Abstract
The malaria parasite sporozoite transmission stage develops and differentiates within parasite oocysts on the Anopheles mosquito midgut. Successful inoculation of the parasite into a mammalian host is critically dependent on the sporozoite's ability to first infect the mosquito salivary glands. Remarkable changes in tissue infection competence are observed as the sporozoites transit from the midgut oocysts to the salivary glands. Our microarray analysis shows that compared to oocyst sporozoites, salivary gland sporozoites upregulate expression of at least 124 unique genes. Conversely, oocyst sporozoites show upregulation of at least 47 genes (upregulated in oocyst sporozoites [UOS genes]) before they infect the salivary glands. Targeted gene deletion of UOS3, encoding a putative transmembrane protein with a thrombospondin repeat that localizes to the sporozoite secretory organelles, rendered oocyst sporozoites unable to infect the mosquito salivary glands but maintained the parasites' liver infection competence. This phenotype demonstrates the significance of differential UOS expression. Thus, the UIS-UOS gene classification provides a framework to elucidate the infectivity and transmission success of Plasmodium sporozoites on a whole-genome scale. Genes identified herein might represent targets for vector-based transmission blocking strategies (UOS genes), as well as strategies that prevent mammalian host infection (UIS genes).
As a vector-borne pathogen, the dispersal success of the malaria parasite Plasmodium relies on its transmission by anopheline mosquitoes. Plasmodium species have effectively exploited the female mosquitoes' need to feed on blood. Ingestion of the parasite-infected blood is followed by fusion between male and female gametes to produce a zygote, which matures into an ookinete. The mobile ookinete penetrates the mosquito midgut and then continues parasite development as an oocyst. Oocysts are lodged between the mosquito midgut epithelium and the basal lamina, which is exposed to the hemolymph-filled mosquito body cavity (reviewed in reference 40). The mature oocyst produces thousands of oocyst sporozoites. Oocyst sporozoites are released into the hemolymph, a process that depends on at least one parasite protease (1) and processing of the circumsporozoite (CS) protein (41). Sporozoites become highly infectious and transmittable to the mammalian host only after they enter the mosquito's salivary glands (reviewed in reference 18). To achieve salivary gland infection, sporozoites must first recognize and attach to the salivary glands. Subsequently, they invade the salivary glands, traverse the gland cells, and finally exit into the secretory cavity (28). During their migration in the mosquito, sporozoites undergo no apparent change in overall morphology. However, the sporozoites released from the oocyst exhibit specific infectivity for the salivary glands but are virtually noninfectious for the mammalian host at this point of development (37). Salivary gland sporozoites gain significant infection capacity for the mammalian liver, but in contrast, they loose infectivity for the salivary glands (36, 37). During the bites of infected mosquitoes, only a few dozen to a few hundred sporozoites are inoculated into a new mammalian host (10, 14). This is sufficient to ensure infection, because each of the highly infective salivary gland-derived sporozoites can initiate development of an intrahepatic liver stage, which can produce more than 10,000 red blood cell-infectious merozoite stages (4, 39). Over the last few years, a better understanding of the sporozoites' complex biology has been achieved and numerous studies have identified proteins involved in various steps required for host infection (reviewed in references 21 and 24).
Using the rodent malaria model parasite Plasmodium berghei, we have previously employed suppression subtractive hybridization (SSH) to identify transcripts that are upregulated in salivary gland sporozoites but are not expressed in oocyst sporozoites (22). This screen identified a set of 30 genes, called UIS (upregulated in infectious sporozoites), that are induced in sporozoites after their transition from the midgut oocysts to the salivary glands. Subsequently, we demonstrated using gene knockouts that two of these genes, UIS3 and UIS4, are indeed not needed for sporozoite salivary gland infection but are critical for the parasites' ability to successfully develop as liver stages (26, 27, 33). Here, using a genome-wide expression screen with the rodent malaria model Plasmodium yoelii, we show that sporozoites exhibit differential expression of a significant part of their transcriptome. Intriguingly, in addition to at least 124 genes that are upregulated in the salivary gland sporozoites, we also found at least 47 genes that are specifically upregulated before salivary gland infection (UOS genes: upregulated in oocyst sporozoites) but downregulated after salivary gland infection. Deletion of one identified UOS gene, UOS3, created mutant parasites that cannot infect the salivary glands but retain liver infection capacity, thus demonstrating the functional significance of UOS gene expression. The identification of comprehensive UIS and UOS gene sets provides a basis to understand the complex processes of differential infectivity control in sporozoites underlying mosquito salivary gland infection and mammalian liver infection.
MATERIALS AND METHODS
Sporozoite preparation.
P. yoelii (17XNL)-infected Anopheles stephensi mosquitoes were used to isolate sporozoites from midguts at day 10 (ooSpzs) and from salivary glands (sgSpzs) at day 15 after an infectious blood meal. Only mosquito cages having at least 70% of mosquitoes infected were kept for further analysis. For microarray experiments, the average number of sporozoites in midguts was 83,011 sporozoites/mosquito and that in salivary glands was 22,635 sporozoites/mosquito. The sporozoites were purified to remove contaminating mosquito tissue over a DEAE cellulose column, resulting in two independent biological replicates of 8 × 106 highly purified sporozoites for each population. The isolation of hemolymph sporozoites was performed as described previously (38).
RNA extraction and T7 RNA amplification.
Total RNA of ooSpzs and sgSpzs was extracted using Trizol reagent (Invitrogen). All samples were digested with DNase I (Invitrogen). The RNA was then subjected to two rounds of linear amplification using the T7-based in vitro transcription system according to the manufacturer′s protocol (Amino Allyl MessageAmp II amplified RNA (aRNA) amplification kit; Ambion). Quality and quantity of aRNA were examined with a high-resolution electrophoresis system, the Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA).
Microarray construction.
P. yoelii spotted microarrays were produced in the Molecular Genomics Core Facility, Drexel University College of Medicine. Arrays contained 65-mer oligonucleotides representing 6,700 open reading frames predicted in the genome of P. yoelii (6).
Preparation of labeled aRNA and microarray hybridization.
For microarray hybridizations, 10 μg of aRNA was coupled with Cy3 or Cy5 (Amersham). The procedure of dye coupling reaction and dye-labeled aRNA purification was followed according to the manufacturer's protocol (Amino Allyl MessageAmp II aRNA amplification kit). The labeled aRNA was fragmented with Ambion's RNA fragmentation reagents for this procedure. The amount of aRNA used for hybridization was 5 μg per microarray. The differentially labeled RNA samples were mixed with 1.6 μl of 5-μg/μl yeast tRNA, 16 μl of 10-μg/μl poly(A) RNA, 9 μl SSC (20×) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.6 μl sodium dodecyl phosphate (20%), and 1.2 μl HEPES (1 M). The hybridization mix was kept at 95°C for 2 min, 42°C for 20 min, and 25°C for 5 min before being added to the microarray. Samples were applied beneath coverslips onto microarray slides. Dual hybridizations in duplicate with both orientations of dye incorporation (dye swaps) were performed in a 60°C water bath for 16 h under a lifter coverslip (Fisher) in hybridization chambers (Corning). The end wells were filled with 20 μl 3× SSC. Microarrays were removed from the hybridization chambers and washed in 1× SSC plus 0. 1% sodium dodecyl phosphate for 2 min at room temperature, 0.2× SSC for 2 min, 0.05× SSC twice for 1 min, and 0.01× SSC for 30 s. Slides were dried by centrifugation for 5 min at 60 × g.
Microarray data analysis.
