Abstract
There are seven SIRT isoforms in mammals, with diverse biological functions including gene regulation, metabolism, and apoptosis. Among them, SIRT3 is the only sirtuin whose increased expression has been shown to correlate with an extended life span in humans. In this study, we examined the role of SIRT3 in murine cardiomyocytes. We found that SIRT3 is a stress-responsive deacetylase and that its increased expression protects myocytes from genotoxic and oxidative stress-mediated cell death. We show that, like human SIRT3, mouse SIRT3 is expressed in two forms, a ∼44-kDa long form and a ∼28-kDa short form. Whereas the long form is localized in the mitochondria, nucleus, and cytoplasm, the short form is localized exclusively in the mitochondria of cardiomyocytes. During stress, SIRT3 levels are increased not only in mitochondria but also in the nuclei of cardiomyocytes. We also identified Ku70 as a new target of SIRT3. SIRT3 physically binds to Ku70 and deacetylates it, and this promotes interaction of Ku70 with the proapoptotic protein Bax. Thus, under stress conditions, increased expression of SIRT3 protects cardiomyocytes, in part by hindering the translocation of Bax to mitochondria. These studies underscore an essential role of SIRT3 in the survival of cardiomyocytes in stress situations.
Mammalian cardiomyocytes are mostly terminally differentiated cells. They lose their proliferation capability soon after birth and undergo hypertrophic growth in response to various stress stimuli (14). During hypertrophy of cardiomyocytes, a continuous growth signal at some point causes cells to malfunction and leads to cell death. As cells die, the workload of the remaining cells increases, which further aggravates this process and eventually leads to heart failure. The molecular mechanism of myocyte death during heart failure is not yet fully understood (15). In regards to the activation of caspases, it has been shown that caspase inhibitors capable of preventing myocyte death improve the functioning of the ischemic heart. However, in many other pathological conditions cell death has been seen occurring independently of caspase activation (6, 13). Therefore, an understanding of the mechanisms regulating cardiomyocyte cell survival and/or death pathways remains an important goal in cardiac cell biology.
SIRT3 is a member of class III of histone deacetylases (HDACs), also called sirtuins (SIRTs). The sirtuin family members are catalytically distinct from other HDACs, as they require NAD for their deacetylase activity (19). SIRTs are implicated in transcriptional silencing, genetic control of aging, and calorie restriction-mediated longevity of organisms ranging from yeasts to humans (19, 29). It has been suggested that the phylogenetically conserved family of SIRT proteins is involved in sensing cellular energy and the redox state (34). During stress, a change in the metabolic state of the cell (NAD/NADH ratio) alters the deacetylase activity of SIRTs, and this has been proposed to influence cell survival (4). Increased cellular NAD content elevates the deacetylase activity of SIRTs, whereas high nicotinamide (NAM) and/or NADH levels inhibit these enzymes (4). The role of SIRT1, a founding member of this family, has been examined in cardiomyocytes, and the results indicated that overexpression of SIRT1 protects cells from oxidative stress-mediated cell death and age-related degeneration of cardiomyocytes (1, 26). In transgenic mice, however, overexpression of SIRT1 had a dose-dependent effect: it protected hearts at low doses but exacerbated stress-mediated cell death and fibrosis at higher doses, indicating that SIRT1 has a very narrow window of activity in the heart (1). Recently, another member of the SIRT family, SIRT7, was shown to protect cardiomyocytes against oxidative stress and inflammatory outbursts (35). Both SIRT1 and SIRT7 have been found to deacetylate p53 in cardiomyocytes, and this mechanism was implicated in the ability of these proteins to create cell resistance to stress (26, 35). The role of other SIRT isoforms in cardiomyocytes is, however, unknown at present.
Seven mammalian SIRT isoforms (SIRT1 to SIRT7) have been identified (24). SIRT1, SIRT6, and SIRT7 were shown to be localized primarily in the nucleus, whereas SIRT2 was present in the cytoplasm and SIRT3, SIRT4, and SIRT5 were present in mitochondria (20). A recent study, however, has shown that human SIRT3 (hSIRT3) is localized not only in mitochondria but also in the nucleus (31). In this study it was pointed out that, during normal cell growth, nuclear SIRT3 enters into mitochondria upon cellular stress, induced by its own overexpression or by UV damage or treatment with etoposide (31).
Initially, it was reported that full-length hSIRT3 (∼44 kDa) is an inert protein and that it is activated inside the mitochondria following deletion of 142 amino acids of the N-terminal segment. This processed form of SIRT3 is approximately 28 kDa and possesses deacetylase activity (32). In a recent study, however, it was shown that full-length hSIRT3 is present in the nucleus and also possesses deacetylase activity and that it is capable of deacetylating H3 and H4 (31). SIRT3 expression has been shown to be activated by calorie restriction, and increased expression of mouse SIRT3 (mSIRT3) in adipocytes was found to be capable of inducing the expression of genes involved in mitochondrial function and biogenesis, thus again linking SIRT3 concentration to the regulation of nuclear gene expression (33). Furthermore, both mitochondrial targeting and nuclear localization signal sequences have been identified in mSIRT3 (22). Within mitochondria SIRT3 was found to be capable of deacetylating and activating the enzyme acetyl coenzyme A-synthetase 2, thus suggesting an important role of the deacetylase in this organelle (12). From these studies, it appears that SIRT3 plays a role both in the nucleus and in the mitochondria depending upon cellular stress (12, 31, 32).
SIRT3-null mice have been created (17). Unlike SIRT1-knockout mice, which die before maturation with defects in the heart and other organs, SIRT3-knockout mice are alive and show no apparent signs of developmental abnormality but display massive acetylation of mitochondrial proteins (1, 17). Based on the results with SIRT3-null mice, it has been proposed that SIRT3 perhaps plays a role under stress conditions but not in the development of the animal (17). Recently, in tumor cells, hSIRT3 was identified as a cell survival factor, which protects cells from genotoxic stress by utilizing the mitochondrial NAD pool; however, little else is currently known about the mechanism of SIRT3-mediated cell protection during stress (36).
This study was designed to examine the role of SIRT3 in cardiomyocytes. We show that SIRT3 is highly expressed in the heart and is localized not only in mitochondria but also in the nucleus and cytoplasm of cardiomyocytes. Overexpression of SIRT3 protects cells against stress-mediated cell death. We also identified Ku70 as a new target of SIRT3. Deacetylation of Ku70 by SIRT3 promotes Ku70/Bax interaction, and this makes cells resistant to Bax-mediated cell damage. These studies demonstrate a role of SIRT3 outside the mitochondria and suggest that it is a survival factor for cardiomyocytes under stress conditions.
MATERIALS AND METHODS
Antibodies used.
The following antibodies and conjugates were used in this study: rabbit anti-SIRT3 (ab56214 [Abcam], AP6242a [Abgent], and PAB-11098 [Orbigen]), goat anti-SIRT3 (sc-49744; Santa Cruz), rabbit anti-Ku70 (AB3742; Chemicon), goat anti-Ku70 (sc-1487; Santa Cruz), rabbit anti-Flag (ab1162; Abcam), rabbit anti-Bax (554104 [BD Biosciences], sc-526 [Santa Cruz], and ab7977 [Abcam]), rabbit antiacetyllysine (06-933; Upstate), mouse antiacetyllysine (Ac-K-103; Cell Signaling), rabbit anti-atrial natriuretic factor (anti-ANF; T-4014; Peninsula Laboratories), goat anti-HSP70 (sc-1060; Santa Cruz), mouse anti-poly(ADP-ribose) (Alx 804220; Alexis), goat anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH; sc-20357, Santa Cruz), mouse antihistones (MAB052; Chemicon), goat anti-RNA polymerase II (sc-5943; Santa Cruz), anti-acetyl-H3 (ab10812; Abcam), anti-alpha-tubulin (sc-8035; Santa Cruz), anti-Flag M2 affinity gel (A2220; Sigma), goat anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase (HRP) (sc-2054; Santa Cruz), donkey anti-mouse IgG-HRP (sc-2096; Santa Cruz), donkey anti-goat IgG-HRP (sc-2056; Santa Cruz), donkey anti-goat IgG-Alexa Fluor 594 (A11058; Invitrogen), donkey anti-rabbit IgG-Alexa Fluor 594 (A21207; Invitrogen), goat antiactin (sc-1616; Santa Cruz), and rabbit anti-Mn superoxide dismutase (anti-MnSOD; 06-984; Millipore). The blocking peptides used in the study were sc-49744 P (Santa Cruz) and BP6242a (Abgent).
Plasmid constructs.
The constructs Flag-SIRT3 (23, 32), Flag-SIRT3-HY (SIRT3 catalytic mutant [mt]) (32), Flag-SIRT1 and His-SIRT3 (23), Flag-Ku70 (30), pYFP and pYFP-Bax (7), and pcDNA-Bax (30) have been described previously. A plasmid (His-SIRT3119-398) encoding a His-tagged 30-kDa fragment of SIRT3 was from the lab of J. M. Denu (12). hSIRT3 adenovirus was from Vector BioLabs, Philadelphia, PA. The mt adenovirus vector, which synthesizes a benign protein, was designed in our lab. Plasmid His-p38d synthesizes a truncated His-tagged p38 having only 54 amino acids; the predicted molecular mass is ∼6 kDa. Plasmid CMV-hSIRT3Δ1-25, which encodes a SIRT3 deletion mt lacking mitochondrial import signal, was generated essentially as described earlier (32).
Cell culture, transfection, and adenovirus infection.
Primary cultures of 2-day-old neonatal rat heart myocytes were carried out using an established procedure described previously (26). All animal protocols were reviewed and approved by the University of Chicago Institutional Animal Care and Use Committee. At 24 to 30 hours after seeding, cells were used for adenovirus infection or immunostaining as described below. For all the adenovirus experiments viruses were used at a multiplicity of infection of 10. Cos7 and HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with penicillin-streptomycin and 10% fetal bovine serum (complete growth medium). Cells were transfected with appropriate plasmids using Superfect transfection reagent (Qiagen) according to the manufacturer's protocol. To generate SIRT3 or SIRT3-HY-overexpressing stable clones, pcDNA.SIRT3 or pcDNA.SIRT3-HY vectors were transfected into HeLa cells and selected with 0.5 mg/ml Geneticin. For the RNA interference experiments, cardiac fibroblasts or HeLa cells were transfected with 200 nM On-Target-plus small interfering RNA (siRNA) specific for hSIRT3 using Dharma-FECT transfection reagents according to the manufacturer's protocol. Similarly, SIRT1 was knocked down in wild-type (wt) SIRT3 or mt SIRT3 stable HeLa cells with 100 nM On-Target-plus siRNA specific for hSIRT1 using Dharma-FECT transfection reagents.
Cell stress experiments.
To serum starve cardiomyocytes, cells were grown in serum-free defined medium (Opti-MEM; Invitrogen). Cells were harvested after 48 h of serum starvation, and SIRT3 expression was analyzed by Western blotting. For inducing genotoxic or oxidative stress in cardiomyocytes, different concentrations of MNNG (N-methyl-N′-nitro-N-nitrosoguanidine) (10, 20, 30, and 40 μM) or H2O2 (20, 40, and 80 μM) were used for 24 h in low-serum medium. Cardiomyocytes were also exposed to 20 μM phenylephrine (PE) or 5 μM angiotensin II (Ang-II) for 48 h in low-serum medium and assayed for SIRT3 expression.
Cell death assays.
