Abstract
The epidermal growth factor receptor (EGFR) family regulates essential biological processes. Various epithelial tumors are linked to EGFR overexpression or expression of variant forms, such as the EGFR1 variant, EGFRvIII. Perturbations in expression of the transcription initiation factor, TATA-binding protein (TBP), alter cellular growth properties. Here we demonstrate that EGFR1 and EGFRvIII, but not HER2, induce TBP expression at a transcriptional level through distinct mechanisms. EGFR1 enhances the phosphorylation and function of Elk-1, recruiting it to the TBP promoter. In contrast, EGFRvIII robustly induces c-jun expression, stimulating recruitment of c-fos/c-jun to an overlapping AP-1 site. Enhancing c-jun expression alone induces TBP promoter activity through the AP-1 site. To determine the underlying mechanism for differences in Elk-1 function and c-jun expression by these receptors, we inhibited the internalization of EGFR1. Persistent EGFR1 cell surface occupancy mimics EGFRvIII-mediated effects on Elk-1 and c-jun and switches the requirement of Elk-1 to AP-1 for TBP promoter induction. Together, these studies define a new molecular mechanism for the regulation of TBP expression. In addition, we identify distinct molecular targets of EGFR1 and EGFRvIII and demonstrate the importance of receptor internalization in distinguishing their specific functions.
The epidermal growth factor receptor (EGFR) family comprises four transmembrane receptor tyrosine kinases, EGFR1 (ErbB1/HER1), HER2 (ErbB2), HER3 (ErbB3), and HER4 (ErbB4). They share a similar kinase domain structure and homology but differ in their extracellular domains and carboxy-terminal tails (48). The EGFRs play vital roles during development and are important regulators of cellular proliferation, survival, and migration. Amplification and overexpression of EGFR1 and HER2 are common in a variety of human cancers. Overexpression of HER2 and EGFR1 usually results from gene amplification, and in the case of EGFR1, overexpression can result in the formation of a variety of genetic mutations. EGFRvIII is the most common genetic variant form of EGFR1 and contains an in-frame deletion of exons 2 to 7, corresponding to amino acids 6 to 273, in the extracellular domain (3). The EGFRvIII-specific deletion results in a novel extracellular domain architecture that mimics an activated receptor unable to bind EGF. In contrast to EGFRvIII, ligand-bound EGFR1 is rapidly endocytosed by clathrin-coated pits (51). After internalization, ligand-EGFR1 complexes traffic through various endosomal compartments where they are either shuttled back to the plasma membrane or degraded in lysosomes. Internalized EGFR1 continues to signal from within the endosomes, and this is thought to modulate the duration, intensity, and specificity of signaling processes (49, 57).
The TATA-binding protein (TBP) is a ubiquitously expressed transcription initiation factor indispensable for cell function. Specific protein-protein interactions allocate TBP for participation in transcription by RNA polymerase I (Pol I), Pol II, or Pol III. The recruitment of TBP to cellular promoters is thought to be a rate-limiting step in the formation of active transcription initiation complexes (41). In higher eukaryotes, TBP expression is tightly regulated at the level of transcription. TBP expression can be induced by the tumor promoter, 12-O-tetradecanoylphorbol-13-acetate, through the activation of protein kinase C (19, 20) and by the expression of oncogenic Ras (29). An examination of growth factor receptors that might regulate TBP expression revealed that EGF-induced activation of EGFR1 stimulates TBP expression, requiring the activation of Ras and all three classes of mitogen-activated protein kinases (MAPKs) (29, 64). Subsequent analysis of the role of the c-jun N-terminal kinases (JNKs) in regulating TBP expression revealed that while JNK1 enhances TBP expression, JNK2 represses it (63). In addition, we recently identified human Maf1 as a negative regulator of TBP expression and showed that it directly represses TBP transcription (30).
Alterations in the cellular amounts of TBP produce both qualitative and quantitative changes in gene expression. Depending on the cell type and context, small increases in TBP expression can induce RNA Pol I- and Pol III-dependent transcription, resulting in increased rRNA and tRNA production (55, 56, 64). Overproduction of RNA Pol III transcripts is observed in a variety of human tumors (8, 9, 58). In addition, recent studies have shown that enhanced RNA Pol III-dependent transcription is required for oncogenic transformation (27, 35). A subset of RNA Pol II-dependent promoters are differentially affected by alterations in cellular TBP concentrations, depending on the promoter architecture (12, 33, 47). TBP-mediated changes in gene expression have dramatic phenotypic consequences. In mouse embryo fibroblasts, alterations in TBP concentrations change cellular proliferation rates (64). Heterozygous disruption of TBP in chicken DT cells results in delayed mitosis (53). In Rat1A fibroblasts, small increases in TBP concentrations do not alter proliferation rates but promote anchorage-independent growth and tumorigenesis (29). Deregulation of TBP expression has biological consequences as well; upregulated expression of TBP is observed in a clinically significant number of human colon tumors compared to matched normal colon epithelium (28). In summary, work implicating the involvement of TBP in proliferation and tumorigenesis underscores the importance of understanding the mechanism by which this central transcription initiation factor is regulated.
EGFRvIII is exclusively expressed in human tumors, and its presence is associated with a more aggressive disease and poorer prognosis. Thus, understanding the molecular functions that differentiate EGFRvIII from EGFR1 will be critical for the development of EGFRvIII-specific therapeutic targets. Here we identify key molecular targets that distinguish the functions of EGFR1 and EGFRvIII. Using mouse NR6 and human U87 cells previously engineered to overexpress either EGFR1 or EGFRvIII, we find that both receptors induce the expression of TBP through activation of the TBP promoter. This contrasts with HER2, which despite its ability to activate Ras signaling does not regulate TBP expression. Surprisingly, however, EGFR1 and EGFRvIII target different transcription factors to the TBP promoter. EGFR1, but not EGFRvIII, induces Elk-1 phosphorylation and its transactivation function and recruits it to a specific Elk-1 binding site within the TBP promoter. In contrast, EGFRvIII more strongly induces c-jun expression and the recruitment of c-fos and c-jun to an AP-1 site that overlaps the Elk-1 binding site. These differences in Elk-1 function and c-jun expression result from differences in the abilities of the receptors to be internalized into clathrin-dependent endosomes. Together, these studies demonstrate a new mechanism by which TBP expression can be regulated and identify key molecular events that distinguish the functions of EGFR1 and EGFRvIII.
MATERIALS AND METHODS
Cell lines.
NR6 stable lines overexpressing EGFR1, EGFRvIII, or HER2 (2, 7); U87 stable lines overexpressing EGFR1 or EGFRvIII (39); and the MCF-7 parental and stable line overexpressing HER2 (7) were cultured in Dulbecco's modified Eagle's medium (4.5 g/liter glucose) supplemented with 10% fetal calf serum. All cell lines were previously characterized.
Immunoblot analysis.
Chemical treatments were performed 16 h prior to protein isolation and 6 to 10 h after serum starvation (0.5% serum). Cell lysates were prepared and subjected to Western blot analysis (100 μg total protein) as previously described (13). The antibodies used were purchased from Cell Signaling (c-fos, c-jun, Elk-1, and phospho-Tyr1173-EGFR1), BD Biosciences (EGFR1 and clathrin heavy chain), Santa Cruz Biotechnology (Elk-1 and phosphotyrosine-immunoglobulin G), Upstate Biotechnology (TBP), and Chemicon (β-actin). The HER2 antibody, 10H8, was provided by J. M. Park et al. (44).
Plasmid DNAs.