Following hybridization and washing, the slides were scanned using a GenePix 4000A laser scanner and the array features (spots) were quantified using the GenePix Pro software program (Axon Instruments Inc.). Array data were analyzed using the R statistical language and environment (http://www.r-project.org), specifically with the software packages from the Bioconductior Project (http://www.bioconductor.org/). To survey the total number of genes detected in sporozoite populations, the feature intensities were first locally background corrected and then divided by the median intensity of negative control spots of the same channels on the same array. The negative controls were spotted with a single oligonucleotide of random sequence. The geometric mean of ratios was calculated for each oligonucleotide signal in each sample across all replicates. To detect differentially expressed genes, data were background corrected and then normalized using the vsn software package, which applies variance-stabilizing transformation (13). Differentially expressed genes were then detected using the RankProd software package (12) at a false-discovery rate of <5%.
Annotations.
Protein domain annotations were done locally using Pfam (release 20) (3) using pfam_scan.pl (http://www.sanger.ac.uk/Users/sgj/code/pfam/scripts/search/pfam_scan.pl). Signal peptides were predicted using the SignalP 3.0 server (5). Only open reading frames with a start codon were considered. Transmembrane domains were predicted using the TMHMM server, v. 2.0 (20). A gene was considered “hypothetical” if the keyword “hypothetical” appeared in its description line. The Plasmodium falciparum orthologs were identified as reciprocal BLAST best hits as described in detail by Tarun et al. (34). We annotated P. yoelii genes using the gene ontology annotations on their P. falciparum orthologs. P. falciparum gene ontology annotation was downloaded from the Gene Ontology Consortium website (http://www.geneontology.org/).
qPCR.
Amplified RNA from purified sgSpzs and ooSpzs (500 ng each) was reversed transcribed with SuperScript II reverse transcriptase according to the manufacturer's protocol (Invitrogen). The resulting cDNA was diluted 1:5 with nuclease-free water. PCR oligonucleotide primers were designed for six UIS genes and for five UOS genes, using the Primer Express software program. Quantitative real-time PCR (qPCR) amplification was done in an AB1 PRISM 7300 real-time PCR cycler (Applied Biosystems, Foster City, CA) using the double-stranded DNA binding probe Sybr green I (Applied Biosystems). Reactions were subjected to one cycle of 10 min at 95°C and 40 cycles of 15 s at 95°C, 1 min at 60°C. qPCR experiments were done in triplicate. The amplicon size for all oligonucleotide primer pairs was kept at ∼90 to 120 bp. PCR fragments were cloned into plasmid pCR2.1 (Invitrogen). Each plasmid construct was used in a 10-fold dilution series (10 copies to 106 copies, each in triplicate) to determine a standard curve. The standard curve plots the threshold value, defined as the cycle number at which the reporter dye fluorescent intensity increases over the background level, over the plasmid copy number. The absolute transcript copy number for each gene is calculated based on the external standard curve. Transcript levels were normalized to a selected gene (PY01511) that showed constitutive expression in the two populations of sporozoites by microarray analysis. Sequences of the oligonucleotide primers used for qPCR experiments are shown in Table 1.
TABLE 1.
PCR oligonucleotide primer sequences
| Function and primera | Sequence |
|---|---|
| Validation of P. yoelii UIS and UOS gene expressionb | |
| PY03047F | AACCACAGATGTAGACCAACCTGAT |
| PY03047R | GGGTTTGTAGCATTTGCTTCATT |
| PY07608F | TCCGAGGCCAATAAGTTATCAAA |
| PY07608R | GGATCTGCAAGGTTGTTATTAATGTAAT |
| PY03831F | GAAATAAGAACAGCAATGGAAAAGC |
| PY03831R | CGTCCTCATCAGTTCTTAGCATATG |
| PY02400F | CAAATGCATTAGATGAAGCTTGCT |
| PY02400R | TTGAGTTTCGACATCTTCAGTTTCTT |
| PY00204F | CTTGCTTGTATGCACCCTGAAG |
| PY00204R | GGTATGGATTTTCGACTGGGC |
| PY03011F | AACCTTTATTCCAATCATGTCTTCCT |
| PY03011R | TGCCTCAATTTTTCACATGCATAAT |
| PY02296F | ACGAAAACAATATAGAGAAACCCAAAC |
| PY02296R | ATTGCTATTTACCGAACTCTCTTTCTTT |
| PY04547F | ATGGAATGGTCCACAAGGTGTT |
| PY04547R | TGTAATAGCTCCATTTTGTGTTGCT |
| PY04986F | GGGTACATGTGATGCTGGCTATAA |
| PY04986R | CCGTGCAAGGTGGCAAA |
| PY07598F | CTAATCCACAAAATCCCAACCAA |
| PY07598R | TCGTTTGTTTCTTGAGTTTTGTCTTC |
| PY03955F | TTCTATTAACCAAGCAGAATGTGATCA |
| PY03955R | GACCAGTCACGAACAAATTGTCTT |
| PY01511F | TGCTTATTCATCATATCCTCATTCG |
| PY01511R | GTCTCGAGGGAAAAGAGAAGTTTTT |
| Construction of uos3 knockout plasmidc | |
| Pr.1 | GGGATATCGCAATGTTAAACAAGCAATATGCTC |
| Pr.2 | TTGCCCTCCTGCAGGTTCGTGGTCTACACTTGTAT |
| Pr.3 | GTGGTTTACCTGCAGGAGGGCAATTTGTATCATATGACC |
| Pr.4 | ATGCGGCCGCGCTGTATAGTTTTTTGAAAGTGGAG |
| UOS3 test For | GGCAATGTTATTTCAGTTTC |
| UOS3 test Rev | TTGCAAAGTGATCCATGTGT |
| Test 1 For | TTGTTACCCTTGTTCTATAATCCAC |
| Test 1 Rev | GCAAGGCGATTAAGTTGGGT |
| Test 2 For | GGCTACGTCCCGCACGGACGAATCCAGATGG |
| Test 2 Rev | TGTACAGGTATACCTTCTTCTACTGTTTTAG |
| Vector test For | AGGGCAATTTGTATCATATGACC |
| Vector test Rev | GCAAGGCGATTAAGTTGGGT |
| CS For | AAGAAGTGTACCATTTTAGTTGTAGCGTCAC |
| CS Rev | CACTACTGGTTGATTCAATTTATTTTGAGCCTC |
| Myc tagging of UOS3 C terminusd | |
| UOS3 For | GCCCGCGGTACACATGCAAAATAAAGCGGATA |
| UOS3 Rev | GGACTAGTTGACCAATCATCATTAACGTAACT |
| UOS3 Test For | GGCAATGTTATTTCAGTTTC |
| UOS3 Test Rev | TTGCAAAGTGATCCATGTGT |
| Test 1 For | ATTAAAGTGGAAAGAGATGC |
| Test 1 Rev | GCAAGGCGATTAAGTTGGGT |
| Test 2 For | GGCTACGTCCCGCACGGACGAATCCAGATGG |
| Test 2 Rev | TGGGTTCGTTACATATTATT |
| Evaluation of P. falciparum UIS and UOS gene expressione | |
| falc_CSF | CAGTGCTATGGAAGTTCGTCAAA |
| falc_CSR | ATACCAATTTTCCTGTTTCCCATAAT |
| falc_TRAPF | TTGTATGCTGATTCTGCATGGG |
| falc_TRAPR | ACATGGAGACCATTCGTCCC |
| falc_UIS3F | AGAAGAACACAACAAAAGGAAGAAACTA |
| falc_UIS3R | TCTTCTCGCGATTTTTTATATCCA |
| falc_UIS4F | TATCTACTGCTGCTGTTGCTTTGG |
| falc_UIS4R | CAGAGTCGGATCCATCATTCAC |
| falc_UIS28F | ACCTACCGAACGTCGACGAA |
| falc_UIS28R | AAATCAGCTGCTTCCCAATTT |
| falc_UIS2F | TGAAGTGTTCGTATCTCCTAATTGT |
| falc_UIS2R | TGTCCGATATCTCCTAACATCATAAT |
| falc_PF14_0467F | GCTTAGTCATTCCAATAGCTGTTCA |
| falc_PF14_0467R | TAATTGGTTGTGCTATATTCTTTGATTGT |
| falc_UOS3F | GATCGTGATGATCGTGCACTT |
| falc_UOS3R | TGTTCGATGAGTTTATGTTGTTTATCT |
| falc_PF14_0471F | TGATATGTACGAATCAAATGAGGATAGT |
| falc_PF14_0471R | AACAGAAAATACGCGAAGATGTCT |
| falc_PFI1105wF | GCAAAAGGTAGTATCGAATGTCTCA |
| falc_PFI1105wR | TTCGTTTTTCTTATTTTGTTGTTCAAC |
| falc_PFE0175cF | CAGGAAAAACAGAAGCATCCAA |
| falc_PFE0175cR | CATTACCAAACGCCTCCAAT |
“F” or “For” in designation indicates forward primer; “R” or “Rev” indicates reverse.