Cardiomyocytes and HeLa cells were treated with MNNG (500 μM), camptothecin (10 μM), or H2O2 (200 to 500 μM) for 6 h. For MNNG, cells were treated with 500 μM MNNG for 10 min and then washed and kept in the complete medium for 3 or 6 h. Cell death was analyzed by fluorescence-activated cell sorting (FACS) by using propidium iodide (PI) staining or Hoechst and PI staining as described before (26, 27). The cell death results were also confirmed by trypan blue cell viability assay (2). The percentages of apoptotic cells were also quantified using the annexin V-phycoerythrin apoptosis detection kit I (BD Pharmingen) after various chemical treatments. HeLa cells stably overexpressing wt SIRT3 or mt SIRT3 or subjected to SIRT3 knockdown were transfected with yellow fluorescent protein (YFP) (0.5 μg), YFP-Bax (0.5 μg), or YFP-Bax (0.5 μg) and Ku70 (1 μg) in each well of a 12-well tissue culture plate. The percentage of YFP-positive cells with apoptotic nuclei was scored 12 h after transfection as previously described (7, 30).
Subcellular fractionation, IP, and Western analyses.
Subcellular protein fractions of mouse hearts were prepared using a Proteoextract subcellular proteome extraction kit (Calbiochem) and the NE-PER nuclear and cytoplasmic extraction kit (Pierce) according to the manufacturer's protocol (10). Western blotting and immunoprecipitation (IP) experiments were done using a standard protocol as described elsewhere (10). Blocking peptide experiments were performed according to manufacturers' protocols.
In vitro acetylation-deacetylation assay.
Unlabeled Flag-Ku70 was in vitro translated using the TNT coupled transcription-translation rabbit reticulocyte lysate kit (Promega) and immunoprecipitated using anti-Flag M2 affinity beads. Beads with bound protein were washed four to five times with radioimmunoprecipitation assay buffer followed by a phosphate-buffered saline (PBS) wash. The final wash was performed in 1× HAT buffer (50 mM Tris, pH 8, 10% glycerol, 0.1 mM EDTA, 1 mM dithiothreitol). A typical acetylation reaction mixture contained 1 μg active p300/CBP-associated factor (PCAF) enzyme (Upstate Biotechnology), 0.3 mM acetyl coenzyme A (Sigma), and 10 mM sodium butyrate in 1× HAT buffer. Reaction mixtures were incubated at 30°C for 2 h on a rotator. For the deacetylation assay, acetylated Flag-Ku70 bound to beads was washed as described above and resuspended in 1× HDAC buffer (50 mM Tris, pH 8, 4 mM MgCl2, 0.2 mM dithiothreitol). The acetylated Flag-Ku70 substrate was either incubated only in 1× HDAC buffer (control) or with equal amounts of His-SIRT3 or His-SIRT3-HY (catalytic mt) with or without NAD (1 mM) in 1× HDAC buffer. His-SIRT3 or His-SIRT3-HY catalytic mt was produced in a prokaryotic expression system as described earlier (23). Reaction mixtures were incubated for 2 to 3 h at 37°C on a rotator. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting.
In vitro protein binding assay.
For in vitro binding assays, 20 μg of His-SIRT3 or His-p38d (a negative control) was captured on nickel-nitrilotriacetic acid beads. Briefly, the plasmids pQE-His-SIRT3-101-399 and pQE-His-p38d were expressed in bacteria and the cells were lysed in a lysis buffer containing 50 mM Tris-HCl (pH 8), 250 mM NaCl, 5 mM imidazole, and protease inhibitor cocktail (Sigma). The cleared supernatant was agitated with nickel-nitrilotriacetic acid resin for 2 h at 4°C. The resin was washed three times with the lysis buffer followed by three washes with PBS containing 1% Triton X-100 (1% PBST) and a final wash with PBS. The washed beads containing His-SIRT3, His-P38d, or empty beads were incubated at 4°C overnight on a rotator with the in vitro-translated [35S]methionine-labeled Ku70 in PBS containing protease inhibitor cocktail. Beads were then washed six times with 1% PBST followed by a rinse in PBS. Bound complexes were resolved by SDS-PAGE and detected by autoradiography. For studying Ku70 and Bax binding, in vitro acetylated or deacetylated Flag-Ku70 was incubated with HeLa cell-CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} buffer extract overnight at 4°C with rotation (30). Flag agarose beads with bound proteins were separated and washed four to five times with CHAPS buffer followed by a PBS rinse. Proteins were resolved by SDS-PAGE and subjected to Western blotting with Bax, Ku70, or acetyllysine antibodies.
Electron microscopy.
Freshly isolated mouse heart ventricular tissue was fixed in 0.2% glutaraldehyde and embedded in L.R. White resin. Postembedding staining of sections was done using primary antibody rabbit anti-SIRT3 (PAB-11098; Orbigen) or nonspecific IgG. Primary antibody staining was detected using goat anti-rabbit IgG conjugated to 10-nm gold beads (Ted Pella Inc.). Transmission electron microscopy and imaging were carried out at the electron microscopy core facility of the University of Chicago.
Immunostaining of cells.
Cardiomyocytes (10,000 to 20,000) plated on 1% laminin-coated coverslips were used for immunostaining. Two-day-old cultures were infected with SIRT3 adenovirus for 24 h and then stained with different agents as described below. Cells were incubated with 400 nM Mitotracker Green FM (M7514; Invitrogen) for 1 h at 37°C in complete medium. The unbound Mitotracker was removed by washing the cells three times with fresh medium for 5 min each. The cells were fixed for 15 min in 4% paraformaldehyde prepared in complete medium at 37°C in an incubator. After three washes with PBS, cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min and then washed again three times with PBS containing 0.05% Triton X-100. Cells were incubated at room temperature for 1 hour in blocking solution (0.05% Triton X-100, 10% donkey serum, 1% bovine serum albumin in 1× PBS) and then with a primary antibody (1:25, diluted in 50% blocking buffer) overnight at 4°C in a humid chamber. Next day, cells were washed three times (10 min for each wash) with PBS containing 0.05% Triton X-100 and 1% bovine serum albumin (PBST) and then incubated for 1 h with appropriate secondary antibody (1:500, diluted in 50% blocking buffer) conjugated with Alexa Fluor 594 (red). Cells were again washed with PBST and mounted in Vectashield hard-set mounting medium with DAPI (4′,6′-diamidino-2-phenylindole) (Vector Laboratories). All microscopy and imaging analyses were done in the digital confocal microscopy core facility of the University of Chicago.
RESULTS
SIRT3 is a stress-responsive factor in cardiomyocytes.
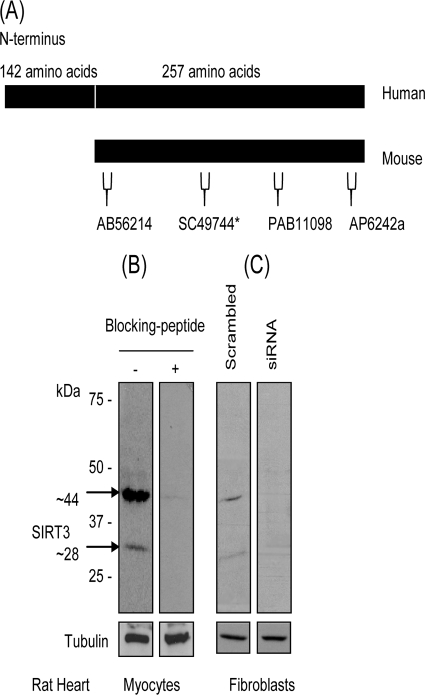
In order to examine SIRT3 function in cardiomyocytes, we first sought to test the specificity of different commercially available antibodies that recognize murine SIRT3. Among six antibodies tested, we found only three (Abgent AP6242a, Orbigen PAB-11098, and Abcam Ab56214) that recognize both the ∼44-kDa long form and ∼28-kDa short form of SIRT3 from rat cardiomyocytes. These three antibodies are specific to three different regions of SIRT3 (Fig. 1A). The specificity of the antibody for both bands was validated by use of a specific blocking peptide, inclusion of which eliminated the reactivity of the antibody (Fig. 1B). We also examined the specificity of antibody by knocking down SIRT3 levels in rat fibroblasts by using SIRT3-specific siRNA. As shown in Fig. 1C, both bands were reduced to nearly nondetectable levels after knocking down SIRT3 levels of fibroblasts, thus again confirming the ability of the antibody to recognize both bands of SIRT3. These antibodies were used in our subsequent experiments.
FIG. 1.
Characterization of antibodies recognizing two forms of SIRT3 from cardiac tissue. (A) Schematic representation of hSIRT3 and mSIRT3 with epitope map of different antibodies used in this study. The asterisk indicates an antibody that was used only for immunostaining of cells. (B) Western blot analysis with SIRT3 antibody (AP6242a) showing two specific bands of SIRT3 in cardiomyocytes, as determined by using a blocking peptide. (C) SIRT3 levels in rat heart fibroblasts were knocked down by using SIRT3-specfic siRNA. Cell lysate was analyzed by Western analysis with the same antibody as that in panel B. Note the reduced levels of both forms of SIRT3 after knockdown of endogenous SIRT3 levels.
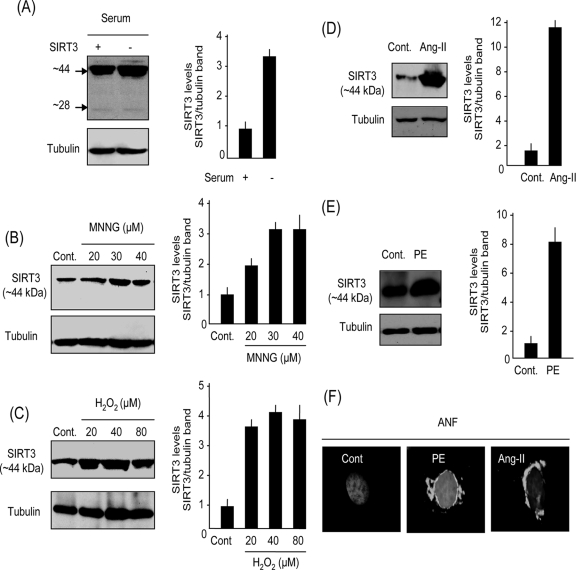
To examine the expression pattern of SIRT3 in cardiomyocytes during stress, we grew primary cultures of cardiomyocytes either in a serum-containing medium or in the serum-free medium for 48 h. We found that cells grown in serum-free medium had two- to threefold-higher levels of expression of the 44-kDa long form of SIRT3 than did controls maintained in serum-containing medium (Fig. 2A). However, we found no change in expression of the 28-kDa short form of SIRT3 in cultures grown with or without serum. To confirm this observation, we treated cells with two other stress stimuli: (i) MNNG, an alkylating agent known to cause genotoxic stress, and (ii) hydrogen peroxide, an established free-radical-generating agent known to induce oxidative stress in cardiomyocytes (26). Cells were treated for 24 h with low doses of MNNG (20 to 40 μM) or H2O2 (20 to 80 μM) which are incapable of inducing cell death. The results obtained from these cultures indicated that both MNNG and H2O2 treatments elevated the levels of long-form SIRT3 in a concentration-dependent manner over those of the nontreated controls (Fig. 2B and C). We then examined the effect of two physiologically relevant cardiac stressors, Ang-II and PE, which are known to induce cell growth (hypertrophy) as well as cell death of cardiomyocytes at higher doses. The stress response of cardiomyocytes to Ang-II and PE treatments was monitored by measurement of ANF release from nuclei (ANF is a marker of cardiomyocyte growth during stress). As shown in Fig. 2D and E, both Ang-II and PE treatments increased the levels of long-form SIRT3 in cardiomyocytes significantly, and this corresponded with the release of ANF from the cell nuclei (Fig. 2F). In this series of experiments, we could not detect a change in the level of the 28-kDa short form of SIRT3 after cellular stress (not shown). These results indicated that the levels of long-form (∼44-kDa) SIRT3 are elevated during mild stress of cardiomyocytes.
