Abstract
CTCF is a zinc finger DNA-binding protein that regulates the epigenetic states of numerous target genes. Using allelic regulation of mouse insulin-like growth factor II (Igf2) as a model, we demonstrate that CTCF binds to the unmethylated maternal allele of the imprinting control region (ICR) in the Igf2/H19 imprinting domain and forms a long-range intrachromosomal loop to interact with the three clustered Igf2 promoters. Polycomb repressive complex 2 is recruited through the interaction of CTCF with Suz12, leading to allele-specific methylation at lysine 27 of histone H3 (H3-K27) and to suppression of the maternal Igf2 promoters. Targeted mutation or deletion of the maternal ICR abolishes this chromatin loop, decreases allelic H3-K27 methylation, and causes loss of Igf2 imprinting. RNA interference knockdown of Suz12 also leads to reactivation of the maternal Igf2 allele and biallelic Igf2 expression. CTCF and Suz12 are coprecipitated from nuclear extracts with antibodies specific for either protein, and they interact with each other in a two-hybrid system. These findings offer insight into general epigenetic mechanisms by which CTCF governs gene expression by orchestrating chromatin loop structures and by serving as a DNA-binding protein scaffold to recruit and bind polycomb repressive complexes.
The transcriptional regulator CCCTC-binding factor (CTCF) is a highly conserved 11-zinc-finger nuclear protein that controls the expression of a number of genes via chromatin insulation or enhancer blocking (for reviews, see references 5, 8, 23, and 28). CTCF silences genes by binding to sites within promoters, silencers, and insulators through the use of different combinations of zinc fingers (20). More than 15,000 CTCF-binding sites have been identified throughout the genome (16).
The role of CTCF as an insulator regulating the imprinting of Igf2 and H19 has been extensively studied. Igf2 and H19 imprinting is directed by epigenetic modifications in the differentially methylated region (DMR) of the imprinting control region (ICR) located between these two adjacent genes (1, 9, 19, 21, 29, 30). The binding of CTCF to the unmethylated maternal ICR creates a physical boundary, blocking the interaction of downstream enhancers with the remote Igf2 promoters and silencing the maternal allele (4, 13, 15). When this ICR is deleted (35) or mutated (32, 34), the maternal Igf2 allele is expressed, leading to biallelic expression. In addition, CTCF has recently been shown to act as a tethering protein, serving as a molecular glue to secure long-range intrachromosomal (17) and interchromosomal (18) interactions.
By chromosome configuration capture (3C) methodology, it has been shown that CTCF participates in the formation of a long-range chromosomal loop to the upstream Igf2 DMRs when it is bound to the maternal ICR (17, 42, 21). This model suggests that CTCF may not only function as a physical insulator but also actively participate in the regulation of the imprinted Igf2 allele. We were interested in learning how CTCF mediates the suppression of three imprinted Igf2 promoters that are located 90 kb upstream of the ICR. We postulated that CTCF mediates the suppression of the three imprinted maternal Igf2 promoters (P1 to P3) by guiding the formation of a suppressor complex around the three promoters.
MATERIALS AND METHODS
Cell lines.
Mouse fibroblast MBW2 cells were cultured from an F1 newborn mouse derived from breeding a Mus spretus male with a C57B/6 female (6). HBF1 human fibroblast cells were cultured from the skin of a human fetus as previously described (14). ICR deletion-containing mouse fibroblasts, kindly provided by M. S. Bartolomei, were cultured from neonates generated from reciprocal crosses of C57BL/6(CAST) with F1 heterozygotes maintained in a C57BL/6 background (35). Fetal liver tissues, kindly provided by P. E. Szabo, were derived from breeding male FVB/NJ.CAST/Ei(N7) and female 129SI/ImJ mice to produce F1 mice that are heterozygous for a mutation in the ICR (34).
Chromosome conformation capture (3C).
MBW2 mouse fibroblast cells derived from an F1 newborn mouse bred from an M. spretus male crossed with a C57B/6 female (6) were used for this study. The 3C assay was performed by a previously described method (7) as modified by Murrell et al. (21). Briefly, 107 MBW2 cells were cross-linked with 2% formaldehyde and lysed with cell lysis buffer (10 mM Tris [pH 8.0], 10 mM NaCl, 0.2% NP-40, protease inhibitors). Nuclei were collected, suspended in 1× restriction enzyme buffer in the presence of 0.3% sodium dodecyl sulfate (SDS), and incubated at 37°C for 1 h. Triton X-100 was then added to a final concentration of 1.8% to sequester the SDS. An aliquot of nuclei (2 × 106) was digested with 800 U of restriction enzyme at 37°C overnight. After stopping the reaction by adding 1.6% SDS and incubating the mixture at 65°C for 20 min, chromatin DNA was diluted with NEB ligation reaction buffer and 2 μg DNA was ligated with 4,000 U of T4 DNA ligase (New England BioLabs) at 16°C for 4 h (final DNA concentration, 2.5 μg/ml). After treatment with 10 mg/ml proteinase K at 65°C overnight to reverse cross-links and with 0.4 μg/ml RNase A for 30 min at 37°C, DNA was extracted with phenol-chloroform, ethanol precipitated, and used for PCR amplification for the ligated DNA products. Information about the PCR primers used in this study is available on request. To distinguish the two parental alleles, DNA was digested with polymorphic restriction enzymes HpaII and DpnII, which distinguishes polymorphisms located in the ICR (Fig. 1A).
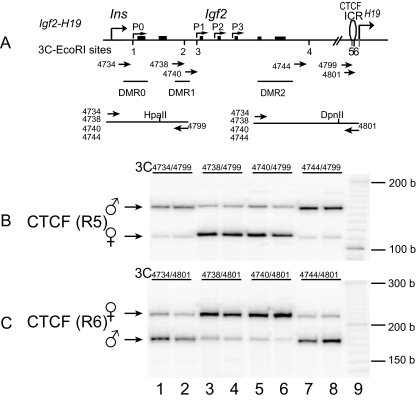
FIG. 1.
Intrachromosomal interaction between the ICR and Igf2 promoters. (A) Schematic presentation of Igf2, H19, DMRs, and EcoRI sites used for 3C assay. The orientation and location of the 3C primers are shown by arrows under each EcoRI restriction site. (B and C) Ligated 3C products between the ICR (EcoRI sites 5 and 6) and EcoRI sites 1 to 4 located up- or downstream of the Igf2 promoters. Chromatin was fixed with formaldehyde, digested with the restriction enzyme EcoRI, and ligated with T4 DNA ligase. The ligated DNA was amplified by PCR with primers covering two EcoRI sites (5 and 6) in the ICR and four EcoRI sites (1 to 4) near the Igf2 promoters. Allele-specific intrachromosomal looping was distinguished by the use of two restriction enzyme polymorphisms (HpaII and DpnII) in the ICR. The ligated intrachromosomal DNA was amplified with the primers in the same orientation to reduce the background. b, bases.
Chromatin immunoprecipitation (ChIP).
