliant Blue G-250 and Western blotting using anti-polyHis antibodies conjugated with horseradish peroxidase (Sigma), as described previously (Olczak et al. 2006).
Statistical analysis
Data expressed as mean ± SD were analyzed using the Student’s t test; P values below 0.05 were considered significant.
Results
Analysis of the HmuY sequences in the P. gingivalis 381 and A7436 strains
Contradictory reports on HmuY prompted us to perform a detailed analysis of this coding region in several P. gingivalis strains. While this work was underway, Lewis et al. (2006) reported that the hmuY gene is absent in the 381 strain. However, using primers specific to the upstream and downstream regions of the hmuY sequence present in the P. gingivalis W83 strain (Table 2S), we successfully amplified and sequenced this genomic fragment located in the P. gingivalis 381 strain. The predicted sequence of the HmuY protein present in the P. gingivalis 381 strain has been deposited in the EMBL Nucleotide Sequence Database under the accession number AM489505. Detailed analysis of the hmuY gene sequence as well as proper annotation showed that the 381 strain’s HmuY protein is almost identical (Fig. 2) to the corresponding protein in other P. gingivalis strains (Nelson et al. 2003; Lewis et al. 2006; Ong et al. unpublished data). In contrast to previously published HmuY sequences or those available in databases (Simpson et al. 1999, 2000; Karunakaran et al. 1997; LANL and TIGR databases), but in agreement with the sequence of the HmuY protein from the P. gingivalis W83 and W50 strains (Lewis et al. 2006; Ong et al. unpublished data), the amino-acid sequence deduced here contained additional 74 amino acids at the N-terminus (Fig. 2). The same extension in HmuY was also present in protein derived from strain A7436 (EMBL accession number AM489506), which we previously used to characterize the truncated version of HmuY protein (Olczak et al. 2006). Also, the hmuY upstream region was found to be almost identical in various P. gingivalis strains except for a few base changes (Nelson et al. 2003; Lewis et al. 2006; and this study, data not shown).
Fig. 2.
Comparison of the amino-acid sequences of P. gingivalis HmuY protein with putative homologues present in the B. fragilis and B. thetaiotaomicron strains. P. gingivalis protein IDs: AAQ66587 (W83) (Nelson et al. 2003; Lewis et al. 2006); CAM31898 and AAF07986 (A7436) (Simpson et al. 1999, 2000; Karunakaran et al. 1997; and this study); ABL74281 (W50) (Ong et al. unpublished data); CAM31897 (381; this study); AAB46901 and AAB46902 (381; peptides derived from hemin-binding protein, HBP) (Kim et al. 1996); AAB25049 (W50; a peptide derived from fibroblast-activating factor, FAF) (Mihara and Holt 1993a). B. fragilis protein IDs: BAD49437 (YCH46) (Kuwahara et al. 2004); BAD47866 (YCH46) (Kuwahara et al. 2004); CAH08403 (NCTC 9343) (Cerdeno-Tarraga et al. 2005); CAH06766 (NCTC 9343) (Cerdeno-Tarraga et al. 2005); AAD56752 (638R) (Comstock et al. 1999). B. thetaiotaomicron protein ID: AAO75604 (VPI-5482) (Xu et al. 2003). The arrow indicates the putative cleavage site for signal peptidase II and the arrowhead the putative cleavage site for signal peptidase I. Asterisks denote positions with identical amino acids (also shown in gray boxes) and a colon those with similar amino acids. Dashes indicate gaps introduced to maximize the alignment
Analysis of the organization of the hmu operon and its transcription products
After the W83 P. gingivalis genome sequence was completed (Nelson et al. 2003) it was obvious that hmuY and hmuR are located in one operon together with four other genes (Fig. 1b). The gene arrangement at this hemin-uptake locus (hmu) seems to be highly conserved among P. gingivalis strains (data not shown). A similar operon organization was also found in B. fragilis and B. thetaiotaomicron strains (data not shown). However, comparison of the deduced amino-acid sequence of the P. gingivalis HmuY protein and a Bacteroides protein homologue encoded by this operon revealed a relatively low level of identity (Fig. 2a). Interestingly, in the genomes of the YCH46, NCTC 9343, and 638 strains of B. fragilis, but not in the genome of B. thetaiotaomicron, we found an ORF far more similar to P. gingivalis hmuY than that present in the hmu operon (Fig. 2b). This ORF in B. fragilis is located upstream of a gene encoding a TonB-dependent outer-membrane receptor similar to P. gingivalis HmuR (~28% identity and ~46% similarity) and downstream of a polysaccharide biosynthesis locus (data not shown). Finally, the genome sequence analysis of other dental pathogens, such as A. actinomycetemcomitans, T. denticola, and P. intermedia, did not reveal the presence of a hmu-like operon or a gene encoding a protein homologous to HmuY.