FIG. 2.
SIRT3 is a stress-responsive factor in cardiomyocytes. (A) Expression levels of SIRT3 in cardiomyocytes grown for 48 h in a medium with serum or defined medium without serum. Note induction of ∼44-kDa band but not 28-kDa band of SIRT3 in serum-free medium. (B) SIRT3 levels in cardiomyocytes treated with vehicle (Cont.) or MNNG at indicated concentrations for 24 h. (C) SIRT3 levels in cardiomyocytes treated with different concentrations of H2O2 for 24 h. (D and E) SIRT3 levels in cardiomyocytes treated with Ang-II (D) (5 μM) or PE (E) (20 μM) for 48 h. (F) Immunostaining of cardiomyocytes for ANF release from nuclei (white), a marker of cellular stress. Positions of nuclei were determined by DAPI staining. Values are means ± standard errors of three to four experiments. SIRT3 antibody AP6242a was used for Western analysis.
SIRT3 is required for survival of cardiomyocytes under stress conditions.
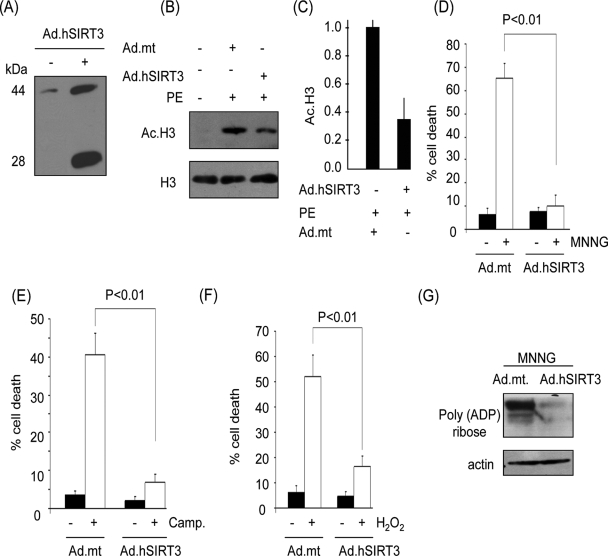
To determine the function of SIRT3 under stress conditions, we transiently expressed hSIRT3 in cardiomyocytes by using an hSIRT3-adenovirus vector (mSIRT3-expressing vectors are not available yet). In a set of preliminary experiments, we first quantified the level of hSIRT3 expression at different doses and different time points post-infection with adenovirus. At a virus dose equivalent to a multiplicity of infection of 10 for 18 h of infection, nearly 5- to 10-fold induction of SIRT3, relative to endogenous levels, was observed, which mimics the stress-mediated upregulation of deacetylase (Fig. 3A). This dose of adenovirus was utilized in subsequent experiments. We also confirmed the enzymatic activity of SIRT3 expressed by the adenovirus vector. Cells were infected with virus synthesizing wt SIRT3 or the mt (benign) protein and then stimulated with PE, a known inducer of cardiomyocyte acetylation (11). We analyzed the acetylation of H3 in these cells and found that the histone acetylation was notably lower in SIRT3-expressing cells than in cells infected with the mt vector (Fig. 3B and C).
FIG. 3.
SIRT3 overexpression protects cardiomyocytes from genotoxic and oxidative stress-mediated cell death. (A) Expression levels of hSIRT3 in cardiomyocytes infected with adenovirus (Ad) vector for 24 h were determined by Western analysis with SIRT3 antibody PAB-11098. Note the presence of both forms of hSIRT3 in adenovirus-infected cells. (B) Deacetylase activity of hSIRT3 in cardiomyocytes. Cells infected with viral vectors, synthesizing the wt SIRT3 or an mt (benign) protein, were treated with PE to induce cellular acetylation. Acetylation of histones was determined by Western analysis. (C) Quantification of H3 deacetylation in SIRT3-overexpressing cells. (D to F) Cardiomyocytes overexpressing hSIRT3 or the mt protein were treated with MNNG (500 μM), camptothecin (Camp.; 10 μM), or H2O2 (200 μM). Cell death was determined 6 h after treatment by Hoechst and PI staining followed by FACS analysis. (G) Western analysis of the cell lysate with a poly(ADP-ribose) antibody, indicating PARP1 activity. Quantitative values are means ± standard errors of five to seven plates of three separate experiments.
To examine the role of SIRT3 in cell death, cardiomyocytes were challenged with a cell-death-inducing dose of MNNG (500 μM). Cell death was monitored by Hoechst and PI staining followed by FACS analysis, as well as by measuring the activity of PARP1, a marker of DNA damage. We found that the MNNG treatment killed almost 60% of cells expressing the mt protein; however, the wt SIRT3-expressing cardiomyocytes were mostly protected (Fig. 3D). The intensity of cell death was also correlated with the magnitude of PARP1 activation, as measured by total poly(ADP)-ribosylation of cellular proteins, thus indicating that SIRT3 has the potential to block the genotoxic stress-mediated cell death (Fig. 3D and G). Similar cell-protective effects of SIRT3 were also noticed when cells were challenged with two other cell-death-inducing agents, H2O2 (200 μM) and camptothecin (10 μM), an alkaloid known to induce DNA damage-mediated cell death (Fig. 3E and F). These results demonstrated that SIRT3 protects cardiomyocytes from genotoxic and oxidative stress-mediated cell death.
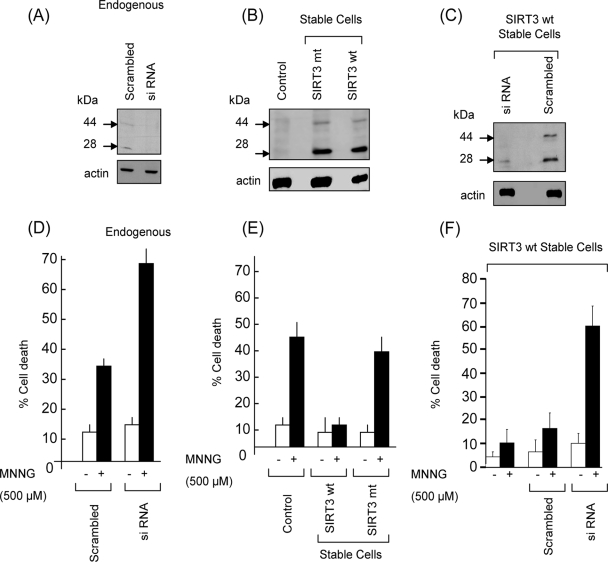
To confirm these results, we examined the role of SIRT3 in HeLa cells. As in cardiomyocytes, we detected two bands of SIRT3 from HeLa cells by using the same antibodies (Fig. 4A). By knocking down SIRT3 levels of HeLa cells, we found that both bands were drastically reduced (Fig. 4A), thus again confirming the specificity of antibody to two forms of SIRT3. We then examined the role of SIRT3 during genotoxic stress of HeLa cells. We found that the SIRT3-knockout cells were twice as sensitive to cell death as were the control cells, thus again suggesting a cell-protective role of SIRT3 during genotoxic stress (Fig. 4D).
FIG. 4.
SIRT3 expression is required for survival of HeLa cells after genotoxic stress. (A) Representative Western blot showing endogenous expression of SIRT3 in control cells and cells subjected to SIRT3 knockout. Note reduced levels of both forms of SIRT3 after siRNA-mediated knockdown. (B) Western blot showing expression of SIRT3 in control and stable HeLa cells. (C) Western blot showing reduced levels of both forms of SIRT3 after siRNA-mediated knockdown in stable cells. (D) HeLa cells (control and endogenous SIRT3 knockdown cells) were treated with MNNG (500 μM) for 10 min and then washed and kept in complete medium for 6 h. Cell death was determined by Hoechst and PI staining followed by FACS analysis. (E) SIRT3 stable cells expressing wt SIRT3 or mt SIRT3 were examined for MNNG-mediated cell death. (F) SIRT3 stable cells were subjected to SIRT3 knockdown by siRNA and then tested for MNNG-mediated cell death. SIRT3 antibody PAB-11098 was used in all these Western blot assays. Values are means ± standard errors of three experiments.
To get further evidence for these findings, we generated two stable HeLa cell lines, one expressing wt hSIRT3 and the other expressing the mt hSIRT3. By Western analysis, we observed that the expression level of SIRT3 in stable cells was nearly fivefold higher than that in the native cells (Fig. 4B). We generated yet another cell line in which the SIRT3 level of the stable cells was knocked down by use of SIRT3-specific siRNA. The SIRT3 levels of knocked-out cells were reduced by >80%, compared to those of controls which received scrambled RNA (Fig. 4C). These cells were then examined for their sensitivity to different cell-death-inducing agents, including MNNG, camptothecin, and H2O2 treatments. We noticed that whereas mt SIRT3-expressing cells were as prone to cell death as were their native counterparts, the wt SIRT3-expressing cells were, in general, resistant to MNNG-induced cell death (Fig. 4E). We obtained similar results by challenging these cells with camptothecin and H2O2. We then examined the response of stable cells subjected to SIRT3 knockdown, and we found that these cells were apparently more susceptible to cell death than were their counterparts expressing high levels of SIRT3 (Fig. 4F). These results together strongly indicated that SIRT3 overexpression renders cells resistant to stress-mediated cell death.
Localization of SIRT3 in cardiomyocytes.
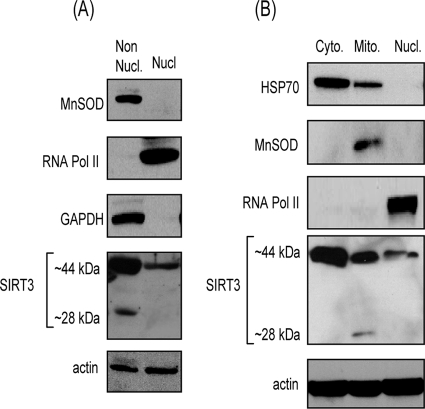
To begin to understand the mechanism of cell protection by SIRT3, we first examined the compartmentalization of the protein in cardiomyocytes. Previous studies carried out with overexpressed SIRT3 have demonstrated contradictory results. Whereas initial studies showed exclusive mitochondrial localization of SIRT3, some recent studies have shown nuclear expression of the deacetylase as well (31, 32). To examine the localization of the endogenous SIRT3 in cardiomyocytes, we generated two subcellular fractions of the adult mouse heart by using a Pierce cell fractionation kit (NE-PER). In this procedure nuclei (nuclear fraction) are separated from other cellular components, which are left behind in the cytoplasmic fraction (nonnuclear fraction). Both fractions were characterized by use of different fraction-specific protein antibodies. MnSOD, a mitochondrial marker, and GAPDH, a cytosolic marker, were present in the nonnuclear fraction, whereas RNA polymerase II was present only in the nuclear fraction, as expected (Fig. 5A). When these fractions were analyzed for SIRT3 expression, we found only the long form (44 kDa) of SIRT3 in the nuclear fraction, whereas both SIRT3 forms (44 and 28 kDa) were present in the cytoplasmic, nonnuclear fraction (Fig. 5A).