Expression plasmids for EGFR1 and EGFRvIII, p-HβAPr-1neo-β-actin-EGFR1 (or EGFRvIII), were provided by S. K. Batra et al. (2). Human wild-type and Elk-1 mutant TBP promoter-luciferase constructs were previously described (16). The AP-1 mutant TBP promoter is described below. The human β3-integrin luciferase reporter gene construct was provided by K. J. Cohen et al. (11). The cytome galovirus (CMV)-β-galactosidase and β-actin-renilla reporter gene constructs were from Amy Lee. The HER2, p-SV2-neo-LTR-HER2, and p-SV2-neo-LTR were provided by J. M. Park et al. (7). Human c-fos-luciferase and Δ56dE-fos mutant luciferase promoter constructs were provided by C. K. Galang et al. (17). RSV-LTR-c-jun and RSV-LTR-c-Fos expression vectors were provided by Ebrahim Zandi.
Site-directed mutagenesis.
Mutagenesis of the −736/+66, −230/+66, and −4500/+66 TBP promoter-luciferase constructs was performed using QuikChange and QuikChange II XL site-directed mutagenesis kits (Stratagene). The AP-1 recognition sequence was changed from GTGACATTAT to the non-recognition sequence GTGATGTTAT (base changes are bold). Mutagenesis was confirmed by sequence analysis (USC/Norris Microchemical Core).
Transient transfections.
Logarithmically growing cells were seeded and then transfected the following day using 3 μg of DNA and 3 μl of F1 reagent (Targeting Systems), following the protocol provided. Twenty-four hours posttransfection, the cells were serum deprived (0.5%). Ten hours after serum deprivation, cells were treated with designated inhibitor (2 μM AG1478 for mouse lines, 10 μM AG1478 for human lines, and 25 μM AG825, 10 μM U0126, 20 μM SP600125, or 10 μM SB202190). Cells were treated with EGF (50 ng/ml) 6 to 10 h after serum deprivation or 1 h after inhibitor treatment.
For transfection of small interfering RNA (siRNA) oligonucleotide sequences, the Targeting Systems siRNA kit was used, following the protocol provided. siRNA transfection occurred 4 h after plasmid DNA transfection. Two days after siRNA transfection, cells were serum deprived (0.5% serum) and harvested the following day. The siRNA oligonucleotide sequences were described previously for Elk-1 (60). ON-TARGETplus duplex c-jun, c-fos, and clathrin siRNAs were purchased from Dharmacon. The Promega protocol was followed for preparing lysates for luciferase and β-galactosidase assays. Changes in promoter activities were calculated based on control (reporter gene plus empty expression vector or untreated or mismatch [mm] siRNA) and normalized to total protein. Standard errors of the mean were calculated using at least three biologically independent sample determinations. β-Galactosidase assays were used to measure transfection efficiency using equal amounts of protein lysate.
EGF internalization assay.
U87-EGFR1 cells transfected with mm siRNA or clathrin siRNA as described above were plated on collagen-coated coverslips for 8 h. Cells were serum starved overnight in medium with 0.5% fetal calf serum and then labeled with 5 μg/ml Texas red-EGF (Molecular Probes) at 4°C for 1 h. Excess EGF was removed by rinsing cells with cold medium. Cells were then warmed to 37°C for 15 min. Cells were fixed in paraformaldehyde and then imaged on a Zeiss LSM 510 confocal laser scanning microscope.
ChIP assay and real-time PCR.
Cells were pretreated with dimethyl sulfoxide or AG1478 for 10 min prior to a 10- to 15-min stimulation with EGF (50 ng/ml). Chromatin immunoprecipitation (ChIP) assays were then performed as described previously (13, 14, 61). Briefly, chromatin was immunoprecipitated with antibody rotating overnight. The DNA region cross-linked to the protein was determined by quantitative PCR (qPCR) using iQ SYBR Green Supermix (Bio-Rad) on an MX3000P system (Stratagene). Primer sequences for TBP promoter regions −119 to +5 and −3315 to −3158 were previously described (30). All threshold cycle (CT) values were normalized to the PCR efficiency using the 1/(2·PCR efficiency)CTcalculation. Normalized CT values for antibody pull-downs were normalized to input using the antibody IP·10/input calculation and immunoglobulin G. Fold changes in promoter occupancy were calculated by setting the level of promoter occupancy of the transcription factor in the cells transfected with mm siRNA and in the absence of EGF treatment to 1.
RESULTS
EGFR1 and EGFRvIII, but not HER2, induce TBP expression.
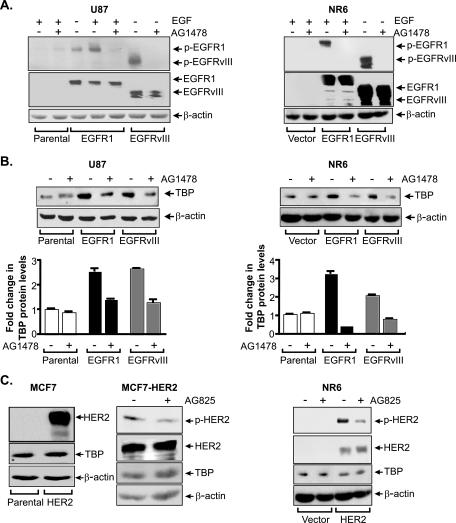
As the activation of EGFR1 was previously shown to induce TBP expression (64), we asked whether variant forms of EGFR1, EGFRvIII, and another member of the EGFR family, HER2, would similarly enhance TBP expression. Stable human glioblastoma U87 and mouse fibroblast NR6 cell lines expressing either EGFR1 or EGFRvIII were examined for receptor expression and activation, which are associated with phosphorylation of the C-terminal tail (Fig. 1A). Immunoblot analysis revealed that neither the vector control U87 nor NR6 cells expressed detectable amounts of EGFR1 or EGFRvIII, while overexpression of EGFR1 and EGFRvIII was detected in the stable lines expressing cDNAs for these receptors. While EGFRvIII was constitutively activated in the U87-EGFRvIII cells, a low level of EGFR1 activation could be further enhanced with treatment of EGF in the U87-EGFR1 cells. Activation of both receptors was inhibited by treatment of the cells with the EGFR-specific inhibitor, AG1478. Analysis of TBP protein levels in these cells revealed that both EGFR1- and EGFRvIII-expressing cells displayed approximately a three- to fourfold increase in TBP amounts compared to that of the control cells (Fig. 1B). Enhanced TBP expression was blocked by inactivation of these receptors. To determine whether HER2 affects TBP expression, NR6 and MCF7 stable cell lines expressing HER2 were examined. Immunoblot analysis demonstrated that neither the vector control MCF7 nor NR6 cells expressed detectable amounts of HER2 (Fig. 1C). HER2 was activated in both HER2 overexpressing cell lines, and the activation of HER2 was blocked with treatment of the HER2-specific inhibitor, AG825; however, there was no effect on TBP expression (Fig. 1C).
FIG. 1.
EGFR1 and EGFRvIII, but not HER2, enhance TBP expression in human and mouse cell lines. (A) Receptor activation in cell lines stably expressing EGFR1 or EGFRvIII. Immunoblot analysis was performed with whole cell lysates (WCL) derived from serum-deprived U87 or NR6 stable cell lines treated as designated using phospho-EGFR1 or EGFR1 antibodies. (B) TBP expression is increased in cell lines stably expressing EGFR1 or EGFRvIII. Immunoblot analysis was performed with WCL as shown in panel A using TBP or β-actin antibodies. Protein levels were quantified by densitometry, and TBP levels were calculated by normalization to β-actin levels. The change was calculated relative to untreated parental control cell lines. (C) TBP expression is not regulated by HER2. Immunoblot analysis was performed with lysates derived from serum-deprived MCF7 parental and MCF7 stable cells overexpressing HER2 or the NR6 stable lines. Lysates were prepared and immunoprecipitated with HER2 antibody and subjected to immunoblot analysis with HER2 or phospho-tyrosine antibody. Direct immunoblot analysis was also performed using TBP and β-actin antibodies.