ChIP assays were performed with a ChIP assay kit (Upstate, Lake Placid, NY) by following the protocol provided by the manufacturer. Briefly, ∼5 million cells were fixed with 1% formaldehyde and then sonicated for 180 s (10 s on and 10 s off) on ice with a Branson sonicator with a 2-mm microtip at 40% output control and 90% duty cycle settings. The sonicated chromatin (0.9 ml) was clarified by centrifugation, aliquoted, and snap-frozen in liquid nitrogen. To perform ChIP, sonicated chromatin (150 μl) was diluted 10-fold and purified with specific antiserum (2 to 5 μl) and protein G-agarose (60 μl). Antibodies to CTCF, Suz12, and dimethyl-H3-K27 (lysine 27 of histone H3) were obtained from Upstate Biotechnology (Waltham, MA). DNA that was released from the bound chromatin after cross-linking reversal and proteinase K treatment was precipitated and diluted in 100 μl of low-TE buffer (1 mM Tris, 0.1 mM EDTA).
PCRs (3 μl under liquid wax) contained 1 μl ChIP (or input) DNA, 0.5 mM appropriate primer pairs, 50 μM deoxynucleotide triphosphate, and 0.2 U KlenTaq I (Ab Peptides, St. Louis, MO). Standard PCR conditions were 95°C for 60 s, followed by 35 cycles of 95°C for 15 s, 65°C for 30 s of annealing, and 72°C for 1 min of extension. All primer sets were tested for the absence of primer-dimer products. To avoid heteroduplex formation that may interfere with restriction enzyme digestion, one primer of each primer pair was end labeled with [γ-32P]ATP. The γ-32P-labeled primer was added to the PCR mixture (1 μl) at the last cycle of amplification. PCR products were checked to exclude PCR allelic bias and digested with 1 U of the appropriate polymorphic restriction enzymes (data not shown) in a total volume of 6 μl for 3 h. The digested products were separated on a 5% polyacrylamide-urea gel and quantified by a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Coimmunoprecipitation (co-IP) of CTCF with Suz12.
Nuclear extracts were prepared by suspending cells in three packed cell volumes of hypotonic buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], protease inhibitors) for 10 min on ice. The cells were homogenized, transferred to new tubes, and centrifuged for 30 min at 10,000 × g. The released nuclei were suspended in half the packed cell volume of low-salt buffer (20 mM HEPES [pH 7.9], 20 mM KCl, 1.5 mM MgCl2, 0.1 mM EDTA, 25% glycerol, 0.2 mM DTT), followed by the dropwise addition of high-salt buffer (20 mM HEPES [pH 7.9], 0.6 M KCl, 1.5 mM MgCl2, 25% glycerol, 0.2 mM DTT, protease inhibitors). The nuclear suspensions were extracted for 30 min at 4°C with gentle agitation and centrifuged for 30 min at 14,000 × g. The supernatants (nuclear extracts) were stored at −80°C in aliquots.
IP was performed with 200 μl of nuclear extract and a 1,000-fold dilution of the preimmune serum, the anti-CTCF antibody (catalog no. 06-917), or the anti-Suz12 antibody (catalog no. 07-379) in IP buffer at 4°C overnight. The reaction mixtures were incubated with protein G-Sepharose beads (Upstate), 60 μl in a 50% suspension in IP buffer, at 4°C for 90 min on a rotator. The immunoprecipitated complexes were washed twice with 10 volumes of lysis buffer and three times with phosphate-buffered saline buffer. The washed beads were incubated with 30 μl of IP buffer and 30 μl of 2× sample buffer 99°C for 5 min. The proteins released from components of the complexes were examined by SDS-polyacrylamide gel electrophoresis (PAGE) and Western immunoblotting.
CTCF nucleotide pull-down assay.
We examined whether the CTCF bound to the Igf2 ICR also bound to Suz12 at CTCF-binding sites in the promoter P2 and promoter P3 regions. We labeled four DNA fragments from these regions. Four micrograms of each biotin-labeled double-stranded DNA fragment was incubated with 300 μg of nuclear proteins for 20 min at room temperature in a binding buffer consisting of 12% glycerol, 12 mM HEPES (pH 7.9), 4 mM Tris (pH 7.9), 150 mM KCl, 1 mM EDTA, 1 mM DTT, and 10 μg of poly(dI-dC) competitor. Following the incubation, 30 μl of streptavidin-agarose beads was added to the reaction mixture, which was then incubated at 4°C for 4 h. Prior to this step, 300 μl of the original streptavidin-agarose bead preparation was preadsorbed with 500 μl of bovine serum albumin (BSA; 1 mg/ml), 50 μg of poly(dI-dC), and 50 μg of sheared salmon sperm DNA for 30 min at 25°C. The beads were washed three times and resuspended in 300 μl of the binding buffer. The protein-DNA-streptavidin-agarose complex was washed three times with binding buffer. Each sample was heated at 95°C for 5 min and loaded onto a recast 4 to 20% Tris-glycine-acrylamide gel (Bio-Rad) for resolution of bound products. After electrophoresis, the gels were transferred to nitrocellulose membranes for immunoblotting to detect CTCF and Suz12.
Western blotting of coimmunoprecipitated CTCF and Suz12 protein.
Detection of the CTCF and Suz12 proteins was done by Western blotting as previously described (41). The protein-DNA-streptavidin-agarose complex was dissolved in 130 mM Tris-Cl (pH 8.0)-20% (vol/vol) glycerol-4.6% (wt/vol) SDS-0.02% bromophenol blue-2% DTT. The proteins were examined by SDS-PAGE and Western immunoblotting with the anti-CTCF and anti-Suz12 antibodies (1:1,000; Upstate, MA) and the ECL detection system (Amersham) by following the instructions of the manufacturer.
CTCF-Suz12 interaction by mammalian two-hybrid assays.
Human CTCF and Suz12 cDNAs (clone identification no. 6821922 and 3982679; OpenBiosystems, Huntsville, AL) were cloned, respectively, into the pACT (transcriptional activation domain) and pBIND (DNA-binding domain) vectors of the CheckMate mammalian two-hybrid system (Promega, Madison, WI). Human skin fibroblast (HBF1) cells were maintained in Dulbecco medium supplemented with 10% fetal calf serum. HBF1 cells at 60% confluence in 96-well plates were transfected with 300 ng of plasmid DNA (pBIND+ACT+pG5-luc) with 0.8 μl of Lipofectamine 2000 (Invitrogen, CA). Luciferase enzyme activity was quantified with a luciferase reporter assay system (Promega, Madison, WI) and measured with an LMax microplate luminometer (Molecular Devices, Sunnyvale, CA).
In vitro CTCF-Suz12 interaction assay with recombinant proteins.
Recombinant CTCF-glutathione S-transferase (GST) protein was purchased from Novus Biologicals, Inc. (Littleton, CO). To prepare recombinant proteins for SUZ12 and CBX2, cDNAs were cloned into TA vector (Invitrogen, CA) and translated by TNT coupled wheat germ extract systems (Promega, Madison, WI) with the SP6 promoter. For the in vitro interaction assay, GST-CTCF (5 μg) was incubated with recombinant SUZ12 or CBX2 or equal concentrations of BSA (negative control) in 100 μl binding buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 2.5 ng/ml BSA, 10 mM EDTA, 0.1% Triton X-100, 1 mM DTT, 10% glycerol) containing glutathione-immobilized paramagnetic particles (MagneGST Protein Purification System; Promega, Madison, WI). After incubation for 3 h at 4°C, the particles were washed three times with washing buffer. Binding proteins were eluted for immunoblotting with anti-Suz12 and anti-CBX2 antibodies.