FIG. 5.
SIRT3 is localized in the cytoplasm and mitochondria as well as nuclei of heart tissue. (A) Nuclear (Nucl.) and nonnuclear (Non Nucl.) fractions of mouse heart were prepared by using the Pierce cell fractionation kit. Fractions were characterized by Western blotting taking GAPDH and MnSOD as nonnuclear markers and RNA polymerase II as a nuclear marker. Both fractions were analyzed for the expression level of SIRT3 by Western blotting. Actin was utilized as a loading control. Note the presence of the (∼44-kDa) long form of SIRT3 in both the nuclear and nonnuclear fractions, while the ∼28-kDa short form is detected only in the nonnuclear fraction. (B) Three subcellular fractions (cytoplasmic, mitochondrial, and nuclear) of a mouse heart were prepared, utilizing the Calbiochem fractionation kit, and characterized by Western blotting using fraction-specific protein antibodies. All three fractions were also analyzed for expression of mSIRT3 by Western blotting. The SIRT3 antibody Ab56214 was used in these blotting assays. Note the presence of ∼44-kDa SIRT3 in all three fractions, whereas the short form (∼28 kDa) of SIRT3 was detected only in the mitochondrial fraction of the heart.
To characterize the location of the 28-kDa form of SIRT3 further, we utilized another cell fractionation procedure (Calbiochem), which separates subcellular fractions based on differences in the solubilities of certain cellular components. By using this procedure, we prepared three different heart fractions; these included the mitochondrial, nuclear, and cytosolic fractions. Again, fractions were characterized by using fraction-specific protein antibodies, as shown in Fig. 5B. Analysis of these fractions for SIRT3 expression revealed that whereas the 44-kDa long form of SIRT3 was present in all three fractions, the 28-kDa short form was detected only in the mitochondrial fraction. Much to our surprise, we also noticed that the 44-kDa form was more abundant in the cytosolic fraction than in the two other fractions. From these results we conclude that the 44-kDa long form of SIRT3 is localized in all three fractions, mitochondria and nucleus, as well as the cytoplasm of cardiomyocytes. The 28-kDa short form of SIRT3 was, however, present only in mitochondria.
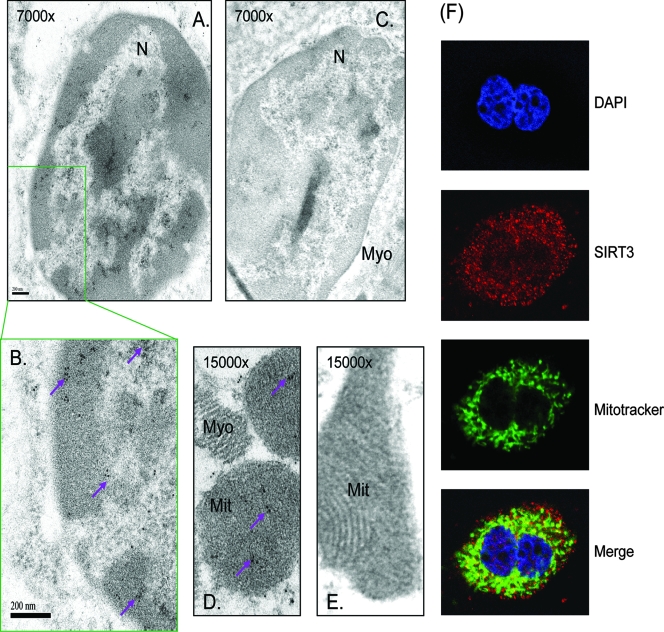
The mitochondrial localization of SIRT3 is well accepted; however, its presence in the nucleus is still being debated. Therefore, to obtain firm evidence for SIRT3 localization in the nucleus, we performed transmission electron microscopy of mouse heart sections stained for endogenous SIRT3. The immunogold particles, as reflected by high-density black dots, depicting SIRT3 were found to be localized in the nucleus as well as in the mitochondria (Fig. 6A, B, and D). These results were validated with another SIRT3-specific antibody as well as by use of different negative controls, i.e., no primary antibody or nonspecific IgG (Fig. 6C and E). These results further substantiated the presence of SIRT3 in the nucleus of cardiomyocytes.
FIG. 6.
Microscopic analysis of heart tissue showing nuclear localization of mSIRT3. (A, B, and D) Representative electron micrographs of mouse heart sections stained with anti-SIRT3 antibody (PAB-11098). Panel B is an enlarged portion of the nucleus shown in panel A. (C and E) Heart sections stained with nonspecific IgG. N, nucleus; Mit, mitochondria; Myo, myofilaments. Numbers at the top of the picture indicate magnifications. High-density black dots (purple arrows) indicate the localization of SIRT3 in the nucleus and mitochondria. (F) Confocal microscopic image of a cardiomyocyte stained with anti-SIRT3 antibody (SC49744) (red). Positions of nuclei and mitochondria were detected by DAPI (blue) and Mitotracker (green) staining, respectively. Note the presence of SIRT3 (red in the merged picture) in the cytoplasm and mitochondria as well as the nuclei of cardiomyocytes.
To get additional evidence for these findings, we carried out confocal microscopic analysis of cultured cardiomyocytes stained for endogenous SIRT3. Mitochondria and nuclei were localized by Mitotracker (green) and DAPI (blue) staining, respectively. As shown in Fig. 6F, we found that the endogenous SIRT3 was highly expressed in the cytoplasm, with relatively smaller amounts also present in the nucleus and mitochondria (see merged picture) of cardiomyocytes. These experiments were repeated with two different anti-SIRT3 antibodies, one raised against an internal region of SIRT3 and the other raised against the C-terminal segment of the protein. The two antibodies gave identical results. The specificity of the antibodies used was confirmed by utilizing two different negative controls, one being SIRT3-blocking peptides and the other being an IgG control. Neither negative control showed SIRT3 staining of cells, thus indicating that the results with the SIRT3 antibody were specific. These results are consistent with the findings of fractionation studies and demonstrated that the endogenous SIRT3 is present not only in mitochondria but also in the cytoplasm and nucleus of cardiomyocytes.
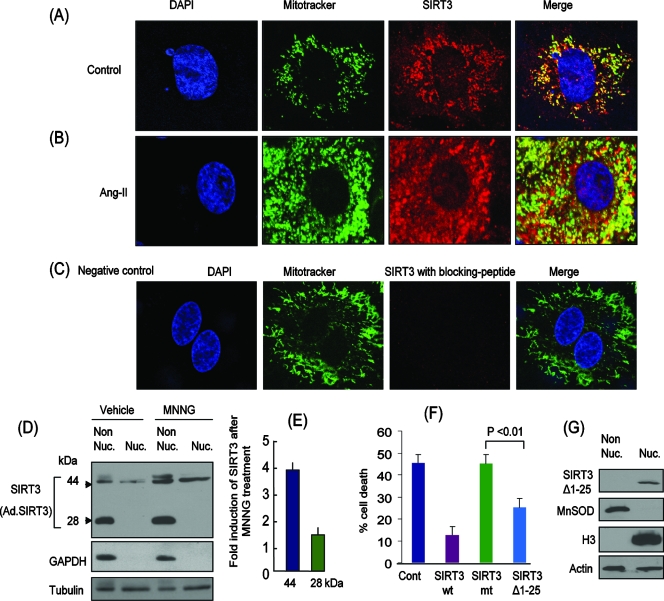
We asked next how stress influences the localization of SIRT3 in cardiomyocytes. For this purpose, we infected cardiomyoctes with ad.hSIRT3 vector, and 24 h later we treated them with Ang-II for another 24 h. Stress of cardiomyocytes was confirmed by release of ANF from the cell nuclei (not shown). On staining of these cells for SIRT3, we found that, in control unstressed cells, SIRT3 was present in the mitochondria and cytoplasm, as well as to a lesser extent in the nucleus, as expected (Fig. 7A). When these cells were stressed with Ang-II, we observed increased levels of SIRT3 in mitochondria, but a notable amount of SIRT3 was also found in the nucleus of the cells (Fig. 7B). To confirm these results, we prepared nuclear and nonnuclear protein fractions of cells and analyzed them for SIRT3 expression. We found that, as with the heart fractions (Fig. 5A), the 44-kDa long form was present both in the nuclear and in the nonnuclear fractions, whereas the 28-kDa short form of SIRT3 was detected only in the nonnuclear fractions. Also, after stress (by MNNG treatment) both forms (44 and 28 kDa) of SIRT3 were notably elevated (Fig. 7D and E). Together, these results indicated that during stress of cardiomyocytes, SIRT3 levels are elevated in the mitochondria as well as in the nucleus of the cell.
FIG. 7.
Overexpressed SIRT3 is localized in the cytoplasm and mitochondria as well as the nuclei of cardiomyocytes. Representative confocal microscopic images of cardiomyocytes infected with ad.hSIRT3 vector and stained with anti-SIRT3 antibody (SC49744) (red). Mitotracker and DAPI staining was utilized for localization of mitochondria and nuclei, respectively. (A) Control cells treated with vehicle. (B) Cells treated with Ang-II (5 μM) for 48 h. Note the induction of SIRT3 in both the mitochondria and nuclei of the cell. (C) Negative control in which a specific blocking peptide was used to validate the specificity of SIRT3 antibody. (D) Cells were overexpressed with ad.hSIRT3 and treated with vehicle or MNNG (500 μM) for 6 h. Subcellular fractions (nuclear and nonnuclear) were prepared and analyzed for the expression of SIRT3 by Western analysis (SIRT3 antibody PAB-11098). GAPDH was utilized as a cytosolic marker, and tubulin was used as a loading control. (E) Quantification of SIRT3 induction after MNNG treatment of cells. Note induction of both the 44- and 28-kDa forms of SIRT3 after genotoxic stress. (F) SIRT3Δ1-25 deletion mt lacking the mitochondrial import signal is effective in protecting cells from MNNG-mediated cell death. HeLa cells were overexpressed with wt SIRT3, mt SIRT3 (catalytically inert), or SIRT3Δ1-25. Cells were treated with MNNG (500 μM), and cell death was monitored 3 h later. Note significantly reduced cell death in SIRT3Δ1-25-expressing cells compared to that of cells overexpressing catalytically inert protein. Cont, control. Values are means ± standard errors of four separate experiments. (G) Localization of SIRT3Δ1-25 deletion mt in the nucleus but not in the nonnuclear fraction.
To understand the role of SIRT3 outside mitochondria, we generated a deletion mt (SIRT3Δ1-25) lacking the mitochondrial import signal (32). This construct was compared with wt SIRT3 and the catalytically inert mt (SIRT3.mt) for their abilities to protect cells from genotoxic stress-mediated cell death. We found that overexpression of SIRT3Δ1-25 was capable of protecting HeLa cells from MNNG-mediated cell death, but to a lesser extent than that of the wt SIRT3 (Fig. 7F). The catalytically inert mt of SIRT3 had no effect on MNNG-mediated cell death. These results thus demonstrated that SIRT3 plays a role outside the mitochondria as well as in protecting cells from stress.
SIRT3 interacts with Ku70 in vitro and in vivo.
