Abstract
The DNA-dependent protein kinase (DNA-PK) complex is a serine/threonine protein kinase comprised of a 469-kDa catalytic subunit (DNA-PKcs) and the DNA binding regulatory heterodimeric (Ku70/Ku86) complex Ku. DNA-PK functions in the nonhomologous end-joining pathway for the repair of DNA double-stranded breaks (DSBs) introduced by either exogenous DNA damage or endogenous processes, such as lymphoid V(D)J recombination. Not surprisingly, mutations in Ku70, Ku86, or DNA-PKcs result in animals that are sensitive to agents that cause DSBs and that are also immune deficient. While these phenotypes have been validated in several model systems, an extension of them to humans has been missing due to the lack of patients with mutations in any one of the three DNA-PK subunits. The worldwide lack of patients suggests that during mammalian evolution this complex has become uniquely essential in primates. This hypothesis was substantiated by the demonstration that functional inactivation of either Ku70 or Ku86 in human somatic cell lines is lethal. Here we report on the functional inactivation of DNA-PKcs in human somatic cells. Surprisingly, DNA-PKcs does not appear to be essential, although the cell line lacking this gene has profound proliferation and genomic stability deficits not observed for other mammalian systems.
The repair of DNA double-stranded breaks (DSBs) is crucial for cell survival, the maintenance of genomic integrity, and the prevention of tumorigenesis (48). DSBs can be caused by exposure to a variety of exogenous agents, including ionizing radiation (IR) and chemotherapeutic agents (reviewed in references 31 and 47), as well as being generated by endogenous cellular mechanisms such as variable (diversity) joining [V(D)J] and class switch recombination, processes required for the maturation of B and T lymphocytes (reviewed in reference 71). Moreover, the terminus of every linear chromosome presents the cell with a naturally occurring double-stranded DNA (dsDNA) end, i.e., a telomere, and this must be specifically regulated in order to ensure the stable maintenance of the genome (reviewed in reference 19). Because of the importance of DNA repair, immune function, and genomic stability for organismal well-being, eukaryotes have evolved at least two major mechanisms for the repair of DSBs: homologous recombination (HR) (reviewed in references 46 and 77) and nonhomologous end joining (NHEJ) (reviewed in reference 47).
In lower eukaryotes, HR, in which large stretches of homology between the donor and the recipient DNAs are required, appears to be the major—and often virtually exclusive—pathway for general DNA DSB repair. In higher eukaryotes, HR also plays very important, if not essential, roles in meiosis, sister chromatid exchange, and the repair of stalled or collapsed DNA replication forks (reviewed in references 46 and 77). Moreover, HR is the mechanism that permits gene targeting, a technology used extensively in research and clinical settings (reviewed in reference 32).
Despite the importance of HR in higher eukaryotes and especially in mammals, the process of NHEJ in which two DNA ends are joined together irrespective of their DNA sequence homology nonetheless predominates. At least seven proteins are required for NHEJ: Ku70, Ku86, DNA-PKcs, LIGIV, XRCC-4, Cernunnos/XLF, and Artemis. The heterodimeric Ku proteins Ku70 and Ku86 (reviewed in reference 33) and DNA-PKcs (the DNA-dependent protein kinase complex catalytic subunit) form a trimer that constitutes the DNA-PK (DNA-dependent protein kinase) complex, whose serine/threonine kinase activity is important for NHEJ (reviewed in reference 52). In addition, the LIGIV complex is composed of LIGIV (DNA ligase IV), XRCC4 (X-ray cross complementing 4), and Cernunnos/XLF (XRCC4-like factor) and is required to seal virtually all DSBs generated during NHEJ (reviewed in reference 67). The seventh factor, the nuclease Artemis, plays a crucial, albeit poorly defined, role in many aspects of NHEJ (56). The importance of the NHEJ genes for human health is emphatically demonstrated by the existence of patients who have mutations of LIGIV (60, 65), Artemis (56), or XLF (10) and who, without exception, present with IR sensitivity, immune deficiency, and/or cancer predisposition.
Given the existence of other NHEJ-deficient patients, it is surprising that patients with mutations for XRCC4, Ku70, Ku86, and DNA-PKcs have yet to be identified or described. In mice, the functional inactivation of XRCC4 results in lethality (24), and if the essential feature of this gene is conserved in humans, this would perforce explain its absence in patients. Similarly, although Ku70 and Ku86 knockout mice are viable (reviewed in reference 33) and a plethora of Ku-null cell lines have been described previously (81), no spontaneous whole animal models defective in either Ku subunit have ever been reported. Moreover, functional inactivation via gene targeting of Ku70 (17) or Ku86 (44) results in nonviable human somatic cell lines. Extrapolation of these observations concerning Ku to humans suggests that the human Ku subunits are essential, and this explains the absence of Ku-deficient patients.
The reason for the lack of DNA-PKcs-deficient patients, however, is not so obvious. Mice made functionally null for DNA-PKcs via gene targeting are viable (8, 23, 72). Moreover, spontaneous mutation of the DNA-PKcs gene has occurred at least four times in three different mammalian species—twice in mice (9, 35), once in a dog (53), and once in a horse (51)—and each time this has resulted in viable animals. Thus, the absence of DNA-PKcs is clearly still compatible with proper development in most mammals. Moreover, a human somatic cell line, MO59J, which harbors a nonsense frameshift mutation in the DNA-PKcs gene and is thus functionally null (5, 43), has been described previously (3). In addition, numerous investigators have attempted to phenocopy the DNA-PKcs-null condition in a variety of human somatic cell lines with pharmacological inhibitors and molecular (e.g., RNA interference, antisense oligonucleotides, etc.) approaches and have generally achieved significant inhibition (reviewed in reference 12). The latter technologies, however, invariably failed to achieve a complete knockdown of DNA-PKcs expression. Moreover, MO59J, which has been used extensively by researchers in the field for the past decade and a half (reviewed in reference 52), is an abnormal cell line. First, since the patient and the tumor from which MO59J was derived were wild type (WT) for DNA-PKcs, the mutation was generated spontaneously during propagation of the glioma in cell culture (3, 43). Second, in addition to being defective for DNA-PKcs, MO59J has defective p53 (4) and diminished ataxia telangiectasia mutated (ATM) (74) expression. Third, these specific DNA repair gene defects are superimposed on an aneuploid (nearly pentaploid) karyotype (5). Consequently, whether the mutation in DNA-PKcs caused these additional abnormalities or whether DNA-PKcs-deficient human somatic cells are viable only under these highly aberrant karyotypic conditions is unknown.
To explore the requirement for DNA-PKcs expression in humans/human somatic cells in more detail, we attempted to disrupt via gene targeting the DNA-PKcs gene in the human adenocarcinoma somatic tissue culture cell line HCT116. While HCT116 is an immortalized and transformed cell line, it is diploid, has a stable karyotype, and is wild type for most DNA repair, DNA checkpoint, and chromosome stability genes (reviewed in reference 32). We describe here the isolation and characterization of HCT116 cell lines that are heterozygous and null for DNA-PKcs expression. These experiments demonstrate that DNA-PKcs is not an essential gene in human somatic cells, and they provide a clear demonstration for a separation of function between Ku and DNA-PKcs. Moreover, the phenotypes of the DNA-PKcs-null cell line, such as profound growth retardation, IR sensitivity, shortened telomeres, and increased genetic instability, collectively shed light on the role of DNA-PKcs in human cells. Last, but nonetheless importantly, these cell lines should provide the field with powerful tools for the characterization of DNA-PK and NHEJ.
MATERIALS AND METHODS
Cell culture.
Human wild-type HCT116 cells were cultured in McCoy's 5A medium containing 10% fetal calf serum, 100 U/ml penicillin, and 50 U/ml streptomycin and supplemented with 2 mM l-glutamine. The cells were incubated at 37°C in a humidified incubator with 5% CO2. All cell lines derived from correct targeting were grown in the presence of 1 mg/ml G418.
Targeting vector construction.
The targeting vectors were constructed utilizing a system described by Kohli et al. (39). Briefly, the left and right homology arms (left HA and right HA, respectively) for the DNA-PKcs exon 81 to 83 targeting vector were constructed by PCR from HCT116 genomic DNA. The primers used to construct the left HA for DNA-PKcs were CS8183P1, 5′-ATACATACGCGGCCGCGTGGCGGGCGCCTGTAATCC-3′, and CS8183P2, 5′-GCTCCAGCTTTTGTTCCCTTTAGGGCAAAGATCACCAAGAATTC-3′. The right HA was constructed using the primers CS8183P3, 5′-CGCCCTATAGTGAGTCGTATTACGCTGTGCTCTCGTGATCCTGGGC-3′, and CS8183P4, 5′-ATACATACGCGGCCGCCAAGCAGACAATCCTAAATAC-3′. The arms were used in a fusion PCR, together with a 4-kb PvuI restriction enzyme fragment containing the drug selection marker. The fusion PCR product was gel purified and ligated to the pAAV backbone by use of NotI restriction enzyme sites to construct the final targeting vector.
Packaging and isolating virus.
The targeting vector (8 μg) was mixed with pAAV-RC and pHelper plasmids (8 μg of each) from an adeno-associated virus (AAV) helper-free system (Stratagene) and was transfected into AAV 293 cells (Invitrogen) by use of Lipofectamine 2000 (Invitrogen). Virus was isolated from the AAV 293 cells 48 h after transfection by use of a freeze-thaw method (39).
Infections.
HCT116 and NALM-6 cells were grown to ∼70 to 80% confluence in six-well tissue culture plates. Fresh medium (1.5 ml) was added to the cells 3 h prior to addition of the virus. The required volume of the virus was added dropwise to the plates. After a 2-h incubation at 37°C, another 1.5 ml of medium was added to the plates. After a further 48-h incubation, the cells were transferred to 96-well plates at dilutions designed to obtain single colonies and placed under selection (1 mg/ml G418).
Isolation of genomic DNA and genomic PCR.
Genomic DNA for PCR screening was isolated using a Puregene DNA purification kit (Gentra Systems). Cells were harvested from confluent wells from a 24-well culture dish. DNA was dissolved in a final volume of 50 μl, 1 μl of which was used in each PCR. For DNA-PKcs+/− targeting events, PCR was carried out at both the 5′ and 3′ sides of the targeted locus. For the 5′ end, a control PCR was performed using the primer set LArmR, 5′-GCTCCAGCTTTTGTTCCCTTTAG-3′, and CS8183P1, 5′-ATACATACGCGGCCGCGTGGCGGGCGCCTGTAATCC-3′ (Fig. 1D). Correct targeting was determined using LArmR and PKcs81-83F1, 5′-CTCATACTTACTATGGATTGTGTGTATATCTACC-3′ (Fig. 1D). For the 3′ end, the control PCR was carried out using the primer set RArmF, 5′-CGCCCTATAGTGAGTCGTATTAC-3′, and CS8183P4, 5′-ATACATACGCGGCCGCCAAGCAGACAATCCTAAATAC-3′ (Fig. 1D). PCR to screen for correctly targeted clones was performed using RArmF and PKcs81-83R1, 5′-CCTCAGCAGTCCCCTTATTTCTCCAA-3′ (Fig. 1D). For the second round of targeting for DNA-PKcs−/− targeting events, PCR was carried out at the 3′ side of the targeted locus. Since the LoxP sites used to remove the neomycin (Neo) cassette from the DNA-PKcs+/− cell line are located upstream and downstream of the vector sequences, a vector-specific primer, pNeDaKOF2, 5′-GTGGCCGAGGAGCAGGACTGAATAAC-3′, as opposed to RArmF, was used (Fig. 1E). For confirmation of DNA-PKcs−/− targeting events, PCR primers that span the targeted locus were used. These include 8183F1, 5′-CTATTTTTCAATGAATTCTTGGTGATCTTTGCC-3′; 8183F2, 5′-TCCTAATTCCAATTCCCCTTTTCTCACTC-3′; 8183R1, 5′-AACGCATGCCCAAAGTCGATCCCG-3′; and 8183R2, 5′-CCTTGCCCAGGATCACGAGAGCACAGC-3′.