RNAi knockdown.
Tree Stealth RNA interference (RNAi) duplexes, purchased from Invitrogen (Carlsbad, CA), were transfected into MBW2 cells with Lipofectamine RNAiMAX complexes by following the manufacturer's reverse transfection protocol. The three RNAi oligonucleotides used were as follows: (i) RNAi 1 (Suz12 MSS225221), UUA UUG GAC AAC UUA CAU CCU UCC U; (ii) RNAi 2 (Suz12 MSS225223), AAU UCA UUA CUG GAA ACU GCC AGG G; (iii) RNAi 3 (Suz12 MSS225222), UAA AUU CUC UUC UUC CUG GAC GAG U). To reduce the concentration of preexisting Suz12 protein that was already incorporated into the chromatin, we harvested the treated cells and repeated the above-described transfection procedure three times. Seventy-two hours following the third transfection, cells were collected for allelic measurement of Igf2 by reverse transcription (RT)-PCR products by using DpnII polymorphism as previously described (6, 40) and for Suz12 Western and H3-K27 methylation analysis.
RESULTS
CTCF orchestrates the allelic intrachromosomal interaction between the ICR and Igf2 promoters.
A mouse fibroblast cell line (MBW2) derived by breeding M. spretus males with C57BL/c females (6) was used to study the allelic interactions with CTCF. We first examined whether CTCF interacts directly with the Igf2 promoters by 3C methodology (17). CTCF-bound DNA in the ICR (EcoRI sites 5 and 6) interacted with the DNA near the imprinted promoters primarily on the maternal allele (EcoRI sites 2 and 3, lanes 3 to 6, Fig. 1), suggesting that CTCF physically interacts with the three suppressed promoters. Similar findings were also confirmed separately in three additional mouse cell lines, including an embryonic stem cell line, and in mouse tissues, including the liver (26). These data are in complete agreement with those reported by Yoon et al. (42), who used a different restriction enzyme system in the 3C assay in reciprocally bred animals, but are slightly different from those reported by Kurukuti et al. (17), possibly due to the different tissues used (42).
We then employed ChIP to map the CTCF-interacting sites around the three Igf2 promoters in detail. In agreement with the 3C data shown in Fig. 1 and as recently reported by Yoon et al. (42), we showed that CTCF primarily interacted with the three Igf2 promoters of the maternal (C57BL/c) allele. This maternal interaction starts gradually from DMR1, with strong interaction around the two major promoters (P2 and P3) (Fig. 2A, lane 4). This allele-specific CTCF interaction in other regions downstream or upstream of the Igf2 promoters was weak or undetectable (data not shown).
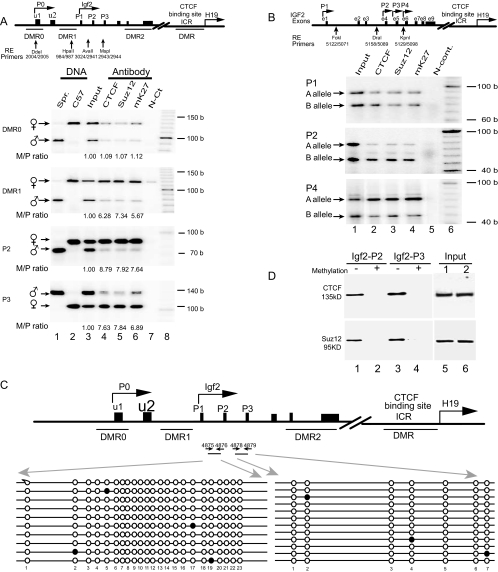
FIG. 2.
Allele-specific ChIP assay across DMR0, DMR1, and the promoter region of Igf2. (A, top) Scheme of the Igf2/H19 imprinting domain. The exons are depicted as solid boxes. DMRs are shown as underlines, and the polymorphic restriction enzyme sites are shown as vertical arrows. The allelic interaction of CTCF with the Igf2 promoters (P0 to P3) and DMRs was identified by using four polymorphic restriction enzymes (Dde1, Hpa2, Ava1, and Msp1) that distinguish M. spretus from C57BL/c. (A, bottom) ChIP of CTCF, Suz12, and dimethylated H3-K27 (mK27) in F1 mouse fibroblasts derived from breeding M. spretus males with C57BL/c females. Cross-linked DNA-protein complexes were immunoprecipitated with antisera against CTCF, Suz12, and dimethyl-H3-K27 (mK27), followed by PCR amplification with specific primers for the DMR0, DMR1, and Igf2 promoters (P1 to P3). Allelic ChIP products were distinguished by polymorphic restriction enzymes (RE). N-Ct lane, negative control (no antibody); input lane, genomic DNA collected before antibody precipitation (positive control). The M/P ratio is the ratio of the maternal to the paternal alleles after normalization with the input DNA. (B) ChIP of CTCF, Suz12, and dimethylated H3-K27 in human fibroblasts. Alleles are labeled A and B because the parental alleles are not known. (C) DNA methylation of CpG dinucleotides at the Igf2 promoters. After sodium bisulfite treatment, genomic DNA fragments were amplified with primers 4875 and 4876 for promoter P2 and primers 4878 and 4879 for promoter P3. Each line represents a single sequenced PCR molecule. Black circles represent methylated CpG dinucleotides, and open circles represent unmethylated CpG dinucleotides. Note the DNA hypomethylation in both promoters. (D) Oligonucleotide pull-down assay for CTCF and Suz12 with Igf2 promoter P2 and P3 DNA fragments labeled with biotin-streptavidin. Wild-type (−) and methylated (+) DNA fragments were end labeled with biotin and incubated with nuclear extracts. After incubation, DNA fragments were pulled down with streptavidin beads. Proteins bound to the DNA fragments were eluted and detected by Western blotting with antibodies directed against CTCF and Suz12. (Input) Aliquots of nuclear proteins, collected before oligonucleotide pull down, were analyzed in parallel with the samples in lanes 1 to 4 and detected by Western blotting. b, bases.
We extended this finding to a human fetal skin-derived fibroblast cell line by showing that there was an allele-specific interaction between CTCF and the imprinted IGF2 promoters (P2 and P4) (Fig. 2B). Human IGF2 promoter P1 is close to the insulin gene and is biallelically expressed (37). As expected, we could not detect any allele-specific interaction between CTCF and this promoter. Thus, this epigenetic mechanism is evolutionarily conserved between human and mouse genes. Using a genome-wide transcription factor-binding location strategy, Kim et al. (16) mapped the CTCF-binding sites in human fibroblasts. Among the identified CTCF-binding sites in the IGF2/H19 imprinting locus, one site was located exactly at human IGF2 promoter P3 and another was downstream of promoter P4, a finding consistent with the involvement of CTCF in the regulation of the IGF2/H19 imprinting domain.
CTCF recruits polycomb repressive complex 2 (PRC2) and induces H3-K27 methylation in Igf2 promoters.