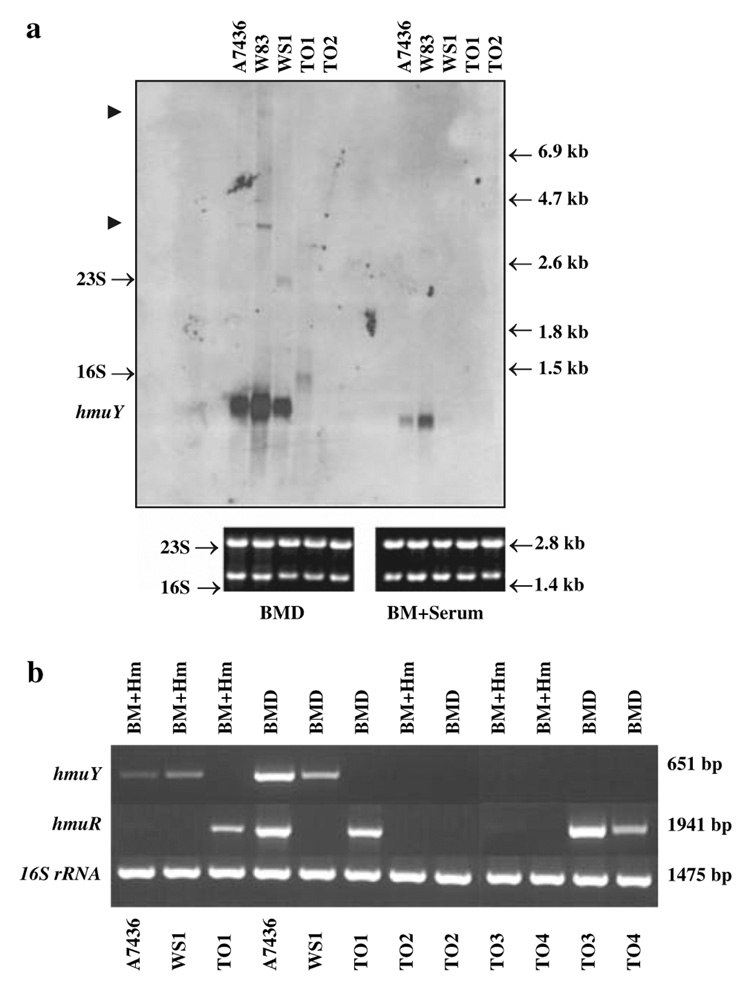
The contradictory published results on the size and expression level of the hmuY transcript (Karunakaran et al. 1997; Simpson et al. 2000; Lewis et al. 2006) prompted us to perform a detailed analysis of this transcription product. Northern blotting performed with a hmuY-specific RNA probe showed the presence of a ~1-kb transcript produced at high levels in bacteria grown under stringent iron/heme-restriction (Fig. 3a). Under these conditions we also observed small amounts of ~3.5 and ~9-kb transcripts produced by both wild-type strains. The size of the mRNA precisely matched the size of hmuY–hmuR and the entire operon polycistronic co-transcriptions, respectively. When starved wild-type P. gingivalis was grown in medium supplemented with human serum, only the ~1-kb hmuY transcript was formed (Fig. 3a), albeit at lower levels compared with the cells grown in iron/heme-restricted medium. We did not find the hmuY transcription product in A7436 and W83 cells grown under hemin-rich conditions (data not shown). Other positive signals seem to be unspecific since they do not correspond to the predicted sizes of the putative co-transcription products or transcription products containing the respective antibiotic cassettes. Using the hmuY-specific probe we did not observe the ~2-kb transcription product reported recently by Lewis et al. (2006). The production of transcripts of other members of the hmu operon is not shown here since those data were presented recently by others (Simpson et al. 1999, 2000; Liu et al. 2004; Lewis et al. 2006).
Fig. 3.
Analysis of the hmuY transcript by Northern blotting (a) and RT-PCR (b). a Total RNA was isolated from P. gingivalis wild-type and mutant strains grown in basal medium supplemented with 160 µM dipirydyl (BMD) or basal medium supplemented with 5% human serum (BM + serum). The hmuY transcript was detected using the hmuY-specific probe labeled with digoxigenin. Putative hmuY–hmuR and the entire operon co-transcription products are shown by arrowheads. Equal amounts of RNA (1.5 µg) were loaded onto formaldehyde aga-rose gels and checked for their purity and integrity. b RT-PCR. Total RNA was isolated from P. gingivalis wild-type and mutant strains. RNA (3 µg) and random hexamers were used for the reverse transcriptase reaction. To perform PCR, specific primers amplifying the entire sequences of the hmuY, hmuR, or the entire hmuY–hmuR region and 1 µl of a cDNA sample as a template were used. Analysis of the expression of 16S rRNA was used as a control. BM + Hm, basal medium supplemented with 1.5 µM hemin; BMD, basal medium supplemented with 160 µM dipirydyl; A7436 and W83, P. gingivalis wild-type strains; WS1, P. gingivalis hmuR::ermF mutant strain; TO1, P. gingivalis hmuY::tetQ mutant strain; TO2, P. gingivalis hmuY::tetQ hmuR::ermF mutant strain; TO3, P. gingivalis in-frame hmuY deletion mutant strain; TO4, P. gingivalis in-frame hmuY deletion mutant strain
Phenotypes of P. gingivalis mutant strains differ from the wild-type strain in heme and hemoprotein utilization
To determine whether HmuY and HmuR are both essential for P. gingivalis growth and survival, we first constructed mutants in the A7436 strain by insertional inactivation of the genes encoding these proteins (Fig. 1a). The respective antibiotic cassettes were inserted into the hmuY and/or hmuR genes such that their reading frames were in the same orientation as the hmu cluster. Since hmuY and hmuR are located at the beginning of the operon, there was concern that insertion of the ermF or tetQ cassettes would have a polar effect, resulting in the lack of transcription of downstream genes. To examine this, transcription of the genes located downstream of hmuY was analyzed by RT-PCR. This analysis showed that hmuR was transcribed in the TO1 strain (Fig. 3b), in keeping with a report that insertion of the ermF or tetQ cassette has no significant polar effect on the expressions of downstream genes in an operon (Limberger et al. 1999).
As determined by RT-PCR, the hmuR transcript was produced at high levels in cells grown under iron/heme-restricted conditions, whereas no transcript or a very low level of the hmuR transcript was produced in the cells grown under iron/heme-rich conditions (Fig. 3b), which is in agreement with previous studies (Simpson et al. 1999, 2000; Karunkaran et al. 1997; Liu et al. 2004). As shown in Fig. 3b, lower levels of the hmuY transcript were observed in the hmuR::ermF mutant strain (WS1) compared with both wild-type strains. In the hmuY mutant (TO1), similar expression of the hmuR transcript was observed regardless of the bacterial growth conditions (Fig. 3b). Since this expression was constitutive and not regulated by iron, we concluded that it could be driven by the tetQ promoter. Therefore, we constructed additional hmuY mutants (Fig. 1a). For this purpose, the internal 300-bp hmuY region was deleted and the ermF cassette was inserted in both directions downstream of the hmuR gene, resulting in the TO3 and TO4 mutant strains, respectively. As examined by RT-PCR, both mutant strains lacked the native hmuY transcript, but produced the hmuR transcript at levels similar to the wild-type strain (Fig. 3b).