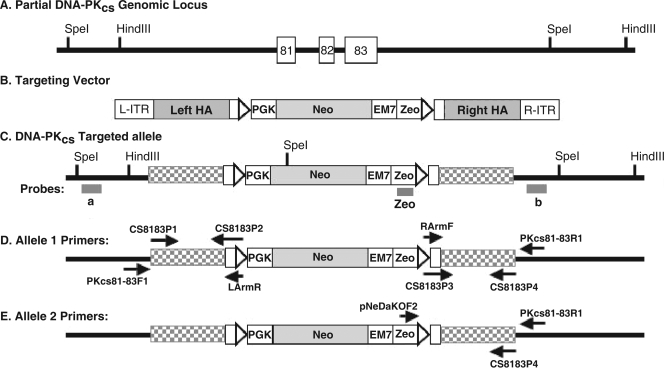
FIG. 1.
Scheme for functional inactivation of the human DNA-PKcs locus. (A) Partial DNA-PKcs genomic locus. Sites for the restriction enzymes HindIII and SpeI are shown. (B) Cartoon of the targeting vector. L-ITR and R-ITR, left and right inverted terminal repeats, respectively; triangles, LoxP sites; PGK, phosphoglycerate kinase eukaryotic promoter; EM7, EM7 prokaryotic promoter; Zeo, Zeo resistance gene; Neo, Neo resistance gene; and left HA and right HA, left and right homology arms, respectively, used to facilitate targeting by HR. (C) Cartoon of a first-round targeted allele. Exons 81 to 83 on one chromosome have been replaced by the targeting vector. a, b, and Zeo are the external probes that were used for Southern blot analysis. A unique SpeI restriction site in the targeting construct is shown. The left HA and right HA are now drawn as checkered rectangles, and L-ITR and R-ITR are lost upon vector integration. (D) Primers used for the diagnostic PCR screening for DNA-PKcs+/− clones. (E) Primers used for the diagnostic second round of targeting PCR screening.
Isolation of genomic DNA and Southern hybridizations.
Chromosomal DNA was prepared, digested, subjected to electrophoresis, and then transferred to a nitrocellulose membrane as described previously (42). The membrane was hybridized with a 5′ probe, a 3′ probe, or an internal Zeomycin (Zeo) probe (Fig. 1C) to detect correct targeting of the PKcs81-83 targeting vector. The 5′ probe corresponded to 634 bp of a region 5′ of the targeting vector and was made by PCR with the primers PKcs81-83.5SF1, 5′-CAATTAACATGTACATTTTAACCACAAGGTTTT-3′, and PKcs81-83.5SR1, 5′-GGGTATTGGAAGTGCCCTGTAAAAAGGCAG-3′, while the 3′ probe corresponded to 644 bp of a region 3′ of the targeting vector and was made by PCR with the primers PKcs81-83.3SF1, 5′-GATTGTATATACCATTATGATCAGCCGGG-3′, and PKcs81-83.3SR1, 5′-CAAGAGAAAACTTTAAGGCTAACCTAAAAGCC-3′. The Zeo probe was obtained from the pNeDaKO Neo vector as a 416-bp AflIII-plus-ApaLI restriction fragment, which includes the entire Zeo coding region. The PCR products were electrophoresed on a 1% agarose gel and gel purified prior to use. A Prime-It II kit (Stratagene) was used to radiolabel the Southern probe with [α-32P]dATP. For the 5′ Southern analysis, genomic DNA was digested with SpeI to generate a parental band of 6,430 bp and a targeted band of 2,883 bp. For the 3′ and internal Southern analyses, genomic DNA was digested with HindIII, which yielded a parental band of 8,631 bp and a targeted band of 10,136 bp.
Whole-cell-extract preparation.
Cells were trypsinized and washed twice with phosphate-buffered saline (PBS). For whole-cell extraction, cells were boiled in DNase buffer (Gibco) for 10 min. The samples were then digested with DNase I (0.1 U/μl; Gibco) for 10 min at 37°C. The samples were finally boiled in 5× sodium dodecyl sulfate buffer (0.225 M Tris-HCl, pH 6.8, 50% [vol/vol] glycerol, 5% sodium dodecyl sulfate, 0.05% bromophenol blue, 0.14 M β-mercaptoethanol).
Nuclear-extract preparation.
Nuclear extracts were prepared according to the protocol supplied with a CelLytic NuCLEAR extraction kit (Sigma). Briefly, cells were collected and washed with cold PBS. The cells were then resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 15 mM CaCl2, 1.5 M sucrose) and kept on ice for 15 min, followed by mixing by inversion. Centrifugation was carried out to obtain a pellet that was then resuspended in extraction buffer (20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 0.42 M NaCl, 0.2 M EDTA, 25% [vol/vol] glycerol). The suspension was agitated for 30 min at 4°C and centrifuged. The supernatant from the final centrifugation step was collected as the nuclear extract.
Immunoblotting.
For immunoblot detection, proteins were subjected to electrophoresis on a 4 to 20% gradient gel (Bio-Rad), electroblotted onto a nitrocellulose membrane, and detected as described previously (28).
DNA end binding assay.
Ku DNA end binding activity was measured by a gel mobility shift assay as described previously (29), with some modifications. A 43-bp dsDNA obtained by annealing two single-stranded oligonucleotides was radiolabeled with [γ-32P]ATP by use of T4 polynucleotide kinase. Radiolabeled DNA (∼4 ng) was incubated with 1 μg of nuclear protein in 15 μl of binding buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 10% glycerol, 200 mM NaCl), with 1 μg of circular TA plasmid (Invitrogen) as the competitor, on ice for 5 min. Electrophoresis was then carried out using a 5% native polyacrylamide gel for 3 h at 130 V. The gel was dried and exposed to X-ray film for ∼15 to 30 min at −80°C.
DNA-PK assay.
Nuclear extracts were incubated on ice for 15 min with preswollen dsDNA-cellulose (Sigma). Nuclear extract (100 μg) was used with each sample. Following incubation on ice, the samples were washed twice in Z′ 0.05 buffer (25 mM HEPES-KOH, 50 mM KCl, 10 mM MgCl2, 20% glycerol, 0.1% Igepal, 1 mM dithiothreitol). After the washing steps, the samples were centrifuged and the precipitate was resuspended in 100 μl Z′ 0.05 buffer. The sample was then incubated at 30°C for 15 min with either a good DNA-PK substrate peptide (EPPLSQEAFADLLKK) or a mutant peptide (EPPLSEQAFADLLKK) and [γ-32P]ATP. The wild-type peptide can be phosphorylated by DNA-PK at the serine residue, while the mutant peptide is not recognized by DNA-PK. Following incubation, polyacrylamide gel electrophoresis was carried out. The gel was vacuum dried and exposed to X-ray film. The amount of phosphorylated peptide was quantified using a phosphorimager.
Cell proliferation assay.
To obtain a growth curve, 4 × 103 HCT116 cells were plated out in each well of a six-well plate in duplicate. Cell numbers were determined using a hemocytometer every day thereafter starting at day 4, utilizing growth media without selection.
Plating efficiency assay.
Parental HCT116, DNA-PKcs+/−, and DNA-PKcs−/− cells were plated at increasing concentrations on six-well plates in triplicate. For each cell line, 100, 200, 300, and 400 cells per well were used. In addition, for DNA-PKcs−/− cells, 1,000 and 2,000 cells per well were included. Cells were allowed to grow for 12 days before the colonies were fixed, stained, and counted and plating efficiency was calculated as follows: (number of colonies/number of cells plated) × 100. Additionally, because of their slow-growth phenotype, 100, 200, 300, and 400 DNA-PKcs−/− cells were plated in triplicate and allowed to grow for 3 weeks.
X-ray survival assay.
For the parental HCT116 and DNA-PKcs+/− cell lines, 300 cells were seeded into each well of a six-well tissue culture plate about 10 to 12 h before irradiation. For the DNA-PKcs−/− cell line, 3,000 cells were plated. Cells were then X irradiated using a 137Cs source at doses of 0, 0.2, 0.5, 1.0, 2.5, and 5.0 Gy. After irradiation, cells were allowed to grow for ∼10 days before the colonies were fixed, stained, and counted and cell survival percentage was calculated (42).
Telomeric TRF analysis.
Terminal restriction fragment (TRF) analysis was performed as described previously (58, 76), utilizing the restriction enzymes AluI and MboI (New England Biolabs). The probe was the telomere-specific oligonucleotide (Operon) (C3TA2)4.
Cytogenetic analysis and telomere FISH.
G-banding cytogenetic and telomeric fluorescent in situ hybridization (FISH) analyses were performed in the Cytogenetics Core Laboratory at the University of Minnesota as described previously (58).
FACS analysis.
Fluorescence-activated cell sorter (FACS) analyses were performed in the Flow Cytometry Core Facility at the University of Minnesota. Briefly, 1 × 106 cells were incubated with 20 μg/ml of propidium iodide in 1× PBS containing 0.1% Triton X-100 and DNA content was determined with a FACSCalibur (Becton Dickinson) instrument. Data were plotted using FlowJo 8.5.2 software; 25,000 events were analyzed per sample.
RT-PCR.
Total RNA was isolated from cells by use of an RNeasy mini kit (Qiagen). Reverse transcription PCRs (RT-PCRs) were performed using a Qiagen LongRange two-step RT-PCR kit. Briefly, 2 μg of total RNA was used as a template for the first-strand cDNA synthesis primed by a DNA-PKcs-specific reverse primer (PKcs84cDNAR, 5′-TCAAAATTTTTCCAATCAAAGGA-3′). The resulting cDNAs were then used in a PCR with PKcs80F (5′-TGAACACCATGTCCCAAGAG-3′) and PKcs84R (5′-GGGCTCCTTGACAAACACATC-3′). The expected size for a DNA-PKcs-specific product that includes exons 81 to 83 is 578 bp, while the deletion of exons 81 to 83 results in a product 214 bp long.
RESULTS
Generation of heterozygous DNA-PKcs+/− HCT116 and NALM-6 cell lines.