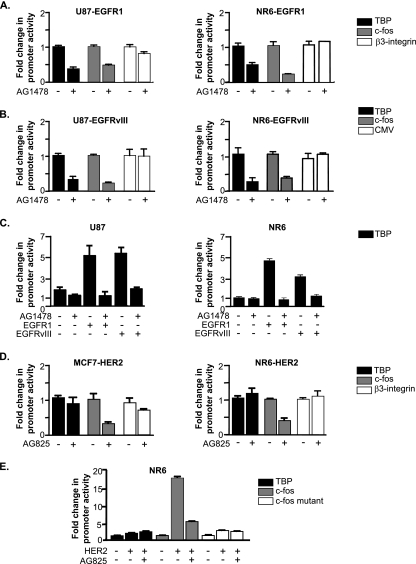
To further address the mechanism by which EGFR1 and EGFRvIII regulate TBP expression, we asked whether these receptors induce transcription of the TBP promoter. The activity of a transfected human TBP promoter construct was measured in the EGFR1- and EGFRvIII-expressing cells in the presence and absence of AG1478 (Fig. 2A and B). Inhibiting activation of either EGFR1 or EGFRvIII reduced TBP promoter activity, as well as that of a known target of these receptors, the c-fos promoter. This is in contrast to the activity of the β3-integrin or CMV promoters that were not affected by altering receptor activation. In addition, transient expression of EGFR1 and EGFRvIII in the parental U87 and NR6 cells resulted in an induction of TBP promoter activity (Fig. 2C). In contrast, TBP promoter activity in the MCF7-HER2 and NR6-HER2 cells was not altered by inactivation of HER2 by AG825 treatment (Fig. 2D). This is distinct from the c-fos promoter, whose activity was decreased by AG825 treatment. Similarly, transient expression of HER2 in the NR6 cells did not induce TBP promoter activity (Fig. 2E). Together, these results indicate that while both activation of EGFR1 and its variant, EGFRvIII, induce cellular TBP expression at a transcriptional level, HER2 does not regulate TBP expression.
FIG. 2.
EGFR1 and EGFRvIII, but not HER2, transcriptionally regulate TBP. (A) EGFR1 transcriptionally regulates TBP. Stable cell lines expressing EGFR1 were transfected with promoter-reporter constructs for TBP, c-fos, or β3-integrin. Cells were then harvested and analyzed for luciferase activity. (B) EGFRvIII transcriptionally regulates TBP. Stable cell lines expressing EGFRvIII were transfected with promoter-reporter constructs for TBP, c-fos, or CMV. Cells were then harvested and analyzed for luciferase activity. (C) Transient expression of EGFR1 or EGFRvIII induces TBP promoter activity. U87 or NR6 cell lines were cotransfected with either empty vector, EGFR1 expression vector, or EGFRvIII expression vector and the TBP promoter construct. Cells were then harvested and analyzed for luciferase activity. (D) HER2 overexpression does not regulate TBP promoter activity. MCF-7 and NR6 stable lines expressing HER2 were transfected with TBP, c-fos, or β3-integrin promoter constructs. Cells were then harvested and analyzed for luciferase activity. (E) Transient expression of HER2 does not induce TBP promoter activity. NR6 cells were cotransfected with empty vector or a HER2 expression vector and TBP, c-fos, or c-fos mutant promoter constructs. Cells were then harvested and analyzed for luciferase activity. For panels A to E, the change was calculated based on normalization to untreated and/or empty vector control for each promoter construct.
EGFR1 and EGFRvIII differentially regulate TBP through the recruitment of Elk-1 and AP-1, respectively, to the TBP promoter.
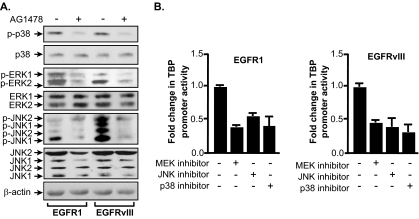
To determine whether EGFR1 and EGFRvIII regulate TBP expression through the same or distinct mechanisms, we first determined the activation states of the three classes of MAPKs in the receptor-expressing cells by measuring their phosphorylation states. Immunoblot analysis revealed that levels of expression of extracellular signal-regulated kinase 1/2 (ERK1/2), p38, and JNK1/2 in the U87-EGFR1 and U87-EGFRvIII cells were comparable (Fig. 3A). All three MAPKs were phosphorylated in the EGFR1- and EGFRvIII-expressing cells. This was receptor activation dependent as treatment with AG1478 diminished the phosphorylation of each MAPK. While the extent of p38 phosphorylation was similar in the U87-EGFR1 and U87-EGFRvIII cells, phospho-ERK amounts were reproducibly higher in the U87-EGFR1 cells compared to those observed in the U87-EGFRvIII cells. In contrast, the amount of phospho-JNK was greater in the U87-EGFRvIII cells compared to that observed in the U87-EGFR1 cells. Thus, while EGFR1 and EGFRvIII activate all three classes of MAPKs, the extent of ERK and JNK activation differs. These results are consistent with previous observations in NIH 3T3 stable cell lines expressing these receptors (1). To determine whether the activation of these MAPKs was required for TBP promoter induction, the cells were treated with chemical inhibitors for ERK, p38, and JNK, and TBP promoter activity was determined (Fig. 3B). Blocking the activation of ERK, p38, or JNK each reduced TBP promoter activity in both the EGFR1 and EGFRvIII cell lines. These results indicate that both these receptors require the activation of all three MAPKs for TBP promoter induction.
FIG. 3.
EGFR1 and EGFRvIII differentially activate MAPKs to induce TBP promoter activity. (A) EGFR1 and EGFRvIII differentially activate ERK1/2 and JNK1/2. Immunoblot analysis was performed using whole cell lysates derived from serum-deprived U87 or NR6 stable cell lines using antibodies against p38, ERK, and JNK as well as phospho-specific antibodies. (B) EGFR1 and EGFRvIII require activation of all three classes of MAPKs to induce TBP promoter activity. U87 stable cell lines were transfected with the wild-type TBP promoter-luciferase construct. Cells were treated as described in Materials and Methods with inhibitors for ERK1/2 (U0126), p38 (SP600125), or JNK1/2 (SB202190) and analyzed for luciferase activity. The change was calculated based on normalization to untreated cells.
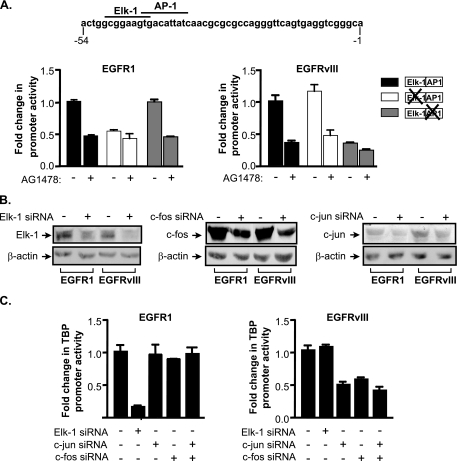
The response elements required for receptor-mediated induction of the TBP promoter were next identified. Mutation of an Elk-1 binding site abrogated EGFR1-mediated, but not EGFRvIII-mediated, enhancement of TBP promoter activity in the U87 stable cell lines (Fig. 4A). In contrast, mutation of an adjacent consensus AP-1 binding site abolished EGFRvIII-mediated, but not EGFR1-mediated, induction of the TBP promoter. To further determine the differential requirement for Elk-1 and AP-1 in driving TBP promoter induction, siRNAs were used to repress the expression of Elk-1, c-jun, or c-fos in these cells (Fig. 4B). Decreased expression of Elk-1 substantially reduced TBP promoter activity in the U87-EGFR1 cells but had no effect on promoter activity in the U87-EGFRvIII cells (Fig. 4C). Conversely, decreased expression of c-jun and/or c-fos had no effect on TBP promoter activity in the U87-EGFR1 cells but reduced TBP promoter activity in the U87-EGFRvIII cells. These results indicate that EGFR1-mediated TBP promoter induction requires Elk-1, whereas EGFRvIII-mediated induction requires c-jun and c-fos.