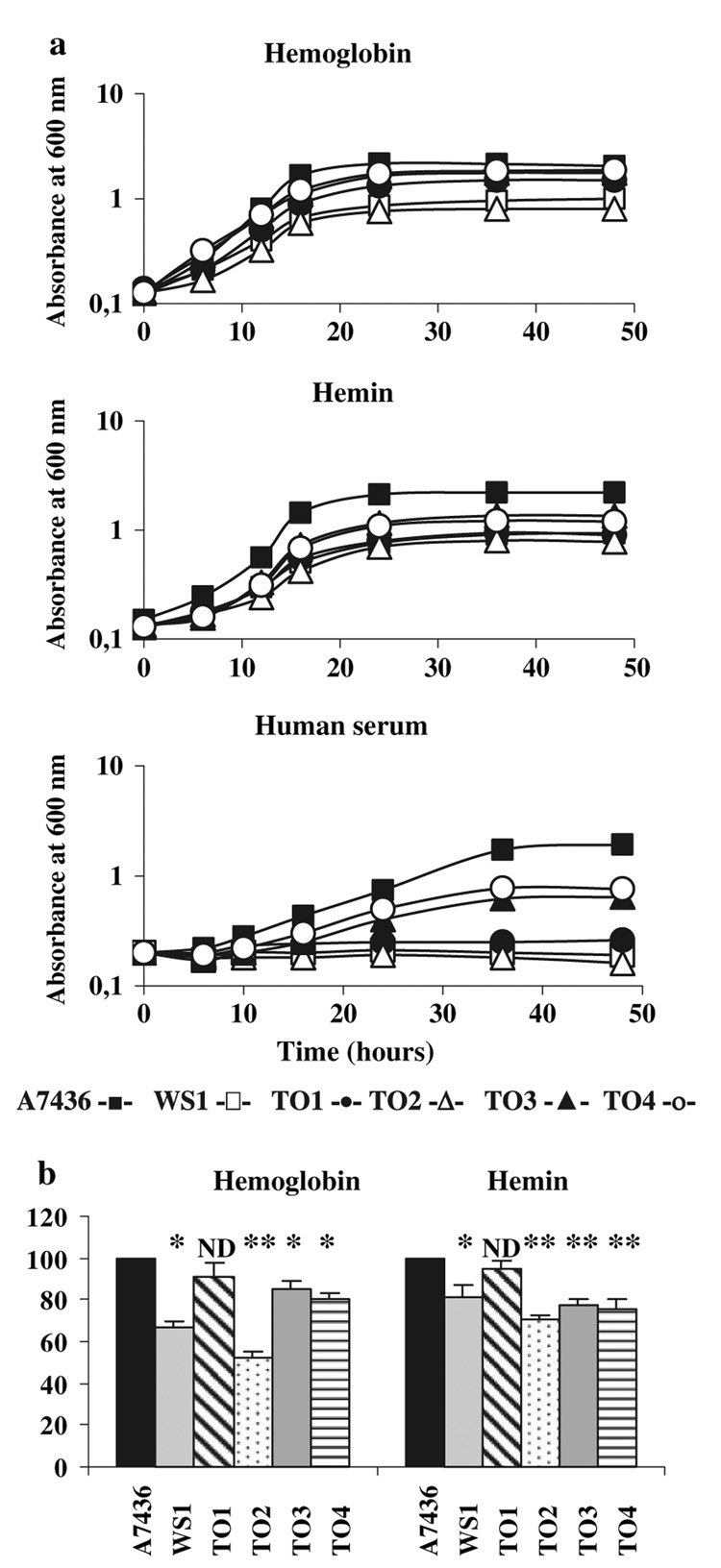
The phenotypes of the mutant strains grown on ABA plates were divergent, suggesting that only the respective genes were inactivated. In contrast to the dark-black colonies of the WS1 hmuR mutant, the TO1, TO3, TO4 hmuY, and TO2 hmuY–hmuR mutants became brown and beige, respectively, after prolonged growth on ABA plates (data not shown). To further characterize the mutant strains, first we examined the ability of all the strains to grow in the presence of various iron/heme sources. When hemin or hemoglobin was used as the sole iron/heme source, we observed defective growth of all the mutant strains (Fig. 4a). Interestingly, all the mutants exhibited decreased or no growth recovery when human serum was used as the sole iron/heme source. The phenotypes of the mutants were further examined by spectrophotometric analysis of their ability to bind hemoglobin or hemin. In comparison to the parental strain and the insertional hmuY mutant (TO1 mutant strain), in-frame hmuY deletion (TO3 and TO4 mutant strains) and double insertional hmuY-hmuR gene inactivation (TO2 mutant strain) resulted in significantly lower hemoglobin and hemin binding (Fig. 4b). The double mutant exhibited the most pronounced decrease in hemoglobin binding of all the mutants.
Fig. 4.
Phenotypic analysis of P. gingivalis A7436 wild-type and mutant strains. a Growth analysis. Starved P. gingivalis cultures were used to inoculate basal medium supplemented with hemoglobin (4 µM), hemin (1.5 µM), or human serum (5%). Bacterial growth was monitored by measuring OD660 at the indicated time points. Five independent experiments were performed and representative growth curves are shown. b Binding of hemoglobin or hemin. Starved P. gingivalis cells were adjusted to an OD660 of 1.0 and incubated with hemoglobin (4 µM) or hemin (10 µM). Hemin or hemoglobin binding to P. gingivalis whole cells was determined by comparing the decrease in the supernatant absorbance at 412 or 380 nm, respectively, of the mutant cells with that of the wild-type strain, which was arbitrarily set at 100%. Five independent experiments, each in triplicate, were performed and the mean ± SD values are shown. *P < 0.05 or **P < 0.005 (mutant strains versus the wild-type strain); ND, no difference. A7436, P. gingivalis wild-type strain; WS1, P. gingivalis hmuR::ermF mutant strain; TO1, P. gingivalis hmuY::tetQ mutant strain; TO2, P. gingivalis hmuY::tetQ hmuR::ermF mutant strain; TO3, P. gingivalis in-frame hmuY deletion mutant strain; TO4, P. gingivalis in-frame hmuY deletion mutant strain
HmuY and HmuR complement an E. coli hemA mutation
An E. coli hemA aroB mutant (EB53) has an enzymatic lesion in the heme biosynthesis pathway (Eberspacher and Braun 1980) and cannot use exogenously supplied heme. The E. coli K-12 outer membrane is impermeable to extra-cellular hemin and its precursor, protoporphyrin IX, and medium supplemented with hemin does not support growth of the mutant cells. We therefore used this mutant to characterize heme utilization through the hmuY–hmuR system by complementation of the heme biosynthesis defect. Initially, complementation analysis was performed in liquid media; however, under these conditions the mutant cells showed a tendency to produce revertants, since some growth in the absence of ALA occurred (data not shown). Therefore, the bacteria were cultured on solid media to minimize the reversion of the hemA mutation in the absence of ALA. The lack of reversion was documented by control plates with LB or M9 medium without ALA and without hemin (data not shown). In our preliminary experiments we found that when membrane-associated HmuR or HmuY were overexpressed under the T7 promoter in ER2566 E. coli and grown on LB plates containing 1 mM IPTG and 10 µM hemin, the cells, especially those expressing HmuY, became brown compared with control cells harboring the pUC18 vector alone (data not shown). This confirmed the ability of HmuR and HmuY to bind heme on the cell surface. When tested for the ability to use the exogenous heme in EB53 E. coli mutant cells, only the cells expressing both HmuY and HmuR under their native promoter grew in the presence of hemin (data not shown), indicating that the proteins mediated an efficient utilization of hemin. Neither HmuY or HmuR alone nor expression of HmuY and HmuR together in E. coli EB53 enabled the cells to use hemoglobin as a heme source (data not shown).