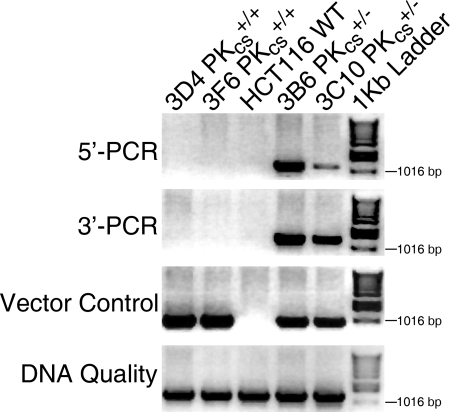
A recombinant AAV (rAAV) was used for the delivery of the targeting vector into the desired cell line (39; also reviewed in references 32 and 78). The targeting vectors contained ∼900-bp-long left and right HAs flanking a Neo resistance selection cassette (Fig. 1B). DNA-PKcs (which is also designated Prkdc, or protein kinase DNA-activated catalytic subunit [55]) is a 469-kDa protein encoded by 87 exons that span an ∼250-kb region (q11) on chromosome 8 (21). Initially, we attempted to disrupt exon 1; however, no correctly targeted clones were recovered from 372 G418-resistant, control-PCR-positive clones analyzed (Table 1 and data not shown). An additional attempt was then made to disrupt exon 10. A total of 421 G418-resistant, control-PCR-positive clones were analyzed, again without identifying a correctly targeted clone (Table 1 and data not shown). Finally, we attempted to disrupt a region at the 3′ end of the locus that contains three tightly clustered exons (81 to 83) (Fig. 1A). The left HA for the DNA-PKcs targeting vector contained sequences 5′ of exon 81, while the right HA contained sequences 3′ of exon 83 (Fig. 1B). The selection cassette was an ∼2.8-kb restriction fragment that contained the phosphoglycerate kinase promoter, the Neo gene, the EM7 promoter, and the Zeo resistance gene (Fig. 1B). The selection cassette was also flanked on either end by LoxP sites. Correct targeting of the endogenous genomic DNA-PKcs locus (Fig. 1A) removes exons 81, 82, and 83 (Fig. 1C) and is predicted to result in a catalytically inactive DNA-PKcs since these exons encode the kinase catalytic domain of DNA-PKcs. All rAAV integration events resulted in the generation of G418-resistant colonies. In order to screen these colonies for correct targeting events, genomic DNA was isolated and diagnostic PCR analyses were performed. Control PCRs were carried out using two sets of primers. RArmF and CS8183P4 (Fig. 1D) were used to confirm the presence of the vector sequence in all G418-resistant clones (Fig. 2, Vector Control), while CS8183P3 and PKcs81-83R1 (Fig. 1D) were used to check for the quality of the isolated DNA and to confirm the presence of DNA in the parental cell line (Fig. 2, DNA Quality). Experimental screening PCR was also carried out, using the following two sets of PCR primers: PKcs81-83F1 and LArmR for the 5′ PCR and RArmF and PKcs81-83R1 for the 3′ PCR. LArmR and RArmF correspond to sequences unique to the targeting vector, while PKcs81-83F1 and PKcs81-83R1 reside externally in the 5′ and 3′ flanking regions, respectively (Fig. 1D). A total of 238 HCT116 clones were screened in this manner, and correct targeting events were observed for three independent clones, for a targeting frequency of 1.3% (Table 1). The correct targeting events in two of the clones (3B6 and 3C10) were confirmed by the 5′ flanking PCR strategy, where both clones showed the expected 1,148-bp PCR band (Fig. 2, 5′-PCR), and by the 3′ flanking PCR strategy, where the correct targeting events resulted in the production of a 1,307-bp band (Fig. 2, 3′-PCR). The PCR bands resulting from correct targeting events were absent from two randomly targeted clones (3D4 and 3F6) as well as the parental HCT116 cell line (Fig. 2).
TABLE 1.
Summary of gene targeting experiments
| Cell line | Gene | Exon(s) | No. of colonies screeneda | No. of correctly targeted clonesb | Targeting frequency (%)c |
|---|---|---|---|---|---|
| NALM-6 (WT) | DNA-PKcs | 81 to 83 | 432 | 1 | 0.2 |
| HCT116 (WT) | DNA-PKcs | 1 | 372 | 0 | 0.0 |
| HCT116 (WT) | DNA-PKcs | 10 | 421 | 0 | 0.0 |
| HCT116 (WT) | DNA-PKcs | 81 to 83 | 238 | 3 | 1.3 |
| DNA-PKcs+/− | DNA-PKcs | 81 to 83 | 638 | 17 | 2.7 |
Drug-resistant clones that were also positive for the internal-control PCR.
Drug-resistant clones that showed correct targeting by PCR.
The targeting frequency is the number of correctly targeted colonies per 100 drug-resistant colonies screened.
FIG. 2.
Identification of DNA-PKcs+/− cell lines. Four diagnostic PCRs were carried out: an experimental 5′ PCR to confirm correct targeting events on the 5′ side, using the primers PKcs81-83F1 and LArmR; an experimental 3′ PCR to confirm correct targeting events from the 3′ side, using the primers RArmF and PKcs81-83R1; a vector control PCR to confirm the presence of the targeting vector in the cell lines, using the primers RArmF and CS8183P4; and a DNA quality PCR to confirm the quality of the genomic DNA preparation, using primers CS8183P3 and PKcs81-83R1. An ethidium bromide-stained agarose gel picture is shown, with a 1-kb marker ladder on the far right. Genomic DNA was isolated from two heterozygous clones (3B6 and 3C10), two randomly targeted clones (3D4 and 3F6), and the WT HCT116 cell line.
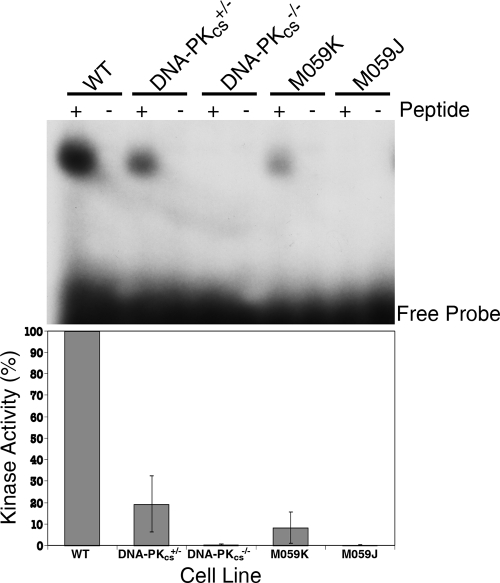
FIG. 5.
DNA-PKcs+/− cells have diminished levels and DNA-PKcs−/− cells have undetectable levels of DNA-PK activity. (Top) DNA-PK activity was assessed by incubating nuclear extracts from the indicated cell lines with [γ-32P]ATP and peptides that either can (+) or cannot (−) be phosphorylated by DNA-PK. After incubation, polyacrylamide gel electrophoresis was performed and the radioactivity incorporated in the peptides was quantitated. The positions of the free probe and the phosphorylated peptides are shown. (Bottom) Phosphorimager quantitation of the kinase assays. Blots similar to that shown in the top panel were quantitated using a phosphorimager. The graph represents the average relative DNA-PK activity levels (±standard deviations) from two experiments.
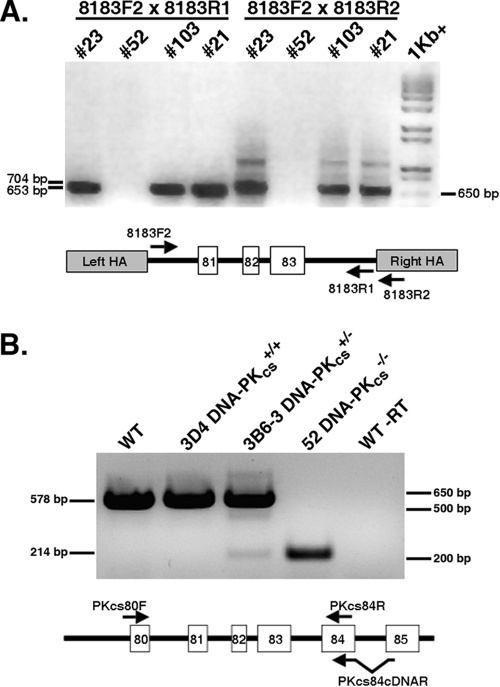
FIG. 3.
Identification of a DNA-PKcs−/− cell line. (A) Two diagnostic PCRs were carried out to confirm the loss of exon 81 to 83 sequences. One primer pair consisted of 8183F2 and 8183R1, and the other primer pair consisted of 8183F2 and 8183R2. Clones no. 21, 23, and 103 represent retargeted clones with one remaining wild-type allele, whereas clone no. 52 represents the null cell line. An ethidium bromide-stained agarose gel picture is shown, with a 1-kb-plus marker ladder on the far right. A cartoon of the PCR strategy is shown below the figure. (B) An RT-PCR analysis was carried out to demonstrate the lack of full-length mRNA in the DNA-PKcs−/− cell line. Total mRNA from the indicated cell lines was reverse transcribed using the primer PKcs84cDNAR or not reverse transcribed (WT −RT). This first-strand cDNA was then used in a PCR with primers PKcs80F and PKcs84R, and the products were identified on an ethidium bromide-stained agarose gel. The predicted full-length product is 578 bp, and a product derived from skipping from exon 80 to 84 should yield a 214-bp product. A cartoon of the RT-PCR strategy is shown below the figure.
Southern hybridization was used to confirm the targeting events. Genomic DNA was isolated and digested with the restriction enzyme SpeI or HindIII. Southern blot analysis was then performed using a 5′ probe (Fig. 1C, probe a), a 3′ probe (Fig. 1C, probe b), or a probe corresponding to the vector (Fig. 1C, probe Zeo). For the 5′ Southern analysis, the appearance of a novel ∼2.8-kb band, caused by the presence of an SpeI restriction enzyme site introduced from the targeting vector, confirmed that a correct targeting event had occurred in 3B6 and 3C10 (see Fig. S1A in the supplemental material). The band was absent, as expected, from DNA isolated from the parental HCT116 cell line and the randomly targeted clones 3D4 and 3F6 (see Fig. S1A in the supplemental material). For the 3′ Southern analysis, a novel ∼10.1-kb HindIII restriction fragment, caused by replacing the 703-bp region containing exons 81 to 83 with 2,208 bp from the targeting vector, confirmed that correct targeting events had occurred on one allele in 3B6 and 3C10 (see Fig. S1B in the supplemental material). This 10.1-kb fragment was absent, as expected, from the parental HCT116 cell line (WT) and the randomly targeted clones 3D4 and 3F6, where only the endogenous 8.6-kb band was observed (see Fig. S1B in the supplemental material). Last, when genomic DNA from the indicated lines was cleaved with HindIII and probed with Zeo, a single band of the predicted size (∼10.1 kb) was observed in the presumptive targeted clones and this band was absent from the parental line (WT) and from the randomly targeted clones (3D4 and 3F6), confirming that only a single viral integration event had occurred in these lines. In summary, PCR and Southern analyses confirmed the isolation of at least two independent DNA-PKcs+/− HCT116 cell lines.
An identical experimental protocol was utilized to disrupt exons 81 to 83 of the DNA-PKcs gene in the B-cell line NALM-6. A single correctly targeted clone (S137) from 432 G418-resistant, internal-control-PCR-positive clones was identified and characterized as described above (Table 1 and data not shown).
Generation of a homozygous DNA-PKcs−/− HCT116 cell line.