To delineate how this long-range CTCF-promoter interaction is involved in suppressing the expression of maternal Igf2, we first examined DNA methylation by sodium bisulfite sequencing (6). As previously reported (10, 31), we found that the Igf2 promoters were normally unmethylated on both alleles (Fig. 2C), thus excluding a role for DNA methylation in promoter suppression. We synthesized and biotin labeled two DNA fragments covering mouse Igf2 promoters P2 and P3 and used them to pull down nuclear proteins. Western blotting confirmed that CTCF bound to wild-type, but not methylated, Igf2 promoter DNAs (Fig. 2D). These data suggest that CTCF may orchestrate the intrachromosomal interaction through self-dimerization or polymerization after binding to both the ICR and the Igf2 promoters. Interestingly, H3-K27 was hypermethylated near the imprinted maternal promoters and was hypomethylated near the expressed paternal promoters (Fig. 2A, lane 6), thus establishing a correlation between CTCF binding and Igf2 promoter silencing by H3-K27 methylation. In support of this finding, Szabo and colleagues recently also demonstrated allele-specific H3-K27 methylation at the maternal Igf2 P2 promoter and Igf2 DMRs (12).
Because Suz12 is an essential component of PRC2 which stimulates H3-K27 methylation (25), we used ChIP to map the binding of Suz12 to the three Igf2 promoters. After precipitation with anti-Suz12 antibodies, DNA was amplified with the same Igf2 promoter primers used for CTCF. Like CTCF, Suz12 also bound specifically to the maternal allele of both of the mouse Igf2 promoters (P2 and P3) (Fig. 2A, lane 5). In the presence of nuclear extract, Suz12 also bound specifically to synthetic oligonucleotides containing the sequences of the major Igf2 promoters, P2 and P3. No binding was seen when the CpGs in these sequences were methylated (Fig. 2D). These data suggest that CTCF binds to DNA and then serves as a scaffold for binding Suz12, leading to H3-K27 methylation and the suppression of the Igf2 promoters.
The direct interactions between Suz12 and CTCF were first examined with a co-IP assay. Nuclear proteins were first precipitated with either CTCF or Suz12 antisera. The precipitated proteins were separated by SDS-PAGE, and Western immunoblotting was used to detect the interaction between CTCF and Suz12. As shown in Fig. 3A, CTCF antibody-precipitated nuclear proteins contain both CTCF and Suz12 (left panel), and Suz12 antiserum precipitated both CTCF and Suz12 from the protein complex (right panel). After washing with high-stringency buffer, the Suz12 signal in the CTCF-immunoprecipitated proteins became weaker (Fig. 3A, lane 3); similarly, the CTCF signal declined in the Suz12 IP under high-stringency conditions (Fig. 3A, lane 7). These data indicate that Suz12 and CTCF physically interact with each other.
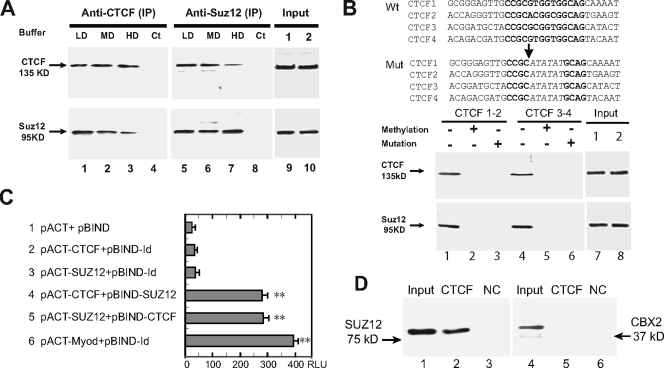
FIG. 3.
(A) Interaction between CTCF and Suz12 as measured by co-IP and Western immunoblot assays. Nuclear proteins were immunoprecipitated, respectively, with anti-CTCF and anti-Suz12 antibodies and washed with detergent buffers. After separation on a 7.5% SDS-polyacrylamide gel, immunoprecipitates were immunodetected with appropriate antibodies to CTCF and Suz12. LD, low-detergent buffer; MD, medium-detergent buffer; HD, high-detergent buffer; Ct, negative IP control with no antibody. (Input) Aliquots of nuclear proteins, collected before CTCF and Suz12 IP, were analyzed in parallel with the samples in lanes 1 to 8 and detected by Western blotting. (B) Oligonucleotide pull down of CTCF and Suz12. At the top are the sequences of the wild-type (Wt) and mutated (Mut) CTCF-binding sites in synthesized oligonucleotide fragments. Consensus CTCF-binding sites are in bold and are partially replaced by ATATAT in mutated oligonucleotides. (Input) Aliquots of nuclear proteins, collected before CTCF oligonucleotide pull down, were analyzed in parallel with the samples in lanes 1 to 6 and detected by Western blotting. At the bottom is Western blotting of CTCF and Suz12 in nuclear proteins pulled down by wild-type (−), methylated (+), and mutated (+) CTCF oligonucleotide fragments. DNA fragments were end labeled with biotin and incubated with nuclear extracts. After incubation, DNA fragments were pulled down with streptavidin beads. Proteins bound to the DNA fragments were eluted and detected with CTCF and Suz12 antibodies. (C) Two-hybrid interactions between CTCF and Suz12. Lane 1 shows background expression of firefly luciferase from the pG5luc vector as determined by cotransfection with the pACT and pBIND vectors, which did not contain CTCF or Suz12, into HBF1 human fibroblast cells. Lanes 2 and 3 show two controls used to determine the background activity of individual CTCF or Suz12 (no interaction). In lanes 4 and 5, The reporter vector was cotransfected with CTCF and SUZ12 in fusion with the VP16 transcription activation domain (pACT constructs) or the GAL4 DNA-binding domain (pBIND constructs). In lane 6, pBIND-Id and pACT-MyoD were used as the positive control encoding two proteins known to interact in vivo. Luciferase activity was measured as relative luminescence units (RLU). *, P < 0.01 compared with the three control groups. The values shown are averages ± standard deviations (n = 6). (D) In vitro binding assay with recombinant proteins. Lanes 1 to 3 show the CTCF-and-Suz12 interaction; lanes 4 to 6 show the CTCF-and-CBX2 interaction. Input, reaction mixture aliquot collected before particle pull down and analyzed in parallel with the samples (CTCF-GST and BSA) by Western blotting. NC, negative control with an equal amount of BSA.
ChIP showed that Suz12 also interacted with the maternal ICR. However, it is difficult to distinguish specific Suz12 binding to the ICR from potential binding of Suz12 to the adjoining paternal H19 promoter region (data not shown). To avoid contamination with the H19 promoter, we synthesized three groups of DNA fragments covering ICR CTCF-binding sites 1 and 2 and sites 3 and 4 and used them to pull down CTCF and Suz12 from extracts of nuclear proteins. If CTCF serves as a scaffold to bind Suz12, methylation or mutation of the CTCF-binding sites in the ICR should abolish the binding of both CTCF and Suz12. The first group of oligonucleotides comprised the unmethylated wild-type CTCF-binding region. In the second and third groups, DNA was either methylated in vitro with SssI DNA methylase or mutated in the conserved CTCF-binding sequences (Fig. 3B). As predicted, both CTCF and Suz12 bound specifically to the wild-type, unmethylated ICR DNA fragments (lanes 1 and 4) but not to the methylated (lanes 2 and 5) or the mutated (lanes 3 and 6) ICR DNA.