Analysis of hmu promoter activity
To examine whether the expression of the P. gingivalis heme-uptake system is regulated by iron, we constructed the transcriptional fusion plasmid and tested its expression in E. coli cells. The plasmid contained the hmu promoter region cloned immediately upstream of a lacZ reporter gene. As a negative control we used the plasmid containing the hmu promoter region only. As shown in Table 1, E. coli cells harboring the phmu–lacZ fusion exhibited higher β-galactosidase activity under iron restriction compared with high-iron conditions, suggesting iron influence on the hmu promoter activity.
Table 1.
Transcriptional analysis of hmu promoter activity
| Construct | β-galactosidase activity (Miller units) |
|
|---|---|---|
| High-iron conditionsa | Low-iron conditionsb | |
| phmu | 9.91 ± 0.52 | 11.93 ± 0.73 |
| phmu–lacZ | 17.52 ± 1.62 | 109.35 ± 1.65 |
High-iron conditions, LB medium supplemented with 100 µM FeCl3
Low-iron conditions, LB medium supplemented with 150 µM dipirydyl. Results are shown as means ± SD values from three independent experiments performed in duplicate
Biochemical analysis of HmuY protein
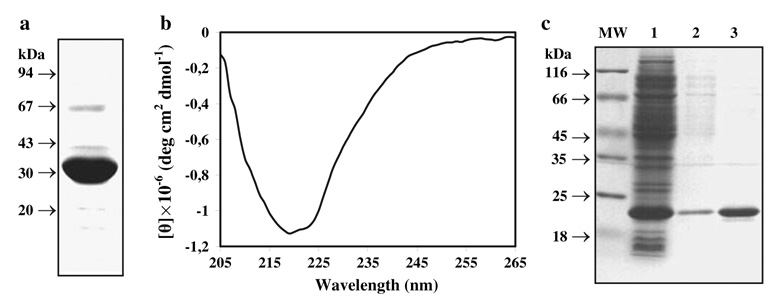
We previously reported on the overexpression, purification, and preliminary characterization of a truncated version of HmuY protein (Olczak et al. 2006). Here we overexpressed and purified under modified conditions a full-length version of the recombinant HmuY protein, containing the C-terminal HSV-His-tag. These improvements made it possible to isolate substantial amounts (>30 mg per liter of the culture) of highly purified protein and facilitated a more detailed protein analysis. In this study, we characterized two HmuY variants: a mature HmuY protein (mHmuY) lacking 21 N-terminal amino acids and a mature processed HmuY (mpHmuY) lacking 25 N-terminal amino acids, which were not found in a native protein purified from P. gingivalis cells (Mihara and Holt 1993a; Lewis et al. 2006). Since both HmuY variants showed identical properties, except for the slower migration of mpHmuY during electrophoresis under native conditions resulting from the different amino-acid composition, in this study we showed results for mpHmuY only. Since there was concern that a His-tag attached to HmuY may interfere with hemin binding, we also overexpressed and purified HmuY without a fusion protein. All the purified recombinant proteins were more than 98% homogenous as determined by densitometry scanning of the Coomassie Brilliant Blue G-250 stained SDS-PAGE gels (Fig. 5a, c). Analysis of the theoretical HmuY secondary structure revealed the presence of ~15% α helix, ~35% extended strand, ~12% β turn, and ~38% random coil. The representative CD spectrum shown in Fig. 5b showed that purified HmuY has a well-ordered structure containing high β structure content.
Fig. 5.
SDS-PAGE and far UV CD analysis of purified recombinant HmuY. a Purified recombinant HmuY with a HSV-His-tag was separated in 12% SDS-PAGE gel and stained with Coomassie Brilliant Blue G-250. All protein bands were visualized with anti-His antibodies, suggesting the presence of a monomer, oligomers, and degradation products (data not shown). b CD analysis of purified recombinant HmuY. c Purification of HmuY variant without a fusion protein. 1 E. coli cell lysate; 2 HmuY purified by ion-exchange chromatography; 3 HmuY re-purified by gel filtration chromatography
Previously, we demonstrated that the truncated version of HmuY protein was overexpressed and purified as apoprotein which bound hemin in vitro (Olczak et al. 2006). Interestingly, we now noticed that full-length mature HmuY, overexpressed at high yield and purified, was colored, indicating that it may be associated with heme. Indeed, UV–vis analysis of the protein samples containing HmuY with a His-tag or HmuY alone displayed a diagnostic absorbance peak characteristic for protein-bound heme (the Soret band) at 411 or 413 nm, respectively (data not shown). Although the ratio of A411/A280 or A413/A280 was low (~0.1), these data suggest that recombinant HmuY can efficiently scavenge in vivo intracellular heme within the E. coli cell. After addition of hemin (iron protoporphyrin IX chloride) to purified HmuY with a His-tag or HmuY alone, the peak at 411 or 413 nm, corresponding to holo-protein, increased in a dose-dependent manner and the absorption maxima at 526 and 559 or 531 and 561 nm, respectively, became clearly visible (data not shown). Further, we attempted to examine the stoichiometry of hemin binding to all variants of HmuY and found that one protein monomer bound one hemin moiety (data not shown). This finding is in agreement with our previous report showing a 1:1 molar ratio of hemin binding to the truncated version of HmuY (Olczak et al. 2006).