In order to construct a DNA-PKcs−/− HCT116 cell line, the DNA-PKcs+/− cell line 3B6-3 (3B6 cells, subclone no. 3) was transiently exposed to Cre recombinase. Cre should excise the internal Neo cassette, which is flanked by LoxP sites, within the integrated targeting vector (Fig. 1B). Eighteen single-cell clones were picked and duplicated into medium containing or lacking G418, and G418-sensitive clones were identified. Correct excision of the neomycin gene was confirmed by the generation of a diagnostic PCR product (data not shown). One (3B6-3 Cre13.1) of several such clones obtained in this fashion was used for a second round of gene targeting with the original targeting vector containing the Neo cassette. Productive infection of 3B6-3 Cre13.1 cells should produce one of three potential outcomes: (i) random targeting (the vast majority of the events); (ii) correct targeting, but of the already inactivated allele from the first round of targeting (i.e., “retargeting”); or (iii) correct targeting of the remaining functional allele to generate the desired null clone. Initially, 17 correctly targeted clones from 638 G418-resistant, internal-control-PCR-positive clones (targeting frequency of 2.7%) were identified using the primers pNeDaKOF2 and PKcs81-83R1 (Fig. 1E, Table 1, and data not shown). A diagnostic PCR strategy was then utilized to distinguish retargeting from null events (Fig. 3A). Strikingly, 16 of the 17 clones were retargeted. This conclusion was substantiated by the fact that although these 16 clones were correctly targeted (data not shown), they still retained exon 81, 82, and 83 sequences (e.g., clones no. 21, 23, and 103) (Fig. 3A and data not shown). A single clone (3B6-3 Cre13.1 no. 52, herein referred to as DNA-PKcs-null or DNA-PKcs−/− cells) lacked exon 81, 82, and 83 sequences (Fig. 3A).
To confirm that no full-length DNA-PKcs mRNA was being produced in the DNA-PKcs-null cells, poly(A)+ mRNA was isolated and first-strand cDNA was generated using a primer (PKcs84cDNAR) that spanned exon 84 and 85 sequences, ensuring that only mRNA, and not genomic DNA, would be amplified (Fig. 3B). This cDNA was then subjected to PCR using primers (PKcs80F and PKcs84R) complementary to sequences located in exons 80 and 84, respectively (Fig. 3B). A 578-bp PCR product corresponding to the region encompassing exons 80 to 84 was produced from mRNA isolated from the parental cell line (WT), a randomly targeted clone (3D4), and a DNA-PKcs+/− cell line (3B6-3) but was absent from the DNA-PKcs−/− cells (Fig. 3B), demonstrating that the DNA-PKcs-null cell line did not produce detectable full-length DNA-PKcs mRNA. A truncated 214-bp product that corresponded to a cDNA template in which exons 81, 82, and 83 had been skipped was detected in the DNA-PKcs+/− and DNA-PKcs−/− cell lines. Since skipping from exon 80 to 84 generates a frameshift within the DNA-PKcs open reading frame that quickly results in a stop codon, this analysis predicted that no functional full-length DNA-PKcs protein would be produced in DNA-PKcs−/− cells.
Primary characterizations.
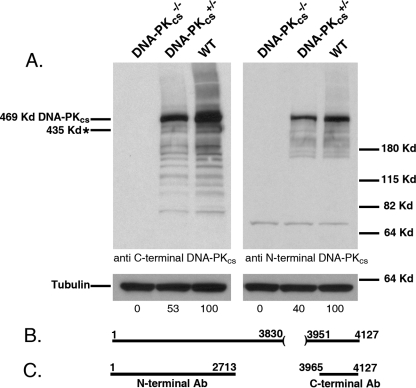
The removal of exons 81 to 83 (which encode amino acids 3831 to 3950) (Fig. 4B) in the C terminus of DNA-PKcs should result in the production of a truncated 435-kDa protein due to the introduction of a premature stop codon regardless of whether exon 80 splices to exon 84 (e.g., Fig. 3B) or whether intron 80 is incorporated into the mature mRNA. The expression levels of the wild-type and the putative truncated DNA-PKcs proteins in the DNA-PKcs+/− and DNA-PKcs−/− cell lines were determined by immunoblot analysis using either a mouse monoclonal antibody generated against the N-terminal 2,713 amino acids of DNA-PKcs or a rabbit polyclonal antibody directed against the C terminus of DNA-PKcs (amino acids 3965 to 4127) (Fig. 4C). The DNA-PKcs+/− clones expressed DNA-PKcs at a level of ∼50% (with α-tubulin as a loading control) of that observed for the parental cells (Fig. 4A), while the DNA-PKcs−/− cells contained no detectable wild-type or truncated protein (Fig. 4A). The complete absence of DNA-PKcs protein in the DNA-PKcs−/− cell line suggested that the expected 435-kDa C-terminally truncated DNA-PKcs protein was highly unstable, a conclusion for which there is ample precedent (38, 53, 62, 79).
FIG. 4.
DNA-PKcs+/− cells have diminished levels and DNA-PKcs−/− cells have nondetectable levels of DNA-PKcs protein. (A) Whole-cell extract was prepared from wild-type HCT116 (WT), clone 3B6-3 no. 23 (DNA-PKcs+/−), and clone no. 52 (DNA-PKcs−/−) cells. The extracts were analyzed by immunoblotting for DNA-PKcs using either a C-terminal or an N-terminal DNA-PKcs antibody. An α-tubulin antibody was used as a loading control. In each panel, single blots that were probed sequentially with the two different antibodies are shown. Phosphorimager quantitation of the DNA-PKcs portion of the blot is shown below the gels. The approximate predicted position of the putative truncated DNA-PKcs protein is shown on the far left (marked with an asterisk). Molecular size markers are shown on the far right. (B) Cartoon of the DNA-PKcs protein and the amino acids that should be missing due to the targeting event (amino acids 3831 to 3950). Note that since the targeting event should generate a nonsense mutation, amino acids 3951 to 4127 should also be missing in the putative truncated protein. (C) Cartoon of the regions of DNA-PKcs that were used in generating the antibodies (Ab).
To confirm that the loss of DNA-PKcs protein resulted in the loss of functional activity in the DNA-PKcs−/− cell line, DNA-PK assays were performed. Nuclear extract was prepared from the cell lines indicated in Fig. 5 and incubated either with a peptide derived from p53 with a consensus phosphorylation site for DNA-PK or with a control peptide in which that site had been mutated (20). The HCT116 DNA-PKcs+/− cell line had diminished (19.3% ± 13.0%, n = 2) DNA-PK activity compared to that of the parental cell line, while the DNA-PKcs−/− cell line had no detectable DNA-PK activity whatsoever (Fig. 5). Extracts from two human glioma cell lines, M059K and M059J, which are known to be positive and negative, respectively, for DNA-PK activity, were used as controls. The activity detected with the control cell lines agreed with published results (43), confirming that the assay was detecting only DNA-PK activity.
Finally, to examine whether the loss of DNA PK activity was due to a defect in the Ku heterodimer, the activity of Ku in this cell line was quantitated based upon Ku's ability to bind to dsDNA by use of an electrophoretic mobility shift assay (64). By use of a radioactive dsDNA probe of 43 bp in length and nuclear extracts prepared from parental (WT), DNA-PKcs+/−, and DNA-PKcs−/− cells, the binding of Ku to the ends of the dsDNA can be quantitated. The DNA-PKcs+/− and the DNA-PKcs−/− cell lines had no obvious defects in Ku DNA end binding activity and, if anything, exhibited an increase in activity respective to that of the wild type (see Fig. S2 in the supplemental material). As expected, extracts prepared from cell lines with reduced Ku70 (clone no. 53 [18]) or Ku86 (clone no. 70-32 [44]) expression showed lower DNA end binding activity. From this experiment, we concluded that the loss of DNA-PK activity in the DNA-PKcs−/− cell line was due to the loss of DNA-PKcs, and not Ku, expression.
In summary, based upon the molecular characterizations (Fig. 2 and 3), the immunoblot and RT-PCR analyses (Fig. 4), and the DNA-PK activity assays (Fig. 5), we concluded that the DNA-PKcs−/− cell line was indeed null for DNA-PKcs expression and activity.
Biological endpoints.
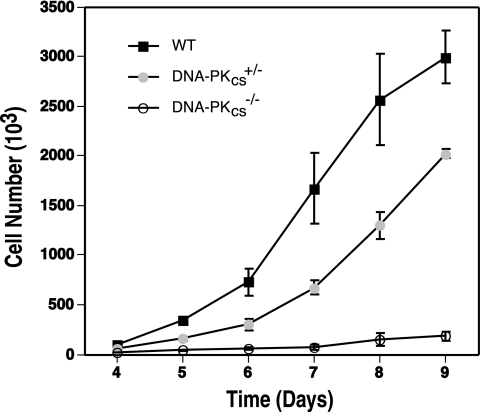
Human HCT116 cells heterozygous for Ku70 (18) and Ku86 (44) display a haploinsufficient growth defect. Moreover, after undergoing a second successful gene targeting event, Ku86-null cell lines grew extremely slowly for 8 to 10 cell divisions before ultimately succumbing. Since Ku functions with DNA-PKcs in the DNA-PK complex, we hypothesized that human cells reduced or deficient for DNA-PKcs might also have a growth defect. This seemed to be a particularly important issue, since this phenotype is not observed with rodents, as only Ku-deficient (45, 59, 61, 82) and not DNA-PKcs-null (8, 23, 35, 72) mice show growth defects. To experimentally quantitate this property, equal numbers of cells for all of the cell lines were plated out in duplicate on day 0 and then the cell number was determined beginning 4 days thereafter for 9 days. Both the DNA-PKcs+/− (clone 3B6-3) and DNA-PKcs−/− cell lines showed a significantly reduced rate of growth compared to that of the parental HCT116 cells (Fig. 6). The doubling time of the 3B6-3 DNA-PKcs+/− cell line was 24.1 (±0.1) h, which was slightly slower than that of the parental line, at 22.8 (±0.4) h. The DNA-PKcs−/− cell line, in contrast, showed a remarkably severe growth defect, with a doubling time of 39.8 (±2.4) h.
FIG. 6.
Growth defects of human somatic cells with reduced and no DNA-PKcs expression. Wild-type (WT), DNA-PKcs+/−, and DNA-PKcs−/− HCT116 cells were seeded on tissue culture plates (4 × 103), and the increase in cell number was determined by counting trypan blue-excluding cells with a hemocytometer at daily intervals. The averages (±standard deviations) of two experiments, each done in duplicate, are shown.
The growth deficit in the DNA-PK-null cell line seemed empirically to be most severe when the cells were subcultured at a low cell density (data not shown). To quantitate this effect, the plating efficiencies for HCT116 WT, DNA-PKcs+/−, and DNA-PKcs−/− cells were determined. HCT116 WT cells had a plating efficiency of 58.6% (±9.5%) (n = 12), while DNA-PKcs+/− and DNA-PKcs−/− cells had efficiencies of 35.4% (±3.9%) (n = 12) and 2.3% (±1.6%) (n = 12), respectively. Thus, DNA-PKcs+/− cell lines are haploinsufficient and DNA-PKcs−/− cell lines are profoundly defective for cell proliferation and survival at a low cell density.
DNA damage sensitivity.