FIG. 4.
EGFR1 and EGFRvIII require different transcription factors to induce TBP promoter activity. (A) TBP promoter induction by EGFR1 requires an Elk-1 binding site, whereas EGFRvIII requires an AP-1 binding site. (Top) The human TBP promoter sequence denoting the Elk-1 and AP-1 sites located between −54 and −1 of transcriptional start site. U87 stable lines were transfected with the designated TBP promoter constructs together with a CMV-driven promoter β-galactosidase reporter construct. Luciferase activity was normalized to β-galactosidase activity, and changes were calculated based on untreated control. (B) Specific downregulation of Elk-1, c-fos, or c-jun expression in EGFR1- and EGFRvIII-expressing U87 cells. Cells were transfected with either mm control siRNA (−) or siRNAs specific for Elk-1, c-fos, or c-jun. Lysates were prepared, and immunoblot analysis was performed using antibodies against Elk-1, c-jun, c-fos, or β-actin. (C) EGFR1 requires Elk-1, whereas EGFRvIII requires c-fos and c-jun for TBP promoter induction. U87 stable cell lines were cotransfected with the TBP promoter and either mm (−), Elk-1, c-jun, or c-fos siRNAs (+). Resultant lysates were analyzed for luciferase activity. Luciferase activities were normalized to total protein levels, and cells transfected with mm siRNA were set to 1. Changes were calculated based on activities in cells transfected with mm siRNA.
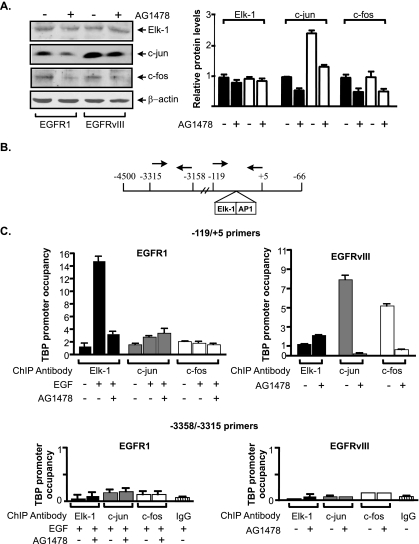
To determine whether Elk-1 and AP-1 were directly involved in regulating TBP promoter activity, we conducted ChIP assays. Immunoblot analysis revealed that comparable levels of Elk-1 and c-fos were expressed in the U87-EGFR1 and U87-EGFRvIII cells (Fig. 5A). However, EGFRvIII induced higher levels of c-jun compared to EGFR1. ChIP analysis was used to interrogate the binding of Elk-1, c-jun, and c-fos to the TBP promoter. Primers were used that span the region of the TBP promoter containing the Elk-1 and AP-1 binding sites (Fig. 5B). Treatment of the U87-EGFR1 cells with EGF induced recruitment of Elk-1 to the TBP promoter, which could be blocked with AG1478 (Fig. 5C, top). Comparatively less c-jun was recruited to the TBP promoter, and its occupancy was not dependent on receptor activation. No detectable binding of c-fos to the TBP promoter was observed. In contrast, little recruitment of Elk-1 to the TBP promoter was observed in the U87-EGFRvIII cells. However, both c-jun and c-fos were found to occupy the promoter in an AG1478-dependent manner. As a control, sequences upstream of the Elk-1 and AP-1 binding sites were examined. Activation of EGFR1 or EGFRvIII did not result in the recruitment of Elk-1, c-jun, or c-fos to this region (Fig. 5C, bottom). These results support the idea that EGFR1 activation results in the selective recruitment of Elk-1 to the TBP promoter, while EGFRvIII activation results in the recruitment of c-jun and c-fos. This also suggests that Elk-1 and c-jun/c-fos are able to independently bind to the TBP promoter and do not appear to cooperate to induce transcription.
FIG. 5.
EGFR1 induces Elk-1 recruitment, whereas EGFRvIII induces c-jun and c-fos recruitment to the TBP promoter. (A) Elk-1, c-jun, and c-fos expression in U87 cells stably expressing EGFR1 or EGFRvIII. (Left) Protein lysates derived from U87 stable cell lines were subjected to immunoblot analysis using Elk-1, c-jun, c-fos, or β-actin antibodies. Three independent experiments were used to quantify the relative levels of each protein by densitometry. (Right) Changes were calculated based on β-actin normalized protein levels in untreated U87-EGFR1 cells. (B) Schematic of the human TBP promoter. The relative location of the PCR primers used for the ChIP analysis is designated with arrows. (C) Elk-1, c-jun, and c-fos differentially occupy the TBP promoter in U87 cells expressing EGFR1 or EGFRvIII. ChIP assays were performed on U87 stable lines using Elk-1, c-jun, or c-fos antibodies. qPCR was performed to quantify the amplified DNA using the primer sets shown in panel B that span the Elk-1 and AP-1 sites (top) or sequences upstream of these binding sites (bottom). The n-fold change in TBP occupancy was calculated based on the untreated control.
EGFR1 and EGFRvIII can be distinguished by their differential effects on Elk-1 and AP-1.
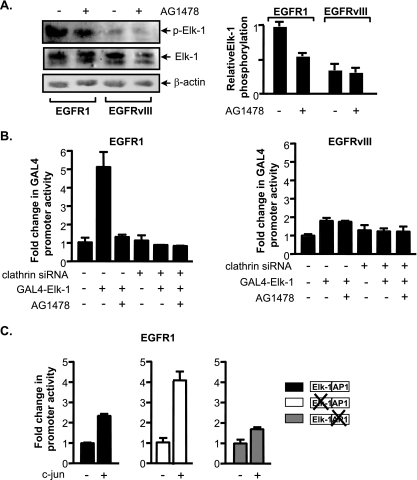
Since EGFR1 and EGFRvIII were found to target distinct transcription components, we sought to identify differences in Elk-1 and AP-1 in these receptor-expressing cells that might contribute to their distinct functions. Since immunoblot analysis revealed that Elk-1 expression did not differ in the EGFR1- and EGFRvIII-expressing cells (Fig. 5A), we determined if the function of Elk-1 might be altered. Transcriptional activation of Elk-1 occurs through its phosphorylation by MAPKs, particularly at serine 383 (21, 25, 34). Immunoblot analysis revealed that while Elk-1 was phosphorylated in both EGFR1- and EGFRvIII-expressing cells, the level of S383 phosphorylation was appreciably higher in the U87-EGFR1 cells than in the U87-EGFRvIII cells (Fig. 6A).
FIG. 6.
EGFR1 and EGFRvIII differentially affect Elk-1 function and c-jun expression. (A) EGFR1, but not EGFRvIII, enhances Elk-1 phosphorylation. (Left) Immunoblot analysis performed on lysates derived from U87 stable lines using antibodies specific for Elk-1 phospho-serine 383, Elk-1, and β-actin antibodies. (Right) For quantification of the relative levels of p-Elk-1, changes (n-fold) were calculated based on β-actin normalized protein levels in untreated U87-EGFR1 using three independent experiments. (B) Elk-1 transactivation function in EGFR1- and EGFRvIII-expressing cells. U87 stable cell lines were cotransfected with a Gal4-responsive promoter-luciferase construct containing a vector and either an Elk-1-Gal4 fusion protein expression vector or an empty vector. All cells were cotransfected with a CMV-driven β-galactosidase reporter construct. Resultant cell lysates were analyzed for luciferase and β-galactosidase activities. Change was calculated based on normalization to luciferase activity in untreated vector control cells transfected with mm siRNA. (C) Increased c-jun expression drives TBP promoter activity through the AP-1 site in EGFR1-expressing cells. Cells were cotransfected with wild-type or mutated TBP promoter constructs, a CMV-driven β-galactosidase reporter construct, and a c-jun expression vector. Resultant lysates were measured for luciferase and β-galactosidase activities. For all experiments, luciferase activities were normalized to β-galactosidase activity and the relative change was determined by setting the luciferase activity with empty vector to 1.