Primers used to validate the expression patterns of selected P. yoelii UIS and UOS genes by quantitative real-time RT-PCR as shown in Fig. 2A.
Primers used to produce the uos3 knockout plasmid in the b3D.DT∧H.∧D vector as shown in Fig. 3.
Primers used to insert coding for the UOS3 C terminus into the b3D.DT∧H.∧D (quadruple Myc) plasmid as shown in Fig. 7.
Primers used to evaluate the expression patterns of selected P. falciparum UIS genes and UOS genes by RT-PCR as shown in Fig. 2A.
Generation of transgenic parasites.
The targeted deletion of UOS3 by gene replacement was done as described in detail by Mikolajczak et al. (23). The sequences of all test primers can be found in Table 1. For the generation of UOS3 tagged with the Myc epitope (UOS3myc), we have introduced a quadruple (4×) Myc tag sequence into the b3D.DT∧H.∧D vector (catalog no. MRA-80 in the MR4-Malaria Research and Reference Reagent Resource Center; http://www.malaria.mr4.org) followed by the 3′ untranslated region of the Plasmodium berghei dihydrofolate reductase gene. The C-terminal fragment of uos3 was cloned into the plasmid in frame and adjacent to the Myc tag. The plasmid was linearized with the BsaBI restriction enzyme, and the selection of transgenic parasites was done as previously described (23). Primer sequences can be found in Table 1.
Microscopy and indirect immunofluorescence assays.
For visualization of whole mosquitoes infected with the red fluorescent protein (RFP)-fluorescent knockout parasites, as well as isolated midguts and salivary glands, a Nikon Eclipse E600 microscope was used and images were processed with the Metamorph software program.
For the indirect immunofluorescence assays, midguts or hemolymph sporozoites were fixed with 2% paraformaldehyde (Sigma), permeabilized with Triton X-100 (0.1%), and incubated with specific antibodies for CS protein (9D3), TRAP, or c-Myc (A-14; Santa Cruz Biotechnology). For fluorescent detection, the secondary antibodies Alexa Fluor 488 and Alexa Fluor 594 (Invitrogen) were used. The images were acquired using the Applied Precision DeltaVision RT microscopy system and its deconvolution software.
RT-PCR analysis of P. falciparum.
OoSpzs and sgSpzs were isolated from P. falciparum-infected A. stephensi mosquitoes at days 10 and 15 postinfection, respectively. Total RNA was extracted using Trizol reagent (Invitrogen). RNA was treated with DNase I (Invitrogen). Oligonucleotide primer sequences used in P. falciparum reverse transcriptase PCR (RT-PCR) are provided in Table 1. PCR conditions used are as follows: 94°C for 5 min; 94°C for 30 s and 55°C for 30 s; 60°C for 30 s (30 cycles); and 60°C for 10 min.
Microarray data accession numbers.
The microarray data reported in this paper have been deposited in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo) under the following identifiers: GSM200758, GSM200759, GSM200764, and GSM200765.
RESULTS
Widespread differential gene expression in sporozoites.
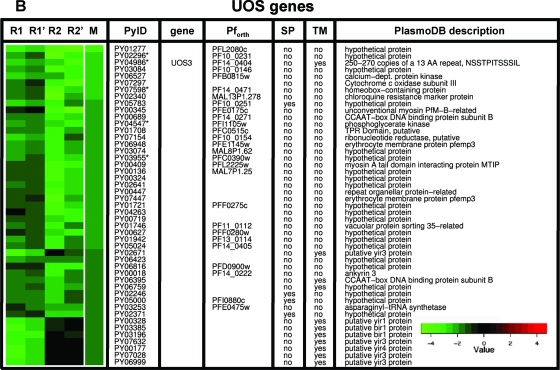
We used an oligonucleotide microarray that was designed based on the annotated open reading frames of P. yoelii to analyze gene expression in two distinct sporozoite populations. The first population (ooSpzs) was isolated from the mosquito midgut oocysts and represented fully mature sporozoites at day 10 after mosquito infection. The second sporozoite population (sgSpzs) was isolated from the mosquito salivary glands at day 15 after infection. RNA was isolated from each purified sporozoite population, amplified/labeled, and hybridized to the array. To identify genes which are differentially expressed between ooSpzs and sgSpzs, we used a nonparametric method based on the analysis of rank product (12). This procedure performs well when only a small number of biological replicates are available. The most strongly upregulated genes have a rank of 1. For each gene, a rank product is calculated as the product of the ranks of the gene in all replicates. Genes with the smallest rank product values are considered the most significant candidates for upregulated genes. A permutation-based estimation procedure can be used to determine the significance level of those rank products, that is, how likely it is to observe a given rank product value or better in a random experiment. Genes which were identified as differentially expressed between ooSpzs and sgSpzs at a false discovery rate of <5% are shown in Fig. 1. One hundred twenty-four genes showed significant upregulation in sgSpzs compared to expression in ooSpzs (Fig. 1A). We compared the set of 124 sgSpz upregulated genes to a set of 30 P. berghei genes, which were previously identified by subtractive cDNA hybridization (UIS genes) (22). Strikingly, only 7 of the 124 genes classified as upregulated by microarray analysis had been identified by the subtractive hybridization screen (UIS1, UIS2, UIS3, UIS4, UIS7, UIS16, and UIS28). UIS3 and UIS4, which are among the most highly ranked differentially expressed genes (Fig. 1A), were indeed shown to have essential functions only in mammalian liver infection (26, 27). Therefore, we identified 117 novel candidate genes which may have roles in mammalian host infection. Thirty-one of the UIS genes encode proteins with putative signal peptides and/or transmembrane domains, indicating that they might enter the sgSpz secretory pathway and therefore might function in sporozoite-mammalian host cell interactions. We also identified 47 genes, which exhibited upregulation in ooSpzs (UOS) (Fig. 1B). Expression of UOS genes is downregulated in sgSpzs. Fifteen UOS genes encode proteins with putative signal peptides and/or transmembrane domains. This indicates that the UOS proteins may enter the ooSpz secretory pathway and that they might have a role in sporozoite salivary gland infection but not in mammalian host infection. In addition, a comparative analysis of our data and recently published P. yoelii sporozoite microarray data from a report on malaria parasite gene expression profiling by Zhou et al. (42) indicated extensive concordance of differential sporozoite gene expression. Out of 47 UOS genes identified by our analysis, 44 genes had expression data in the data set of Zhou et al. and 77% (34/44) of those UOS genes were upregulated in the data set of Zhou et al.. Out of the 124 UIS genes identified by our analysis, 82 genes had expression data according to Zhou et al. and 87% (71/82) of those genes were upregulated in the data set of Zhou et al..