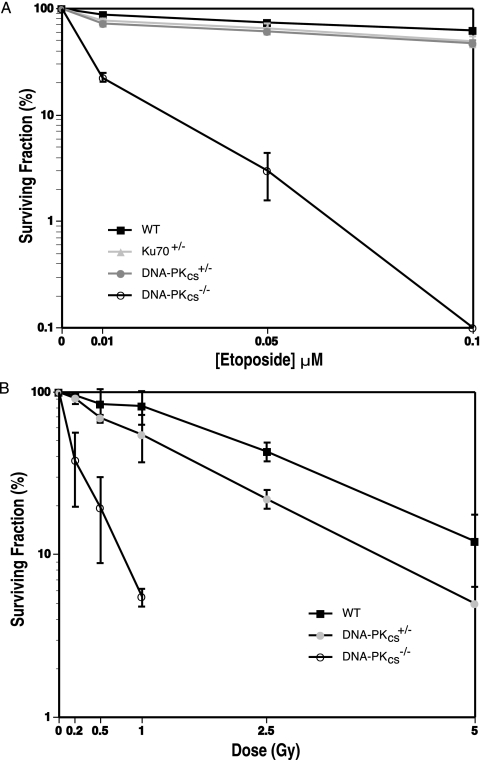
A universal role for DNA-PKcs is in DNA DSB repair, and lowered levels of DNA-PKcs would be expected to sensitize cells to DNA-damaging agents. Indeed, DNA-PKcs+/− cells were slightly sensitive to etoposide (Fig. 7A), a topoisomerase II poison and a potent inducer of DNA DSBs (54). The DNA-PKcs+/− cells were as sensitive as HCT116 Ku70+/− cells, a cell line known to be sensitive to agents causing DSBs (18). In contrast to these heterozygous cell lines, the DNA-PKcs−/− cells (even when plated at a 10-fold-higher concentration to accommodate the plating efficiency issue described above) were more than an order of magnitude more sensitive to etoposide than the parental cells (Fig. 7A). To confirm these studies, the sensitivity of the cell lines to IR was determined. The D37 (the dose required to reduce survival to 37%) for the parental cell line was 3.0 Gy, the D37 for the DNA-PKcs+/− line was 1.8 Gy, and the D37 for the DNA-PKcs−/− line was 0.2 Gy (Fig. 7B). Thus, DNA-PKcs-reduced and -deficient cells had slight and profound sensitivities, respectively, to agents that cause DNA DSBs.
FIG. 7.
DNA-PK-deficient cells are sensitive to DNA-damaging agents. (A) Etoposide sensitivity. For the parental HCT116 (WT) and Ku70+/− cells, 300 cells were seeded on tissue culture plates in duplicate and exposed to the indicated levels of etoposide. For DNA-PKcs+/− and DNA-PKcs−/− cells, 1,000 and 3,000 cells were used, respectively. Cells surviving to form colonies of at least 50 cells after 10 or 15 (DNA-PKcs−/− cells) days were scored. (B) IR sensitivity. For HCT116 WT and DNA-PKcs+/− cells, 300 cells were seeded on tissue culture plates in duplicate and X irradiated at the indicated doses. For DNA-PKcs−/− cells, 3,000 cells were treated similarly. Cells surviving to form colonies of at least 50 cells after ∼10 days were scored. Both profiles show the averages (±standard deviations) of at least two experiments.
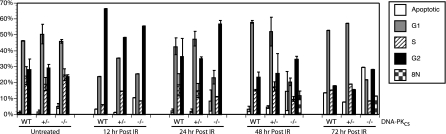
To extend this analysis, the effect of IR on cell cycle progression and checkpoint function in the mutant cell lines was examined. Asynchronous populations of the parental (WT), DNA-PKcs+/−, and DNA-PKcs−/− cells were either left untreated or exposed to 5 Gy of IR and then analyzed by FACS analysis at various times postirradiation (Fig. 8). Untreated populations of the three cell lines were virtually indistinguishable by FACS, with the exception of a slightly higher fraction of apoptotic cells in the DNA-PKcs−/− population. The responses of WT and DNA-PKcs+/− cells to IR were the same. Both cell lines showed a strong G2 arrest at 12 h postirradiation and then appeared to resume cell cycle progression such that by 72 h postirradiation the only obvious change from the untreated profiles was a higher fraction of apoptotic cells and a corresponding reduction in the G2 population. In contrast, DNA-PKcs−/− cells, although they also showed an initial G2 arrest, never appeared to recover from the IR exposure. In particular, a significant fraction of the cells appeared to become aneuploid within 48 h postirradiation, and by 72 h 45% of the total population had become either apoptotic or aneuploid (Fig. 8). From these experiments, we concluded that all of the cell lines initially respond normally to IR exposure. This conclusion was supported by the observation that all three cell lines showed robust phosphorylation of p53 at serine 15 following IR exposure (see Fig. S3 in the supplemental material), suggesting that the upstream checkpoint kinases were intact. We also concluded that although DNA-PKcs−/− cells initially respond normally to IR, they are severely impaired in exiting from a G2 arrest and instead engage either a cell death (apoptosis) or an aberrant mitotic (aneuploidy) program.
FIG. 8.
Cell cycle profiles of DNA-PKcs+/− and DNA-PKcs−/− cells following IR exposure. Parental HCT116 (WT), DNA-PKcs+/− (+/−), and DNA-PKcs−/− (−/−) cells were either left untreated or exposed to 5 Gy of IR and then analyzed for cell cycle distribution by propidium iodide staining and FACS analysis at the indicated times postirradiation. Error bars represent standard deviations. 8N, aneuploid.
Telomeric defects.
In humans, the inactivation of even one allele of either Ku70 or Ku86 generally leads to shortened telomeres (18, 34, 58), although this is not observed for all human cell lines (18, 75). To test whether this observation was also true for the HCT116 DNA-PKcs+/− and DNA-PKcs−/− cell lines, a TRF assay (50) was performed. Telomeric DNA, which is made up of only a T2AG3 repetitive sequence, is impervious to digestion by any restriction enzyme. Therefore, when genomic DNA is completely digested with frequent cutting restriction enzymes like AluI and MboI, the telomeric DNA remains largely intact while the remaining chromosomal DNA is reduced to small fragments. The telomeric DNA can then be detected with a radioactive d(C3TA2)3 probe in a Southern blotting procedure.
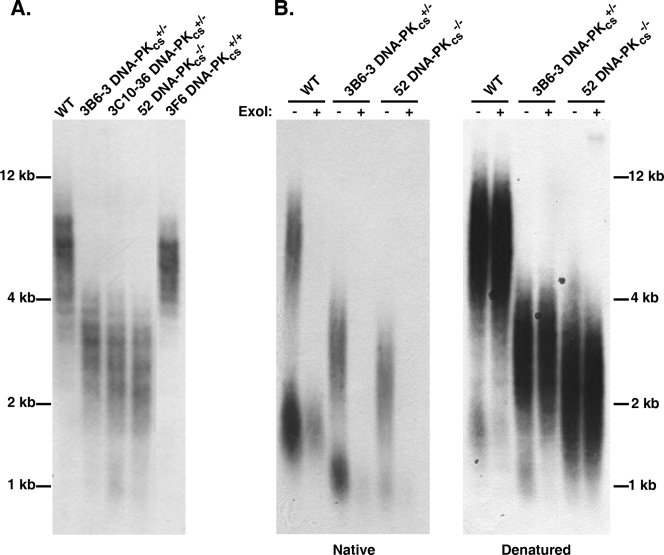
TRF analysis of DNA-PKcs+/− and DNA-PKcs−/− cells revealed a severe shortening of their telomeres similar to that observed for Ku70+/− and Ku86+/− cells, cell lines with documented telomere shortening (58). The wild-type HCT116 cells and the randomly targeted clone 3F6 showed telomeres ranging from ∼3 to 8 kb in length, with average sizes of 6.2 kb (±0.4 kb) (n = 2) and 5.7 kb (±0.3 kb) (n = 2), respectively. In contrast, the DNA-PKcs+/− clones 3B6 and 3C10 and the DNA-PKcs−/− cells had TRFs ranging from only 1 to 4 kb, with averages of 3.0 kb (±0.5 kb) (n = 2), 2.6 kb (±0.3 kb) (n = 2), and 2.6 kb (±0.1 kb) (n = 2), respectively (Fig. 9A). This profound telomeric shortening is likely causally linked to the reduction in DNA-PKcs expression, since when randomly targeted HCT116 clones were expanded from single cells, only ∼1 in 10 clones showed spontaneously shortened telomeres whereas the other ∼90% of the clones had telomeres that were either of the parental size or larger (see Fig. S4 in the supplemental material).
FIG. 9.
Telomere shortening in human DNA-PKcs+/− and DNA-PKcs−/− cell lines. (A) Genomic DNA was purified from the indicated cell lines, digested to completion with AluI and MboI, and then subjected to TRF Southern blot analysis under denaturing conditions with a (C3TA2)3 5′-end-radiolabeled oligonucleotide probe. Approximate molecular size markers are shown on the far left. (B) DNA-PKcs mutant cell lines have slightly elongated G overhangs. A modified TRF analysis in which one-half of each indicated sample was first exposed to exonuclease I (+ExoI) to remove any telomeric G overhangs was carried out. The samples were subsequently electrophoresed onto an agarose gel under nondenaturing conditions (Native) and subjected to an in-gel hybridization procedure with the (C3TA2)3 5′-end-radiolabeled oligonucleotide probe. This gel was subsequently denatured (Denatured) and rehybridized with the telomeric (C3TA2)3 5′-end-radiolabeled oligonucleotide probe. Molecular size markers are indicated on the far right.
Mutations in Ku affect the length of the G-strand telomeric overhang in Saccharomyces cerevisiae (7, 27) and Arabidopsis thaliana (66). Moreover, we have demonstrated previously that reduced Ku86 expression in HCT116 cells leads to slightly elongated G-strand overhangs (18, 58). To assess the length of the G-strand overhang in human DNA-PKcs mutant cells, we utilized a TRF analysis combined with nondenaturing gel electrophoresis (50). In a native (nondenaturing) gel, the single-stranded G overhang is the only substrate capable of hybridizing to the probe (Fig. 9B, “−” ExoI lanes). This interpretation was confirmed by prior treatment of the samples with exonuclease I (50), a single-stranded exonuclease, which removed the overhangs and eliminated the hybridization signal (Fig. 9B, “+” ExoI lanes). The total amount of telomeric DNA in each lane was quantitated by subsequently denaturing the gel and rehybridizing it with the probe (Fig. 9B). The ratio of the signal obtained from the native gel (i.e., a function of the length of the G overhang only) versus that obtained from the denatured gel (i.e., a function of the total telomere length) was arbitrarily set to 1.0 for the HCT116 cell line. Experimentally, the ratio for the 3B6 DNA-PKcs heterozygous clone was 2.9 (±0.3) (n = 2) while that for the DNA-PKcs-null clone was 1.5 (±0.3) (n = 2). Together, these experiments suggest that a deficiency in DNA-PKcs, like that of Ku86 (58), while causing overall telomere shortening, results in more telomeres with G-strand overhangs and/or telomeres with slightly longer G-strand overhangs.