The functional consequence of this difference in Elk-1 phosphorylation was examined using a GAL4-Elk-1 reporter assay (5). We tested the ability of an expressed protein containing the GAL4 DNA binding domain fused to the Elk-1 transactivation domain to activate a GAL4 reporter promoter (Fig. 6B). GAL4-Elk-1 markedly induced the GAL4 reporter promoter in EGF-stimulated U87-EGFR1 cells. Blocking EGFR1 activation inhibited induction of the reporter. In contrast, expression of GAL4-Elk-1 in the U87-EGFRvIII cells failed to induce GAL4 reporter promoter activity. Together, these results demonstrate that EGFR1 is capable of inducing phosphorylation of Elk-1, allowing its recruitment to the promoter and transcription activation. In contrast, EGFRvIII does not induce Elk-1 phosphorylation and its activation.
We next examined potential differences in AP-1 that might contribute to its preferential use by the TBP promoter in the U87-EGFRvIII cells compared to the U87-EGFR1 cells. Since U87-EGFRvIII cells express a relatively higher level of c-jun than the U87-EGFR1 cells (Fig. 5A), this suggested that c-jun might be limiting for the formation of AP-1 complexes in the U87-EGFR1 cells. Therefore, c-jun expression was ectopically increased in the U87-EGFR1 cells, and the activities of the wild-type and mutated TBP promoter-reporter constructs were examined (Fig. 6C). Increased expression of c-jun resulted in stimulation of the wild-type TBP promoter, but a more substantial induction of the TBP promoter was observed when the Elk-1 site was mutated. However, the TBP promoter lacking a functional AP-1 site was not induced. These results indicate that while c-jun is fully functional in the EGFR1-expressing cells, it is limiting for TBP promoter activity. When c-jun expression is increased in these cells, EGFR1-mediated TBP promoter induction alternatively requires the AP-1 site instead of the Elk-1 site. These results indicate that increased expression of c-jun alone can drive transcription of the TBP promoter through the AP-1 response element in EGFR1-expressing cells.
Reduced internalization of EGFR1 mimics EGFRvIII-mediated effects on Elk-1 and c-jun.
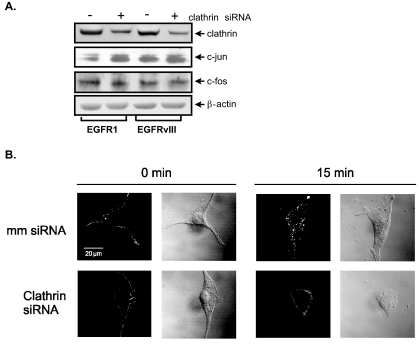
To determine the underlying mechanisms that distinguish EGFR1 and EGFRvIII signaling to the TBP promoter, we considered that a major difference between these receptors is that EGFR1 undergoes rapid ligand-mediated internalization and recycling through clathrin-coated pits (54, 65), whereas EGFRvIII does not (22, 23). We asked whether inhibiting the internalization of EGFR1, mimicking the prolonged cell surface occupancy of EGFRvIII, would alter Elk-1 function and c-jun expression. Immunoblot analysis showed that clathrin-specific siRNA downregulated clathrin expression in both U87-EGFR1 and U87-EGFRvIII cells by approximately threefold (Fig. 7A). This reduction in clathrin was sufficient to prevent internalization of EGF-bound EGFR1 (Fig. 7B). Decreased clathrin expression resulted in an increase in c-jun expression in the U87-EGFR1 cells, corresponding to the levels of c-jun observed in the U87-EGFRvIII cells (Fig. 7A). We next assessed potential changes in Elk-1 function when clathrin expression was repressed (Fig. 6B). The ability of GAL-4-Elk-1 to induce the GAL4 reporter promoter in U87-EGFR1 cells was abrogated when clathrin expression was decreased. In contrast, the Elk-1-dependent promoter was not induced in the U87-EGFRvIII cells and decreased clathrin expression had no effect on promoter activity. These results indicate that decreased clathrin expression in U87-EGFR1 cells results in an increase in c-jun expression and a loss of Elk-1 transactivation activity. These results support the idea that inhibiting internalization of EGFR1 results in altered EGFR1 signaling that mimics that of EGFRvIII.
FIG. 7.
Decreased clathrin expression increases c-jun expression and inhibits EGFR1 internalization in U87-EGFR1 cells. (A) Decreased clathrin expression in U87-EGFR1 cells increases c-jun expression. U87 stable lines were transfected with either clathrin siRNA or mm control siRNA. Immunoblot analysis was performed using the resultant cell lysates and antibodies against clathrin, c-jun, c-fos, and β-actin. (B) Decreased clathrin expression in U87-EGFR1 cells inhibits EGFR1 internalization. Cells transfected with mm siRNA (top row) or clathrin siRNA (bottom row) were labeled with Texas red-EGF. Labeled cells were then fixed in paraformaldehyde and imaged by confocal microscopy. Fluorescent images are shown next to differential interference contrast images for a representative cell for each treatment/time point.
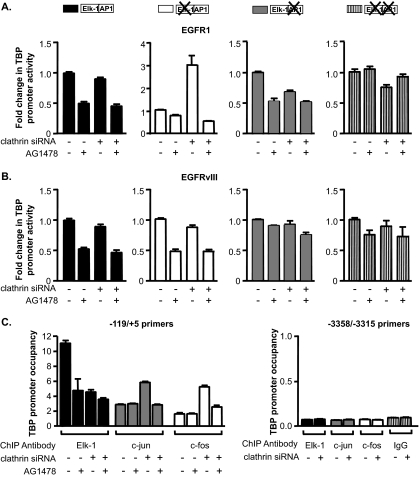
We next determined whether the changes in Elk-1 transactivation activity and c-jun expression in the U87-EGFR1 cells observed upon decreased clathrin expression would result in changes in the utilization of these factors by the TBP promoter. Various TBP promoter reporter constructs were tested in the U87-EGFR1 cells transfected with either a control siRNA or clathrin siRNA. No change in wild-type TBP promoter activity was observed upon decreased clathrin expression in either the U87-EGFR1 or U87-EGFRvIII cells (Fig. 8A and B). As expected, the promoter containing the mutated Elk-1 site did not respond to EGFR1; however, reduction of clathrin expression resulted in an increase in promoter activity, which could be abolished with AG1478 (Fig. 8A). These results suggest that by reducing the internalization of EGFR1, TBP promoter activity is now regulated through the AP-1 site. Consistent with these results, mutation of the AP-1 site did not prevent EGFR1-mediated induction of the promoter. However, downregulation of clathrin resulted in dependence for the AP-1 site, as the promoter lacking this site could no longer be induced by EGFR1. Mutation of both the Elk-1 and AP-1 sites resulted in an inability of the TBP promoter to be stimulated by EGFR1, independent of clathrin levels. Examination of TBP promoter activity in the U87-EGFRvIII cells revealed that decreasing the levels of clathrin did not affect the ability of EGFRvIII to induce TBP promoter activity through the AP-1 site (Fig. 8B). Furthermore, ChIP assays revealed that decreased clathrin expression in the U87-EGRF1 cells resulted in reduced Elk-1 recruitment to the TBP promoter, with a concomitant increase in both c-jun and c-fos recruitment (Fig. 8C). Together, these results demonstrate that the differential effect that EGFR1 and EGFRvIII have on Elk-1 and c-jun is a result of the abilities of these receptors to be internalized.
FIG. 8.