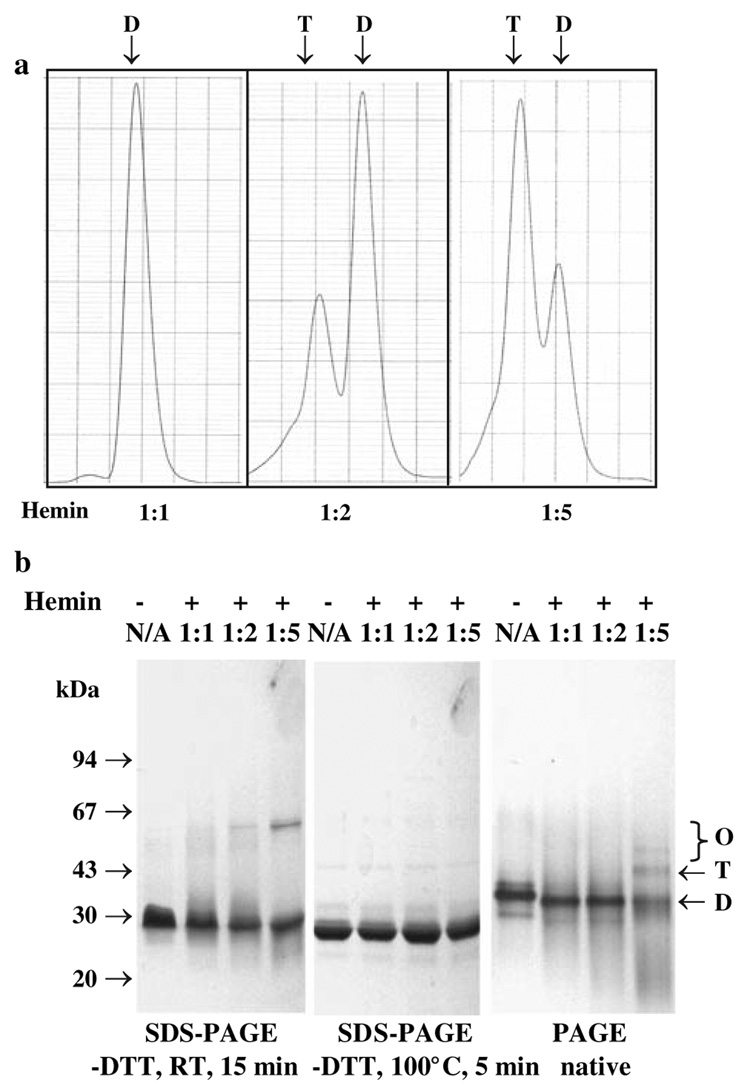
Preliminary experiments suggested that HmuY may form dimeric or higher-order oligomeric structures (Olczak et al. 2006; Lewis et al. 2006). Therefore in this study the oligomeric state of HmuY with a His-tag or HmuY alone was examined by analytical gel filtration chromatography. The gel filtration analysis revealed that both variants of HmuY eluted as dimers, and Fig. 6a shows a typical separation pattern. Upon addition of hemin to the protein samples, tetramers were formed. When the HmuY protein was analyzed by PAGE and SDS-PAGE, a tendency to form oligomers in the presence of hemin was also observed (Fig. 6c). It is also worth noting that the formation of dimers, tetramers and oligomers by HmuY, as judged by gel filtration and electrophoresis, was independent of the presence of DTT, suggesting that disulfide bonds are not involved in the multimerization of recombinant HmuY.
Fig. 6.
Determination of the HmuY oligomeric state. a Analytical gel filtration chromatography. Protein separation was carried out using Superdex 200 HR 10/30 with 130 µg of HmuY in 20 mM sodium phosphate buffer, pH 7.4, containing 250 mM NaCl. Typical profiles are shown. b SDS-PAGE and PAGE. Separation of HmuY by electrophoresis was performed in 12% denaturing or native gels and visualized by staining with Coomassie Brilliant Blue G-250. Hemin was added to HmuY to obtain 1:1, 1:2, or 1:5 molar ratios. N/A, hemin not added to the purified HmuY sample. D Dimer, T tetramer, O oligomers
Discussion
As part of our ongoing studies we further analyzed heme uptake in P. gingivalis by HmuY and HmuR proteins. Previously, we showed that HmuY binds hemin in vitro (Olczak et al. 2006). We proposed that the heme–HmuY interaction has rather a transient character, since the protein was purified in a heme-free form. Conversely, here we have demonstrated that the full-length HmuY purifies as a mixture of apo- and holo-protein and the protein binds hemin in vitro with high affinity. In the likely scenario in vivo, HmuY may bind hemin at the cell surface, serving as an initial heme receptor. In the following step, heme is passed from HmuY to the HmuR protein, which is the TonB-dependent outer-membrane heme transporter. A similar mechanism was described in Neisseria meningitidis, where HpuA and HpuB act as a complex involved in the internalization of heme present in hemoglobin (Lewis et al. 1997). The arrangement of the first two genes at the hmu locus is also analogous to that of the lactoferrin and transferrin binding systems in Neisseria and Haemophilus species, respectively (Gray-Owen and Schryvers 1996). Indeed, analysis of the mutant phenotypes after prolonged growth of P. gingivalis cells on ABA plates argues for the presence of a similar mechanism in P. gingivalis. The colonies of the WS1 mutant lacking the functional HmuR transporter become dark black, suggesting that HmuY protein binds heme, but in the absence of HmuR heme cannot be efficiently transported into the cell and the mutant shows retarded growth compared with the parental strain. The hmuY mutants also show growth retardation, but in this case it is due to the inefficient binding of hemin, illustrated by beige or, at best, brownish color of colonies on blood agar. Despite this functional similarity of the HmuY/HmuR system to the Neisseria and Haemophilus heme-acquisition systems, HmuY is a unique protein and occurs only in some species of the Bacteroidetes phylum.
Although HmuY binds heme, it does not possess a typical heme binding motif. It has been recently reported that the cofacial hemes may be enclosed by a pocket formed by a dimer of a putative Campylobacter jejuni ChaN lipoprotein composed of two protein monomers, allowing the heme dimer to serve as a bridge linking two protein molecules (Chan et al. 2006). This lipoprotein is thought to associate with the outer membrane and interact with the outer-membrane receptor ChaR. It is likely that a similar mechanism may occur in P. gingivalis. Previously, we suggested that purified HmuY exists in a dimeric form which could participate in heme binding (Olczak et al. 2006), and this study supported our hypothesis. The recombinant full-length mature form of HmuY can bind hemin very eYciently, although it purifies from E. coli cell extract only partially saturated with heme. When the Soret band of HmuY with a His-tag and HmuY alone were examined, only a slight shift was observed, suggesting that a His-tag does not significantly interfere with hemin binding. Interestingly, being a dimer at low hemin concentrations, both variants of HmuY form a tetramer in the presence of an excess of hemin. It is likely that this property could be a common trait of this novel protein family.