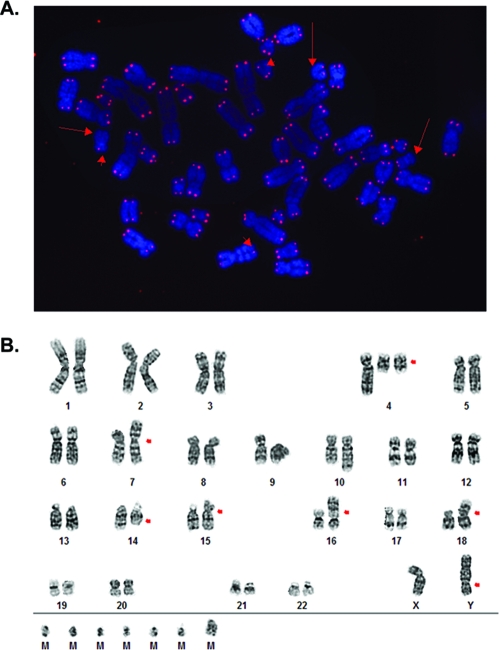
To independently confirm the telomere defect observed by TRF analysis, the parental HCT116, DNA-PKcs+/−, and DNA-PKcs−/− cell lines were arrested in metaphase, hybridized with a protein:nucleic acid telomere probe, and analyzed by FISH. Sixty-seven of the 7,168 chromatid ends examined in the parental HCT116 cells failed to hybridize to the probe (0.93% signal-free ends). In DNA-PKcs+/− cells, 82 of 3,684 telomeric ends examined were completely devoid of any telomeric signal (2.23% signal-free ends). Thus, an additional 1.30% of the telomeres in DNA-PKcs+/− cells were either entirely missing or so short that they could no longer be detected by this probe. This phenotype was exacerbated in the DNA-PKcs-null cell line, where 410 signal-free ends were detected from 11,020 chromatid ends analyzed (3.72%) (Fig. 10A). Thus, although the DNA-PKcs-null cell line had a TRF pattern indistinguishable from that of the DNA-PKcs+/− cell line, it appeared to have a more severe telomere length maintenance-defective phenotype.
FIG. 10.
Signal-free telomere ends and genomic instability in DNA-PKcs−/− cell lines. (A) FISH analysis with a telomere-specific Cy3-(C3TA2)3 protein-nucleic acid probe. Telomeres are seen as red dots, and metaphase chromosomes are stained blue. The arrows point to some of the chromosome ends where no discernible hybridization was detected. (B) G-band karyotype. The red arrows show instances of obvious aberrant chromosomal abnormalities. The translocations on chromosomes 16 and 18 originate from the parental cell line. M is a “Mars” or marker chromosome and is presumed to represent a chromosome fragment.
Genomic (in)stability.
Defects in telomere length maintenance have been shown to correlate with defects in genomic stability (reviewed in reference 48), although the majority of these studies have been carried out with nonhuman systems. In the few human somatic cell lines characterized to date for these two phenotypes, this correlation seems less absolute (18). To see whether the short/signalless telomere phenotype in the DNA-PK-defective cell lines correlated with genomic instability, the lines were cytogenetically analyzed by G banding. In these studies, the HCT116 parental cells had a slightly higher level of genetic instability than we (58) or others (1, 49) have previously reported, with 11.8% (14/120) of all metaphases examined containing some aberrant gross chromosomal rearrangements (GCRs), including translocations, duplications, or deletions. Nonetheless, the HCT116 DNA-PKcs+/− cell line showed a higher level of instability, as 30.0% (12/40) of the metaphases contained GCRs. Strikingly, the instability of the DNA-PKcs−/− cell line was extremely elevated, with 15/20 (75.0%) of the metaphases containing GCRs, including several with a high frequency of fragmented chromosomes (Fig. 10B; also see Fig. S5 in the supplemental material). Thus, the loss of one allele of DNA-PKcs appeared to result in increases in telomere shortening/uncapping and genomic instability, and these phenotypes were greatly accentuated by the loss of the second DNA-PKcs allele.
DISCUSSION
There has never been a human patient with a mutation in the catalytic subunit of the DNA-PK complex described, a surprising finding since the gene is not essential in many other vertebrate model systems (reviewed in reference 52). To experimentally cope with this dearth, researchers have relied upon inhibitors (some more specific than others) or RNA interference or have turned to a single human cell line, M059J. This cell line was isolated and characterized in 1993 (3). In the intervening 15 years, M059J has been a useful tool in numerous DNA repair studies. Despite this success, M059J and, correspondingly, its utility have been limited by the aberrant genetic constellation of the line. In addition to the mutation in DNA-PKcs, the cell line has mutations in p53 (4) and ATM (74) and has a highly aneuploid karyotype (5). Moreover, because of the spontaneous nature of the mutation it has been unclear whether the loss of DNA-PKcs was responsible for and/or whether the absence of DNA-PKcs could be tolerated only in such an aberrant genetic background. Given that both Ku subunits, which constitute the DNA binding portion of the DNA-PK complex, are essential in diploid human somatic cell lines (17, 44), it seemed likely that DNA-PKcs would also prove to be essential. Much to our surprise, we report here the isolation of a new human somatic cell line that appears to be null for DNA-PKcs.
The null status of this line was confirmed at the DNA (Fig. 2 and 3), protein and mRNA (Fig. 4), and functional (Fig. 5 to 8) levels. Importantly, because a graded series of isogenic lines progressing from WT to heterozygous to null now exists, a number of important conclusions can safely be drawn. First, the loss of even one allele of DNA-PKcs results in growth retardation, and the loss of both DNA-PKcs alleles results in a severe proliferation deficit. This is identical to the phenotype of human somatic cell lines heterozygous (18, 44) or null (44) for Ku. This is an important conclusion because, in the mouse, mutations only in Ku70 or in Ku86 (reviewed in reference 33) and not in DNA-PKcs (8, 9, 23, 35, 72) result in growth deficits. Thus, in this regard, human somatic cell lines are different from mice. Second, the loss of DNA-PKcs alleles appears to correlate with telomere shortening and genomic instability. TRF analysis demonstrated that the DNA-PKcs+/− and DNA-PKcs−/− cell lines had shortened telomeres with slightly elongated G overhangs, comparable to those observed for human Ku heterozygous cell lines (18, 58). Although it was anticipated that the DNA-PKcs−/− cell line would have even shorter telomeres than the DNA-PKcs+/− cell lines, this was not the case as judged by the TRF analysis (Fig. 9). This may be misleading, however, as the length observed for these cell lines appears to define a minimum telomere length for HCT116 cell lines that is compatible with viability (unpublished data). Consistent with this interpretation, FISH analysis using a telomere-specific probe to identify signal-free ends suggested that the telomere length maintenance defect was indeed significantly worse for the DNA-PKcs−/− cell line than for the DNA-PKcs+/− lines (Fig. 10A). Although the relationship between telomere shortening and genomic instability with regard to mutations of the Ku subunits is confusing (18), in the case of DNA-PKcs there appears to be a strong correlation. Thus, DNA-PKcs+/− lines had more signal-free telomeric ends than the parental HCT116 cell line and they were less stable. Similarly, the DNA-PKcs−/− cell line had even more signal-free telomeric ends than the DNA-PKcs+/− lines and they were correspondingly more unstable. Again, in this regard, human somatic cell lines are different from mice, as telomeres from DNA-PKcs-null mice have been reported to be slightly shorter (16), unchanged (15, 25, 26), or even longer (30) and to play a more important role in regulating telomeric fusions (6, 26). Third, we conclude that the regulation of p53 and cell cycle arrests are essentially normal and intact, respectively, in DNA-PKcs-deficient cell lines. Human cell lines heterozygous for either Ku70 (18) or Ku86 (44) show spontaneously elevated levels of the DNA damage regulator p53. Similarly, the DNA-PKcs+/− and DNA-PKcs−/− cell lines also have spontaneously elevated levels of activated p53 (see Fig. S3 in the supplemental material, nonirradiated lanes). We hypothesize that these DNA-PK-reduced/deficient cell lines have activated p53 due to telomere dysfunction and/or increased spontaneous DSB formation. Importantly, however, the cell lines nonetheless showed robust activation of p53 at serine 15 and initial normal checkpoint responses following IR exposure: the number of cells actively replicating decreased within 12 h and a large portion of the cells arrested in G2 within 24 h (Fig. 8). At this juncture, however, the fates of the cell lines diverged. The DNA-PKcs+/− line appeared to recover and the majority of the population again became asynchronous within 48 to 72 h (Fig. 8). In contrast, the DNA-PKcs-null cells became either polyploid or apoptotic. Thus, while DNA-PK activity is not required for a G2 checkpoint (2), it does appear to be required for cells to exit from G2 arrest. This is consistent with a bevy of previous reports (2, 13, 40, 41, 57, 59). In summary, in at least three regards—growth, telomere maintenance/genomic stability, and p53 activation/cell cycle checkpoints—the DNA-PKcs-reduced/deficient cell lines described here provide powerful tools to gain insight into how human cells cope with DNA damage.
Although we by no means wish to downplay the potential utility of the DNA-PKcs−/− cell line, it should still be stressed that there is nonetheless a strong selective bias against being null for DNA-PKcs. This is empirically evident in the vastly reduced doubling time of the DNA-PKcs−/− cell line (Fig. 6) and its extremely low plating efficiency. Such characteristics would mean that in a mixed population, DNA-PKcs−/− cells would quickly become outcompeted by either the DNA-PKcs+/− lines or the parental cells. Furthermore, the great asymmetry observed in the second round of gene targeting, in which 16 of 17 events were retargetings and only a single null event occurred, also speaks to this point. There is overwhelming evidence to support the view that targeting vectors integrate at the paternal and maternal alleles without bias. In an experiment designed to disrupt β-catenin, 11 second-round targeting events occurred on the functional allele and 15 on the inactive allele (36). Similarly, in two independent studies involving Ki-ras, four of seven (70) and 9 of 15 (37) targeting events occurred on the functional allele. Moreover, in rAAV-mediated targeting studies, 10 of 19 targeting events for COL1A1 occurred on the wild-type allele and 9 of 19 on the mutant allele (11). Because of this lack of bias in the targeting methodology, the striking disequilibrium of 16 out of 17 retargeting events is strong evidence that many DNA-PKcs−/− cell clones died or grew so slowly that they were not recovered in the experimental time frame. Nonetheless, the isolation of the DNA-PKcs−/− cell clone reported here demonstrates that human somatic cells can tolerate the loss of DNA-PKcs better than they can tolerate the loss of Ku. We presume this is because Ku contains a dsDNA end binding activity (see Fig. S2 in the supplemental material) that appears to be unique in mammals whereas DNA-PKcs is one of six members of a related family of phosphatidylinositol 3-kinase-related protein kinases (PIKKs). Presumably, one of the other PIKK family members can substitute, at low efficiency, for the absence of DNA-PKcs in our DNA-PKcs-null cell line.
It is worth noting that our initial knockout attempts, in which N-terminally located exons were targeted, were uniformly unsuccessful. The fact that exon 1 could not be targeted can perhaps be explained by the close proximity of DNA-PKcs exon 1 to the promoter for MCM4, an essential DNA replication factor (68). It is less clear why attempts to target exon 10 were unsuccessful. Although some of the mouse knockouts involved functional inactivation of N-terminally located exons (23, 35), almost all of the naturally occurring spontaneous mutations in DNA-PKcs occur exclusively in the C-terminal half of the protein and generally in the extreme C terminus (9, 14, 22, 63, 69, 80). This circumstantial evidence suggests that the N-terminal portion of DNA-PKcs may actually encode the essential region of the protein and that perhaps small, generally undetectable amounts of a C-terminally truncated protein may keep the cells/animals alive. Intriguingly, TEL2 (hClk2/TELO2), an essential gene, was recently shown to regulate the stability of all six PIKKs and to interact with the HEAT (huntingtin, elongation factor 3, alpha regulatory subunit of protein phosphatase 2A, and target of rapamycin 1) repeats found in the N-terminal region of DNA-PKcs and the other five PIKKs (73). If the stability between DNA-PKcs and TEL2 were reciprocal, this might explain a requirement for the N terminus of DNA-PKcs. With this hypothesis in mind, we have searched hard for even hints of N-terminally truncated DNA-PKcs protein, but these attempts have been uniformly unsuccessful.