Inhibition of EGFR1 internalization mimics EGFRvIII-mediated effects on TBP promoter induction. (A) Decreased clathrin expression in U87-EGFR1 cells alters TBP promoter induction through the AP-1 site. Cells were transfected with either mm or clathrin-specific siRNAs and cotransfected with TBP-luciferase and CMV-β-galactosidase promoter-reporter constructs. Cell lysates were analyzed for luciferase and β-galactosidase activities. Luciferase activity levels were normalized to β-galactosidase activity, and changes were calculated based on untreated cells transfected with mm siRNA. (B) Decreased clathrin expression does not affect EGFRvIII-mediated induction of TBP promoter activity through the AP-1 site. Experiments were conducted as described in panel A using U87-EGFRvIII cells. (C) Decreased clathrin expression in U87-EGFR1 cells increases the recruitment of c-jun and c-fos while decreasing the recruitment of Elk-1 to the TBP promoter. ChIP assays were performed on cells transfected with either mm (−) or clathrin-specific siRNA using Elk-1, c-jun, and c-fos antibodies. qPCR was performed to quantify the amplified DNA using the primer sets as described for Fig. 5. The n-fold change in TBP occupancy was calculated based on untreated mm control.
DISCUSSION
EGFRvIII plays an important role in a variety of human cancers, including those of the brain, breast, lung, ovary, and prostate (37). Studies indicate that tumors expressing EGFRvIII are linked to radiation and chemotherapeutic resistance, and these cancers exhibit a more aggressive disease and poorer prognosis (24, 38). In addition, EGFRvIII is more tumorigenic than the wild-type receptor (40). The enhanced tumorigenicity of EGFRvIII has been attributed to its inability to be downregulated due its low rate of endocytosis compared to that of EGFR1, which is rapidly internalized through clathrin-coated vesicles (23). While extensive studies have determined the molecular mechanisms that define the dramatic differences in the endocytosis rates of EGFR1 and EGFRvIII, how this ultimately affects their molecular and biological functions remained unclear. Our studies have identified distinct molecular functions of these receptors that are dictated by their abilities to undergo internalization.
EGFR1 induces Elk-1 phosphorylation and transcriptional activation, and this requires clathrin-dependent internalization of the receptor. Accordingly, EGFRvIII expression does not lead to enhancement of Elk-1 phosphorylation and function. EGFR1 has been shown to produce a transient but potent induction of ERK-1, whereas EGFRvIII displays a much weaker induction of ERK-1 (1, 37). As ERK-1 is responsible for phosphorylating Elk-1 at serine 383 (26), it is likely that differences in the activation of ERK-1 by these receptors determine whether Elk-1 is phosphorylated. Recent studies have revealed that trafficking of activated EGFR1 through endosomes controls the spatial and temporal regulation of MAPK signaling (52). Continuous signaling of EGFR1 from late endosomes resulted in sustained ERK-1 activation and hyperactivation of Elk-1. Together, these results support the idea that transcriptional activation of Elk-1 is mediated through EGFR1 signaling from endosomes. Thus, regulation of EGFR1 internalization and endosomal trafficking is an important mechanism for determining EGFR1 targets. Interestingly, recent studies have shown that expression of the androgen receptor in prostate cancer cells reduces clathrin-mediated endocytosis of EGFR1, which could regulate EGFR1 signaling responses (4).
While the activation of both EGFR1 and EGFRvIII induces c-jun expression, we consistently observed a more pronounced increase in c-jun levels in the EGFRvIII-expressing cells. This is consistent with the observation that EGFRvIII induces a more potent activation of JNK than EGFR1 (1, Fig. 3A). Although EGFR1-mediated TBP promoter induction requires Elk-1 activation and its recruitment to the promoter, enhanced expression of c-jun in these cells was able to switch dependence from the Elk-1 response element to the AP-1 binding site for promoter induction. These results suggest that the inability of c-jun to be recruited to the TBP promoter in the EGFR1-activated cells is not because it is functionally impaired, but rather that c-jun is limiting for TBP promoter activity. Upon enhanced expression of c-jun, it can be effectively used to drive TBP promoter activity, even in the presence of transcriptionally competent Elk-1. Importantly, the increased expression of c-jun and its recruitment to the TBP promoter observed in the EGFRvIII-expressing cells can be reproduced in the EGFR1-expressing cells by inhibiting its internalization. Thus, EGFRvIII signaling, or prolonged EGFR1 signaling at the cell surface, results in a greater enhancement of c-jun expression and its recruitment to the TBP promoter. As sustained ERK1/2 activation has been shown to increase AP-1 DNA binding activity (6), it is also possible that the duration of ERK1/2 activation determines the extent to which c-jun levels are induced.
Our results have uncovered a new pathway by which TBP expression is regulated. Increased cellular levels of c-jun induce TBP promoter activity through the recruitment of c-jun and c-fos to an AP-1 binding site that overlaps the Elk-1 response element. Composite promoters containing overlapping Ets and AP-1 binding sites have been found in many oncogenic responsive genes (17, 31). Studies have established a strong link between activation of these composite promoters and Ras transformation (17, 18). Consistent with these findings, Ras-mediated increases in TBP expression are required for Ras transforming activity (28). Analysis of certain promoters containing tandem Ets and AP-1 binding sites revealed that only a subset of Ets proteins cooperatively binds DNA with c-jun/c-fos complexes to direct gene expression (31). Our results support the idea that Elk-1 and c-jun/c-fos bind independently to stimulate TBP promoter activity. Moreover, mutation of one of these DNA binding sites appears to enhance binding of the other protein to its cognate site, suggesting that the binding of Elk-1 or c-jun/c-fos may exclude the binding of the other. Given our results, it is likely that the cellular concentrations of c-jun and functional Elk-1 determine which binding site within the promoter is used for regulating TBP expression. It will be interesting to identify other cellular contexts, in addition to EGFR activation, by which c-jun or Elk-1 is preferentially used to drive TBP expression. The ability of TBP to be independently regulated by these two key transcription factors ensures that TBP expression is induced under a wide variety of conditions and regulatory events.
These studies, and our previous work, support the idea that TBP and c-jun positively regulate each other's expression. While c-jun levels modulate TBP promoter activity, TBP also transcriptionally regulates c-jun expression, which then dictates the proliferation rates of mouse embryo fibroblasts (63). Overexpression of c-jun has been shown to induce oncogenesis (50), and AP-1 activity is deregulated in a large variety of human tumors (15, 36, 42, 59). AP-1 regulates a program of gene expression that induces invasion in both human and mouse fibroblasts (43). Furthermore, increased expression of c-jun is required for tumor angiogenesis (62). The metalloproteinases, which are produced by endothelial cells and are required for the switch to the angiogenic phenotype in tumor models, are c-jun and c-fos targets (43). Interestingly, analysis of primary glioblastoma multiforme (GBM) tumor subtypes revealed that increased metalloproteinase 9 activation is strongly correlated with EGFRvIII expression, occurring in 83% of the analyzed EGFRvIII-immunopositive tumors but in none of the EGFRvIII-negative tumors (10). Expression of EGFRvIII induces the motility and invasive properties of GBM (32). These results, together with our current findings, support the idea that EGFRvIII-mediated increases in c-jun expression contribute to the enhanced invasive properties of GBMs expressing EGFRvIII.
Our studies have demonstrated that the EGFRs can be distinguished by their functions in regulating expression of the central transcription initiation factor, TBP. EGFR1 and its mutant variant, EGFRvIII, regulate TBP through distinct pathways, whereas HER2 does not affect TBP expression. While our previous studies have shown that expression of oncogenic Ras induces TBP expression (29) and HER2 activates Ras signaling (45, 46), HER2 fails to regulate TBP expression. Whether this is a consequence of HER2-specific differences in the activation of signaling pathways downstream of Ras, or due to the activation of other pathways that negate the effects of HER2 on Ras signaling required for regulation of TBP expression, remains to be determined.