FIG. 1.
Heat map of genes differentially expressed between sgSpzs and ooSpzs. A set of 124 upregulated genes (A) was identified when RNA isolated from sgSpzs was compared to that from ooSpzs (UIS genes). Forty-seven downregulated genes (B) were also identified in sgSpzs compared with ooSpzs (UOS genes). In each row, repeated measurements of the log2 ratios of gene expression levels in sgSpzs to those in ooSpzs (sgSpz/ooSpz) for the same gene are shown. In each heat map, the replicated hybridizations are shown in the first four columns: two biological replicates (R1 and R2) and dye swaps of each biological replicate (R1′ and R2′). The fifth column is the mean for four replicates. Differentially expressed genes were selected using a rank-based algorithm with a false-discovery rate of <5%. The genes verified by qPCR are labeled with an asterisk. Previously identified UIS and S genes are indicated; PyID, P. yoelii gene identifier; gene, common name; Pforth, P. falciparum ortholog; SP, signal peptide; TM, transmembrane domain.
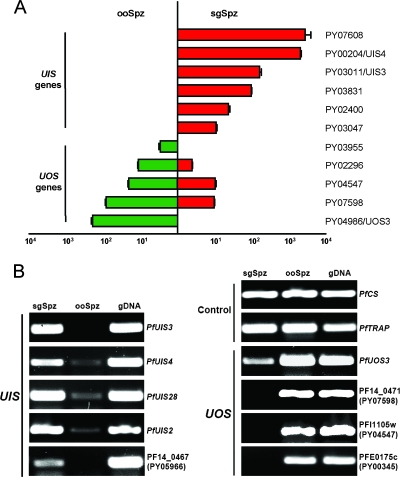
To further assess the validity of differential gene expression data obtained for ooSpzs and sgSpzs, six UIS genes and five UOS genes were selected and their differential transcript abundance was assessed by qPCR (Fig. 2A). Expression levels were normalized against a gene (PY01511) that showed constitutive expression in ooSpzs and sgSpzs in the microarray analysis (data not shown). Normalized and absolute qPCR expression data did not show significant deviations, indicating that the ooSpz RNA and sgSpz RNA template quantities were approximately equivalent (data not shown). The qPCR results obtained for the six UIS genes showed high transcript abundance in sgSpzs, but transcripts were virtually undetectable in ooSpzs (Fig. 2A). The qPCR analysis of UOS genes revealed variable degrees of transcript abundance among the genes (Fig. 2A). The genes PY03955 and PY04986 exhibited an extreme downregulation of transcript abundance in sgSpzs. However, transcripts for the genes PY07598, PY02296, and PY04547 were still detected in sgSpzs, albeit at much lower level than that in ooSpzs. Together, the qPCR data obtained for 11 differentially expressed genes are in agreement with data obtained by microarray hybridizations.
FIG. 2.
Differential expression profile of UIS genes and UOS genes in Plasmodium yoelii and Plasmodium falciparum. (A) Quantitative real-time RT-PCR with RNA from P. yoelii sporozoites from either the salivary gland or the midgut as templates using gene-specific oligonucleotide primer pairs for each UIS gene or UOS gene. The transcript quantity is presented as the number of copies (± standard deviation) in comparison with an external standard curve generated with gene-specific plasmids. Each experiment was done in triplicate. (B) Differential gene expression in P. falciparum sporozoites. RT-PCR analysis was used to verify that differential gene expression between sgSpzs and ooSpzs also occurs in P. falciparum. PCR products of the expected amplicon sizes were amplified for all tested genes. P. falciparum genomic DNA (gDNA) was used as a PCR control. As an expression control, CS and TRAP gene expression is detected in ooSpzs and sgSpzs. The P. yoelii orthologs are shown in parentheses when only a PF gene identification number but no common name is available.
Differential sporozoite gene expression in P. falciparum.
We next investigated if differential sporozoite gene expression also occurs in the human malaria parasite P. falciparum. Eleven gene orthologs, PfCS, PfTRAP, PfUIS3, PfUIS4, PfUIS28, PfUIS2, PF14_0467 (PY05966 ortholog), PfUOS3, PF14_0471 (PY007598 ortholog), PFl1105w (PY04547 ortholog), and PFE0175c (PY00345 ortholog), were selected for RT-PCR analysis of ooSpz and sgSpz RNA (Fig. 2B). Constitutively expressed CS and TRAP were tested as a control and indeed showed similar expression in ooSpzs and sgSpzs. PfUIS3, PfUIS4, PfUIS28, PfUIS2, and PF14_0467 showed preferential expression in sgSpzs, and PfUOS3, PF14_0471, PFl1105w, and PFE0175c showed significant downregulation in sgSpzs. Thus, the observed sporozoite transcript abundance patterns of the tested P. falciparum genes are similar to those observed for the respective P. yoelii orthologs, establishing that differential gene expression also occurs in P. falciparum sporozoites.
UOS3 is essential for salivary gland invasion.
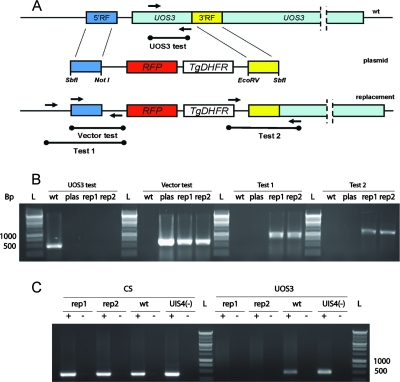
One UOS gene, UOS3 (PY04986), exhibited significant differential expression in sporozoites. UOS3 expression is high in ooSpzs (Fig. 2A) but low in sgSpzs, which suggests a role of this gene in salivary gland infection. The gene was previously identified as a pre-erythrocytic stage-specific gene in an SSH screen of P. yoelii sgSpzs versus blood-stage merozoites (designated S6 [sporozoite-specific gene 6]) (15). UOS3 encoded a 2,690-amino-acid protein with a 47-amino-acid TRAP-type cytoplasmic domain (15, 16). Interestingly, a close evaluation of the N-terminal domain of UOS3 also revealed a partially conserved thrombospondin repeat (TSR) domain containing an N-terminal 177WSXW180 tetrapeptide and a C-terminal cluster of positively charged residues (187RQRRK191). The key residues are well conserved between P. yoelli UOS3 and its P. falciparum ortholog (data not shown). Based on its predicted structure and the observed expression profile, we postulated that UOS3 is involved in salivary gland infection. To test this, we deleted the gene by double-crossover homologous recombination (Fig. 3). Two clonal lines of knockout parasites were isolated from the transfected and drug-selected parental population and used in the subsequent experiments (Fig. 3). The gene deletion strategy also introduced an RFP cassette into the knockout parasite to create a uos3− rfp line. This allowed for direct visualization of the uos3− parasites. uos3− rfp parasites did not exhibit any apparent defects in asexual blood-stage replication (data not shown). In addition, the morphology of male and female gametocytes in thin infected-blood smears and male gamete exflagellation in wet mounts of infected blood were indistinguishable from those of P. yoelii wild-type (WT) parasites (data not shown). Anopheles stephensi mosquitoes were infected with of uos3− rfp parasites by blood feeding on infected mice. uos3− rfp mosquito infections exhibited normal oocyst development (Fig. 4A and B). Strikingly, however, at day 12 postinfection, fluorescence microscopy observation detected few sporozoites associated with the salivary glands of uos3− rfp parasite-infected mosquitoes (Fig. 4A and 5F to H). In contrast, salivary glands were heavily infected with uis4− rfp sporozoites (Fig. 4B and 5C to E), as expected from previous work that showed no defect in the salivary gland infection for this knockout (23, 26, 33). Direct quantification of ooSpzs and sgSpzs confirmed these observations (Fig. 6). Sporozoite numbers derived from the infected midguts at day 10 after the infected-blood meal were similar between uos3− rfp and uis4− rfp parasites (Fig. 6). In contrast, salivary gland sporozoite numbers for uos3− rfp at day 14 after the infected meal were dramatically reduced (∼90% reduction) compared to uis4− rfp sporozoite numbers (Fig. 6). Fluorescence microscopy observation of uos3− rfp parasite-infected salivary glands (Fig. 5F to J) suggested that the sporozoites were mainly attached to the glands but did not localize to the interior of the gland. To test the hypothesis that uos3− rfp sporozoites cannot infect the glands and as a consequence cannot reach the salivary gland ducts, we performed natural bite experiments where uos3− rfp parasite-infected mosquitoes (day 14 postinfection) were allowed to feed on naive BALB/c mice. The exposed mice did not develop blood-stage parasitemia (monitored until day 10 postinfection) (Table 2). Control experimental mice exposed to WT-infected mosquitoes developed blood-stage parasitemia at day 3. To test whether this lack of infection was caused by a defect in liver infection, we injected 105 oocyst-derived or 2 × 104 hemolymph-derived uos3− rfp sporozoites intravenously into mice. All mice developed blood-stage parasitemia on the same day (day 4) as the WT-sporozoite control-injected mice (Table 2). Together the data show that UOS3 is critical for sporozoite salivary gland infection but is not important for infection of the mammalian host.