In conclusion, we have demonstrated that human adenocarcinoma HCT116 cell lines heterozygous for DNA-PKcs are haploinsufficient and that, under cell culture conditions, DNA-PKcs is nonessential. Because not all cell types may have the same requirements for NHEJ, we are currently performing the same targeting strategy with NALM-6 and BJ cells immortalized with telomerase to determine whether they are viable as well when null for DNA-PKcs.
Supplementary Material
Acknowledgments
The FISH and G-banding cytogenetic analyses were performed by LeAnn Oseth in the Cytogenetics Core Laboratory at the University of Minnesota, with support from Comprehensive Cancer Center NIH grant no. P30 CA077598-09. This work has been supported in part by grants HL079559 and GM069576 from the NIH to E.A.H.
We thank Anja-Katrin Bielinsky and members of our laboratory for their helpful comments on the manuscript. We are indebted to Bert Vogelstein (The Johns Hopkins University, Baltimore, MD) and the many members of his laboratory who were very generous with their reagents and advice.
Footnotes
Published ahead of print on 18 August 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abdel-Rahman, W. M., K. Katsura, W. Rens, P. A. Gorman, D. Sheer, D. Bicknell, W. F. Bodmer, M. J. Arends, A. H. Wyllie, and P. A. Edwards. 2001. Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement. Proc. Natl. Acad. Sci. USA 982538-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allalunis-Turner, J., G. M. Barron, and R. S. Day III. 1997. Intact G2-phase checkpoint in cells of a human cell line lacking DNA-dependent protein kinase activity. Radiat. Res. 147284-287. [PubMed] [Google Scholar]
- 3.Allalunis-Turner, M. J., G. M. Barron, R. S. Day III, K. D. Dobler, and R. Mirzayans. 1993. Isolation of two cell lines from a human malignant glioma specimen differing in sensitivity to radiation and chemotherapeutic drugs. Radiat. Res. 134349-354. [PubMed] [Google Scholar]
- 4.Anderson, C. W., and M. J. Allalunis-Turner. 2000. Human TP53 from the malignant glioma-derived cell lines M059J and M059K has a cancer-associated mutation in exon 8. Radiat. Res. 154473-476. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, C. W., J. J. Dunn, P. I. Freimuth, A. M. Galloway, and M. J. Allalunis-Turner. 2001. Frameshift mutation in PRKDC, the gene for DNA-PKcs, in the DNA repair-defective, human, glioma-derived cell line M059J. Radiat. Res. 1562-9. [DOI] [PubMed] [Google Scholar]
- 6.Bailey, S. M., M. A. Brenneman, J. Halbrook, J. A. Nickoloff, R. L. Ullrich, and E. H. Goodwin. 2004. The kinase activity of DNA-PK is required to protect mammalian telomeres. DNA Repair 3225-233. [DOI] [PubMed] [Google Scholar]
- 7.Bertuch, A. A., and V. Lundblad. 2003. The Ku heterodimer performs separable activities at double-strand breaks and chromosome termini. Mol. Cell. Biol. 238202-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogue, M. A., C. Jhappan, and D. B. Roth. 1998. Analysis of variable (diversity) joining recombination in DNA-dependent protein kinase (DNA-PK)-deficient mice reveals DNA-PK-independent pathways for both signal and coding joint formation. Proc. Natl. Acad. Sci. USA 9515559-15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosma, G. C., R. P. Custer, and M. J. Bosma. 1983. A severe combined immunodeficiency mutation in the mouse. Nature 301527-530. [DOI] [PubMed] [Google Scholar]
- 10.Buck, D., L. Malivert, R. de Chasseval, A. Barraud, M. C. Fondaneche, O. Sanal, A. Plebani, J. L. Stephan, M. Hufnagel, F. le Deist, A. Fischer, A. Durandy, J. P. de Villartay, and P. Revy. 2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124287-299. [DOI] [PubMed] [Google Scholar]
- 11.Chamberlain, J. R., U. Schwarze, P. R. Wang, R. K. Hirata, K. D. Hankenson, J. M. Pace, R. A. Underwood, K. M. Song, M. Sussman, P. H. Byers, and D. W. Russell. 2004. Gene targeting in stem cells from individuals with osteogenesis imperfecta. Science 3031198-1201. [DOI] [PubMed] [Google Scholar]
- 12.Collis, S. J., T. L. DeWeese, P. A. Jeggo, and A. R. Parker. 2005. The life and death of DNA-PK. Oncogene 24949-961. [DOI] [PubMed] [Google Scholar]
- 13.Darroudi, F., and A. T. Natarajan. 1987. Cytological characterization of Chinese hamster ovary X-ray-sensitive mutant cells xrs 5 and xrs 6. I. Induction of chromosomal aberrations by X-irradiation and its modulation with 3-aminobenzamide and caffeine. Mutat. Res. 177133-148. [DOI] [PubMed] [Google Scholar]
- 14.Ding, Q., L. Bramble, V. Yuzbasiyan-Gurkan, T. Bell, and K. Meek. 2002. DNA-PKcs mutations in dogs and horses: allele frequency and association with neoplasia. Gene 283263-269. [DOI] [PubMed] [Google Scholar]
- 15.Espejel, S., S. Franco, A. Sgura, D. Gae, S. M. Bailey, G. E. Taccioli, and M. A. Blasco. 2002. Functional interaction between DNA-PKcs and telomerase in telomere length maintenance. EMBO J. 216275-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espejel, S., M. Martin, P. Klatt, J. Martin-Caballero, J. M. Flores, and M. A. Blasco. 2004. Shorter telomeres, accelerated ageing and increased lymphoma in DNA-PKcs-deficient mice. EMBO Rep. 5503-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fattah, F. J., N. F. Lichter, K. R. Fattah, S. Oh, and E. A. Hendrickson. 2008. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proc. Natl. Acad. Sci. USA 1058703-8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fattah, K. R., B. L. Ruis, and E. A. Hendrickson. 2008. Mutations to Ku reveal differences in human somatic cell lines. DNA Repair 7762-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira, M. G., K. M. Miller, and J. P. Cooper. 2004. Indecent exposure: when telomeres become uncapped. Mol. Cell 137-18. [DOI] [PubMed] [Google Scholar]
- 20.Finnie, N. J., T. M. Gottlieb, T. Blunt, P. A. Jeggo, and S. P. Jackson. 1995. DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 92320-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujimori, A., R. Araki, R. Fukumura, T. Saito, M. Mori, K. Mita, K. Tatsumi, and M. Abe. 1997. The murine DNA-PKcs gene consists of 86 exons dispersed in more than 250 kb. Genomics 45194-199. [DOI] [PubMed] [Google Scholar]
- 22.Fukumura, R., R. Araki, A. Fujimori, M. Mori, T. Saito, F. Watanabe, M. Sarashi, H. Itsukaichi, K. Eguchi-Kasai, K. Sato, K. Tatsumi, and M. Abe. 1998. Murine cell line SX9 bearing a mutation in the DNA-PKcs gene exhibits aberrant V(D)J recombination not only in the coding joint but also in the signal joint. J. Biol. Chem. 27313058-13064. [DOI] [PubMed] [Google Scholar]
- 23.Gao, Y., J. Chaudhuri, C. Zhu, L. Davidson, D. T. Weaver, and F. W. Alt. 1998. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for Ku in V(D)J recombination. Immunity 9367-376. [DOI] [PubMed] [Google Scholar]
- 24.Gao, Y., Y. Sun, K. M. Frank, P. Dikkes, Y. Fujiwara, K. J. Seidl, J. M. Sekiguchi, G. A. Rathbun, W. Swat, J. Wang, R. T. Bronson, B. A. Malynn, M. Bryans, C. Zhu, J. Chaudhuri, L. Davidson, R. Ferrini, T. Stamato, S. H. Orkin, M. E. Greenberg, and F. W. Alt. 1998. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 95891-902. [DOI] [PubMed] [Google Scholar]
- 25.Gilley, D., H. Tanaka, M. P. Hande, A. Kurimasa, G. C. Li, M. Oshimura, and D. J. Chen. 2001. DNA-PKcs is critical for telomere capping. Proc. Natl. Acad. Sci. USA 9815084-15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goytisolo, F. A., E. Samper, S. Edmonson, G. E. Taccioli, and M. A. Blasco. 2001. The absence of the DNA-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol. Cell. Biol. 213642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gravel, S., M. Larrivee, P. Labrecque, and R. J. Wellinger. 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280741-744. [DOI] [PubMed] [Google Scholar]
- 28.Han, Z., T. H. Carter, W. H. Reeves, J. H. Wyche, and E. A. Hendrickson. 1996. DNA-dependent protein kinase is a target of a CPP32-like apoptotic protease. J. Biol. Chem. 27125035-25040. [DOI] [PubMed] [Google Scholar]
- 29.Han, Z., C. Johnston, W. H. Reeves, T. Carter, J. H. Wyche, and E. A. Hendrickson. 1996. Characterization of a Ku86 variant protein that results in altered DNA binding and diminished DNA-dependent protein kinase activity. J. Biol. Chem. 27114098-14104. [DOI] [PubMed] [Google Scholar]
- 30.Hande, P., P. Slijepcevic, A. Silver, S. Bouffler, P. van Buul, P. Bryant, and P. Lansdorp. 1999. Elongated telomeres in scid mice. Genomics 56221-223. [DOI] [PubMed] [Google Scholar]
- 31.Hefferin, M. L., and A. E. Tomkinson. 2005. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair 4639-648. [DOI] [PubMed] [Google Scholar]
- 32.Hendrickson, E. A. 2008. Gene targeting in human somatic cells, p. 509-525. In P. M. Conn (ed.), Source book of models for biomedical research. Humana Press, Inc., Totowa, NJ.
- 33.Hendrickson, E. A., J. L. Huffman, and J. A. Tainer. 2006. Structural aspects of Ku and the DNA-dependent protein kinase complex, p. 629-684. In W. Seide, Y. W. Kow, and P. Doetsch (ed.), DNA damage recognition. Taylor and Francis Group, New York, NY.