Acknowledgments
We thank Sandra Schmidt for helpful advice. Microscopy was performed by the Cell and Tissue Imaging Core of the USC Center for Liver Diseases, NIH P30 DK048522.
This work was supported by NIH CA74138 to D.L.J.
Footnotes
Published ahead of print on 18 August 2008.
REFERENCES
- 1.Antonyak, M. A., D. K. Moscatello, and A. J. Wong. 1998. Constitutive activation of c-Jun N-terminal kinase by a mutant epidermal growth factor receptor. J. Biol. Chem. 2732817-2822. [DOI] [PubMed] [Google Scholar]
- 2.Batra, S. K., S. Castelino-Prabhu, C. J. Wikstrand, X. Zhu, P. A. Humphrey, H. S. Friedman, and D. D. Bigner. 1995. Epidermal growth factor ligand-independent, unregulated, cell-transforming potential of a naturally occurring human mutant EGFRvIII gene. Cell Growth Differ. 61251-1259. [PubMed] [Google Scholar]
- 3.Bigner, S. H., P. A. Humphrey, A. J. Wong, B. Vogelstein, J. Mark, H. S. Friedman, and D. D. Bigner. 1990. Characterization of the epidermal growth factor receptor in human glioma cell lines and xenografts. Cancer Res. 508017-8022. [PubMed] [Google Scholar]
- 4.Bonaccorsi, L., D. Nosi, M. Muratori, L. Formigli, G. Forti, and E. Baldi. 2007. Altered endocytosis of epidermal growth factor receptor in androgen receptor positive prostate cancer cell lines. J. Mol. Endocrinol. 3851-66. [DOI] [PubMed] [Google Scholar]
- 5.Cantin, G. T., J. L. Stevens, and A. J. Berk. 2003. Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc. Natl. Acad. Sci. USA 10012003-12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalmers, C. J., R. Gilley, H. N. March, K. Balmanno, and S. J. Cook. 2007. The duration of ERK1/2 activity determines the activation of c-Fos and Fra-1 and the composition and quantitative transcriptional output of AP-1. Cell. Signal. 19695-704. [DOI] [PubMed] [Google Scholar]
- 7.Chazin, V. R., M. Kaleko, A. D. Miller, and D. J. Slamon. 1992. Transformation mediated by the human HER-2 gene independent of the epidermal growth factor receptor. Oncogene 71859-1866. [PubMed] [Google Scholar]
- 8.Chen, W., W. Bocker, J. Brosius, and H. Tiedge. 1997. Expression of neural BC200 RNA in human tumours. J. Pathol. 183345-351. [DOI] [PubMed] [Google Scholar]
- 9.Chen, W., J. Heierhorst, J. Brosius, and H. Tiedge. 1997. Expression of neural BC1 RNA: induction in murine tumours. Eur. J. Cancer 33288-292. [DOI] [PubMed] [Google Scholar]
- 10.Choe, G., J. K. Park, L. Jouben-Steele, T. J. Kremen, L. M. Liau, H. V. Vinters, T. F. Cloughesy, and P. S. Mischel. 2002. Active matrix metalloproteinase 9 expression is associated with primary glioblastoma subtype. Clin. Cancer Res. 82894-2901. [PubMed] [Google Scholar]
- 11.Cohen, K. J., J. S. Hanna, J. E. Prescott, and C. V. Dang. 1996. Transformation by the Bmi-1 oncoprotein correlates with its subnuclear localization but not its transcriptional suppression activity. Mol. Cell. Biol. 165527-5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colgan, J., and J. L. Manley. 1992. TFIID can be rate limiting in vivo for TATA-containing, but not TATA-lacking, RNA polymerase II promoters. Genes Dev. 6304-315. [DOI] [PubMed] [Google Scholar]
- 13.Crighton, D., A. Woiwode, C. Zhang, N. Mandavia, J. P. Morton, L. J. Warnock, J. Milner, R. J. White, and D. L. Johnson. 2003. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J. 222810-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Cristofano, A., B. Pesce, C. Cordon-Cardo, and P. P. Pandolfi. 1998. Pten is essential for embryonic development and tumour suppression. Nat. Genet. 19348-355. [DOI] [PubMed] [Google Scholar]
- 15.Eferl, R., and E. F. Wagner. 2003. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3859-868. [DOI] [PubMed] [Google Scholar]
- 16.Foulds, C. E., and D. K. Hawley. 1997. Analysis of the human TATA binding protein promoter and identification of an Ets site critical for activity. Nucleic Acids Res. 252485-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galang, C. K., C. J. Der, and C. A. Hauser. 1994. Oncogenic Ras can induce transcriptional activation through a variety of promoter elements, including tandem c-Ets-2 binding sites. Oncogene 92913-2921. [PubMed] [Google Scholar]
- 18.Galang, C. K., J. Garcia-Ramirez, P. A. Solski, J. K. Westwick, C. J. Der, N. N. Neznanov, R. G. Oshima, and C. A. Hauser. 1996. Oncogenic Neu/ErbB-2 increases Ets, AP-1, and NF-κB-dependent gene expression, and inhibiting Ets activation blocks Neu-mediated cellular transformation. J. Biol. Chem. 2717992-7998. [DOI] [PubMed] [Google Scholar]
- 19.Garber, M., S. Panchanathan, R. S. Fan, and D. L. Johnson. 1991. The phorbol ester, 12-O-tetradecanoylphorbol-13-acetate, induces specific transcription by RNA polymerase III in Drosophila Schneider cells. J. Biol. Chem. 26620598-20601. [PubMed] [Google Scholar]
- 20.Garber, M. E., A. Vilalta, and D. L. Johnson. 1994. Induction of Drosophila RNA polymerase III gene expression by the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) is mediated by transcription factor IIIB. Mol. Cell. Biol. 14339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gille, H., T. Strahl, and P. E. Shaw. 1995. Activation of ternary complex factor Elk-1 by stress-activated protein kinases. Curr. Biol. 51191-1200. [DOI] [PubMed] [Google Scholar]
- 22.Han, W., T. Zhang, H. Yu, J. G. Foulke, and C. K. Tang. 2006. Hypophosphorylation of residue Y1045 leads to defective downregulation of EGFRvIII. Cancer Biol. Ther. 51361-1368. [DOI] [PubMed] [Google Scholar]
- 23.Huang, H. S., M. Nagane, C. K. Klingbeil, H. Lin, R. Nishikawa, X. D. Ji, C. M. Huang, G. N. Gill, H. S. Wiley, and W. K. Cavenee. 1997. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J. Biol. Chem. 2722927-2935. [DOI] [PubMed] [Google Scholar]
- 24.Huang, P. H., A. Mukasa, R. Bonavia, R. A. Flynn, Z. E. Brewer, W. K. Cavenee, F. B. Furnari, and F. M. White. 2007. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc. Natl. Acad. Sci. USA 10412867-12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janknecht, R., W. H. Ernst, V. Pingoud, and A. Nordheim. 1993. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 125097-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janknecht, R., and T. Hunter. 1997. Convergence of MAP kinase pathways on the ternary complex factor Sap-1a. EMBO J. 161620-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, S. A., L. Dubeau, and D. L. Johnson. 2008. Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J. Biol. Chem. 28319184-19191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, S. A., L. Dubeau, M. Kawalek, A. Dervan, A. H. Schonthal, C. V. Dang, and D. L. Johnson. 2003. Increased expression of TATA-binding protein, the central transcription factor, can contribute to oncogenesis. Mol. Cell. Biol. 233043-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, S. A., N. Mandavia, H. D. Wang, and D. L. Johnson. 2000. Transcriptional regulation of the TATA-binding protein by Ras cellular signaling. Mol. Cell. Biol. 205000-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, S. S., C. Zhang, J. Fromm, I. M. Willis, and D. L. Johnson. 2007. Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol. Cell 26367-379. [DOI] [PubMed] [Google Scholar]