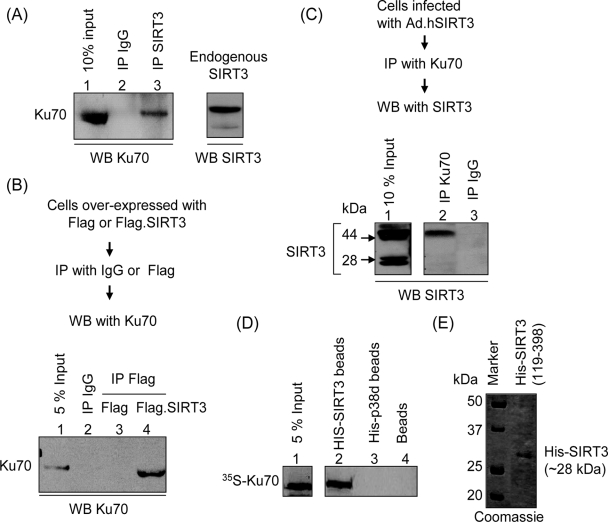
To delineate the mechanism of cell-protective effects of SIRT3, we searched for its partner proteins. We immunoprecipitated SIRT3 from cardiomyocyte lysate and looked for pull-down of partner proteins in the precipitate. We found that Ku70, a nuclear protein, was successfully pulled down with SIRT3 but not with the IgG negative control (Fig. 8A). To confirm this observation, we overexpressed Cos7 cells with plasmids expressing either Flag-SIRT3 or the Flag tag alone. Cell lysate was prepared and subjected to IP with the Flag antibody or IgG. The resulting beads were analyzed by Western analysis with anti-Ku70 antibody. As shown in Fig. 8B, Ku70 was coprecipitated with the Flag antibody from Flag.SIRT3-expressing cells but not from cells expressing the Flag tag alone. Also, no Ku70 was pulled down with IgG, which served as another negative control (Fig. 8B).
FIG. 8.
SIRT3 interacts with Ku70 in vitro and in vivo. (A) Endogenous SIRT3 interacts with Ku70. Cardiomyocyte lysate was subjected to IP with either nonspecific IgG or specific anti-SIRT3 antibody (AP6242a). Resulting beads were analyzed by Western blotting (WB) with anti-Ku70 antibody. (B) Cos7 cells were induced to overexpress with the Flag tag or the Flag-tagged SIRT3. Cell lysate was subjected to IP with either nonspecific IgG-conjugated beads or anti-Flag M2 agarose beads. Precipitated beads were analyzed by Western blotting with anti-Ku70 antibody. (C) Cells infected with ad.hSIRT3 vector were subjected to IP with Ku70 antibody, and the resulting beads were analyzed by Western blotting with anti-SIRT3 antibody (PAB11098). Note the presence of both forms of SIRT3 in the input lane, but only the long form of SIRT3 was pulled down by Ku70 in this assay. (D) In vitro protein binding assay. In vitro-synthesized [35S]methionine-labeled Ku70 was incubated with beads containing His-tagged 28-kDa SIRT3, His-tagged p38d, or nickel beads alone. His-tagged proteins were precipitated as nickel resin beads, and bound proteins were analyzed by SDS-PAGE. (E) Picture of a Coomassie blue-stained gel showing synthesis of the His-tagged 28-kDa form of SIRT3 from plasmid (His-SIRT3119-398).
To obtain further evidence for a SIRT3/Ku70 interaction, we performed an inverse experiment in which SIRT3 was intended to be pulled down by Ku70. Cardiomyocytes infected with ad.hSIRT3 vector were subjected to IP with IgG or Ku70 antibody, and the resulting beads were analyzed by Western analysis by use of an anti-SIRT3 antibody. As mentioned before, in SIRT3-overexpressing cells both 44-kDa and 28-kDa forms of SIRT3 were readily detectable. In some experiments these forms appear as doublets (Fig. 8C, lane 1). The exact reason for these doublets is not known to us at present. In co-IP experiments, we found that only the 44-kDa long form of SIRT3 was pulled down with Ku70, and not the 28-kDa short form of the deacetylase. These experiments were repeated with SIRT3-expressing stable HeLa cells, and similar results were obtained.
The lack of interaction of the 28-kDa form of SIRT3 with Ku70 raised the question of whether this is due to the inability of the short form of SIRT3 to bind to Ku70 or is related to localization of this form in the mitochondria, where Ku70 is not found. To address this issue, we examined the interaction of the 28-kDa form of SIRT3 with Ku70 under in vitro assay conditions. A plasmid (His-SIRT3119-398) encoding the His-tagged 28-kDa form of SIRT3 was primed to synthesize the protein in vitro (Fig. 8E). Another plasmid synthesizing His-p38d was utilized as a negative control. For the binding assay, His-tagged proteins were incubated with in vitro-synthesized [35S]methionine-labeled Ku70. His-tagged proteins were precipitated as nickel resin beads, and bound proteins were analyzed by SDS-PAGE. As shown in Fig. 8D, 35S-labeled Ku70 was successfully pulled down by His-SIRT3 but not by His-p38d or resin beads alone. These results thus demonstrated that (i) Ku70 physically binds to SIRT3 and (ii) the 28-kDa short form of SIRT3 is capable of interacting with Ku70.
SIRT3 deacetylates Ku70 under in vitro and in vivo assay conditions.
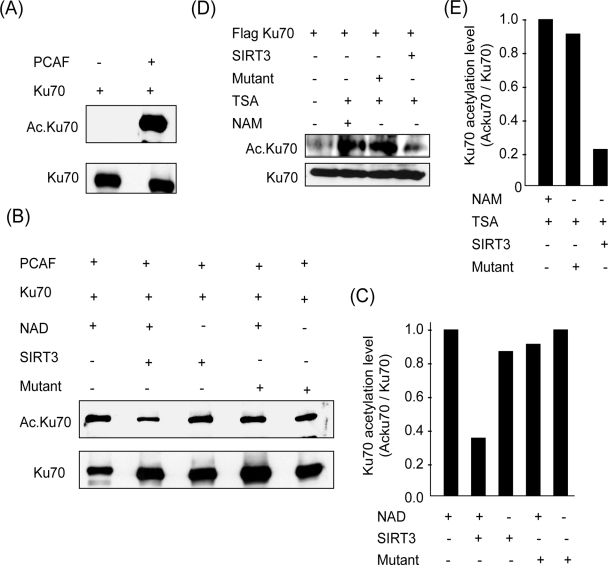
Previous studies have shown that Ku70 is an acetylated protein (7). Our results showing an association of Ku70 with SIRT3 suggested that Ku70 may be a deacetylation target of SIRT3. To test this possibility, we studied deacetylation of Ku70 by SIRT3 under in vitro assay conditions. Ku70 was synthesized in vitro and incubated with PCAF in an acetylation reaction buffer. We found, by Western analysis, that Ku70 was highly acetylated in the presence of PCAF (Fig. 9A). To test the deacetylation of Ku70 by SIRT3, we first tested the ability of in vitro-synthesized SIRT3 to deacetylate Ku70. We found, however, no appreciable deacetylation of Ku70 in this experiment (data not shown).
FIG. 9.
SIRT3 deacetylates Ku70 under in vitro and in vivo assay conditions. (A) In an acetylation buffer Flag.Ku70 was incubated with PCAF. Acetylation of protein was determined by Western blotting with antiacetyllysine antibody. (B) Deacetylation of Ku70 by SIRT3 in vitro. Flag.Ku70 was acetylated in vitro with PACF, and it was precipitated with Flag M2 beads. Acetylated Flag.Ku70 was then incubated with beads containing wt or mt SIRT3 in a deacetylation buffer with or without NAD. Both wt and mt SIRT3 were immunoprecipitated from stable HeLa cells. (C) Quantification of Ku70 deacetylation by SIRT3. (D) In vivo deacetylation of Ku70 by SIRT3. Stable cells expressing wt or mt SIRT3 were induced to overexpress with Flag.Ku70 and treated with NAM (10 mM for 24 h) and/or TSA (5 μM for 6 h) as indicated. Flag.Ku70 was immunoprecipitated, and the level of acetylation was analyzed by probing with antiacetyllysine antibody. (E) Quantification of Ku70 deacetylation by SIRT3 in vivo. Values are means of three experiments.
We then examined the deacetylase activity of in vivo-synthesized SIRT3. Cells were induced to overexpress protein with the use of plasmids synthesizing wt Flag.SIRT3 (Flag tag at the C-terminal end) or the mt. Proteins were immunoprecipitated by use of a Flag-specific antibody, and the resulting beads were incubated with the acetylated Ku70 in a deacetylation buffer with or without NAD. We found that beads containing wt SIRT3 had the ability to deacetylate Ku70 in an NAD-dependent manner. However, the SIRT3 mt, having only one point mutation which eliminates its deacetylase activity, was incapable of deacetylating Ku70, even when NAD was present in the buffer (Fig. 9B and C). These results thus demonstrated that SIRT3 has the ability to deacetylate Ku70.
We then studied the SIRT3-mediated deacetylation of Ku70 in vivo. Stable HeLa cells overexpressing wt or mt SIRT3 were transfected with a Flag.Ku70-expressing plasmid and treated with trichostatin A (TSA), a class I and II HDAC inhibitor, or with NAM, a class III HDAC inhibitor. Flag.Ku70 was immunoprecipitated from these cells, and the acetylation of the protein was analyzed by Western analysis by use of an antiacetyllysine antibody. As shown in Fig. 9D and E, Ku70 was highly acetylated in cells treated with the HDAC inhibitors TSA and NAM compared to nontreated controls. When Ku70 was analyzed from SIRT3-expressing cells, we found that it was substantially deacetylated in these cells but not in cells expressing the mt SIRT3. These data thus indicated that SIRT3 is one of the major regulators of Ku70 acetylation in vivo.
SIRT3 blocks Ku70 acetylation under stress conditions.
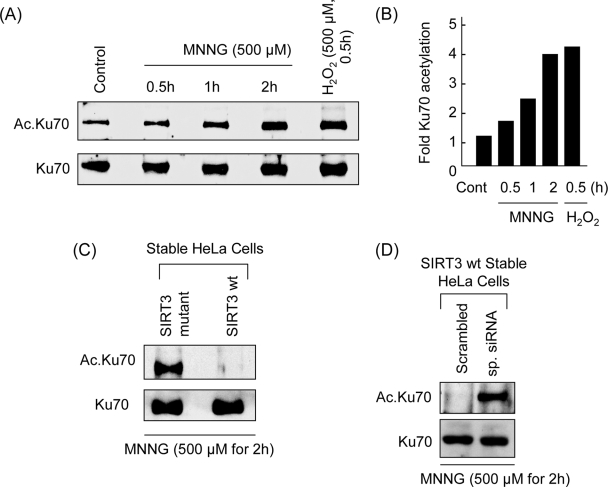
Acetylation of Ku70 increases after DNA damage and stress-induced apoptosis (7, 8). To test whether SIRT3 was capable of blocking Ku70 acetylation under stress conditions, we performed a time course analysis of Ku70 acetylation after MNNG and H2O2 treatment of cells. Cardiomyocytes as well as HeLa cells were treated with a cell-death-inducing dose (500 μM) of MNNG or H2O2. Cells were harvested at different time points after treatment, and Ku70 was immunoprecipitated from the lysate, which was then examined by Western analysis by use of an antiacetyllysine antibody. As shown in Fig. 10A and B, the acetylation of Ku70 was elevated in a time-dependent manner, reaching maximum level at 2 h of MNNG treatment and at 30 min of H2O2 treatment of cells. These results thus indicated that Ku70 acetylation is elevated during genotoxic and oxidative stress of the cells.