The physical interaction between Suz12 and CTCF was confirmed with the CheckMate mammalian two-hybrid system (Promega, Madison, WI). Human CTCF and Suz12 cDNAs were cloned in frame with VP16 in a pACT (transcriptional activation domain) vector and with GAL4 in a pBIND (DNA-binding domain) vector and then cotransfected with a pG5luc reporter vector into human fibroblast cell line HBF1. Neither CTCF nor Suz12 alone activated luciferase activity in the pG5luc vector (Fig. 3C, lanes 2 and 3). However, when pACT-CTCF and pBIND-Suz12 were cotransfected, luciferase activity was significantly enhanced (Fig. 3C, lane 4). Similarly, cotransfection of pACT-Suz12 and pBIND-CTCF also transactivated the reporter gene (Fig. 3C, lane 5). These data demonstrate that CTCF directly interacts with Suz12. In a similar set of experiments, replacing Suz12 with Ezh2, the enzymatic component of PRC2 that methylates H-K27, also led to activation of the reporter gene in this two-hybrid system (data not shown), further demonstrating the requirement for a complete PRC2 complex for CTCF interaction and silencing of the maternal Igf2 allele.
To determine whether CTCF interacts with Suz12 in a direct or indirect manner, we synthesized and purified Suz12 protein with a wheat in vitro translation system and incubated it with recombinant CTCF-GST. After incubation, the CTCF-Suz12 complex was pulled down by glutathione particles and the presence of Suz12 in the interacting complex was detected by Western blotting with anti-Suz12 antibody. We showed a direct interaction between CTCF and Suz12 in this in vitro binding assay (Fig. 3D, lane 2). However, by using the same strategy we could not pull down recombinant CBX2, a component of the PRC1 complex (lane 5), suggesting that CTCF may not interact with CBX2 in a direct manner.
Genomic deletion or mutation of the ICR abolishes the CTCF-Suz12 interaction and induces demethylation of H3-K27 in Igf2 promoters.
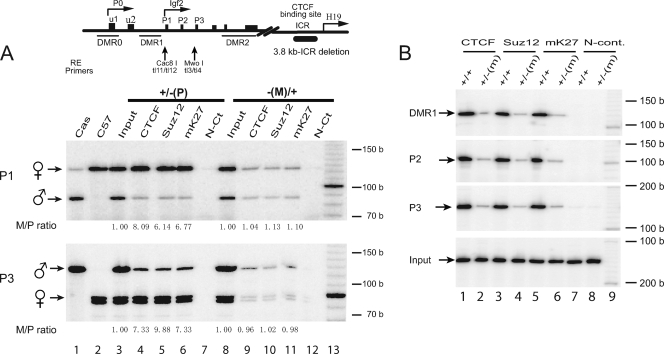
We further examined the CTCF-Suz12 interaction by using transgenic mouse models that harbored a mutation in the CTCF-binding sites in the ICR (34) or a deletion of the ICR (35). In both models, CTCF did not bind to the maternal ICR, leading to loss of Igf2 imprinting (32, 35). In the ICR deletion-containing models, two fibroblast cell lines were cultured from the skin of neonates generated from reciprocal crosses of C57BL/6(CAST) and F1 ICR deletion-containing heterozygotes maintained in a C57BL/6 background (35). As shown in Fig. 4A, CTCF and Suz12 specifically interacted with the maternal promoters (P1 and P3) (lanes 4 and 5) only when the ICR deletion was paternally inherited and Igf2 imprinting was maintained. Correspondingly, H3-K27 was hypermethylated specifically in the maternal Igf2 promoters. The allele-specific promoter interaction and H3-K27 methylation, however, were lost (Fig. 4A, lanes 9 to 11) when the ICR deletion was maternally inherited, leading to biallelic Igf2 expression (35).
FIG. 4.
ChIP of CTCF, Suz12, and methylated H3-K27 in transgenic mouse skin tissues. (A) ICR deletion model. Mouse fibroblasts, kindly provided by M. S. Bartolomei, were cultured from neonates carrying a 3.8-kb deletion of the ICR (35). These mice were generated by reciprocal crosses of C57BL/6(CAST) with F1 ICR heterozygotes maintained in a C57BL/6 background. Heterozygous fetuses inherit either a maternal [−(M)/+] or a paternal [+/−(P)] ICR deletion. Allelic ChIP products were distinguished by polymorphic restriction enzymes Cac81 and Mwo1. N-Ct, negative control (no antibody). b, bases. (B) ICR mutation model. Fetal liver tissues, kindly provided by P. E. Szabo, were derived by breeding male FVB/NJ.CAST/Ei(N7) and female 129SI/ImJ mice to produce F1 mice that are heterozygous for a mutation in the ICR (34). Wild-type (+/+) mice carry both alleles from strain CAST/Ei. −(M)/+ mice carry the maternally inherited mutated ICR from strain 129SI/ImJ (129) and the paternally inherited ICR from strain CAST/Ei. Since the wild-type mice are homozygous, the parental alleles cannot be distinguished. N-cont., negative control (no antibody). b, bases.
In the model in which ICR is mutated (34), the ChIP assay was performed with liver from fetuses derived by breeding FVB/NJ.CAST/Ei(N7) males with 129SI/ImJ females heterozygous for the mutation of the ICR. As expected, there was no association of CTCF (Fig. 4B, lane 2) and Suz12 (Fig. 4B, lane 4) with the Igf2 promoters in the livers of the mice in which the ICR was maternally mutated. In confirmation of the report by Han et al. (12), H3-K27 became hypomethylated near the promoters (Fig. 4B, lane 6), indicating that Suz12 is an essential component of the promoter suppression complex mediated by CTCF.
RNAi knockdown of Suz12 releases the suppression of the imprinted Igf2 maternal allele.
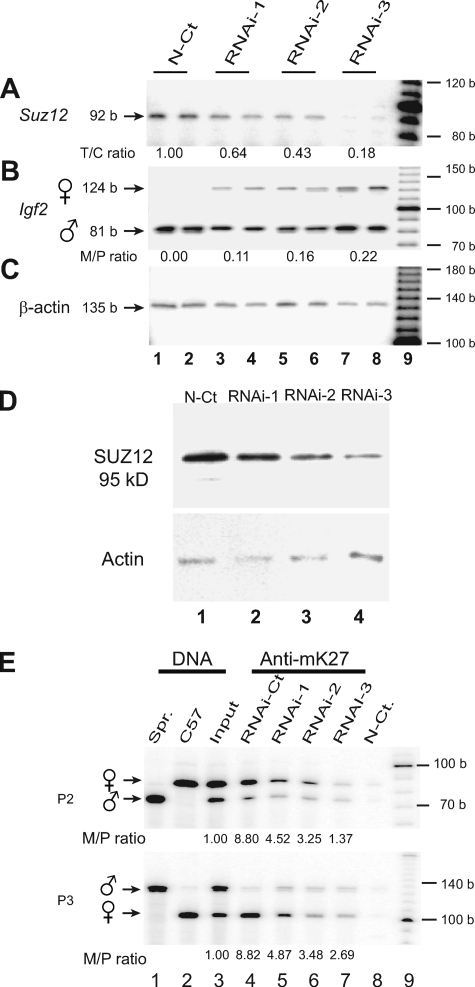
To determine the role of PRC2 proteins in the imprinted expression of Igf2, we used RNAi to knock down Suz12 at both the mRNA and protein levels (Fig. 5A and D). The previously suppressed maternal Igf2 allele was expressed to an extent that was inversely correlated with Suz12 mRNA levels. With very low levels of Suz12 expression, Igf2 was biallelically expressed (Fig. 5B).