Other investigators showed that, in contrast to the hmuR gene, which was downregulated in iron/heme-rich media (Karunakaran et al. 1997; Liu et al. 2004; Simpson et al. 2000), the hmuY gene was upregulated under these conditions (Karunakaran et al. 1997). Our results are in contrast to these studies and demonstrate that under iron/heme-restricted conditions the hmuY transcript is produced with high efficiency. We also found that in addition to the single hmuY transcript, during iron/heme starvation P. gingivalis A7436 and W83 cells co-transcribe the hmuY–hmuR gene couple as well as the entire operon, albeit at a very low level. Such a process might be involved in the regulation of HmuY and HmuR levels in the cell and requires further investigation. Similar regulation has been observed for polycistronic puf mRNA in Rhodobacter capsulatus (Heck et al. 2000) and dicistronic pocA-pfl mRNA in E. coli (Sawers 2005), where the processing and differential decay of primary transcripts were observed. Since hmuY and hmuR are part of a transcriptional unit under the control of one promoter and hmuY should be co-transcribed together with other genes in the hmu operon, the presence of the single dominating transcript could result from the processing of the primary transcript. Lewis et al. (2006) suggested that secondary mRNA structures may be responsible for the differences in the size and level of hmu transcripts. However, no analysis supporting such a hypothesis was presented.
All these data prompted us to further investigate the putative role of both HmuY and HmuR proteins using mutational analysis. In contrast to data obtained from P. gingivalis grown in the presence of serum, all the other differences observed between the wild-type and mutant strains grown in the presence of hemin or hemoglobin as well as differences determined in hemin- or hemoglobin-binding experiments were moderate. Although these two genes, encoded in one operon, seem to be very important for P. gingivalis heme utilization, the disruption of hmuY alone, hmuR alone, or both hmuY and hmuR did not totally abrogate hemin or hemoglobin utilization. It is well known that P. gingivalis expresses additional iron/heme utilization systems and the mutants’ growth may be promoted by the presence of other proteins (Nelson et al. 2003; Olczak et al. 2005). Literature data and the results presented in this study suggest that various iron/heme systems respond to different levels of iron/heme in the environment. Some operons may require lower iron/heme levels for expression than others. HmuY–HmuR cooperation is probably required at early stages of P. gingivalis growth under the low iron/heme conditions typical of healthy gingiva or early periodontitis development when heme is not easily available.
Bacterial genes are often organized in operons to coordinate the production of proteins engaged in the same metabolic or regulatory pathways. The most commonly described system of iron/heme uptake consists of the heme-specific TonB-dependent receptor, a periplasmic iron/heme-binding protein, an ATPase, and a permease which delivers heme to the cytoplasm. Although similar genetic organization of heme-uptake systems has been described in many Gram-negative bacteria, their transcriptional organization and gene regulation are still unclear. In addition, the completion of the genome sequencing of increasing numbers of species has shown that the operons of heme uptake often exhibit different organization. An example is the hmu cluster found in P. gingivalis. It seems that this operon consists of a lipoprotein associated with the outer membrane (HmuY), a TonB-dependent outer membrane receptor (HmuR), a protein belonging to the CobN/Mg chelatase family, a putative ATPase, a protein belonging to the MotA/TolQ/ExbB family, and a putative protein without an assigned function. Such an organization is different from that typically found in other Gram-negative bacteria. Recently, Lewis et al. (2006) defined the hmu cluster according to a typical iron-uptake locus and suggested that HmuS may function as a cobalamin biosynthesis (CobN)/ magnesium chelatase, HmuT and HmuU may function as permeases, and HmuV may be localized in the inner membrane. This prediction needs to be experimentally verified since already now it seems that at least some of these proteins may play different roles from those previously proposed. It seems that a protein assigned a CobN/magnesium chelatase function may be involved in P. gingivalis in vitamin B12 synthesis (Rodionov et al. 2003). Components of vitamin B12 transporters are usually encoded by clusters of co-localized genes that are regulated by the B12 element. Although such an element was found upstream of the P. gingivalis hmu operon, it is likely that this operon may be involved in the transport of various metalloporphyrins rather than in cyanocobalamin biosynthesis. Our previous results showing binding of metalloporphyrins to the HmuR receptor (Olczak et al. 2001) supports this hypothesis. Another protein with different function may be the MotA/ TolQ/ExbB homologue, defined by Lewis et al. (2006) as a permease.
An arrangement of genes similar to that of the P. gingivalis hmu operon was found in the B. fragilis and B. thetaiotaomicron species of the Bacteroidetes phylum, suggesting that this gene cluster plays an important role in these heme-requiring bacteria. It is also worth noting that an additional HmuY-like protein was found in B. fragilis strains encoded in a transcriptional unit with a HmuR homologue. As more microbial genomes have been sequenced, it is apparent that many species may express more than one mechanism involved in heme uptake. Analysis of several P. gingivalis strains and isolates showed a substantial extent of recombination (Frandsen et al. 2001; Koehler et al. 2003). Apparently, dental plaque may constitute a favorable environment for horizontal gene transfer and recombination between different bacteria. In keeping with this, it has recently been shown that the natural competence for DNA uptake increases when bacteria are grown in plaque-like biofilms (Wang et al. 2002). Frequent recombination between strains is likely to facilitate the spread of favorable traits, such as heme-utilization mechanisms.
Heme-uptake systems negatively regulated by iron/heme have been described for many Gram-negative bacteria (Stojiljkovic and Hantke 1992; Letoffe et al. 1994; Mills and Payne 1995; Thompson et al. 1999; Ochsner et al. 2000). The iron-dependent control in these bacteria is usually mediated through the Fur protein. Based on the literature data (Simpson et al. 2000; Liu et al. 2004; Lewis et al. 2006) and the results presented here it is clear that the transcription of hmuY and hmuR is regulated by iron. However, we and others (Lewis et al. 2006) did not find any matches to the Fur consensus binding sequence in the hmu promoter region. It has been recently reported that the heme-binding protein HbpA in Pasteurella multocida is regulated by iron and heme by a Fur-independent mechanism (Garrido et al. 2003). Heme-uptake systems which are regulated by iron but do not contain a sequence homologous to the Fur box in the promoter region have also been identified in other bacteria (Nienaber et al. 2001). It is also likely that other factors in addition to iron or heme regulate the expressions of these genes. Recently, the regulation of iron/heme-uptake systems by the luxS product was described in P. gingivalis and other bacteria (James et al. 2006; Burgess et al. 2002; Chung et al. 2001; Fong et al. 2001). Thus the mechanism of the iron regulation of hmuY and hmuR transcription needs further studies, which are in progress in our laboratories.