FIG. 10.
SIRT3 blocks Ku70 acetylation during stress. (A) Ku70 was acetylated during genotoxic and oxidative stress. Cells were transiently expressed with Flag-Ku70 and treated with MNNG (500 μM) or H2O2 (500 μM). Cells were harvested at indicated time intervals after treatment, and Flag.Ku70 was immunoprecipitated. Flag.Ku70 beads were analyzed by Western blotting using antiacetyllysine and anti-Ku70 antibodies. (B) Quantitative representation of Ku70 acetylation by MNNG and H2O2 treatment of cells. Values are means of three separate experiments. (C) HeLa cells stably expressing either wt or mt SIRT3 were treated with MNNG for 2 h. The endogenous Ku70 was immunoprecipitated and analyzed by Western blotting as in panel A. (D) Knockdown of SIRT3 increases Ku70 acetylation during genotoxic stress. SIRT3 was knocked down from SIRT3-expressing stable cells by use of SIRT3-specific siRNA. Scrambled RNA was used as a negative control. Cells were treated with MNNG for 2 h, and the level of Ku70 acetylation was determined by Western blotting.
After determining the specific time course of Ku70 acetylation following stress, we studied the role of SIRT3 in blocking Ku70 acetylation. For this purpose, we treated stable HeLa cells expressing wt SIRT3 or the mt with 500 μM MNNG for 2 h. As shown in Fig. 10C, MNNG treatment significantly increased the acetylation of Ku70 in the mt-expressing cells but not in the wt SIRT3-expressing cells. To strengthen this finding, we also examined the acetylation of Ku70 in stably SIRT3-expressing cells subjected to siRNA-mediated knockdown of SIRT3 levels. As shown in Fig. 10D, MNNG treatment induced Ku70 acetylation in the cells that received specific SIRT3 siRNA but not the scrambled RNA. Collectively, theses experiments demonstrated that SIRT3 is capable of blocking Ku70 acetylation during oxidative and genotoxic stress of the cells.
SIRT3 prevents Bax-mediated apoptosis by deacetylating Ku70.
Translocation of the Bax protein from the cytoplasm to mitochondria is known to initiate stress-induced apoptosis. Under physiologic conditions, Bax remains in association with Ku70 (7, 30). During stress, Ku70 is acetylated, which diminishes its ability to bind to Bax. The released Bax enters into mitochondria and initiates the process of apoptosis (7). The HDACs capable of deacetylating Ku70 (such as SIRT1) have been shown to enhance Ku70/Bax binding and prevent cell death (8). We therefore speculated that SIRT3 might protect cells by promoting the binding of Ku70 to Bax and hence blocking the Bax translocation to mitochondria.
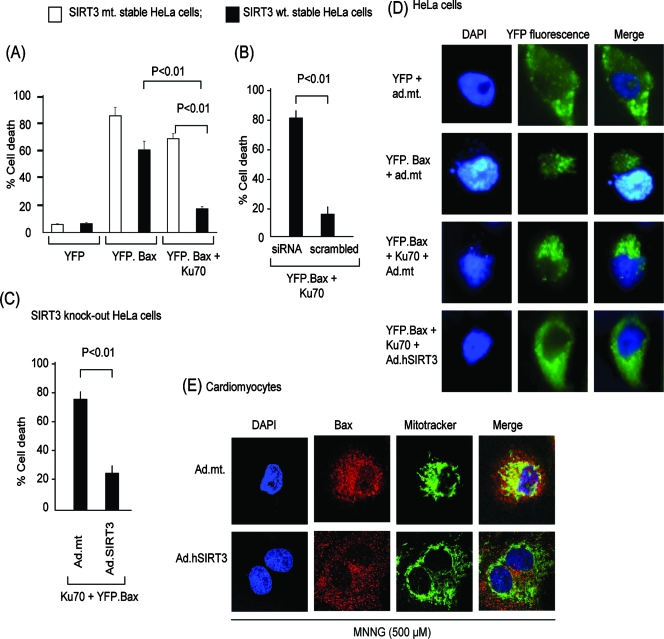
To test this hypothesis, we employed a well-characterized assay system in which stable HeLa cells were transfected with plasmids synthesizing the Bax tagged with YFP (YFP.Bax) and/or Ku70. Cell death was monitored by scoring YFP-positive cells which had a fragmented nucleus, a common marker of apoptosis. As shown in Fig. 11A, stably wt SIRT3-expressing cells overexpressing Bax alone (YFP.Bax) had a significantly higher rate of cell death than did those expressing Bax and Ku70 together. In contrast, stable cells expressing the mt SIRT3 were not resistant to Bax-mediated cell death even when Bax and Ku70 were combined.
FIG. 11.
SIRT3 prevents Bax-mediated apoptosis by deacetylating Ku70. (A) HeLa cells stably expressing wt SIRT3 (black bars) or mt SIRT3 (white bars) were transfected with plasmids synthesizing YFP, YFP.Bax, or YFP.Bax and Ku70 together. The percentage of YFP-positive cells (yellow fluorescence) with apoptotic (fragmented) nuclei was scored 12 h after transfection. Values are the averages of three experiments; during each experiment >200 cells were scored. (B) HeLa cells stably expressing wt SIRT3 were given SIRT3-specific siRNA or scrambled RNA. We observed >80% reduction of the SIRT3 levels in cells to which siRNA was added, as shown in Fig. 4C. These cells were then transfected with plasmids encoding YFP.Bax and Ku70. Cell death was scored 12 h posttransfection. Values are the averages of four experiments. (C) Salvage of wt SIRT3 levels protects cells from Bax-mediated cell death. SIRT3-knockdown HeLa cells were infected with ad.hSIRT3 or the mt vector. After 12 h of virus infection, cells were transfected with YFP.Bax and Ku70 plasmids together. The percentage of YFP-positive cells with apoptotic nuclei was scored 12 h after transfection. Values are the means of four experiments with >200 cells scored in each plate. Nearly 80% of SIRT3 levels were recovered in these cells, as verified by Western blotting (not shown). (D) Representative picture of HeLa cells subjected to Bax-mediated apoptosis. Cells were infected with ad.hSIRT3 or the mt (ad.mt) vector. Twenty-four hours after viral infection, cells were transfected with different plasmids as indicated. The pictures of cells were taken 12 h posttransfection. Note the presence of fragmented apoptotic nuclei in cells transfected with YFP.Bax and YFP.Bax plus Ku70 (middle panels) but not in SIRT3-overexpressing cells (bottom panels). (E) SIRT3 blocks localization of Bax to mitochondria during genotoxic stress. Cardiomyocytes infected with ad.hSIRT3 or the mt (ad.mt) vectors were treated with MNNG for 6 h. Cells were stained with anti-Bax antibody (red). DAPI (blue) staining and Mitotracker (green) staining were utilized as markers of nuclei and mitochondria, respectively. Note shrunken nuclei in upper panels (ad.mt) depicting cell death. In these cells, Bax was totally merged with Mitotracker green (yellow in merged image) representing mitochondrial localization of Bax. In the lower panels, cells overexpressing wt SIRT3 and having well-preserved nuclei showed no Bax translocation to mitochondria (no yellow in merged image).
This experiment was repeated in cells subjected to siRNA-mediated knockout of SIRT3. As expected, stably SIRT3-expressing cells that received scrambled RNA were resistant to cell death when Bax and Ku70 were coexpressed, but the other cells, in which SIRT3 levels were markedly reduced by siRNA, were not resistant (Fig. 11B). To provide further support for these findings, we performed a SIRT3 supplementation experiment, in which SIRT3-knockout cells were infected with adenovirus vectors expressing the wt SIRT3 or the mt protein. Twelve hours post-viral infection, cells were transfected with YFP-Bax and Ku70, and the percentage of cells that underwent apoptosis was scored 12 h later. The cells that received mt vector exhibited significantly higher numbers of cell deaths than did cells infected with the wt SIRT3-expressing vector (Fig. 11C).
To confirm that translocation of Bax in mitochondria was indeed prevented after overexpression of SIRT3, we examined the Bax localization by using fluorescence microscopy. HeLa cells infected with viral vectors were transfected with plasmids encoding YFP, YFP.Bax, and/or Ku70 in different combinations. As shown in Fig. 11D, in SIRT3-expressing cells Bax fluorescence was mostly uniform and was seen mainly in the cytoplasm (lower panel), whereas in the mt protein-expressing cells, Bax fluorescence was noted to have a punctate pattern, accumulated close to fragmented nuclei, and overlapped with Mitotracker staining of the cells (not shown), thus suggesting that SIRT3 overexpression prevented the mitochondrial accumulation of Bax.
These experiments were repeated in MNNG-treated cardiomyocytes, where translocation of the endogenous Bax was examined after treatment of cells with MNNG. As shown in Fig. 11E, cardiomyocytes infected with the mt vector and showing signs of cell death (fragmented and/or shrunken nuclei) had neatly overlapped Bax (red) staining with Mitotracker green staining (see yellow in merged image). However, a similar overlap of Bax and Mitotracker staining was not seen in cells overexpressing the wt SIRT3 and having well-preserved nuclei. These results thus demonstrated that SIRT3 expression blocks translocation of Bax into mitochondria under stress conditions.
To add weight to these findings, we then examined Bax binding to Ku70 in the absence or presence of SIRT3. Flag-tagged Ku70 was synthesized and acetylated by PCAF under in vitro assay conditions. Acetylated Ku70 was subjected to deacetylation by incubation with beads containing in vivo-synthesized wt SIRT3, mt SIRT3, or SIRT1, which was used as a positive control. Following the deacetylation reaction, Flag.Ku70 beads were separated and incubated overnight with lysates of normal HeLa cells. Beads were removed from the reaction mix, and the binding of Bax to Ku70 was determined by Western blotting. As expected, we found strong binding of Bax to Ku70 subjected to deacetylation by SIRT3 or SIRT1, but not with the SIRT3 mt (Fig. 12A, lanes 2 to 4). As a negative control, we also tested the ability of SIRT3 to bind Bax and found no direct binding between these two proteins (not shown). To support these findings further, we tested the ability of acetylated Ku70 to bind Bax, and we found that acetylation of Ku70 completely blocked its binding to Bax (Fig. 12A, lane 1). Together, these results provided strong evidence that SIRT3 prevents cell death by deacetylating Ku70 and hence promoting Ku70/Bax association.
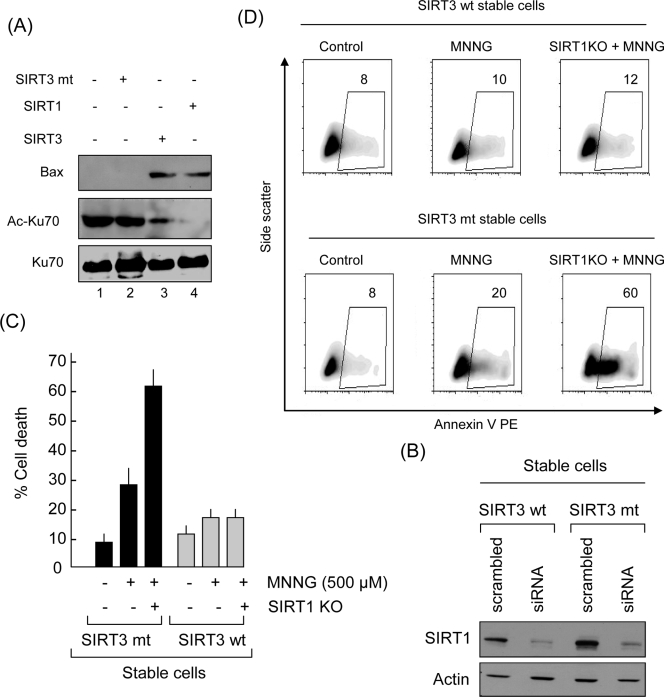
FIG. 12.