FIG. 5.
Loss of Igf2 imprinting induced by RNAi knockdown of Suz12. (A) Quantitation of Suz12 mRNA by RT-PCR in Suz12 knockdown cells. The T/C ratio is the ratio of Suz12 mRNA in RNAi-treated cells to that in control cells (N-Ct). (B) Igf2 imprinting in Suz12 knockdown cells with polymorphic restriction enzyme DpnII. The M/P ratio is the ratio of Igf2 mRNA from the maternal allele to that from the paternal allele. (C) Measurement of β-actin mRNA serves as the PCR control. Lanes 1 and 2, stealth RNAi control; lanes 3 to 8, RNAi-treated cells; lane 9, 100-bp DNA molecular size marker. Three Suz12 RNAi duplex oligonucleotides that target distinct locations of Suz12 were separately transfected into MBW2 cells that maintain normal Igf2 imprinting (6). As the Suz12 RNAi oligonucleotides are rapidly degraded and the preexisting Suz12 protein has a relatively long half-life, we transfected each group of MBW2 cells three times with Suz12 RNAi oligonucleotides. RNAi-1 to -3 are three individual stable clones with duplicated RT-PCR measurements. (D) Suz12 Western blotting in RNAi knockdown fibroblasts. Suz12 was knocked down by three different RNAi oligonucleotides, and equal amounts of proteins were used for detection by Western blotting with β-actin as the internal control. (E) H3-K27 ChIP assay of Igf2 promoters P2 and P3 in Suz12 knockdown fibroblasts. Experimental conditions are the same as in Fig. 2A. N-Ct., control without Suz12 RNAi. Spr., M. spretus; b, bases.
To demonstrate the importance of Suz12 in regulating Igf2 allelic expression, we also examined whether Suz12 knockdown affects H3-K27 methylation at the Igf2 promoters. In control cells, the maternal allele of Igf2 promoters P2 and P3 was hypermethylated (Fig. 5E, lane 4). However, in Suz12 knockdown cells, H3-K27 became hypomethylated, in concordance with the activation of the normally suppressed maternal allele. These data suggest that Suz12 is a critical component of the complex regulating H3-K27 methylation and Igf2 promoter activity.
DISCUSSION
Igf2 and H19 are two tightly coordinated yet reciprocally imprinted genes controlled by the same enhancers utilizing an “enhancer competition” mechanism (3). The finding of allelic-specific binding of CTCF to an ICR between H19 and Igf2 (4, 13) further extended our understanding of how Igf2/H19 reciprocal imprinting is regulated. The CTCF insulator marks the boundary in the ICR that is differentially methylated on the parental alleles. CTCF binds to the unmethylated maternal CTCF DMR and insulates the Igf2 promoter from the remote enhancer downstream of H19. Allelic methylation of the paternal ICR, however, abrogates the binding of CTCF and thus allows the exclusive expression of H19 from the maternal allele and Igf2 from the paternal allele (2, 9, 19, 38, 39). This insulator model is supported by the observation that deletion (35) or mutation (34) of the CTCF DMR relaxes the normally silent maternal allele of Igf2.
In human tumors where IGF2 is biallelically expressed (24, 32, 35), the CTCF model predicts that both parental alleles are methylated and no CTCF insulation occurs at either allele, leading to loss of IGF2 imprinting. Indeed, maternally transmitted microdeletion of two CTCF sites in the ICR results in biallelic IGF2 expression and H19 silencing in Beckwith-Wiedemann syndrome (33). Using nuclear transfer, we previously showed that aberrant IGF2 imprinting in human tumor cells was repaired by the imprinting machinery in the normal fibroblast cytoplasm, leading to monoallelic expression of IGF2 in the reconstructed tumor cybrids or hybrids. However, this epigenetic resetting of IGF2 imprinting in tumors was not accompanied by any changes in DNA methylation at well-known DMRs (DMR0, ICR, and kvDMR1). Neither did we observe an alteration in DNA methylation in human fibroblasts, in which IGF2 is biallelically expressed after treatment with cycloheximide. These findings suggested that alterations of IGF2 imprinting in tumors may not necessarily be accompanied by changes in DNA methylation in known ICRs but may be related to the inactivation of other, unknown, imprinting factors.
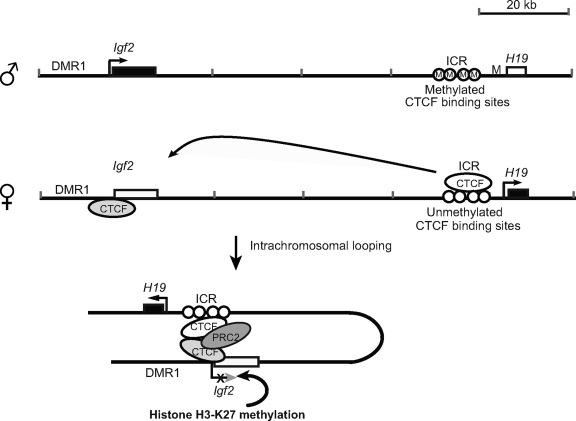
The present study has provided a more detailed delineation of how CTCF specifically regulates Igf2 imprinting. The data are consistent with a model in which CTCF binds to the unmethylated maternal ICR and hinges the ICR to the Igf2 promoters by self-dimerization to form a unique intrachromosomal loop. Through the direct interaction of Suz12 with DNA-bound CTCF, the PRC2 complex is recruited specifically to the maternal promoters, where it methylates H3-K27, leading to the formation of a repressive chromatin state around the maternal Igf2 promoters. CTCF cannot bind to the methylated paternal ICR, and thus, there is no scaffold to secure PRC2 to that site (Fig. 6) (4, 13). In the absence of PRC2 complex binding, the paternal Igf2 promoters are able to access the downstream enhancers and are transcribed in an allele-specific manner.
FIG. 6.
Simplified model of regulation of Igf2 imprinting by the CTCF-mediated PRC2-Igf2 promoter suppressive complex. CTCF binds to the unmethylated ICR and the Igf2 promoters on the maternal allele and forms a long-distance intrachromosomal loop through CTCF dimerization. CTCF recruits PRC2 via Suz12, resulting in methylated H3-K27 and inactive chromatin around the Igf2 promoters. On the paternal allele, the methylated ICR does not bind CTCF and the CTCF-PRC2 suppression complex cannot be formed, resulting in unmethylated H3-K27 in the active Igf2 promoters. Similarly, CTCF-mediated ICR-promoter looping is abolished when the CTCF binding site in the ICR is deleted or mutated. Loss of H3-K9 methylation at the maternal promoters leads to loss of Igf2 imprinting.