- 34.Jaco, I., P. Munoz, and M. A. Blasco. 2004. Role of human Ku86 in telomere length maintenance and telomere capping. Cancer Res. 647271-7278. [DOI] [PubMed] [Google Scholar]
- 35.Jhappan, C., H. C. Morse III, R. D. Fleischmann, M. M. Gottesman, and G. Merlino. 1997. DNA-PKcs: a T-cell tumour suppressor encoded at the mouse scid locus. Nat. Genet. 17483-486. [DOI] [PubMed] [Google Scholar]
- 36.Kim, J. S., H. Crooks, T. Dracheva, T. G. Nishanian, B. Singh, J. Jen, and T. Waldman. 2002. Oncogenic beta-catenin is required for bone morphogenetic protein 4 expression in human cancer cells. Cancer Res. 622744-2748. [PubMed] [Google Scholar]
- 37.Kim, J. S., C. Lee, A. Foxworth, and T. Waldman. 2004. B-Raf is dispensable for K-Ras-mediated oncogenesis in human cancer cells. Cancer Res. 641932-1937. [DOI] [PubMed] [Google Scholar]
- 38.Kirchgessner, C. U., C. K. Patil, J. W. Evans, C. A. Cuomo, L. M. Fried, T. Carter, M. A. Oettinger, and J. M. Brown. 1995. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science 2671178-1183. [DOI] [PubMed] [Google Scholar]
- 39.Kohli, M., C. Rago, C. Lengauer, K. W. Kinzler, and B. Vogelstein. 2004. Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses. Nucleic Acids Res. 32e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, S. E., R. A. Mitchell, A. Cheng, and E. A. Hendrickson. 1997. Evidence for DNA-PK-dependent and -independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol. Cell. Biol. 171425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94399-409. [DOI] [PubMed] [Google Scholar]
- 42.Lee, S. E., C. R. Pulaski, D. M. He, D. M. Benjamin, M. Voss, J. Um, and E. A. Hendrickson. 1995. Isolation of mammalian cell mutants that are X-ray sensitive, impaired in DNA double-strand break repair and defective for V(D)J recombination. Mutat. Res. 336279-291. [DOI] [PubMed] [Google Scholar]
- 43.Lees-Miller, S. P., R. Godbout, D. W. Chan, M. Weinfeld, R. S. Day III, G. M. Barron, and J. Allalunis-Turner. 1995. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science 2671183-1185. [DOI] [PubMed] [Google Scholar]
- 44.Li, G., C. Nelsen, and E. A. Hendrickson. 2002. Ku86 is essential in human somatic cells. Proc. Natl. Acad. Sci. USA 99832-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li, G. C., H. Ouyang, X. Li, H. Nagasawa, J. B. Little, D. J. Chen, C. C. Ling, Z. Fuks, and C. Cordon-Cardo. 1998. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol. Cell 21-8. [DOI] [PubMed] [Google Scholar]
- 46.Li, X., and W. D. Heyer. 2008. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 1899-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lieber, M. R., Y. Ma, Y. Kefei, U. Pannicke, and K. Schwarz. 2006. The mechanism of vertebrate nonhomologous DNA end joining and its role in immune system gene rearrangements, p. 609-627. In W. Seide, Y. W. Kow, and P. Doetsch (ed.), DNA damage recognition. Taylor and Francis Group, New York, NY.
- 48.Maser, R. S., and R. A. DePinho. 2002. Connecting chromosomes, crisis, and cancer. Science 297565-569. [DOI] [PubMed] [Google Scholar]
- 49.Masramon, L., M. Ribas, P. Cifuentes, R. Arribas, F. Garcia, J. Egozcue, M. A. Peinado, and R. Miro. 2000. Cytogenetic characterization of two colon cell lines by using conventional G-banding, comparative genomic hybridization, and whole chromosome painting. Cancer Genet. Cytogenet. 12117-21. [DOI] [PubMed] [Google Scholar]
- 50.McElligott, R., and R. J. Wellinger. 1997. The terminal DNA structure of mammalian chromosomes. EMBO J. 163705-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGuire, T. C., and M. J. Poppie. 1973. Hypogammaglobulinemia and thymic hypoplasia in horses: a primary combined immunodeficiency disorder. Infect. Immunol. 8272-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meek, K., S. Gupta, D. A. Ramsden, and S. P. Lees-Miller. 2004. The DNA-dependent protein kinase: the director at the end. Immunol. Rev. 200132-141. [DOI] [PubMed] [Google Scholar]
- 53.Meek, K., L. Kienker, C. Dallas, W. Wang, M. J. Dark, P. J. Venta, M. L. Huie, R. Hirschhorn, and T. Bell. 2001. SCID in Jack Russell terriers: a new animal model of DNA-PKcs deficiency. J. Immunol. 1672142-2150. [DOI] [PubMed] [Google Scholar]
- 54.Montecucco, A., and G. Biamonti. 2007. Cellular response to etoposide treatment. Cancer Lett. 2529-18. [DOI] [PubMed] [Google Scholar]
- 55.Morozov, V. E., M. Falzon, C. W. Anderson, and E. L. Kuff. 1994. DNA-dependent protein kinase is activated by nicks and larger single-stranded gaps. J. Biol. Chem. 26916684-16688. [PubMed] [Google Scholar]
- 56.Moshous, D., I. Callebaut, R. de Chasseval, B. Corneo, M. Cavazzana-Calvo, F. Le Deist, I. Tezcan, O. Sanal, Y. Bertrand, N. Philippe, A. Fischer, and J. P. de Villartay. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105177-186. [DOI] [PubMed] [Google Scholar]
- 57.Munoz, P., M. Z. Zdzienicka, J. M. Blanchard, and J. Piette. 1998. Hypersensitivity of Ku-deficient cells toward the DNA topoisomerase II inhibitor ICRF-193 suggests a novel role for Ku antigen during the G2 and M phases of the cell cycle. Mol. Cell. Biol. 185797-5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Myung, K., G. Ghosh, F. J. Fattah, G. Li, H. Kim, A. Dutia, E. Pak, S. Smith, and E. A. Hendrickson. 2004. Regulation of telomere length and suppression of genomic instability in human somatic cells by Ku86. Mol. Cell. Biol. 245050-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nussenzweig, A., C. Chen, V. da Costa Soares, M. Sanchez, K. Sokol, M. C. Nussenzweig, and G. C. Li. 1996. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 382551-555. [DOI] [PubMed] [Google Scholar]
- 60.O'Driscoll, M., K. M. Cerosaletti, P. M. Girard, Y. Dai, M. Stumm, B. Kysela, B. Hirsch, A. Gennery, S. E. Palmer, J. Seidel, R. A. Gatti, R. Varon, M. A. Oettinger, H. Neitzel, P. A. Jeggo, and P. Concannon. 2001. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol. Cell 81175-1185. [DOI] [PubMed] [Google Scholar]
- 61.Ouyang, H., A. Nussenzweig, A. Kurimasa, V. C. Soares, X. Li, C. Cordon-Cardo, W. Li, N. Cheong, M. Nussenzweig, G. Iliakis, D. J. Chen, and G. C. Li. 1997. Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination in vivo. J. Exp. Med. 186921-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peterson, S. R., A. Kurimasa, M. Oshimura, W. S. Dynan, E. M. Bradbury, and D. J. Chen. 1995. Loss of the 350kD catalytic subunit of the DNA dependent protein kinase in DNA double-strand break repair mutant mammalian cells. Proc. Natl. Acad. Sci. USA 923171-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Priestley, A., H. J. Beamish, D. Gell, A. G. Amatucci, M. C. Muhlmann-Diaz, B. K. Singleton, G. C. Smith, T. Blunt, L. C. Schalkwyk, J. S. Bedford, S. P. Jackson, P. A. Jeggo, and G. E. Taccioli. 1998. Molecular and biochemical characterisation of DNA-dependent protein kinase-defective rodent mutant irs-20. Nucleic Acids Res. 261965-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rathmell, W. K., and G. Chu. 1994. A DNA end-binding factor involved in double-strand break repair and V(D)J recombination. Mol. Cell. Biol. 144741-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Riballo, E., S. E. Critchlow, S.-H. Teo, A. J. Doherty, A. Priestley, B. Broughton, B. Kysela, H. Beamish, N. Plowman, C. F. Arlett, A. R. Lehmann, S. J. Jackson, and P. A. Jeggo. 1999. Identification of a defect in DNA ligase IV in a radiosensitive leukemia patient. Curr. Biol. 9699-702. [DOI] [PubMed] [Google Scholar]
- 66.Riha, K., and D. E. Shippen. 2003. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc. Natl. Acad. Sci. USA 100611-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sekiguchi, J. M., and D. O. Ferguson. 2006. DNA double-strand break repair: a relentless hunt uncovers new prey. Cell 124260-262. [DOI] [PubMed] [Google Scholar]
- 68.Shima, N., A. Alcaraz, I. Liachko, T. R. Buske, C. A. Andrews, R. J. Munroe, S. A. Hartford, B. K. Tye, and J. C. Schimenti. 2007. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat. Genet. 3993-98. [DOI] [PubMed] [Google Scholar]
- 69.Shin, E. K., L. E. Perryman, and K. Meek. 1997. A kinase-negative mutation of DNA-PKcs in equine SCID results in defective coding and signal joint formation. J. Immunol. 1583565-3569. [PubMed] [Google Scholar]
- 70.Shirasawa, S., M. Furuse, N. Yokoyama, and T. Sasazuki. 1993. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science 26085-88. [DOI] [PubMed] [Google Scholar]
- 71.Soulas-Sprauel, P., P. Rivera-Munoz, L. Malivert, G. Le Guyader, V. Abramowski, P. Revy, and J. P. de Villartay. 2007. V(D)J and immunoglobulin class switch recombinations: a paradigm to study the regulation of DNA end-joining. Oncogene 267780-7791. [DOI] [PubMed] [Google Scholar]
- 72.Taccioli, G. E., A. G. Amatucci, H. J. Beamish, D. Gell, X. H. Xiang, M. I. Torres Arzayus, A. Priestley, S. P. Jackson, A. Marshak Rothstein, P. A. Jeggo, and V. L. Herrera. 1998. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity 9355-366. [DOI] [PubMed] [Google Scholar]
- 73.Takai, H., R. C. Wang, K. K. Takai, H. Yang, and T. de Lange. 2007. Tel2 regulates the stability of PI3K-related protein kinases. Cell 1311248-1259. [DOI] [PubMed] [Google Scholar]
- 74.Tsuchida, R., T. Yamada, M. Takagi, A. Shimada, C. Ishioka, Y. Katsuki, T. Igarashi, L. Chessa, D. Delia, H. Teraoka, and S. Mizutani. 2002. Detection of ATM gene mutation in human glioma cell line M059J by a rapid frameshift/stop codon assay in yeast. Radiat. Res. 158195-201. [DOI] [PubMed] [Google Scholar]
- 75.Uegaki, K., N. Adachi, S. So, S. Iiizumi, and H. Koyama. 2006. Heterozygous inactivation of human Ku70/Ku86 heterodimer does not affect cell growth, double-strand break repair, or genome integrity. DNA Repair 5303-311. [DOI] [PubMed] [Google Scholar]
- 76.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92401-413. [DOI] [PubMed] [Google Scholar]
- 77.van Veelen, L., J. Wesoly, and R. Kanaar. 2006. Biochemical and cellular aspects of homologous recombination, p. 581-607. In W. Seide, Y. W. Kow, and P. Doetsch (ed.), DNA damage recognition. Taylor and Francis Group, New York, NY.
- 78.Vasileva, A., and R. Jessberger. 2005. Precise hit: adeno-associated virus in gene targeting. Nat. Rev. Microbiol. 3837-847. [DOI] [PubMed] [Google Scholar]
- 79.Wiler, R., R. Leber, B. B. Moore, L. F. VanDyk, L. E. Perryman, and K. Meek. 1995. Equine severe combined immunodeficiency: a defect in V(D)J recombination and DNA-dependent protein kinase activity. Proc. Natl. Acad. Sci. USA 9211485-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Woods, T., W. Wang, E. Convery, A. Errami, M. Z. Zdzienicka, and K. Meek. 2002. A single amino acid substitution in DNA-PKcs explains the novel phenotype of the CHO mutant, XR-C2. Nucleic Acids Res. 305120-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zdzienicka, M. Z. 1999. Mammalian X-ray-sensitive mutants which are defective in non-homologous (illegitimate) DNA double-strand break repair. Biochimie 81107-116. [DOI] [PubMed] [Google Scholar]
- 82.Zhu, C., M. A. Bogue, D. S. Lim, P. Hasty, and D. B. Roth. 1996. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell 86379-389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.