Redundant function of SIRT3 and SIRT1 of protecting cells from apoptosis. (A) SIRT3- and SIRT1-mediated deacetylation of Ku70 enhances its binding to Bax. Flag-Ku70 was synthesized in vitro, captured on Flag beads, and then acetylated by PCAF. Acetylated Flag.Ku70 was subjected to deacetylation by SIRT1, SIRT3, or the mt protein. The acetylated and deacetylated Flag.Ku70 was then incubated with HeLa cell extract in CHAPS buffer overnight. Flag.Ku70 beads were separated from the extract and tested for Bax coprecipitation. Beads were also tested for the acetylation status of Ku70 by Western blotting using appropriate antibodies. Note that Bax was pulled down by Ku70 that was deacetylated by either SIRT1 or SIRT3 but not by the acetylated Ku70. (B) SIRT1 was knocked out (KO) from wt SIRT3- or mt SIRT3-expressing stable cell lines, using SIRT1-specific siRNA. The blot shows Western analysis of SIRT1 expression levels in control and KO cells. (C) SIRT1 KO sensitized mt SIRT3-expressing cells, but not the wt SIRT3-expressing cells, to MNNG-mediated cell death. Values are means ± standard errors of four experiments. (D) Representative density blots of FACS analysis of cells treated with MNNG (500 μM). Cell death was monitored 3 h later.
Since both SIRT3 and SIRT1 are capable of deacetylating Ku70 and enhancing Ku70-Bax interaction, we asked whether both have redundant activities to protect cells during stress. To test this possibility, we knocked out SIRT1 from wt SIRT3- and mt SIRT3-expressing stable HeLa cells and then tested them for their sensitivity to MNNG-mediated cell death (Fig. 12B). We found that wt SIRT3-expressing cells were mostly resistant to cell death even when SIRT1 was knocked out. In contrast, mt SIRT3-expressing cells became overly sensitive to MNNG-mediated cell death once SIRT1 was knocked out (Fig. 12C and D). These results thus indicated that both SIRT3 and SIRT1 have redundant functions to protect cells under stress conditions.
DISCUSSION
SIRT3 is highly expressed in the heart (17, 33). In this study, we found that SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from genotoxic and oxidative stress-mediated cell damage. By utilizing different approaches such as cellular fractionation and immunostaining of endogenous as well as ectopically expressed SIRT3, we show for the first time that both the full-length form (long form) and the short form of mSIRT3 are present in cardiomyocytes. Whereas the ∼28-kDa short form of SIRT3 was localized exclusively in mitochondria, the long form (∼44 kDa) was detected in mitochondria, the nucleus, and the cytoplasm. During stress, cardiomyocyte SIRT3 levels were elevated not only in mitochondria but also in the nucleus. Another important finding of this study is that we identified Ku70, a nuclear protein, as a deacetylase target of SIRT3. Thus, our data demonstrate that, in stressed cardiomyocytes, SIRT3 plays a role in both the mitochondrial and the nuclear compartments of the cell.
Expression of hSIRT3 and mSIRT3.
hSIRT3 exists in two different forms, a full-length protein (∼44 kDa) and a processed form in which the N-terminal 142 amino acids are removed (∼28 kDa) (9, 31, 32). The sequence of mSIRT3 cDNA encoding full-length (44-kDa) protein has not been published yet. However, based on sequence conservation, it has been predicted that mSIRT3 starts at the methionine residue, equivalent to human methionine-143 (31, 33, 38), which would result in a protein product of ∼28 kDa (short form). In a recent report, however, Cooper and Spelbrink disputed these reports and predicted that the full-length mSIRT3 is a homologue of hSIRT3 (9). This form of mSIRT3 has an N-terminal stretch of 75 to 77 amino acids prior to the methionine-143 position of hSIRT3. The predicted molecular mass of the full-length (long-form) mSIRT3 is ∼39 kDa, and the short form is ∼28 kDa, which is similar to the mass of hSIRT3 (9). In our study, we observed that the long form of mSIRT3 is about ∼44 kDa and the short form is ∼28 kDa by Western analysis. These results were repeated with three different antibodies, raised against the N-terminal region, the C-terminal region, and an internal region of SIRT3, and identical results were obtained. We searched previous publications with which to compare our findings, but interestingly, although many reports have published results for expression and localization of mSIRT3, no report thus far has given the molecular weight of mSIRT3. Therefore, to know the specificity of mSIRT3 bands detected in our analysis, we relied on the blocking peptide and knockout experiments. Results obtained from these experiments confirmed that both the ∼44-kDa and ∼28-kDa bands observed from cardiac tissue are specific for SIRT3. The difference in the predicted and observed molecular masses of mSIRT3 is most likely related to posttranslational modification of SIRT3, as has been shown in the case of SIRT1 (37).
Localization of SIRT3 in cardiomyocytes.
The subcellular localization of SIRT3 is also controversial. Initial studies carried out with overexpressed hSIRT3 have shown exclusive mitochondrial localization of both the full-length and processed forms of the protein (25, 32). In contrast, a recent study has shown that the full-length hSIRT3 is a nuclear protein and that it is processed inside the nucleus to yield a protein that is capable of mitochondrial import (31). This report has also shown that the full-length nuclear hSIRT3 is catalytically active, contradicting the previous belief that processing of hSIRT3 inside mitochondria is necessary for its activation (31, 32). Another recent study has put forward further evidence showing that both hSIRT3 and mSIRT3 proteins are expressed exclusively in mitochondria (9, 17). Our results, obtained from cardiomyocytes and whole-heart tissue, however, disagree with this report and show that the ∼28-kDa short form of mSIRT3 is present only in mitochondria, whereas the full-length long form (44 kDa) is detected not only in the mitochondria but also in the nucleus as well as in the cytoplasm. Previous reports showing exclusive mitochondrial localization of SIRT3 have argued that the detection of the short form of SIRT3 (28 kDa) in the nuclear fraction, as seen by others, is likely to have originated from mitochondrial contamination of the nuclear pellet (9). This argument does not apply to our findings, as we found no trace of the 28-kDa form of SIRT3 in the nuclear or cytoplasmic fractions, suggesting that this form of mSIRT3 is localized exclusively in the mitochondria of cardiomyocytes. In our protein binding experiments we found that, although the short form of SIRT3 was capable of binding to Ku70 in vitro, it was not pulled down by Ku70 from the in vivo binding assay, thus again supporting the observation that the 28-kDa short form of SIRT3 is localized exclusively in mitochondria, where Ku70 is not expressed. In addition, our results showing deacetylation of H3 by hSIRT3 overexpression and the ability of a SIRT3 deletion mt (SIRT3Δ1-25), which lacks the mitochondrial import signal, to protect cells from MNNG-mediated cell death indicated that the protein is catalytically active outside the mitochondria, which is in agreement with one report (31) but not with the other (32).
There could be several explanations for the observed differences between our study and others. (i) In most of the previous studies, SIRT3 expression was analyzed in SIRT3-overexpressing cell lines, where endogenous SIRT3 levels are extremely low (9, 25, 31, 32). Overexpression of SIRT3 mimics a stress-like situation resulting in predominant expression of the processed form of the deacetylase, as we also observed in our cardiomyocyte cultures (Fig. 3) (31). (ii) In many previous studies, conclusions were drawn from use of a single antibody, while we know that many commercial antibodies do not recognize both forms of SIRT3 (9, 17). (iii) Tissue specificities of the two forms of SIRT3 might also account for the observed differences in different studies.
Previous studies carried out for tissue distribution of SIRT3 have shown relatively higher levels of the deacetylase in highly active tissues such as brain, heart, kidney, liver, and adipose tissues (17). It should be noted that in stress situations cardiomyocytes, which are incapable of dividing, behave very differently than do other cells, competent in proliferation. Cardiomyocytes develop hypertrophy during stress, which is accompanied by a large increase in the number of mitochondria (18). A recent study has shown that the constitutive expression of endogenous mSIRT3 upregulates the expression of mitochondrion-related genes, such as those for PGC1α, UCP1, cytochrome c oxidase subunits II and IV, and ATP synthetase, in adipocytes (33). As we know that PGC1α is a critical regulator of proteins involved in mitochondrial function and biogenesis, it is likely that the elevated levels of nuclear SIRT3, as seen in our study, have a role in controlling mitochondrial propagation in cardiomyocytes under stress conditions (16). This function of SIRT3 in cardiomyocytes, however, needs to be formally demonstrated. Another important observation in this study is that we found a considerable amount of mSIRT3 in the cytoplasm of cardiomyocytes, where no trace of nuclear or mitochondrial contamination could be detected. This was confirmed both in the myocardial tissue and in cultured cardiomyocytes, with identical results. Although we have no explanation for this finding at present, it is possible that in cardiomyocytes SIRT3 targets other organelles in addition to mitochondria. Studies are under way in our laboratory to explore this possibility.
Ku70, a target of SIRT3-mediated deacetylation.
In this study, we have identified Ku70 as a new target of SIRT3 deacetylase. Because Ku70 is not found in mitochondria, identifying this protein as a SIRT3 target again supports our finding of nonmitochondrial localization of SIRT3 in cardiomyocytes (30). Increased oxidative stress and DNA damage have been proposed as key mechanisms leading to cell death of cardiomyocytes during stress and aging (6). We used MNNG and H2O2 in this study to induce DNA damage and oxidative stress-mediated cell death. This type of cell death is controlled by PARP1 activation, which results in poly(ADP)-ribosylation of key DNA repair proteins at the expense of NAD (21, 26, 39). It has been shown that in conditions where DNA damage is limited (e.g., by MNNG treatment at 20 to 40 μM) this physiological machinery could repair the injury and the cell could survive. However, when DNA damage is extensive (e.g., by 500 μM MNNG treatment), overactivation of PARP1 occurs and this activates calpain, a Ca2+-dependent protease (21). Downstream steps from the gain of function of calpain include activation of Bax and cleavage of mitochondrial apoptosis-inducing factor (AIF) to generate an active form of the enzyme, tAIF (21). Activated Bax translocates from the cytosol to the mitochondria, where it facilitates the release of tAIF from the mitochondria to the cytosol. Once liberated in the cytosol, tAIF translocates to the nucleus, where it generates DNA breaks and chromatin condensation, resulting in cell death (21). It has also been reported that ablation of Bax prevents both tAIF release and DNA damage-induced cell death, thus documenting that Bax translocation to mitochondria is indispensable for oxidative and genotoxic stress-mediated cell death (21).