Our data thus demonstrate that CTCF is more than a physical insulator that passively blocks the access of the Igf2 promoters to the H19 enhancers. Rather, it is an active participant in the control of allelic expression of Igf2. First, it orchestrates long-distance communication via an intrachromosomal loop between the Igf2 promoters and the H19 ICR (Fig. 1) (42, 21). While most imprinted genes are allelically methylated or marked in the promoter region to guide allelic expression of the gene, none of the Igf2 promoters are differentially methylated (Fig. 2C) (10, 31) and they are thus not distinguishable by the cellular transcription machinery. Instead, CTCF provides the signal by binding to the unmethylated maternal ICR and delivering the parental allele-specific imprinting message to the remote Igf2 promoters by intrachromosomal looping. Deletion or mutation of the ICR abolishes this intrachromosomal looping and blocks this long-range communication (42). Second, CTCF recruits the PRC2 complex via the direct interaction with Suz12 (Fig. 3D). The maternal allele thus contains a PRC2-CTCF-ICR-loop-Igf2 promoter region that forms a chromatin complex that serves as the substrate for Ezh2-mediated H3-K27 methylation. This change in histone methylation ultimately leads to the suppression of the maternal Igf2 promoters.
Han et al. (12) also provided data to support our model. Using a quantitative ChIP-single-nucleotide primer extension assay, they found that ICR-CTCF binding was essential for the maternal allele-specific repressing mark H3-K27 methylation at the Igf2 DMRs, including the P2 promoter. Point mutations in the ICR resulted in complete reorganization of chromatin at the Igf2/H19 imprinting domain. In conjunction with the data presented here, we now have a clearer picture of the mechanisms by which CTCF specifically coordinates the allelic expression of the remote Igf2 promoters after binding to the ICR. It is especially interesting that CTCF functions as a “second messenger” of the imprinting signal in the regulation of Igf2 allelic expression. Unlike Igf2r and many other imprinted genes, Igf2 promoters do not carry the parent-specific DNA methylation mark (Fig. 2C) (10, 31). Rather, the imprinting control signal is located in the ICR that is 90 kb away from the Igf2 promoters. By intrachromosomal looping, CTCF delivers the parent-specific methylation signal in the ICR to the remote Igf2 promoters, where it then recruits PRC2, which methylates H3-K27 and leads to gene silencing (Fig. 6).
When we performed an in vitro binding assay with synthetic Igf2 P2 and P3 DNA fragments, it appeared that CTCF was able to bind directly to Igf2 promoters P2 and P3 (Fig. 2D). Similarly, in wild-type cells, the ChIP assay also detected the binding of CTCF to the maternal Igf2 promoters (Fig. 2A). However, the ability of CTCF to bind to the Igf2 promoters is lost in the maternal ICR deletion-containing animals (Fig. 4A), suggesting that the ICR-mediated intrachromosomal structure is also essential for CTCF to bind to the Igf2 promoter in cells as opposed to naked DNA. It is not clear how the maternal ICR deletion affects CTCF-Igf2 promoter binding. It is possible that prior binding of CTCF to the maternal ICR is required to institute CTCF-Igf2 promoter interactions. Deletion of the ICR may significantly alter the local chromatin structure and thus decrease the binding of CTCF to the Igf2 promoters. Further delineation of the specific Igf2 promoter sequences to which CTCF binds may help address this intrachromosomal regulation.
CTCF regulates many genes by binding to promoters, enhancers, and silencers. The CTCF-PRC2 complex may play a role in the regulation of many other genes that are regulated by CTCF, such as c-myc, β-globin, amyloid β-protein precursor, and X-inactivated genes. We believe that this previously unrecognized mechanism may also be broadly applied to numerous genes that are controlled by CTCF in the genome. Recently, we showed that CTCF mediates an interchromosomal colocalization between the Igf2/H19 ICR on mouse chromosome 7 and Wsb1/Nf1 on mouse chromosome 11 (18). Inhibition of CTCF synthesis or genomic deletion of the maternal CTCF-binding sites in the ICR abrogates this interchromosomal association and alters the allelic expression of Igf2. It would be of great interest to investigate whether such interchromosomal associations of CTCF are also accompanied by the recruitment of PRC2 proteins. Loss of IGF2 imprinting is a hallmark of many human tumors (11, 22, 27); it is associated with the loss of activity of a non-CTCF trans-imprinting factor(s) (6) and, in some cases, with aberrant methylation of the ICR (36). It will be important to determine whether PRC2 serves as a putative imprinting factor that is either inactivated or mutated in tumors in which IGF2 imprinting has been lost.
In summary, we demonstrate for the first time that CTCF acts as a unique imprinting message carrier in coordinating allelic expression in the Igf2/H19 domain. It delivers the parent-specific methylation signal in the ICR to the remote Igf2 promoters that do not carry any imprinting marks. CTCF recruits PRC2 after anchoring the ICR to Igf2 promoters by long-distance intrachromosomal looping. PRC2 causes allele-specific methylation at H3-K27, leading to suppression of the maternal Igf2 promoters (Fig. 6). Thus, CTCF governs allelic gene expression of Igf2 by serving as a DNA-binding protein scaffold to recruit and bind PRCs.
Acknowledgments
We are indebted to M. S. Bartolomei, Piroska E. Szabo, and Jeffrey R. Mann for providing us with tissues and cell lines that are mutated at CTCF-binding sites of the ICR.
This study was supported by a Department of Defense grant (W81XWH-04-1-0597) and an NIH SBIR grant (R43 CA86664-01) to J.F.H., an NIH grant (DK36054) to A.R.H., and the Research Service of the Department of Veterans Affairs.
Footnotes
Published ahead of print on 28 July 2008.
REFERENCES
- 1.Arney, K. L. 2003. H19 and Igf2—enhancing the confusion? Trends Genet. 1917-23. [DOI] [PubMed] [Google Scholar]
- 2.Bartolomei, M. S. 2003. Epigenetics: role of germ cell imprinting. Adv. Exp. Med. Biol. 518239-245. [DOI] [PubMed] [Google Scholar]
- 3.Bartolomei, M. S., A. L. Webber, M. E. Brunkow, and S. M. Tilghman. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 71663-1673. [DOI] [PubMed] [Google Scholar]
- 4.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405482-485. [DOI] [PubMed] [Google Scholar]
- 5.Bell, A. C., A. G. West, and G. Felsenfeld. 2001. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science 291447-450. [DOI] [PubMed] [Google Scholar]
- 6.Chen, H. L., T. Li, X. W. Qiu, J. Wu, J. Q. Ling, Z. H. Sun, W. Wang, W. Chen, A. Hou, T. H. Vu, A. R. Hoffman, and J. F. Hu. 2006. Correction of aberrant imprinting of IGF2 in human tumors by nuclear transfer-induced epigenetic reprogramming. EMBO J. 255329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dekker, J., K. Rippe, M. Dekker, and N. Kleckner. 2002. Capturing chromosome conformation. Science 2951306-1311. [DOI] [PubMed] [Google Scholar]