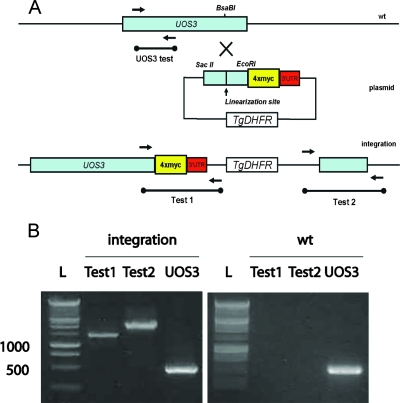
FIG. 3.
Targeted gene disruption strategy for UOS3. The gene was disrupted by deleting the N terminus of UOS3 with a plasmid carrying the Toxoplasma gondii dihydrofolate reductase (TgDHFR) and an RFP cassette by homologous recombination. (A) A graphical representation of the homologous replacement is shown. (B) Gene replacement analysis was preformed on genomic DNA of two uos3− rfp clones (rep1 and rep2), the wild type (wt), and a uis4− clone with primers as shown in panel A. “Test 1” and “Test 2” genomic PCR confirmed uos gene-specific replacement by double homologous recombination. uos3 gene disruption was confirmed by “UOS3 test” genomic PCR, which showed no amplification with rep1 and rep2. Bp, base pairs; rep, uos3 replacement parasites; plas, plasmid; L, DNA ladder. (C) Total RNA was isolated from uos3− rfp (rep1 and rep2), wild-type (wt), and uis4− rfp oocyst sporozoites, and cDNA was generated and amplified for 35 cycles for detection of UOS3 and CS expression (primers used are listed in Table 1). No transcript for UOS3 is detected for the rep1 and rep2 knockout sporozoites.
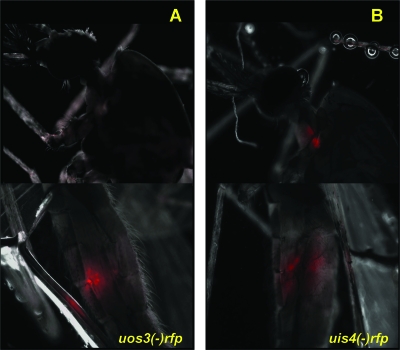
FIG. 4.
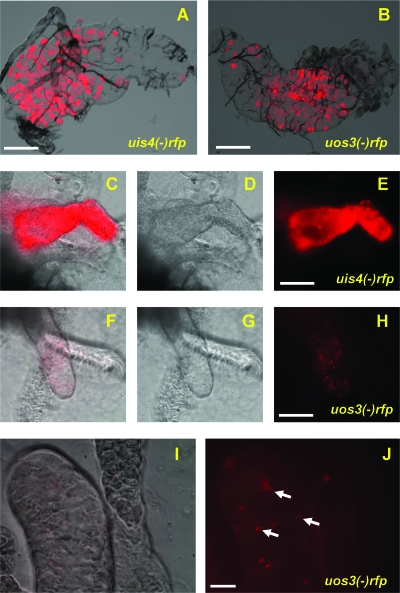
Whole-body imaging of uos3− rfp sporozoites shows a defect in salivary gland infection. Mosquitoes infected with uos3− rfp parasites (A) or uis4− rfp parasites (B) were visualized at day 12 postinfection by fluorescence microscopy (magnification, ×40) in whole mosquitoes. The upper panels show localization to the salivary glands of intact mosquitoes, and the lower panels show localization to the midguts of intact mosquitoes. Mosquito midgut infections appear similar, but uos3− rfp infections are not detected in salivary glands, whereas uis4− rfp infections can be easily observed.
FIG. 5.
uos3− rfp sporozoites do not infect salivary glands efficiently. Fluorescence microscopy analysis of isolated infected mosquito organs is shown. uis4− rfp parasites (A) and uos3− rfp parasites (B) show comparable numbers of oocysts at day 10 after the blood meal. Scale bar, 250 μm. (C to E) A salivary gland lobe heavily infected (day 14) with uis4− rfp sporozoites is shown. (F to H) Salivary glands of uos3− rfp parasite-infected mosquitoes show only a small number of sporozoites associated with the gland. Scale bar, 75 μm. (I and J) Higher-magnification image of a uos3− rfp parasite-infected salivary gland lobe. Overlay of differential interference contrast and the red fluorescent image is shown in panel I and the red fluorescent image in panel J. Few uos3− rfp sporozoites are observed associated with the salivary glands. Scale bar, 30 μm.
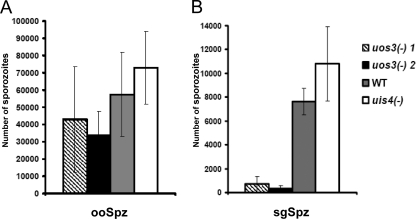
FIG. 6.
Quantification of sporozoite infection. (A) A similar number (∼30) of mosquito midguts infected with uos3− rfp clone 1, uos3− rfp clone 2, wild-type (WT), or uis4− rfp parasites were dissected, and the numbers of ooSpzs were compared between groups. (B) Salivary glands (∼30) were dissected from mosquitoes infected with the same parasites as in panel A. Numbers of sgSpzs show an approximate 90% reduction in uos3− rfp parasite-infected mosquitoes compared to those in wild-type- or uis4− rfp sporozoite-infected mosquitoes. The numbers were collected from three independent mosquito infections.
TABLE 2.