In the quiescent state, Bax normally remains associated with Ku70 (30). Acetylation of Ku70 during cellular stress or inhibition of HDACs promotes Bax-mediated apoptosis, as acetylated Ku70 is incapable of binding and sequestering Bax (7). Deacetylation of Ku70 by SIRT1 has been shown to cause sequestration of Bax away from mitochondria (8). We presumed that SIRT3 may follow the same pathway to protect cells from oxidative and genotoxic stress. Experiments carried out for testing binding and deacetylation of Ku70 confirmed that it is, in fact, as good a target of SIRT3 as was reported for SIRT1 (8). By using a highly specific assay of Bax-mediated cell death, we found that SIRT3 prevented cell death in cooperation with Ku70. This finding suggested that SIRT3 and Ku70 might work together to modulate the susceptibility of cells to stress-induced apoptosis, as has been reported in the case of SIRT1-mediated cell protection (8). By knocking out SIRT1, we found that both SIRT3 and SIRT1 have redundant functions to protect cells from genotoxic stress-mediated cell death. It is likely that in vivo SIRT3 and SIRT1, though working through the same mechanism, are activated by different stress stimuli in different cell types or to different magnitudes of stimuli. They may also have additive effects to protect cells from severely adverse conditions. Based on these findings and our unreported data, we believe that, in addition to mitochondrial proteins, there are other targets which also come into play for SIRT3-mediated cell protection. Our studies are in agreement with a recent report in which Yang et al. have shown that stress of cardiomyocytes (by serum withdrawal) induces expression of Nampt, an enzyme of NAD biosynthesis, and that the Nampt-mediated cell protection requires SIRT3 (36).
Among the different members of the sirtuin family, SIRT3 is the only protein whose increased expression has been directly linked with an extended life span in humans (3, 28). Furthermore, in aging hematopoietic stem cells, a notably reduced level of SIRT3 has been reported, suggesting that SIRT3 might function to maintain the cellular repair process and delay aging (5). Our data presented in this study provide the first evidence that cardiomyocytes are made resistant to stress-mediated cell death by SIRT3 overexpression. If a similar effect of SIRT3 is observed also in other tissues such as brain, liver, and kidney where it is highly expressed, this will suggest that SIRT3 might be a key regulator of cell defense mechanisms during stress, which become weak and dysregulated with aging. Future studies directed toward examining the beneficial effect of SIRT3 at the whole-organ level, where cells are subjected to pathological stresses, should be able to shed more light on its effect on the longevity of the organism.
Acknowledgments
We thank the following investigators for their generous gift of plasmids used in this study: E. Verdin, S. Matsuyama, D. A. Sinclair, and J. M. Denu. We also thank H. Spelbrink, University of Tampere, Finland, for sharing unpublished information on mSIRT3 expression patterns. We also thank C. Labno and S. Bond for their technical assistance in microscopic analyses and A. Isbatan for his help in tissue culture studies.
This study was partially supported by NIH grants RO1 HL-68083, HL-77788, and HL-83423.
Footnotes
Published ahead of print on 18 August 2008.
REFERENCES
- 1.Alcendor, R. R., S. Gao, P. Zhai, D. Zablocki, E. Holle, X. Yu, B. Tian, T. Wagner, S. F. Vatner, and J. Sadoshima. 2007. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ. Res. 1001512-1521. [DOI] [PubMed] [Google Scholar]
- 2.Altman, S. A., L. Randers, and G. Rao. 1993. Comparison of trypan blue dye exclusion and fluorometric assays for mammalian cell viability determinations. Biotechnol. Prog. 9671-674. [DOI] [PubMed] [Google Scholar]
- 3.Bellizzi, D., G. Rose, P. Cavalcante, G. Covello, S. Dato, F. De Rango, V. Greco, M. Maggiolini, E. Feraco, V. Mari, C. Franceschi, G. Passarino, and G. De Benedictis. 2005. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics 85258-263. [DOI] [PubMed] [Google Scholar]
- 4.Blander, G., and L. Guarente. 2004. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73417-435. [DOI] [PubMed] [Google Scholar]
- 5.Chambers, S. M., C. A. Shaw, C. Gatza, C. J. Fisk, L. A. Donehower, and M. A. Goodell. 2007. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 5e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clerk, A., S. M. Cole, T. E. Cullingford, J. G. Harrison, M. Jormakka, and D. M. Valks. 2003. Regulation of cardiac myocyte cell death. Pharmacol. Ther. 97223-261. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, H. Y., S. Lavu, K. J. Bitterman, B. Hekking, T. A. Imahiyerobo, C. Miller, R. Frye, H. Ploegh, B. M. Kessler, and D. A. Sinclair. 2004. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol. Cell 13627-638. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, H. Y., C. Miller, K. J. Bitterman, N. R. Wall, B. Hekking, B. Kessler, K. T. Howitz, M. Gorospe, R. de Cabo, and D. A. Sinclair. 2004. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305390-392. [DOI] [PubMed] [Google Scholar]
- 9.Cooper, H. M., and J. N. Spelbrink. 2008. The human Sirt3 protein deacetylase is exclusively mitochondrial. Biochem. J. 411279-285. [DOI] [PubMed] [Google Scholar]
- 10.Gupta, M., S. Samant, S. Smith, and S. Shroff. 2008. HDAC4 and PCAF bind to cardiac sarcomeres and play a role in regulating myofilament contractile activity. J. Biol. Chem. 28310135-10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gusterson, R., B. Brar, D. Faulkes, A. Giordano, J. Chrivia, and D. Latchman. 2002. The transcriptional co-activators CBP and p300 are activated via phenylephrine through the p42/p44 MAPK cascade. J. Biol. Chem. 2772517-2524. [DOI] [PubMed] [Google Scholar]
- 12.Hallows, W. C., S. Lee, and J. M. Denu. 2006. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. USA 10310230-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayakawa, Y., M. Chandra, W. Miao, J. Shirani, J. H. Brown, G. W. Dorn II, R. C. Armstrong, and R. N. Kitsis. 2003. Inhibition of cardiac myocyte apoptosis improves cardiac function and abolishes mortality in the peripartum cardiomyopathy of Galpha(q) transgenic mice. Circulation 1083036-3041. [DOI] [PubMed] [Google Scholar]
- 14.Heineke, J., and J. D. Molkentin. 2006. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 7589-600. [DOI] [PubMed] [Google Scholar]
- 15.Kang, P. M., and S. Izumo. 2000. Apoptosis and heart failure: a critical review of the literature. Circ. Res. 861107-1113. [DOI] [PubMed] [Google Scholar]
- 16.Lagouge, M., C. Argmann, Z. Gerhart-Hines, H. Meziane, C. Lerin, F. Daussin, N. Messadeq, J. Milne, P. Lambert, P. Elliott, B. Geny, M. Laakso, P. Puigserver, and J. Auwerx. 2006. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 1271109-1122. [DOI] [PubMed] [Google Scholar]
- 17.Lombard, D. B., F. W. Alt, H. L. Cheng, J. Bunkenborg, R. S. Streeper, R. Mostoslavsky, J. Kim, G. Yancopoulos, D. Valenzuela, A. Murphy, Y. Yang, Y. Chen, M. D. Hirschey, R. T. Bronson, M. Haigis, L. P. Guarente, R. V. Farese, Jr., S. Weissman, E. Verdin, and B. Schwer. 2007. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 278807-8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meerson, F. Z., T. A. Zaletayeva, S. S. Lagutchev, and M. G. Pshennikova. 1964. Structure and mass of mitochondria in the process of compensatory hyperfunction and hypertrophy of the heart. Exp. Cell Res. 36568-578. [DOI] [PubMed] [Google Scholar]
- 19.Michan, S., and D. Sinclair. 2007. Sirtuins in mammals: insights into their biological function. Biochem. J. 4041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michishita, E., J. Y. Park, J. M. Burneskis, J. C. Barrett, and I. Horikawa. 2005. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 164623-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moubarak, R. S., V. J. Yuste, C. Artus, A. Bouharrour, P. A. Greer, J. Menissier-de Murcia, and S. A. Susin. 2007. Sequential activation of poly(ADP-ribose) polymerase 1, calpains, and Bax is essential in apoptosis-inducing factor-mediated programmed necrosis. Mol. Cell. Biol. 274844-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura, Y., M. Ogura, D. Tanaka, and N. Inagaki. 2008. Localization of mouse mitochondrial SIRT proteins: shift of SIRT3 to nucleus by co-expression with SIRT5. Biochem. Biophys. Res. Commun. 366174-179. [DOI] [PubMed] [Google Scholar]
- 23.North, B. J., B. Schwer, N. Ahuja, B. Marshall, and E. Verdin. 2005. Preparation of enzymatically active recombinant class III protein deacetylases. Methods 36338-345. [DOI] [PubMed] [Google Scholar]
- 24.North, B. J., and E. Verdin. 2004. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onyango, P., I. Celic, J. M. McCaffery, J. D. Boeke, and A. P. Feinberg. 2002. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc. Natl. Acad. Sci. USA 9913653-13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pillai, J. B., A. Isbatan, S. Imai, and M. P. Gupta. 2005. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J. Biol. Chem. 28043121-43130. [DOI] [PubMed] [Google Scholar]
- 27.Riccardi, C., and I. Nicoletti. 2006. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat. Protoc. 11458-1461. [DOI] [PubMed] [Google Scholar]
- 28.Rose, G., S. Dato, K. Altomare, D. Bellizzi, S. Garasto, V. Greco, G. Passarino, E. Feraco, V. Mari, C. Barbi, M. BonaFe, C. Franceschi, Q. Tan, S. Boiko, A. I. Yashin, and G. De Benedictis. 2003. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp. Gerontol. 381065-1070. [DOI] [PubMed] [Google Scholar]
- 29.Saunders, L. R., and E. Verdin. 2007. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene 265489-5504. [DOI] [PubMed] [Google Scholar]
- 30.Sawada, M., W. Sun, P. Hayes, K. Leskov, D. A. Boothman, and S. Matsuyama. 2003. Ku70 suppresses the apoptotic translocation of Bax to mitochondria. Nat. Cell Biol. 5320-329. [DOI] [PubMed] [Google Scholar]
- 31.Scher, M. B., A. Vaquero, and D. Reinberg. 2007. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 21920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwer, B., B. J. North, R. A. Frye, M. Ott, and E. Verdin. 2002. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 158647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi, T., F. Wang, E. Stieren, and Q. Tong. 2005. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J. Biol. Chem. 28013560-13567. [DOI] [PubMed] [Google Scholar]
- 34.Smith, J. 2002. Human Sir2 and the ‘silencing’ of p53 activity. Trends Cell Biol. 12404-406. [DOI] [PubMed] [Google Scholar]
- 35.Vakhrusheva, O., C. Smolka, P. Gajawada, S. Kostin, T. Boettger, T. Kubin, and E. Bober. 2008. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 102703-710. [DOI] [PubMed] [Google Scholar]
- 36.Yang, H., T. Yang, J. A. Baur, E. Perez, T. Matsui, J. J. Carmona, D. W. Lamming, N. C. Souza-Pinto, V. A. Bohr, A. Rosenzweig, R. de Cabo, A. A. Sauve, and D. A. Sinclair. 2007. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell 1301095-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, Y., W. Fu, J. Chen, N. Olashaw, X. Zhang, S. V. Nicosia, K. Bhalla, and W. Bai. 2007. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat. Cell Biol. 91253-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, Y. H., Y. H. Chen, C. Y. Zhang, M. A. Nimmakayalu, D. C. Ward, and S. Weissman. 2000. Cloning and characterization of two mouse genes with homology to the yeast Sir2 gene. Genomics 69355-369. [DOI] [PubMed] [Google Scholar]
- 39.Yu, S. W., H. Wang, M. F. Poitras, C. Coombs, W. J. Bowers, H. J. Federoff, G. G. Poirier, T. M. Dawson, and V. L. Dawson. 2002. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297259-263. [DOI] [PubMed] [Google Scholar]