- 8.Dunn, K. L., and J. R. Davie. 2003. The many roles of the transcriptional regulator CTCF. Biochem. Cell Biol. 81161-167. [DOI] [PubMed] [Google Scholar]
- 9.Engel, N., and M. S. Bartolomei. 2003. Mechanisms of insulator function in gene regulation and genomic imprinting. Int. Rev. Cytol. 23289-127. [DOI] [PubMed] [Google Scholar]
- 10.Feil, R., J. Walter, N. D. Allen, and W. Reik. 1994. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development 1202933-2943. [DOI] [PubMed] [Google Scholar]
- 11.Feinberg, A. P. 1993. Genomic imprinting and gene activation in cancer. Nat. Genet. 4110-113. [DOI] [PubMed] [Google Scholar]
- 12.Han, L., D. H. Lee, and P. E. Szabo. 2008. CTCF is the master organizer of domain-wide allele-specific chromatin at the H19/Igf2 imprinted region. Mol. Cell. Biol. 281124-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405486-489. [DOI] [PubMed] [Google Scholar]
- 14.Hu, J. F., P. H. Nguyen, N. V. Pham, T. H. Vu, and A. R. Hoffman. 1997. Modulation of Igf2 genomic imprinting in mice induced by 5-azacytidine, an inhibitor of DNA methylation. Mol. Endocrinol. 111891-1898. [DOI] [PubMed] [Google Scholar]
- 15.Kanduri, C., V. Pant, D. Loukinov, E. Pugacheva, C. F. Qi, A. Wolffe, R. Ohlsson, and V. V. Lobanenkov. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10853-856. [DOI] [PubMed] [Google Scholar]
- 16.Kim, T. H., Z. K. Abdullaev, A. D. Smith, K. A. Ching, D. I. Loukinov, R. D. Green, M. Q. Zhang, V. V. Lobanenkov, and B. Ren. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 1281231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurukuti, S., V. K. Tiwari, G. Tavoosidana, E. Pugacheva, A. Murrell, Z. Zhao, V. Lobanenkov, W. Reik, and R. Ohlsson. 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. USA 10310684-10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ling, J. Q., T. Li, J. F. Hu, T. H. Vu, H. L. Chen, X. W. Qiu, A. M. Cherry, and A. R. Hoffman. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312269-272. [DOI] [PubMed] [Google Scholar]
- 19.Mann, J. R., P. E. Szabo, M. R. Reed, and J. Singer-Sam. 2000. Methylated DNA sequences in genomic imprinting. Crit. Rev. Eukaryot. Gene Expr. 10241-257. [DOI] [PubMed] [Google Scholar]
- 20.Mukhopadhyay, R., W. Yu, J. Whitehead, J. Xu, M. Lezcano, S. Pack, C. Kanduri, M. Kanduri, V. Ginjala, A. Vostrov, W. Quitschke, I. Chernukhin, E. Klenova, V. Lobanenkov, and R. Ohlsson. 2004. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 141594-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murrell, A., S. Heeson, and W. Reik. 2004. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 36889-893. [DOI] [PubMed] [Google Scholar]
- 22.Ogawa, O., M. R. Eccles, J. Szeto, L. A. McNoe, K. Yun, M. A. Maw, P. J. Smith, and A. E. Reeve. 1993. Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms' tumour. Nature 362749-751. [DOI] [PubMed] [Google Scholar]
- 23.Ohlsson, R., R. Renkawitz, and V. Lobanenkov. 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 17520-527. [DOI] [PubMed] [Google Scholar]
- 24.Pant, V., S. Kurukuti, E. Pugacheva, S. Shamsuddin, P. Mariano, R. Renkawitz, E. Klenova, V. Lobanenkov, and R. Ohlsson. 2004. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol. Cell. Biol. 243497-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasini, D., A. P. Bracken, M. R. Jensen, E. Lazzerini Denchi, and K. Helin. 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 234061-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu, X., T. H. Vu, Q. Lu, J. Q. Ling, T. Li, A. Hou, S. K. Wang, H. L. Chen, J. F. Hu, and A. R. Hoffman. 2008. A complex deoxyribonucleic acid looping configuration associated with the silencing of the maternal igf2 allele. Mol. Endocrinol. 221476-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rainier, S., L. A. Johnson, C. J. Dobry, A. J. Ping, P. E. Grundy, and A. P. Feinberg. 1993. Relaxation of imprinted genes in human cancer. Nature 362747-749. [DOI] [PubMed] [Google Scholar]
- 28.Recillas-Targa, F., I. A. De La Rosa-Velazquez, E. Soto-Reyes, and L. Benitez-Bribiesca. 2006. Epigenetic boundaries of tumour suppressor gene promoters: the CTCF connection and its role in carcinogenesis. J. Cell. Mol. Med. 10554-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reik, W., M. Constancia, W. Dean, K. Davies, L. Bowden, A. Murrell, R. Feil, J. Walter, and G. Kelsey. 2000. Igf2 imprinting in development and disease. Int. J. Dev. Biol. 44145-150. [PubMed] [Google Scholar]
- 30.Sasaki, H., K. Ishihara, and R. Kato. 2000. Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J. Biochem. (Tokyo) 127711-715. [DOI] [PubMed] [Google Scholar]
- 31.Sasaki, H., P. A. Jones, J. R. Chaillet, S. A. Ferguson, S. C. Barton, W. Reik, and M. A. Surani. 1992. Parental imprinting: potentially active chromatin of the repressed maternal allele of the mouse insulin-like growth factor II (Igf2) gene. Genes Dev. 61843-1856. [DOI] [PubMed] [Google Scholar]
- 32.Schoenherr, C. J., J. M. Levorse, and S. M. Tilghman. 2003. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 3366-69. [DOI] [PubMed] [Google Scholar]
- 33.Sparago, A., F. Cerrato, M. Vernucci, G. B. Ferrero, M. C. Silengo, and A. Riccio. 2004. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nat. Genet. 36958-960. [DOI] [PubMed] [Google Scholar]
- 34.Szabó, P. E., S.-H. Tang, F. J. Silva, W. M. Tsark, and J. R. Mann. 2004. Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol. Cell. Biol. 244791-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thorvaldsen, J. L., K. L. Duran, and M. S. Bartolomei. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 123693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulaner, G. A., Y. Yang, J. F. Hu, T. Li, T. H. Vu, and A. R. Hoffman. 2003. CTCF binding at the insulin-like growth factor-II (IGF2)/H19 imprinting control region is insufficient to regulate IGF2/H19 expression in human tissues. Endocrinology 1444420-4426. [DOI] [PubMed] [Google Scholar]
- 37.Vu, T. H., and A. R. Hoffman. 1994. Promoter-specific imprinting of the human insulin-like growth factor-II gene. Nature 371714-717. [DOI] [PubMed] [Google Scholar]
- 38.West, A. G., M. Gaszner, and G. Felsenfeld. 2002. Insulators: many functions, many mechanisms. Genes Dev. 16271-288. [DOI] [PubMed] [Google Scholar]
- 39.Wolffe, A. P. 2000. Transcriptional control: imprinting insulation. Curr. Biol. 10R463-R465. [DOI] [PubMed] [Google Scholar]
- 40.Yang, Y., J. F. Hu, G. Ulner, T. Li, X. Yao, T. H. Vu, and A. R. Hoffman. 2003. Epigenetic regulation of Igf2/H19 imprinting at CTCF insulator binding sites. J. Cell. Biochem. 901038-1055. [DOI] [PubMed] [Google Scholar]
- 41.Yao, X. M., J. F. Hu, M. Daniels, T. Li, Y. W. Yang, Z. H. Li, T. H. Vu, and A. R. Hoffman. 2004. Epigenetic regulation of the taxol resistance associated gene (TRAG-3) in human tumors. Cancer Genet. Cytogenet. 1511-13. [DOI] [PubMed] [Google Scholar]
- 42.Yoon, Y. S., S. Jeong, Q. Rong, K. Y. Park, J. H. Chung, and K. Pfeifer. 2007. Analysis of the H19ICR insulator. Mol. Cell. Biol. 273499-34510. [DOI] [PMC free article] [PubMed] [Google Scholar]