- 31.Kim, S., C. T. Denny, and R. Wisdom. 2006. Cooperative DNA binding with AP-1 proteins is required for transformation by EWS-Ets fusion proteins. Mol. Cell. Biol. 262467-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lal, A., C. A. Glazer, H. M. Martinson, H. S. Friedman, G. E. Archer, J. H. Sampson, and G. J. Riggins. 2002. Mutant epidermal growth factor receptor up-regulates molecular effectors of tumor invasion. Cancer Res. 623335-3339. [PubMed] [Google Scholar]
- 33.Majello, B., G. Napolitano, and L. Lania. 1998. Recruitment of the TATA-binding protein to the HIV-1 promoter is a limiting step for Tat transactivation. AIDS 121957-1964. [DOI] [PubMed] [Google Scholar]
- 34.Marais, R., J. Wynne, and R. Treisman. 1993. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 73381-393. [DOI] [PubMed] [Google Scholar]
- 35.Marshall, L., N. S. Kenneth, and R. J. White. 2008. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell 13378-89. [DOI] [PubMed] [Google Scholar]
- 36.Milde-Langosch, K. 2005. The Fos family of transcription factors and their role in tumourigenesis. Eur. J. Cancer 412449-2461. [DOI] [PubMed] [Google Scholar]
- 37.Moscatello, D. K., R. B. Montgomery, P. Sundareshan, H. McDanel, M. Y. Wong, and A. J. Wong. 1996. Transformational and altered signal transduction by a naturally occurring mutant EGF receptor. Oncogene 1385-96. [PubMed] [Google Scholar]
- 38.Nagane, M., F. Coufal, H. Lin, O. Bogler, W. K. Cavenee, and H. J. Huang. 1996. A common mutant epidermal growth factor receptor confers enhanced tumorigenicity on human glioblastoma cells by increasing proliferation and reducing apoptosis. Cancer Res. 565079-5086. [PubMed] [Google Scholar]
- 39.Nagane, M., A. Levitzki, A. Gazit, W. K. Cavenee, and H. J. Huang. 1998. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc. Natl. Acad. Sci. USA 955724-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishikawa, R., X. D. Ji, R. C. Harmon, C. S. Lazar, G. N. Gill, W. K. Cavenee, and H. J. Huang. 1994. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc. Natl. Acad. Sci. USA 917727-7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 102657-2683. [DOI] [PubMed] [Google Scholar]
- 42.Ozanne, B. W., L. McGarry, H. J. Spence, I. Johnston, J. Winnie, L. Meagher, and G. Stapleton. 2000. Transcriptional regulation of cell invasion: AP-1 regulation of a multigenic invasion programme. Eur. J. Cancer 361640-1648. [DOI] [PubMed] [Google Scholar]
- 43.Ozanne, B. W., H. J. Spence, L. C. McGarry, and R. F. Hennigan. 2007. Transcription factors control invasion: AP-1 the first among equals. Oncogene 261-10. [DOI] [PubMed] [Google Scholar]
- 44.Park, J. M., X. Yang, J. J. Park, O. W. Press, and M. F. Press. 1999. Assessment of novel anti-p185HER-2 monoclonal antibodies for internalization-dependent therapies. Hybridoma 18487-495. [DOI] [PubMed] [Google Scholar]
- 45.Reese, D. M., and D. J. Slamon. 1997. HER-2/neu signal transduction in human breast and ovarian cancer. Stem Cells 151-8. [DOI] [PubMed] [Google Scholar]
- 46.Rubin, I., and Y. Yarden. 2001. The basic biology of HER2. Ann. Oncol. 12(Suppl. 1)S3-S8. [DOI] [PubMed] [Google Scholar]
- 47.Sadovsky, Y., P. Webb, G. Lopez, J. D. Baxter, P. M. Fitzpatrick, E. Gizang-Ginsberg, V. Cavailles, M. G. Parker, and P. J. Kushner. 1995. Transcriptional activators differ in their responses to overexpression of TATA-box-binding protein. Mol. Cell. Biol. 151554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlessinger, J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103211-225. [DOI] [PubMed] [Google Scholar]
- 49.Schmidt, M. H. H., F. B. Furnari, W. K. Cavenee, and O. Bögler. 2003. Epidermal growth factor receptor signaling intensity determines intracellular protein interactions, ubiquitination, and internalization. Proc. Natl. Acad. Sci. USA 1006505-6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schütte, J., J. D. Minna, and M. J. Birrer. 1989. Deregulated expression of human c-jun transforms primary rat embryo cells in cooperation with an activated c-Ha-ras gene and transforms Rat-1a cells as a single gene. Proc. Natl. Acad. Sci. USA 862257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sorkin, A., and M. Von Zastrow. 2002. Signal transduction and endocytosis: close encounters of many kinds. Nat. Rev. Mol. Cell Biol. 3600-614. [DOI] [PubMed] [Google Scholar]
- 52.Taub, N., D. Teis, H. L. Ebner, M. W. Hess, and L. A. Huber. 2007. Late endosomal traffic of the epidermal growth factor receptor ensures spatial and temporal fidelity of MAPK signaling. Mol. Biol. Cell 184698-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Um, M., J. Yamauchi, S. Kato, and J. L. Manley. 2001. Heterozygous disruption of the TATA-binding protein gene in DT40 cells causes reduced cdc25B phosphatase expression and delayed mitosis. Mol. Cell. Biol. 212435-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vieira, A. V., C. Lamaze, and S. L. Schmid. 1996. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science 2742086-2089. [DOI] [PubMed] [Google Scholar]
- 55.Wang, H. D., A. Trivedi, and D. L. Johnson. 1997. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol. Cell. Biol. 176838-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, H. D., A. Trivedi, and D. L. Johnson. 1998. Regulation of RNA polymerase I-dependent promoters by the hepatitis B virus X protein via activated Ras and TATA-binding protein. Mol. Cell. Biol. 187086-7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wells, A., J. B. Welsh, C. S. Lazar, H. S. Wiley, G. N. Gill, and M. G. Rosenfeld. 1990. Ligand-induced transformation by a noninternalizing epidermal growth factor receptor. Science 247962-964. [DOI] [PubMed] [Google Scholar]
- 58.Winter, A. G., G. Sourvinos, S. J. Allison, K. Tosh, P. H. Scott, D. A. Spandidos, and R. J. White. 2000. RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumors. Proc. Natl. Acad. Sci. USA 9712619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wisdom, R. 1999. AP-1: one switch for many signals. Exp. Cell Res. 253180-185. [DOI] [PubMed] [Google Scholar]
- 60.Yang, S. H., and A. D. Sharrocks. 2004. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 13611-617. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, C., L. Comai, and D. L. Johnson. 2005. PTEN represses RNA polymerase I transcription by disrupting the SL1 complex. Mol. Cell. Biol. 256899-6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, G., C. R. Dass, E. Sumithran, N. Di Girolamo, L. Q. Sun, and L. M. Khachigian. 2004. Effect of deoxyribozymes targeting c-Jun on solid tumor growth and angiogenesis in rodents. J. Natl. Cancer Inst. 96683-696. [DOI] [PubMed] [Google Scholar]
- 63.Zhong, S., J. Fromm, and D. L. Johnson. 2007. TBP is differentially regulated by c-Jun N-terminal kinase 1 (JNK1) and JNK2 through Elk-1, controlling c-Jun expression and cell proliferation. Mol. Cell. Biol. 2754-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong, S., C. Zhang, and D. L. Johnson. 2004. Epidermal growth factor enhances cellular TATA binding protein levels and induces RNA polymerase I- and III-dependent gene activity. Mol. Cell. Biol. 245119-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zwang, Y., and Y. Yarden. 2006. p38 MAP kinase mediates stress-induced internalization of EGFR: implications for cancer chemotherapy. EMBO J. 254195-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]