Infectivity of uos3− sporozoites via intravenous injection or natural bite
| Expt (no. of sporozoites used)a | No. of mice used/no. of parasitemic mice (prepatency) for indicated type of sporozoite
|
|
|---|---|---|
| uos3− | WT | |
| ooSpz injection (1 × 105) | 4/4 (4) | 4/4 (4) |
| heSpz injection (2 × 104) | 4/4 (4) | 4/4 (4) |
| Mosquito bite (10 mos./mousec) | 4/0b | 4/4 (3) |
ooSpz, oocyst sporozoites; heSpz, hemolymph sporozoites; mos., mosquitoes.
Experiment performed twice (average oocyst sporozoite loads in the mosquitoes used: 76,000 and 178,000, respectively).
Mosquitoes infected with uos3− or WT sporozoites were allowed to feed on mice for 8 min.
Localization of UOS3 in oocyst and hemolymph sporozoites.
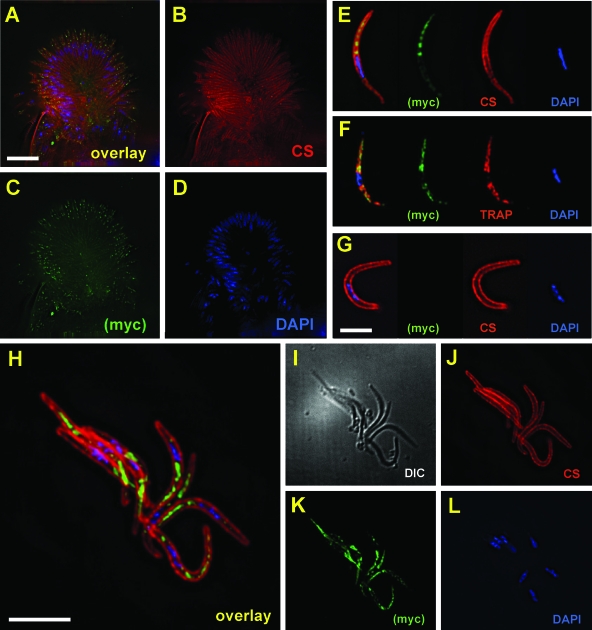
To further characterize UOS3, we investigated its localization in oocyst and hemolymph sporozoites. A quadruple c-Myc tag was fused to the C terminus of UOS3 using a genetic insertion strategy (Fig. 7). The insertion resulted in expression of a UOS3myc chimeric protein under the control of the endogenous UOS3 5′ upstream DNA region. Anti-Myc antibody staining of UOS3myc oocysts at day 10 postinfection revealed that UOS3 localized to the apical end of oocyst sporozoites that bud from the oocysts (Fig. 8A to D). A similar localization of UOS3 was observed in UOS3myc hemolymph sporozoites (Fig. 8E to L). Interestingly, simultaneous staining of UOS3myc and TRAP, a known micronemal protein, showed only a partial overlap in localization (Fig. 8F). The localization and the punctuate appearance of UOS3myc distribution suggested that the protein is a part of the apical invasive organelles and are compatible with its role in the process of salivary gland infection.
FIG. 7.
Myc epitope-tagging strategy for UOS3. (A) Graphical representation of the tagging strategy. To epitope tag UOS3, a quadruple Myc tag sequence (4×myc) was introduced into the b3D.DT∧H.∧D vector. 4×myc is followed by the 3′ untranslated region (3′UTR) of the Plasmodium berghei DHFR gene (∼1,000 bp). The region of UOS3 (∼1,400 bp) corresponding to the C terminus was cloned in frame (without the stop codon) with the 4×myc tag. The plasmid was linearized for parasite transfection at the BsaBI restriction site. (B) 4×myc tag integration analysis was performed on genomic DNA from the parental population of parasites transfected with the 4×myc plasmid (integration) and wild-type (wt) parasites (negative control) using primers as indicated in panel A.
FIG. 8.
Localization of UOS3 in oocyst and hemolymph sporozoites. UOS3 was tagged with the quadruple Myc epitope (UOS3myc) and used in immunofluorescence localization studies. (A to D) At day 10 postinfection, a midgut oocyst with developing sporozoites (A, overlay) was stained for CS protein (B, red), Myc (C, green), and 4′,6′-diamidino-2-phenylindole (DAPI)-DNA content (D, blue). Scale bar, 10 μm. UOS3 localizes to the apical ends of the emerging sporozoites. (E) A UOS3myc hemolymph sporozoite stained for Myc (green), CS protein (red), and DAPI-DNA content (blue). (F) A UOS3myc hemolymph sporozoite stained for Myc (green), TRAP (red), and DAPI-DNA (blue). UOS3 shows internal granular staining that does not colocalize with CS but shows partial overlap with TRAP localization. (G) As a control, wild-type sporozoites were stained with the Myc antibody (green), CS antibody (red), and DAPI-DNA (blue) to show specificity of the Myc antibody. No Myc staining was observed. H to L) UOS3myc hemolymph sporozoites (H, overlay; I, differential interference contrast image) stained with CS antibody (J, red), Myc antibody (K, green), and DAPI (L, blue) show that UOS3myc preferentially localizes to one end of the sporozoites. Scale bar, 5 μm.
DISCUSSION
Malaria parasite sporozoites provide a unique system to study the gene expression programs that regulate parasite-stage-specific interactions with mosquito and mammalian host tissues (17). To this end, we performed transcriptional profiling of ooSpzs and sgSpzs of the rodent malaria parasite P. yoelii using oligonucleotide microarrays that cover all annotated P. yoelii open reading frames (6). Comparing the expression profiles of ooSpzs and sgSpzs on a genome-wide scale, we found that ∼10% of the genes exhibit differential expression in sporozoites. At a <5% false discovery rate, sgSpzs upregulate expression of 124 UIS genes. Our previous work with P. berghei using SSH provided the first evidence for differential gene expression in sgSpzs (22). Surprisingly, the set of 30 UIS genes identified by SSH and the 124 UIS genes identified herein overlap only for 7 genes. This finding may be explained by the fact that the SSH expression screen (8) was done with normalized cDNA populations of ooSpzs and sgSpzs (22). Thus, the screen probably detected small quantitative differences in low-abundance transcripts, which are not considered significant using the microarray analysis with the described cutoff criteria. Conversely, genes that show substantial differential regulation in the microarray analysis may have remained undetected by SSH because this technique is not quantitative and exhaustive and relied on sequencing of a limited number of cDNAs (22).
Of the 124 UIS genes identified by our analysis, 31 encode putative secreted and/or membrane-anchored proteins. Thus, these proteins are likely to be involved in parasite-host tissue interactions during mammalian host infection. UIS3 (PY03011) and UIS4 (PY00204) localize to the secretory organelles of sgSpzs (15). These proteins are also present in the liver-stage parasitophorous vacuole and are essential for early liver-stage development only (23, 26, 27, 33).
A new candidate sgSpz invasion-related protein is the gene product of PY01499. This putative protein exhibits a domain architecture that is similar to the structure of TRAP (thrombospondin-related anonymous protein) (30), including a thrombospondin repeat, a von Willebrand factor-like A-domain, and a TRAP-type conserved cytoplasmic domain. Indeed, a recent study showed that genetic deletion of the P. berghei ortholog of PY01499, named TLP, results in a decreased capacity for cell traversal by sgSpzs and reduced infectivity of sgSpzs in vivo (25). Some of the predicted secreted UIS proteins bear putative enzymatic domains, which may imply a role in manipulation of the mammalian host environment by the parasite. For example, PY07137 encodes a putative secreted lipase domain that is similar to the class 3 lipases, which are not related to any of the known lipase families of eukaryotic lipases (11).