In conclusion, our data show that the utilization of heme alone or complexed to hemoproteins in P. gingivalis is dependent on the HmuY and HmuR proteins. It seems that the strategies by which bacteria respond to iron alone or iron present in the heme moiety may be more diverse than once thought. It is likely that the HmuY could be the first reported member of a new class of proteins in Porphyromonas and other Bacteroidetes species involved in a novel mechanism of heme acquisition, a function being exerted in conjunction with HmuR, the outer-membrane heme transporter. Detailed biochemical and structural analysis of P. gingivalis HmuY in the presence of heme is required to establish its function in heme uptake. Further understanding of the HmuY function also requires the identification of its interacting partners in the bacterial cell. These studies are now being carried out in our laboratories
Acknowledgments
Dr. C.A. Genco (Section of infectious diseases, Boston University School of Medicine, USA) is gratefully acknowledged for giving T.O. the opportunity to continue studies on P. gingivalis heme utilization. The authors thank Dr. Klaus Hantke (University of Tübingen, Germany) for generously supplying us with the E. coli K-12 EB53 strain. This work was supported in part by grants nos. 3 P05A 113 24 and N401 029 32/0742 from the Department of Scientific Research, Ministry of Science and Higher Education, Poland (T.O.) and NIH grant (DE 09761), USA (J.P.). J.P. is a Subsydium Profesorskie award recipient from the Foundation for Polish Science (Warsaw, Poland). Part of the data was presented at the FEBS Congress and the 9th IUBMB Conference (Budapest, Olczak et al. 2005, FEBS J 272S1:15).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00203-007-0309-7) contains supplementary material, which is available to authorized users.
Contributor Information
Teresa Olczak, Laboratory of Biochemistry, Faculty of Biotechnology, University of Wroclaw, Tamka 2, 50-137 Wroclaw, Poland e-mail: Teresa.Olczak@biotech.uni.wroc.pl.
Aneta Sroka, Department of Microbiology, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Gronostajowa 7, 30-387 Krakow, Poland.
Jan Potempa, Department of Microbiology, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Gronostajowa 7, 30-387 Krakow, Poland.
Mariusz Olczak, Laboratory of Biochemistry, Faculty of Biotechnology, University of Wroclaw, Tamka 2, 50-137 Wroclaw, Poland.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Boeekmann B. The SWISS-PROT protein sequence data bank. Nucleic Acids Res. 1991;19:2247–2249. doi: 10.1093/nar/19.suppl.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess NA, Kirke DF, Williams P, Winzer K, Hardie KR, Meyers NL, Aduse-Opoku J, Curtis MA, Camara M. LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates protease and haemagglutinin activities but is not essential for virulence. Microbiology. 2002;148:763–772. doi: 10.1099/00221287-148-3-763. [DOI] [PubMed] [Google Scholar]
- Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, Barron A, Clark L, Corton C, Doggett J, Holden MT, Larke N, Line A, Lord A, Norbertczak H, Ormond D, Price C, Rabbinowitsch E, Woodward J, Barrell B, Parkhill J. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- Chan AC, Lelj-Garolla B, Rosell F, Pedersen KA, Mauk AG, Murphy ME. Cofacial heme binding is linked to dimerization by a bacterial heme transport protein. J Mol Biol. 2006;362:1108–1119. doi: 10.1016/j.jmb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Chung WO, Park Y, Lamont RJ, McNab R, Barbieri B, Demuth DR. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J Bacteriol. 2001;183:3903–3909. doi: 10.1128/JB.183.13.3903-3909.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock LE, Coyne MJ, Tzianabos AO, Kasper DL. Inter-strain variation of the polysaccharide B biosynthesis locus of Bacteroides fragilis: characterization of the region from strain 638R. J Bacteriol. 1999;181:6192–6196. doi: 10.1128/jb.181.19.6192-6196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberspacher B, Braun V. The involvement of cytochromes in the uptake of ferrichrome by Escherichia coli K-12. FEMS Microbiol Lett. 1980;7:61–64. [Google Scholar]
- Fong KP, Chung WO, Lamont RJ, Demuth DR. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect Immun. 2001;69:7625–7634. doi: 10.1128/IAI.69.12.7625-7634.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen EV, Poulson K, Curtis MA, Kilian M. Evidence of recombination in Porphyromonas gingivalis and random distribution of putative virulence markers. Infect Immun. 2001;69:4479–4485. doi: 10.1128/IAI.69.7.4479-4485.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido ME, Bosch M, Medina R, Bigas A, Llagostera M, Perez de Rozas AM, Badiola I, Barbe J. fur-independent regulation of the Pasteurella multocida hbpA gene encoding a haemin-binding protein. Microbiology. 2003;149:2273–2281. doi: 10.1099/mic.0.26370-0. [DOI] [PubMed] [Google Scholar]
- Genco CA, Dixon DW. Emerging strategies in microbial heme capture. Mol Microbiol. 2001;391:1–11. doi: 10.1046/j.1365-2958.2001.02231.x. [DOI] [PubMed] [Google Scholar]
- Geourjon C, Deleage G. Significant improvement in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- Gray-Owen SD, Schryvers AS. Bacterial transferrin and lacto-ferrin receptors. Trends Microbiol. 1996;4:185–191. doi: 10.1016/0966-842x(96)10025-1. [DOI] [PubMed] [Google Scholar]
- James CE, Hasegawa Y, Park Y, Yeung V, Tribble GD, Kuboniwa M, Demuth DR, Lamont RJ. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect Immun. 2006;74:3834–3844. doi: 10.1128/IAI.01768-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Juncker AS, Willenbrock H, von Heijne G, Nielsen H, Brunak S, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck C, Balzer A, Fuhrmann O, Klug G. Initial events in the degradation of the polycistronic puf mRNA in Rhodobacter capsulatus and consequences for further processing steps. Mol Microbiol. 2000;35:90–100. doi: 10.1046/j.1365-2958.2000.01679.x. [DOI] [PubMed] [Google Scholar]
- Karunakaran T, Madden T, Kuramitsu H. Isolation and characterization of a hemin-regulated gene, hemR, from Porphyromonas gingivalis. J Bacteriol. 1997;179:1898–1908. doi: 10.1128/jb.179.6.1898-1908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Chu L, Holt SC. Isolation and characterization of a hemin-binding cell envelope protein from Porphyromonas gingivalis. Microb Pathog. 1996;21:65–70. doi: 10.1006/mpat.1996.0043. [DOI] [PubMed] [Google Scholar]
- Koehler A, Karch H, Beikler T, Flemmig TF, Suerbaum S, Schmidt H. Multilocus sequence analysis of Porphyromonas gingivalis indicates frequent recombination. Microbiology. 2003;149:2407–2415. doi: 10.1099/mic.0.26267-0. [DOI] [PubMed] [Google Scholar]
- Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, Kuhara S, Hattori M, Hayashi T, Ohnishi Y. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci USA. 2004;101:14919–14924. doi: 10.1073/pnas.0404172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letoffee S, Ghigo JM, Wandersman C. Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc Natl Acad Sci USA. 1994;91:9876–9880. doi: 10.1073/pnas.91.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LA, Gray E, Wang YP, Roe BA, Dyer DW. Molecular characterization of hpuAB, the haemoglobin–haptoglobin-utilization operon of Neisseria meningitidis. Mol Microbiol. 1997;23:737–749. doi: 10.1046/j.1365-2958.1997.2501619.x. [DOI] [PubMed] [Google Scholar]
- Lewis JP, Plata K, Fan Y, Rosato A, Anaya C. Transcriptional organization, regulation and role of the Porphyromonas gingivalis W83 hmu haemin-uptake locus. Microbiology. 2006;152:3367–3382. doi: 10.1099/mic.0.29011-0. [DOI] [PubMed] [Google Scholar]
- Limberger RJ, Slivienski LL, Izard J, Samsonoff WA. Insertional inactivation of Treponema denticola tap1 results in a non-motile mutant with elongated flagellar hooks. J Bacteriol. 1999;181:3743–3750. doi: 10.1128/jb.181.12.3743-3750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sroka A, Potempa J, Genco CA. Coordinate expression of the Porphyromonas gingivalis lysine-specific gingipain proteinase, Kgp, arginine-specific gingipain proteinase, RgpA, and the heme/hemoglobin receptor, HmuR. Biol Chem. 2004;385:1049–1057. doi: 10.1515/BC.2004.136. [DOI] [PubMed] [Google Scholar]
- Liu X, Olczak T, Guo HC, Dixon DW, Genco CA. Identification of essential amino acid residues required for hemoprotein utilization in the Porphyromonas gingivalis heme receptor HmuR. Infect Immun. 2006;74:1222–1232. doi: 10.1128/IAI.74.2.1222-1232.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara J, Holt SC. Purification and characterization of fibroblast-activating factor isolated from Porphyromonas gingivalis W50. Infect Immun. 1993a;61:588–595. doi: 10.1128/iai.61.2.588-595.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara J, Holt SC. Modulation of growth and function of human gingival fibroblast-activating factor derived from Porphyromonas gingivalis W50. Infect Immun. 1993b;61:596–601. doi: 10.1128/iai.61.2.596-601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara J, Yoneda T, Holt SC. Role of Porphyromonas gingivalis-derived fibroblast-activating factor in bone resorption. Infect Immun. 1993;61:3562–3564. doi: 10.1128/iai.61.8.3562-3564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor: Cold Springer Harbor Laboratory Press; 1972. [Google Scholar]
- Mills M, Payne SM. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J Bacteriol. 1995;177:3004–3009. doi: 10.1128/jb.177.11.3004-3009.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, Daugherty SC, Dodson RJ, Durkin AS, Gwinn M, Haft DH, Kolonay JF, Nelson WC, Mason T, Tallon L, Gray J, Granger D, Tettelin H, Dong H, Galvin JL, Duncan MJ, Dewhirst FE, Fraser CM. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–5601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nienaber A, Hennecke H, Fischer HM. Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum. Mol Microbiol. 2001;41:787–800. doi: 10.1046/j.1365-2958.2001.02555.x. [DOI] [PubMed] [Google Scholar]
- Ochsner UA, Johnson Z, Vasil ML. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology. 2000;146:185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- Olczak T. Heme receptor HmuR from Porphyromonas gingivalis: towards a further understanding of heme uptake. Arch Microbiol. 2006;186:393–402. doi: 10.1007/s00203-006-0151-3. [DOI] [PubMed] [Google Scholar]
- Olczak T, Dixon DW, Genco CA. Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins. J Bacteriol. 2001;183:5599–5608. doi: 10.1128/JB.183.19.5599-5608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olczak T, Simpson W, Liu X, Genco CA. Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiol Rev. 2005;29:119–144. doi: 10.1016/j.femsre.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Olczak T, Siudeja K, Olczak M. Purification and initial characterization of a novel Porphyromonas gingivalis HmuY protein expressed in Escherichia coli and insect cells. Protein Expr Purif. 2006;49:299–306. doi: 10.1016/j.pep.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Pept Sci. 2003;4:397–407. doi: 10.2174/1389203033487036. [DOI] [PubMed] [Google Scholar]
- Rasmussen JL, Odelson DA, Macrina FL. Complete nucleotide sequence and transcription of ermF, a macrolide-lincosamide-streptogramin B resistance determinant from Bacteroides fragilis. J Bacteriol. 1986;168:523–533. doi: 10.1128/jb.168.2.523-533.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in Prokaryotes. J Biol Chem. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- Sawers RG. Evidence for novel processing of the anaerobically inducible dicistronic focA-pfl mRNA transcript in Escherichia coli. Mol Microbiol. 2005;58:1441–1453. doi: 10.1111/j.1365-2958.2005.04915.x. [DOI] [PubMed] [Google Scholar]
- Simpson W, Wang CY, Bond V, Potempa J, Mikolajczyk-Pawlinska J, Travis J, Genco CA. Transposition of the endogenous insertion sequence element IS1126 modulates gingipain expression in Porphyromonas gingivalis. Infect Immun. 1999;67:5012–5020. doi: 10.1128/iai.67.10.5012-5020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson W, Olczak T, Genco CA. Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J Bacteriol. 2000;182:5737–5748. doi: 10.1128/jb.182.20.5737-5748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson W, Olczak T, Genco CA. Lysine-specific gingipain K and heme/hemoglobin receptor HmuR are involved in heme utilization in Porphyromonas gingivalis. Acta Biochim Pol. 2004;51:253–262. [PubMed] [Google Scholar]
- Stojiljkovic I, Hantke K. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 1992;11:4359–4376. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JM, Jones HA, Perry RD. Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect Immun. 1999;67:3879–3892. doi: 10.1128/iai.67.8.3879-3892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BY, Chi B, Kuramitsu HK. Genetic exchange between Treponema denticola and Streptomyces gordonii in biofilms. Oral Microbiol Immunol. 2002;17:108–112. doi: 10.1046/j.0902-0055.2001.00001.x. [DOI] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Zor T, Selinger Z. Linearization of the Bradford protein assay increases its sensitivity: theoretical and experimental studies. Anal Biochem. 1996;236:302–308. doi: 10.1006/abio.1996.0171. [DOI] [PubMed] [Google Scholar]