Conversely, we found that 47 UOS genes show significant upregulation in ooSpzs compared to results for sgSpzs. A number of these genes exhibit extreme differential expression, and qPCR measurements presented herein confirmed the results obtained by microarray. The identification of UOS genes will provide important information to further a detailed understanding of the molecular events prior to or involved in salivary gland infection. In support of the importance of differential UOS expression, we analyzed UOS3/S6. The presence of a thrombospondin repeat-like domain and a TRAP-type cytoplasmic domain in UOS3 suggested that the protein might have invasive properties during mosquito salivary gland infection. Indeed, targeted deletion of UOS3 resulted in a dramatic reduction of sgSpz loads. The sporozoite defect likely resides in either salivary gland attachment or salivary gland cell traversal to reach the salivary ducts, for the reason that a natural bite experiment with uos3− parasite-infected mosquitoes did not result in blood-stage parasitemia. However, UOS3 has no apparent function in mammalian liver infection, because intravenous injection of uos3− rfp hemolymph sporozoites as well as ooSpzs resulted in blood-stage infection in mice. Thus, the function of UOS3 is specific for salivary gland invasion and is not as broad as has been seen, for example, for TRAP. TRAP deletion affects gliding motility, salivary gland invasion, and liver infectivity (32). In addition to TRAP, four additional proteins are currently implicated in salivary gland infection: CSP (31), MAEBL (19), and PCRM1 and PCRM2 (35). Nevertheless, based on our analysis, these genes are not UOS genes and indeed only PCRM2 function appeared restricted to salivary gland infection (29, 35). It will be of importance to understand whether the above-mentioned proteins act independently or together in complexes with UOS3 in salivary gland infection.
Interestingly, we detected that members of the yir multigene family are upregulated in the ooSpzs (Fig. 1B). This seems surprising since yir genes have been shown to be expressed on the surfaces of infected red blood cells and are thought to play a role in antigenic variation (7). However, a recent report on transcriptional regulation of the yir multigene family revealed that there are distinct groups of yir genes showing limited expression during the asexual blood stage of P. yoelii (9). yir genes expressed in mosquito stages of the parasite might have a role in escaping the mosquitoes' innate immune response.
The identification of comprehensive UIS and UOS gene sets described herein will now allow the functional mapping of each gene to the distinct steps in the journey of sporozoites from oocysts to the mammalian liver. The factors responsible for differential gene expression in sporozoites, however, remain unknown and require future investigation. Recently sporozoite SAP1 (asparagine-rich protein 1) has been described to function as a selective factor controlling the expression of infectivity-associated parasite genes in salivary gland sporozoites, such as UIS3 and UIS4 (2). Further studies of SAP1 functional properties should provide an insight into how regulation of gene expression in sporozoites is achieved.
Additionally, in this report we provide unprecedented evidence that differential sporozoite gene expression also occurs in the most deadly human malaria parasite, P. falciparum. Thus, the study of differential gene expression of important sporozoite virulence factors in rodent malaria models will give critical information about human malaria infection. This may allow the exploitation of UOS proteins as targets for vector-based transmission-blocking strategies. Conversely, UIS proteins are targets for strategies that interfere with initial steps of transmission and mammalian liver infection.
Acknowledgments
This work was funded by a grant from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health Initiative and an SBRI institutional grant to S.H.I.K. Design and construction of the P. yoelii microarray were supported by the National Institutes of Health (to L.W.B.).
A potential conflict of interest is as follows: S.H.I.K. is an inventor listed on U.S. patent no. 7,22,179, U.S. patent no. 7,261,884, and international patent application PCT/US2004/043023, each titled “Live genetically attenuated malaria vaccine.”
Footnotes
Published ahead of print on 18 August 2008.
REFERENCES
- 1.Aly, A. S., and K. Matuschewski. 2005. A malarial cysteine protease is necessary for Plasmodium sporozoite egress from oocysts. J. Exp. Med. 202225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aly, A. S., S. A. Mikolajczak, H. S. Rivera, N. Camargo, V. Jacobs-Lorena, M. Labaied, I. Coppens, and S. H. Kappe. 2008. Targeted deletion of SAP1 abolishes the expression of infectivity factors necessary for successful malaria parasite liver infection. Mol. Microbiol. 69152-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belmonte, M., T. R. Jones, M. Lu, R. Arcilla, T. Smalls, A. Belmonte, J. Rosenbloom, D. J. Carucci, and M. Sedegah. 2003. The infectivity of Plasmodium yoelii in different strains of mice. J. Parasitol. 89602-603. [DOI] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 6.Carlton, J. M., S. V. Angiuoli, B. B. Suh, T. W. Kooij, M. Pertea, J. C. Silva, M. D. Ermolaeva, J. E. Allen, J. D. Selengut, H. L. Koo, J. D. Peterson, M. Pop, D. S. Kosack, M. F. Shumway, S. L. Bidwell, S. J. Shallom, S. E. Van Aken, S. B. Riedmuller, T. V. Feldblyum, J. K. Cho, J. Quackenbush, M. Sedegah, A. Shoaibi, L. M. Cummings, L. Florens, J. R. Yates, J. D. Raine, R. E. Sinden, M. A. Harris, D. A. Cunningham, P. R. Preiser, L. W. Bergman, A. B. Vaidya, L. H. Van Lin, C. J. Janse, A. P. Waters, H. O. Smith, O. R. White, S. L. Salzberg, J. C. Venter, C. M. Fraser, S. L. Hoffman, M. J. Gardner, and D. J. Carucci. 2002. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 419512-519. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham, D. A., W. Jarra, S. Koernig, J. Fonager, D. Fernandez-Reyes, J. E. Blythe, C. Waller, P. R. Preiser, and J. Langhorne. 2005. Host immunity modulates transcriptional changes in a multigene family (yir) of rodent malaria. Mol. Microbiol. 58636-647. [DOI] [PubMed] [Google Scholar]
- 8.Diatchenko, L., S. Lukyanov, Y. F. Lau, and P. D. Siebert. 1999. Suppression subtractive hybridization: a versatile method for identifying differentially expressed genes. Methods Enzymol. 303349-380. [DOI] [PubMed] [Google Scholar]
- 9.Fonager, J., D. Cunningham, W. Jarra, S. Koernig, A. A. Henneman, J. Langhorne, and P. Preiser. 2007. Transcription and alternative splicing in the yir multigene family of the malaria parasite Plasmodium y. yoelii: identification of motifs suggesting epigenetic and post-transcriptional control of RNA expression. Mol. Biochem. Parasitol. 1561-11. [DOI] [PubMed] [Google Scholar]
- 10.Frischknecht, F., P. Baldacci, B. Martin, C. Zimmer, S. Thiberge, J. C. Olivo-Marin, S. L. Shorte, and R. Menard. 2004. Imaging movement of malaria parasites during transmission by Anopheles mosquitoes. Cell Microbiol. 6687-694. [DOI] [PubMed] [Google Scholar]
- 11.Hide, W. A., L. Chan, and W. H. Li. 1992. Structure and evolution of the lipase superfamily. J. Lipid Res. 33167-178. [PubMed] [Google Scholar]
- 12.Hong, F., R. Breitling, C. W. McEntee, B. S. Wittner, J. L. Nemhauser, and J. Chory. 2006. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 222825-2827. [DOI] [PubMed] [Google Scholar]
- 13.Huber, W., A. von Heydebreck, H. Sultmann, A. Poustka, and M. Vingron. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl. 1)S96-S104. [DOI] [PubMed] [Google Scholar]
- 14.Jin, Y., C. Kebaier, and J. Vanderberg. 2007. Direct microscopic quantification of dynamics of Plasmodium berghei sporozoite transmission from mosquitoes to mice. Infect. Immun. 755532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser, K., K. Matuschewski, N. Camargo, J. Ross, and S. H. Kappe. 2004. Differential transcriptome profiling identifies Plasmodium genes encoding pre-erythrocytic stage-specific proteins. Mol. Microbiol. 511221-1232. [DOI] [PubMed] [Google Scholar]
- 16.Kappe, S., T. Bruderer, S. Gantt, H. Fujioka, V. Nussenzweig, and R. Ménard. 1999. Conservation of a gliding motility and cell invasion machinery in apicomplexan parasites. J. Cell Biol. 147937-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kappe, S. H., C. A. Buscaglia, and V. Nussenzweig. 2004. Plasmodium sporozoite molecular cell biology. Annu. Rev. Cell Dev. Biol. 2029-59. [DOI] [PubMed] [Google Scholar]
- 18.Kappe, S. H., K. Kaiser, and K. Matuschewski. 2003. The Plasmodium sporozoite journey: a rite of passage. Trends Parasitol. 19135-143. [DOI] [PubMed] [Google Scholar]
- 19.Kariu, T., M. Yuda, K. Yano, and Y. Chinzei. 2002. MAEBL is essential for malarial infection of the mosquito salivary gland. J. Exp. Med. 1951317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305567-580. [DOI] [PubMed] [Google Scholar]
- 21.Matuschewski, K. 2006. Getting infectious: formation and maturation of Plasmodium sporozoites in the Anopheles vector. Cell Microbiol. 81547-1556. [DOI] [PubMed] [Google Scholar]
- 22.Matuschewski, K., J. Ross, S. M. Brown, K. Kaiser, V. Nussenzweig, and S. H. Kappe. 2002. Infectivity-associated changes in the transcriptional repertoire of the malaria parasite sporozoite stage. J. Biol. Chem. 27741948-41953. [DOI] [PubMed] [Google Scholar]
- 23.Mikolajczak, S. A., A. S. Aly, R. F. Dumpit, A. M. Vaughan, and S. H. Kappe. 2008. An efficient strategy for gene targeting and phenotypic assessment in the Plasmodium yoelii rodent malaria model. Mol. Biochem. Parasitol. 158213-216. [DOI] [PubMed] [Google Scholar]
- 24.Mikolajczak, S. A., and S. H. Kappe. 2006. A clash to conquer: the malaria parasite liver infection. Mol. Microbiol. 621499-1506. [DOI] [PubMed] [Google Scholar]
- 25.Moreira, C. K., T. J. Templeton, C. Lavazec, R. E. Hayward, C. V. Hobbs, H. Kroeze, C. J. Janse, A. P. Waters, P. Sinnis, and A. Coppi. 2008. The Plasmodium TRAP/MIC2 family member, TRAP-like protein (TLP), is involved in tissue traversal by sporozoites. Cell Microbiol. 101505-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller, A. K., N. Camargo, K. Kaiser, C. Andorfer, U. Frevert, K. Matuschewski, and S. H. Kappe. 2005. Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc. Natl. Acad. Sci. USA 1023022-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller, A. K., M. Labaied, S. H. Kappe, and K. Matuschewski. 2005. Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature 433164-167. [DOI] [PubMed] [Google Scholar]
- 28.Pimenta, P. F., M. Touray, and L. Miller. 1994. The journey of malaria sporozoites in the mosquito salivary gland. J. Eukaryot. Microbiol. 41608-624. [DOI] [PubMed] [Google Scholar]
- 29.Preiser, P., L. Renia, N. Singh, B. Balu, W. Jarra, T. Voza, O. Kaneko, P. Blair, M. Torii, I. Landau, and J. H. Adams. 2004. Antibodies against MAEBL ligand domains M1 and M2 inhibit sporozoite development in vitro. Infect. Immun. 723604-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robson, K. J., J. R. Hall, M. W. Jennings, T. J. Harris, K. Marsh, C. I. Newbold, V. E. Tate, and D. J. Weatherall. 1988. A highly conserved amino-acid sequence in thrombospondin, properdin and in proteins from sporozoites and blood stages of a human malaria parasite. Nature 33579-82. [DOI] [PubMed] [Google Scholar]
- 31.Sidjanski, S. P., J. P. Vanderberg, and P. Sinnis. 1997. Anopheles stephensi salivary glands bear receptors for region I of the circumsporozoite protein of Plasmodium falciparum. Mol. Biochem. Parasitol. 9033-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sultan, A. A., V. Thathy, U. Frevert, K. J. Robson, A. Crisanti, V. Nussenzweig, R. S. Nussenzweig, and R. Menard. 1997. TRAP is necessary for gliding motility and infectivity of plasmodium sporozoites. Cell 90511-522. [DOI] [PubMed] [Google Scholar]
- 33.Tarun, A. S., R. F. Dumpit, N. Camargo, M. Labaied, P. Liu, A. Takagi, R. Wang, and S. H. I. Kappe. 2007. Protracted sterile protection with Plasmodium yoelii pre-erythrocytic GAP malaria vaccines is independent of detectable liver stage persistence and is mediated by CD8+ T cells. J. Infect. Dis. 196608-616. [DOI] [PubMed] [Google Scholar]
- 34.Tarun, A. S., X. Peng, R. F. Dumpit, Y. Ogata, H. Silva-Rivera, N. Camargo, T. M. Daly, L. W. Bergman, and S. H. Kappe. 2008. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc. Natl. Acad. Sci. USA 105305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J., D. Fernandez-Reyes, L. Sharling, S. G. Moore, W. M. Eling, S. A. Kyes, C. I. Newbold, F. C. Kafatos, C. J. Janse, and A. P. Waters. 2007. Plasmodium cysteine repeat modular proteins 1-4: complex proteins with roles throughout the malaria parasite life cycle. Cell Microbiol. 91466-1480. [DOI] [PubMed] [Google Scholar]
- 36.Touray, M. G., A. Warburg, A. Laughinghouse, A. U. Krettli, and L. H. Miller. 1992. Developmentally regulated infectivity of malaria sporozoites for mosquito salivary glands and the vertebrate host. J. Exp. Med. 1751607-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanderberg, J. P. 1975. Development of infectivity by the Plasmodium berghei sporozoite. J. Parasitol. 6143-50. [PubMed] [Google Scholar]
- 38.Vanderberg, J. P. 1974. Studies on the motility of Plasmodium sporozoites. J. Protozool. 21527-537. [DOI] [PubMed] [Google Scholar]
- 39.Verhage, D. F., D. S. Telgt, J. T. Bousema, C. C. Hermsen, G. J. van Gemert, J. W. van der Meer, and R. W. Sauerwein. 2005. Clinical outcome of experimental human malaria induced by Plasmodium falciparum-infected mosquitoes. Neth. J. Med. 6352-58. [PubMed] [Google Scholar]
- 40.Vlachou, D., T. Schlegelmilch, E. Runn, A. Mendes, and F. C. Kafatos. 2006. The developmental migration of Plasmodium in mosquitoes. Curr. Opin. Genet. Dev. 16384-391. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Q., H. Fujioka, and V. Nussenzweig. 2005. Exit of Plasmodium sporozoites from oocysts is an active process that involves the circumsporozoite protein. PLoS Pathog. 1e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, Y., V. Ramachandran, K. A. Kumar, S. Westenberger, P. Refour, B. Zhou, F. Li, J. A. Young, K. Chen, D. Plouffe, K. Henson, V. Nussenzweig, J. Carlton, J. M. Vinetz, M. T. Duraisingh, and E. A. Winzeler. 2008. Evidence-based annotation of the malaria parasite's genome using comparative expression profiling. PLoS ONE 3e1570. [DOI] [PMC free article] [PubMed] [Google Scholar]